ABSTRACT
The evolutionarily conserved Target of Rapamycin (TOR) complex-2 (TORC2) is an essential regulator of plasma membrane homeostasis in budding yeast (Saccharomyces cerevisiae). In this yeast, TORC2 phosphorylates and activates the effector protein kinase Ypk1 and its paralog Ypk2. These protein kinases, in turn, carry out all the crucial functions of TORC2 by phosphorylating and thereby controlling the activity of at least a dozen downstream substrates. A previously uncharacterized interplay between the Rab5 GTPases and TORC2 signaling was uncovered through analysis of a newly suspected Ypk1 target. Muk1, one of two guanine nucleotide exchange factors for the Rab5 GTPases, was found to be a physiologically relevant Ypk1 substrate; and, genetic analysis indicates that Ypk1-mediated phosphorylation activates the guanine nucleotide exchange activity of Muk1. Second, it was demonstrated both in vivo and in vitro that the GTP-bound state of the Rab5 GTPase Vps21/Ypt51 physically associates with TORC2 and acts as a direct positive effector required for full TORC2 activity. These interrelationships provide a self-reinforcing control circuit for sustained up-regulation of TORC2-Ypk1 signaling. In this overview, we summarize the experimental basis of these findings, their implications, and speculate as to the molecular basis for Rab5-mediated TORC2 activation.
KEYWORDS: Growth control, protein kinase, phosphorylation, plasma membrane, endocytosis, guanine nucleotide, mutants
Background and overview
In organisms from yeasts to humans, essential aspects of cellular physiology are coordinated by the actions of two, evolutionarily conserved, multi-component protein kinase complexes in which the catalytic subunit is the large Target of Rapamycin (TOR) polypeptide [1–4]. These two TOR-containing complexes – TOR complex 1 (TORC1) and TOR complex 2 (TORC2) – are spatially and functionally distinct. Active TORC1 resides on the cytosolic surface of the lysosome in animal cells and on the surface of its counterpart, the vacuole, in yeast (Saccharomyces cerevisiae), whereas active TORC2 resides mainly on the plasma membrane (PM). Animal cells express a single TOR protein that populates both complexes; in yeast, however, there are two TOR paralogs, Tor1 and Tor2. TORC1 is functional when it contains either Tor1 or Tor2 as its catalytic subunit; but, to be functional, TORC2 must contain Tor2 as its catalytic subunit [5,6]. Hence, a tor1∆ mutant is viable, but a tor2∆ mutant is not [7,8]. The activity of TORC1 can be acutely inhibited by rapamycin and related compounds [9], whereas TORC2 is largely immune to these inhibitors [10], which made analysis of TORC1 function more readily accessible than analysis of TORC2 function. However, the demonstration that the primary essential function of TORC2 is to phosphorylate two, paralogous, AGC-family protein kinases, Ypk1 and Ypk2, which then execute all of the critical downstream functions, has dramatically increased our understanding of the physiological roles of TORC2 [11].
As with other AGC kinases, the basal activity of Ypk1 requires phosphorylation of a conserved Thr residue (T504) in its activation loop within its catalytic domain. This modification is installed by two paralogous protein kinases, Pkh1 and Pkh2 [12,13], which are stably-associated components of the protein coats of PM invaginations called eisosomes [14,15]. Basal activity and stability of Ypk1 also requires its phosphorylation at S644, which lies within a conserved sequence (dubbed the “turn motif”) located downstream of its kinase homology domain within a C-terminal regulatory domain [16]. This modification is installed by TORC2, which is also largely PM-associated [17–21]. Under certain stressful conditions that stimulate TORC2-mediated phosphorylation of Ypk1, such as sphingolipid limitation [22], heat stress [23], hypotonic conditions [24,25], and acetic acid stress [26], TORC2 further elevates Ypk1 activity by phosphorylating four additional sites in its C-terminal regulatory domain, paramount among them is T662, which lies within another conserved sequence (dubbed the “hydrophobic motif”) in the C-terminal domain [13,16]. Phosphorylation of Ypk1 at these locations further enhances both its activity and stability. Under other stressful conditions, such as hypertonic shock [27,28], treatments that damage the cell wall [19], and treatments that decrease “membrane tension” [29], TORC2-mediated phosphorylation of Ypk1 is dramatically reduced.
S. cerevisiae TORC2 comprises four essential core subunits (Avo1, Avo3, Lst8, and Tor2) [30], two classes of non-essential peripherally-associated subunits (Avo2 and Bit61 and its paralog Bit2) [3,31,32], and two, essential ancillary subunits (Slm1 and Slm2) that undergo dynamic shuttling between the eisosomes and TORC2 [24,25]. The tertiary fold of the kinase domain of the catalytic subunit Tor2 is stabilized by its tight association with the β-propeller protein Lst8 (which also binds to Tor1). Tor2 is also intimately entwined with Avo1 and Avo3 [33] to form a dimeric rhombohedral complex [31], creating the scaffold onto which the other TORC2 components dock.
Based on a cryo-EM-derived structure of S. cerevisiae TORC2 [31], Avo1 appears to be located in close proximity to the active site of the Tor2-Lst8 complex. Furthermore, convincing biochemical evidence shows that a sequence in Avo1 shared with its Schizosaccharomyces pombe ortholog Sin1 and its mammalian counterpart (mSIN1), designated the “conserved region in the middle” (CRIM), is the sequence element that binds the corresponding Ypk1 orthologs in these organisms, Gad8 [34] and both SGK1 [35] and AKT1 [36], and presents them to the TOR kinase for phosphorylation. Therefore, by analogy, Ypk1 is likely to be recognized as a substrate for TORC2 by its binding to the CRIM element in Avo1. In this regard, although slm1∆ slm2∆ cells are inviable, fusion of the PtdIns4,5P2-binding PH domain of Slm1 [37,38] to Ypk1 restores viability to slm1∆ slm2∆ cells [24], suggesting that, normally, one function of the Slm1 proteins is to promote, somehow, the Avo1-mediated recognition of Ypk1 by TORC2 at the PM.
Muk1 emerges as a substrate for Ypk1
Various approaches have been used to identify physiologically relevant substrates of the TORC2-Ypk1 signaling axis, including genetic methods [12], biochemical analysis [22,39], chemogenetic strategies [40–42], a genome-wide candidate screen [43], and global phosphoproteomics [44]. As summarized in a recent comprehensive review [11], among the thoroughly validated direct substrates of Ypk1 identified from these studies are: (a) two protein kinases (Fpk1 and Fpk2) whose role is to phosphorylate and thereby stimulate both PM- (Dnf1 and Dnf2) and trans-Golgi- (Dnf3) localized aminoglycerophospholipid flippases; Ypk1-mediated phosphorylation inhibits Fpk1 and Fpk2; (b) one of two glycerol-3P dehydrogenase isoforms (Gpd1) whose role is to supply sn-glycerol-3P for both glycerophospholipid synthesis and glycerol production; Ypk1-mediated phosphorylation inhibits Gpd1; (c) the major aquaglycerolporin (Fps1), which is a channel required for glycerol efflux; Ypk1-mediated phosphorylation of Fps1 is required to keep this channel in its open state; (d) two, endoplasmic reticulum (ER)-localized tetraspanins (Orm1 and Orm2) whose role is to inhibit the enzyme that catalyzes the first-committed step in sphingolipid biosynthesis; Ypk1-mediated phosphorylation blocks the inhibitory functions of Orm1 and Orm2, thereby stimulating metabolic flux into the sphingolipid pathway; (e) the heterodimeric ceramide synthase (Lac1-Lag1) required for generation of sphingolipids; Ypk1-mediated phosphorylation stimulates the activity of Lac1-Lag1, thereby further promoting production of sphingolipids; (f) at least two α-arrestins (Art3/Aly2 and Art4/Rod1), which are adaptors necessary to target the HECT domain ubiquitin ligase Rsp3 to specific integral membrane protein clients to promote their endocytosis; Ypk1-mediated phosphorylation inhibits the function of these proteins; and (g) two paralogous StARkin domain–containing proteins (Ysp2/Lam2/Ltc4 and its paralog Lam4/Ltc3) located at PM-endoplasmic reticulum (ER) contact sites, which participate in the retrograde movement of ergosterol from the PM to the ER; Ypk1-mediated phosphorylation inhibits the ability of these proteins to promote this retrograde sterol transport.
This cohort of substrates demonstrates that TORC2-Ypk1 signaling is essential for growth and viability because it controls virtually every aspect of PM homeostasis – from sphingolipid and glycerolipid production, to PM lipid asymmetry, to the concentration of an intracellular osmolyte (glycerol), to PM protein composition, to sterol content. In a global screen for candidate Ypk1 targets conducted by our laboratory [43], we also identified Muk1, one of two Rab5-specific guanine nucleotide exchange factors (GEFs) encoded in the S. cerevisiae genome (the other is Vps9), among potential targets of Ypk1. We confirmed recently that Muk1 is indeed a bona fide substrate of Ypk1 [45]. We demonstrated that Muk1 is phosphorylated in a Ypk1-dependent manner both in vivo and in vitro and, under either condition, is phosphorylated by Ypk1 at its two consensus Ypk1 phospho-acceptor motifs (RSRSSSG and RPRRSSS). Moreover, using three different phenotypic screens in vivo, we found that a corresponding Muk1(6A) mutant was a total loss-of-function allele, whereas the cognate phosphomimetic allele, Muk1(6E), was somewhat hyperactive compared to wild-type Muk1.
These results predicted that stimulation of TORC2-Ypk1 signaling should result in a Muk1-dependent increase in the pool of active (GTP-bound) Rab5 in the cells. In this regard, we were intrigued by the possibility that one or more of the three Rab5 GTPases encoded in S. cerevisiae genome (Ypt51/Vps21, Ypt52, and Ypt53) might serve as a direct modulator of TORC2 function, based on two precedents. First, the function of the other TOR-containing complex, TORC1, requires its interaction with two other classes of small GTPases, both RHEB [46,47] and RAGs [48,49]. Second, it was reported, largely on the basis of genetic findings, that GTP-bound Ryh1 (a small GTPase that most closely resembles human Rab6 and its S. cerevisiae ortholog Ypt6) stimulates TORC2 in fission yeast [50].
Rab5 GTPases emerge as regulators of TORC2
For reasons explained just above, we examined whether Rab5 function influenced TORC2 activity, as assessed by monitoring the TORC2-mediated phosphorylation and activation of Ypk1. Strikingly, we found, first, that yeast lacking its two Rab5 GEFs (Muk1 and Vps9) or its three Rab5 paralogs (Vps21/Ypt51, Ypt52 and Ypt53) or overexpressing Msb3, a Rab5-directed GTPase-activating protein (GAP), all exhibited pronounced reduction in TORC2-mediated phosphorylation and activation of Ypk1 [45]. Second, we found that cells lacking both Rab5 GEFs and cells lacking all three Rab5 paralogs were much more sensitive than otherwise isogenic WT control cells to the growth-inhibitory action of aureobasidin A (AbA), an antibiotic that blocks sphingolipid biosynthesis [45]. We had demonstrated previously that upregulation of Ypk1 activity via its TORC2-dependent phosphorylation is required for cells to survive inhibition of sphingolipid biosynthesis by another antibiotic (myriocin) [16,22]. Collectively, these results demonstrated that cells that cannot generate active Rab5 exhibit both biochemical and physiological behavior consistent with inefficient TORC2-mediated phosphorylation of Ypk1.
In further support of the conclusion that active Rab5 stimulates TORC2 phosphorylation of Ypk1, we found that expression of Vps21(Q66L), a GTP hydrolysis-defective (so-called “GTP-locked”) variant, restored readily detectable TORC2-mediated phosphorylation of Ypk1 in vps21∆ ypt52∆ ypt53∆ triple mutant cells, whereas expression of Vps21(S21L), a variant (so-called “GDP-locked”) that preferentially binds GDP, did not [45]. We focused on Vps21 because it is constitutively expressed, is the most abundant yeast Rab5 isoform, has been shown to play the major role in endocytic vesicle trafficking, and, when absent, causes more pronounced phenotypes as compared to loss of either of the other two Rab5 GTPases. Once loaded with GTP, Vps21(Q66L) persists in its active state and has been shown to block endocytosis in yeast cells [51,52], suggesting that the ability of Vps21(Q66L) to promote TORC2-dependent phosphorylation of Ypk1 is distinct from its role in promoting early endosome formation and recycling.
To begin to explore the mechanism by which GTP-bound Rab5 supports robust-TORC2-mediated phosphorylation of Ypk1, we first examined whether there might be some more indirect reason for the observed impairment in TORC2-Ypk1 signaling in cells that cannot generate active Rab5.
Given that Rab5 GTPases are important in endocytic vesicle internalization, and have also been implicated in recycling of endosomes back to the PM, it seemed possible that cells deficient in GTP-bound Rab5 might have decreased TORC2 activity simply due to altered TORC2 localization. However, when we analyzed the subcellular distribution of TORC2 by fluorescence microscopy, we saw no marked change in the amount of TORC2 located at the cell cortex between wild-type cells and muk1Δ vps9Δ double mutant cells. Thus, TORC2 did not appear to be grossly mislocalized in the absence of active Rab5. It also seemed possible that, in cells deficient in GTP-bound Rab5, Ypk1 might not be accessible to the cell cortex at all. However, in contrast to its poor phosphorylation by TORC2, there was no difference in activation loop phosphorylation of Ypk1 by eisosome-associated Pkh1 in WT and muk1Δ vps9Δ double mutant cells. Also, using pull-down assays, there was no difference in the amount of Ypk1 associated with Avo1 between wild-type and muk1Δ vps9Δ double mutant cells, indicating that the lack of efficient phosphorylation by TORC2 in cells deficient in GTP-Rab5 could not be attributed to lack of encounter between TORC2 and Ypk1. Another possibility, given that phosphorylation and activation of certain other AGC family protein kinases requires association of their regulatory domains with other classes of small GTPases [53,54], it seemed possible that a conformation change induced in Ypk1 by its binding of GTP-loaded Rab5 might be necessary for it to be competent for phosphorylation by TORC2. However, as judged by co-immunoprecipitation, we could detect no interaction between Ypk1 and GTP-bound Vps21. Taken together, these findings led us to test whether there is a direct physical interaction between Vps21 and TORC2.
Indeed, in marked contrast to the lack of interaction between Vps21 and Ypk1, association of Vps21 with Tor2 was readily detectable by co-immunoprecipitation under a variety of different expression conditions and strain backgrounds. Moreover, consistent with our in vivo results, in in vitro kinase assays using a purified recombinant substrate [E. coli MalE (maltose-binding protein)-Ypk1(603–680)] that contains all of the TORC2 sites in Ypk1, we found that the specific activity of TORC2 isolated from vsp21∆ ypt52∆ double mutant cells was reproducibly lower than that of an equivalent amount of TORC2 purified from wild-type cells. Most strikingly, the ability of TORC2 isolated from vsp21∆ ypt52∆ cells to phosphorylate MalE-Ypk1(603–680) could be markedly stimulated by the addition of purified recombinant Vps21 loaded with GTPγS, but not by yeast Rab7 (Ypt7) loaded with GTPγS. Thus, a GTP-bound Rab5 appears to be a direct activator of TORC2 kinase function.
Given our evidence that GTP-bound Rab5 stimulates TORC2 function, and given that we showed that the Rab5-specific GEF Muk1 is activated in a TORC2-dependent and Ypk1-mediated manner, one expected outcome of these interrelationships is that, once TORC2-Ypk1 signaling has been initiated, more GTP-bound Rab5 should be generated. In essence, therefore, this control circuit imposes self-reinforcing positive feedback to sustain TORC2-Ypk1 signaling. Our unexpected discovery that Rab5 function is necessary to support maximal TORC2 function reveals a previously unappreciated new connection between the vesicle trafficking machinery and the control of PM homeostasis by TORC2-Ypk1 signaling.
Implications of Rab5-dependent regulation of TORC2
Demonstration of the link between Rab5 GTPases and TORC2 raises numerous important mechanistic questions about how and where Rab5 GTPases influence TORC2 function.
How?
One intriguing possibility for how Vps21 GTPase could serve as a direct positive activator of the kinase activity of Tor2 in yeast TORC2 is that it could act in a manner analogous to how the RHEB GTPase activates mTOR in mTORC1 [47]. Structural analysis [46] has revealed that, in its GTP-bound state, RHEB occupies a pocket constituted by the M- and N-HEAT repeats and FAT domain of mTOR, thereby inducing rearrangements of key elements important for stabilizing the catalytically-competent state of mTOR. As one means to assess whether yeast Tor2 in TORC2 might contain a similar binding pocket, we used SWISS-MODEL [55] to build a homology map (Figure 1) in which we threaded the sequence of yeast Tor2 onto that of mTOR in the structure of RHEB-activated mTORC1 (PDB: 6BCU). Superimposition of our homology model of Tor2 onto mTOR in RHEB-bound mTORC1 structure reveals a very similar cavity that could readily accommodate a small GTPase. Additionally, many of the residues in mTOR that make contact with RHEB are conserved in Tor2 and adopt a similar orientation in the binding pocket in our model (Figure 1).
Figure 1.
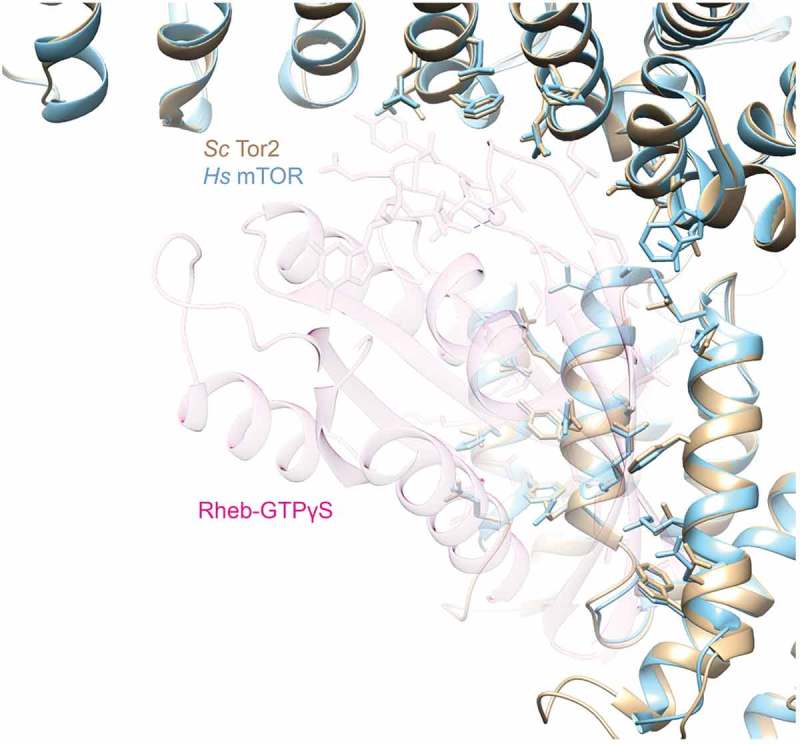
Saccharomyces cerevisiae Tor2 modeled onto human mTOR in the RHEB-binding pocket in mTORC1.
A homology model of ScTor2 (tan) built with SWISS-MODEL [55] based on the confirmation of mTOR (blue) in the cryo-EM-derived structure of mTORC1 occupied by GTPγS-bound RHEB (faint fuschia) (PDB: 6BCU) [46]. Side chains in human mTOR that make intermolecular contacts with RHEB (blue) and the corresponding side chains in yeast Tor2 (tan) are shown in stick representation.
To further explore this possibility, we compared the crystal structure of GTP-bound Vps21 to the structure of GTP-bound RHEB. Despite the divergence in the amino acid sequences between these two Ras super-family members, we found that their GTPase folds adopt a nearly identical shape (Figure 2). Hence, GTP-bound Vps21 is clearly sufficiently compact that it could occupy the analogous pocket in Tor2 in TORC2 as GTP-bound RHEB occupies in mTOR in mTORC1. However, how could Tor2 in TORC2 distinguish GTP-bound Rab5 from any other small GTPase? In this regard, and as with other effectors modulated by small GTPases, it is primarily residues in the Switch I and Switch II loops of RHEB that make contact with the RHEB-binding pocket in mTOR in mTORC1. Notably, the superimposition of Vps21 and RHEB reveals that the most major structural differences between these two GTPases lie in their respective Switch II regions (Figure 2). It is not hard to imagine, therefore, that the distinct subunits that define TORC2 force Tor2 into a conformation that is optimal for making specific contacts with the unique geometry of the Switch II residues in Vps21, thereby stabilizing Tor2 in its most catalytically-competent state.
Figure 2.
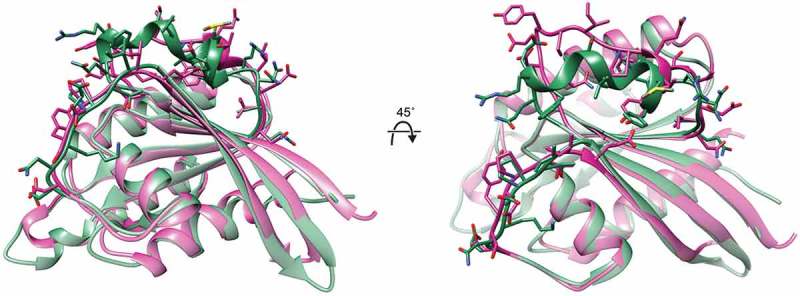
Comparison of Saccharomyces cerevisiae Vps21 to Homo sapiens RHEB.
Crystal structure (1.48 Å resolution) of yeast Vps21 (green) bound to the non-hydrolyzable GTP analog GppNHp (PDB: 1EK0) [83] was aligned with the crystal structure (2.65 Å resolution) of GppNHp-bound RHEB (fuschia) (PDB: 1XTR) [84] by superimposing the GTP-binding fold in each of these two GTPase, revealing the distinct conformation of the Switch II regions (top) in these proteins. Side chains in the Switch I and Switch II loops in both GTPases are shown in stick representation.
In this same regard, in a screen of Drosophila small GTPases [56], it was found that overexpression of activated Rab5 strongly inhibited amino acid- or Rag GTPase-stimulated mTORC1 activity, a process that requires RHEB [57], but did not inhibit mTORC1 stimulation by activated RHEB itself [56]. At the time, it was not known how to interpret these observations. In light of our findings, however, a likely explanation for these prior results is that Rab5, if overexpressed, can associate with the RHEB-binding pocket in mTOR in mTORC1, but does so non-productively (i.e. it cannot stimulate mTOR in this context), and thus under these conditions Rab5 serves, in essence, as a simple competitive inhibitor of RHEB-mediated mTORC1 activation. By contrast, when activated RHEB is overexpressed, it is able to out-compete Rab5.
Where?
Localization of TORC2 in S. cerevisiae to discrete puncta residing at the cell cortex has been well documented, based on analysis of fixed cells by indirect immunofluorescence [20] or immuno-gold EM [6], or examination of live cells expressing fluorescently-tagged derivatives of Tor2 [18,19,21] or TORC2-specific subunits that associate tightly with Tor2 [17,19,21]. In addition, the effector protein kinases (Ypk1 and Ypk2) directly phosphorylated by TORC2 are, as mentioned earlier, also obligatorily phosphorylated by other protein kinases (Pkh1 and Pkh2) that are tightly associated with the PM-anchored eisosomes. Moreover, the PM in yeast is highly enriched in PtdIns4,5P2 compared to any other cellular membrane [29,58] and both the core TORC2 subunit Avo1 [17,59,60] and the more dynamically associated TORC2 subunits Slm1 and Slm2 [24,37,38] contain PtdIns4,5P2-binding PH domains. Furthermore, maintenance of an adequate PtdIns4,5P2 level is necessary for TORC2 activity [61]. These observations are at least consistent with the conclusion that, in the main, the TORC2 cortical puncta are docked on the PM.
The three yeast Rab5 GTPases (Vps21/Ypt51, Ypt52, and Ypt53) [62] are involved at an early stage of the endocytic pathway and, akin to their mammalian Rab5 counterparts [63,64], are therefore located primarly on intracellular endosomal vesicles prior to their fusion and delivery to the multivesicular body [65–67]. Correspondingly, fluorescently-tagged derivatives of the three yeast Rab5 members exhibit punctate staining on vesicular structures within the cell that co-localize with well-documented endocytic cargo [68,69]. Interestingly, however, for GFP-Vps21, a significant fraction of the fluorescent structures are found at the cell periphery on, or in very close apposition to, the PM, whereas GFP-Ypt52 and GFP-Ypt53 generally occupy more interior locations [70].
There is additional evidence that Rab5 GTPases can be associated with the PM. For example, experiments to localize Rab5 in mammalian cells also exhibited some staining on the PM [71]. When heterologously expressed in animal cells, Vps21 colocalizes with the endogenous Rab5 and can stimulate Rab5-specific endocytic events [72], indicating that, in addition to their C-terminal prenylation [73], the yeast and vertebrate proteins must share other conserved localization determinants. In addition, using a bimolecular fluorescence complementation assay, interaction between Vps21 and its specific GAP Msb3 was detected only at patches on the PM [74], indicating again that at least a fraction of the GTP-bound pool of Vps21 resides at the PM. In this context, it is important to note that the phenotypes of cells devoid of each of the three yeast Rab5 isoforms differ significantly. In addition to severe defects in endocytic vesicle trafficking, cells lacking Vps21 also exhibit an obvious defect in growth rate, whereas cells lacking either Ypt52 or Ypt53 (or both) have much milder phenotypes. Although the relative levels of each of these proteins may contribute to the differential effects observed in their absence [62], these results are also consistent with Vps21 having a role in promoting the function of TORC2, a central growth regulator, distinct from its function in vesicle trafficking. In agreement with that proposal, in our study [45], we observed that in cells devoid of all three Rab5 paralogs TORC2-mediated phosphorylation of Ypk1 could be stimulated by expression of a GTP-locked Vps21 mutant, which (as already mentioned above) is unable to support vesicle trafficking. It will be important to test GTP-locked variants of Ypt52 and Ypt53 in the same way to determine whether it is exclusively Vps21 that associates with and acts to activate TORC2.
It is also possible that TORC2 association with and activation by Vps21 might occur on endosomes. In subcellular fractionation studies showing that TORC2 is a protein complex peripherally associated with membranes, Tor2 co-fractionated with PM markers, but also with a second fraction that appeared to represent vesicular structures [20]. Similarly, localization of endogenous Tor2 at the ultrastructural level using immuno-gold EM labeling revealed gold particle clusters at the PM, but also gold particles associated with internal membranous structures that resembled vesicles of the endocytic pathway and were distinct from the vacuole [6]. In this regard, we found that in cells lacking the two Rab5 GEFs (Vps9 and Muk1) that the amount of TORC2 localized at the PM, as judged by monitoring a core TORC2 subunit (Avo3-GFP), is not reduced compared to otherwise isogenic wild-type cells. We reached a similar conclusion using Tor2-mNeonGreen in cells lacking all three Rab5 GTPases (FM Roelants, this laboratory, unpublished data). These observations indicate that PM association of TORC2 does not require Rab5 function. However, we have not carefully analyzed whether the absence of the Rab5 GEFs or the Rab5 GTPases themselves markedly diminishes the minor pool of TORC2 that appears to be associated with internal vesicles. In this regard, there is other evidence that TORC2 may be distributed among spatially distinct populations.
It has been inferred from recent studies in mammalian cells of mTORC2-mediated phosphorylation of one of its effector protein kinases, AKT/PKB, that mTORC2 can act at discrete subcellular locations. AKT has a PH domain specific for binding PtdIns(3,4,5)P3 generated by growth factor-activated PtdIns 3-kinase (PI3K); however, there is some controversy about where and whether lipid binding is necessary to induce a conformational change in AKT that makes it competent to be phosphorylated on its activation loop by the upstream activating kinase PDK1 before it is competent to be subject to further stimulation via its mTORC2-mediated phosphorylation [75–77]. In any event, using a chemically-inducible dimerization system to recruit Akt to endosomes, it was observed that AKT becomes phosphorylated at its mTORC2 sites when tethered in this fashion, arguing that there must be mTORC2 present on the same endosomes [78]. Further, it was purported that mTORC2-dependent activation of Akt at the PM can be uncoupled from the need for PI3K-generated PtdIns(3,4,5)P3, whereas the endosomally-associated mTORC2, it is claimed, requires PI3K generation of PtdIns(3,4,5)P3 [79], suggesting, if true, that these spatially distinct pools may be differentially regulated. However, in none of these studies was the role of Rab5 in this putative endosomal pool of mTORC2 examined. However, in a recent study of mouse hippocampal pyramidal neurons, AKT was found in association with Rab5-positive endosomes, but whether TORC2 was also present was not addressed [80]. To further complicate matters as to which GTPase(s) may regulate TORC2, and where, it was recently reported that, in mammalian cells, oncogenic variants of RAS, a PM-anchored GTPase [81], associate with mTORC2, as assessed by proximity-dependent biotin labeling [82].
Hypothetically, if there are distinct pools of TORC2 with differing requirements for activation, such a scenario could provide a means to adjust the level of TORC2 activity to meet the physiological needs of the cell in the most effective manner. In yeast, for Ypk1 that has been phosphorylated by Pkh1 on its activation loop, all that is required for cell survival under steady-state growth conditions in rich medium with glucose as the carbon source is TORC2-dependent phosphorylation of Ypk1 on a single site (S644 in the turn motif). However, if the cells are subjected to any significant stress, especially limitation for sphingolipids, TORC2-mediated phosphorylation of T662 and additional residues in the carboxy-terminal tail of Ypk1 occurs and modification of these residues is essential for yeast cell survival under these stressful conditions [13,16,22]. That phosphorylation at these additional resides is not required under basal conditions, suggests that TORC2 activity provides a graded mechanism to fine-tune the level of Ypk1 activation. Furthermore, C-terminal phosphorylation of Ypk1 by TORC2 not only enhances its specific activity [22], but also stabilizes Ypk1 against degradation [16], thereby maintaining both its active conformation and prolonging the duration of its activated state. Thus, our demonstration [45] that TORC2-Ypk1 signaling also stimulates the generation of GTP-bound Vps21 and that this Rab5 is, in turn, a direct activator of TORC2, provides a positive feedback mechanism to further sustain TORC2-Ypk1 signaling, once initiated. This control circuit also provides a means for TORC2 to serve as both a sensor and a regulator of the rate of endocytic vesicle trafficking. Moreover, this Rab5-mediated stimulation could exist to up-regulate TORC2 activity and hence Ypk1 signaling at the PM or, potentially, to provide the means for localized TORC2-dependent Ypk1-mediated phosphorylation of specific substrates located on or nearby early endosomes.
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM21841];National Institute of General Medical Sciences [GM07232];University of California [CRCC Predoctoral Fellowship];University of California Berkeley [Summer Research Grant].
Acknowledgments
This work was supported by NIH Predoctoral Traineeship GM07232, by a Summer Research Grant from the University of California Berkeley Graduate Division, and by a Cancer Research Coordinating Committee Fellowship from University of California Systemwide (all to M.N.L.) and by NIH R01 Research Grant GM21841 (to J.T.). We thank Dr. Françoise M. Roelants for careful reading of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Tatebe H, Shiozaki K.. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules. 2017;7:E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Saxton RA, Sabatini DM.. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Eltschinger S, Loewith R.. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016;26:148–159. [DOI] [PubMed] [Google Scholar]
- [4].González A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. Embo J. 2017;36:397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. [DOI] [PubMed] [Google Scholar]
- [6].Wedaman KP, Reinke A, Anderson S, et al. Tor kinases are in distinct membrane-associated protein complexes Saccharomyces Cerevisiae. Mol Biol Cell. 2003;14:1204–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kunz J, Henriquez R, Schneider U, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. [DOI] [PubMed] [Google Scholar]
- [8].Helliwell SB, Wagner P, Kunz J, et al. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dowling RJ, Topisirovic I, Fonseca BD, et al. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta. 2010;1804:433–439. [DOI] [PubMed] [Google Scholar]
- [10].Gaubitz C, Oliveira TM, Prouteau M, et al. Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol Cell. 2015;58:977–988. [DOI] [PubMed] [Google Scholar]
- [11].Roelants FM, Leskoske KL, Martinez Marshall MN, et al. The TORC2-dependent signaling network in the yeast Saccharomyces cerevisiae. Biomolecules. 2017;7:E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roelants FM, Torrance PD, Bezman N, et al. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol Biol Cell. 2002;13:3005–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roelants FM, Torrance PD, Thorner J. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology. 2004;150:3289–3304. [DOI] [PubMed] [Google Scholar]
- [14].Foderaro JE, Douglas LM, Konopka JB. MCC/eisosomes regulate cell wall synthesis and stress responses in fungi. J Fungi (Basel). 2017;3:E61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Walther TC, Brickner JH, Aguilar PS, et al. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. [DOI] [PubMed] [Google Scholar]
- [16].Leskoske KL, Roelants FM, Martinez Marshall MN, et al. The stress-sensing TORC2 complex activates yeast AGC-family protein kinase Ypk1 at multiple novel sites. Genetics. 2017;207:179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sturgill TW, Cohen A, Diefenbacher M, et al. TOR1 and TOR2 have distinct locations in live cells. Eukaryot Cell. 2008;7:1819–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Leskoske KL, Roelants FM, Emmerstorfer-Augustin A, et al. Phosphorylation by the stress-activated MAPK Slt2 down-regulates the yeast TOR complex 2. Genes Dev. 2018;32:1576–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kunz J, Schneider U, Howald I, et al. HEAT repeats mediate plasma membrane localization of Tor2p in yeast. J Biol Chem. 2000;275:37011–37020. [DOI] [PubMed] [Google Scholar]
- [21].Martinez Marshall MN, Emmerstorfer-Augustin A, Leskoske KL, et al. Analysis of the roles of phosphatidylinositol-4,5-bisphosphate and individual subunits in assembly, localization and function of Saccharomyces cerevisiae target of rapamycin complex 2. Mol Biol Cell. 2019. Published online: 2019 Apr 10. doi: 10.1091/mbc.E18-10-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Roelants FM, Breslow DK, Muir A, et al. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis Saccharomyces Cerevisiae. Proc Natl Acad Sci USA. 2011;108:19222–19227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sun Y, Miao Y, Yamane Y, et al. Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol Biol Cell. 2012;23:2388–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Niles BJ, Mogri H, Hill A, et al. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA. 2012;109:1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Berchtold D, Piccolis M, Chiaruttini N, et al. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol. 2012;14:542–547. [DOI] [PubMed] [Google Scholar]
- [26].Guerreiro JF, Muir A, Ramachandran S, et al. Sphingolipid biosynthesis upregulation by TOR complex 2-Ypk1 signaling during yeast adaptive response to acetic acid stress. Biochem J. 2016;473:4311–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee YJ, Jeschke GR, Roelants FM, et al. Reciprocal phosphorylation of yeast glycerol-3-phosphate dehydrogenases in adaptation to distinct types of stress. Mol Cell Biol. 2012;32:4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Muir A, Roelants FM, Timmons G, et al. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. Elife. 2015;4:e09336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Riggi M, Niewola-Staszkowska K, Chiaruttini N, et al. Decrease in plasma membrane tension triggers PtdIns(4,5)P2 phase separation to inactivate TORC2. Nat Cell Biol. 2018;20:1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Karuppasamy M, Kusmider B, Oliveira TM, et al. Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2. Nat Commun. 2017;8:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gaubitz C, Prouteau M, Kusmider B, et al. TORC2 structure and function. Trends Biochem Sci. 2016;41:532–545. [DOI] [PubMed] [Google Scholar]
- [33].Wullschleger S, Loewith R, Oppliger W, et al. Molecular organization of target of rapamycin complex 2. J Biol Chem. 2005;280:30697–30704. [DOI] [PubMed] [Google Scholar]
- [34].Tatebe H, Murayama S, Yonekura T, et al. Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. Elife. 2017;6:e19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Casamayor A, Torrance PD, Kobayashi T, et al. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. [DOI] [PubMed] [Google Scholar]
- [36].Yang Q, Inoki K, Ikenoue T, et al. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tabuchi M, Audhya A, Parsons AB, et al. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol Cell Biol. 2006;26:5861–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fadri M, Daquinag A, Wang S, et al. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol Biol Cell. 2005;16:1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Roelants FM, Baltz AG, Trott AE, et al. A protein kinase network regulates the function of aminophospholipid flippases. Proc Natl Acad Sci USA. 2010;107:34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rispal D, Eltschinger S, Stahl M, et al. Target of rapamycin complex 2 regulates actin polarization and endocytosis via multiple pathways. J Biol Chem. 2015;290:14963–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kliegman JI, Fiedler D, Ryan CJ, et al. Chemical genetics of rapamycin-insensitive TORC2 in S. Cerevisiae Cell Rep. 2013;5:1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bourgoint C, Rispal D, Berti M, et al. Target of rapamycin complex 2-dependent phosphorylation of the coat protein Pan1 by Akl1 controls endocytosis dynamics. Saccharomyces Cerevisiae J Biol Chem. 2018;293:12043–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Muir A, Ramachandran S, Roelants FM, et al. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. Elife. 2014;3:e03779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fröhlich F, Olson DK, Christiano R, et al. Proteomic and phosphoproteomic analyses of yeast reveal the global cellular response to sphingolipid depletion. Proteomics. 2016;16:2759–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Locke MN, Thorner J. Rab5 GTPases are required for optimal TORC2 function. J Cell Biol. 2019;218:961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yang H, Jiang X, Li B, et al. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature. 2017;552:368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Avruch J, Long X, Lin Y, et al. Activation of mTORC1 in two steps: rheb-GTP activation of catalytic function and increased binding of substrates to raptor. Biochem Soc Trans. 2009;37:223–226. [DOI] [PubMed] [Google Scholar]
- [48].Nicastro R, Sardu A, Panchaud N, et al. The architecture of the Rag GTPase signaling network. Biomolecules. 2017;7:E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sancak Y, Sabatini DM. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem Soc Trans. 2009;37:289–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tatebe H, Morigasaki S, Murayama S, et al. Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr Biol. 2010;20:1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Plemel RL, Lobingier BT, Brett CL, et al. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lo SY, Brett CL, Plemel RL, et al. Intrinsic tethering activity of endosomal Rab proteins. Nat Struct Mol Biol. 2012;19:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pearce LR, Komander D, Alessi DR. The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol. 2010;11:9–22. [DOI] [PubMed] [Google Scholar]
- [54].Leroux AE, Schulze JO, Biondi RM. AGC kinases, mechanisms of regulation and innovative drug development. Semin Cancer Biol. 2018;48:1–17. [DOI] [PubMed] [Google Scholar]
- [55].Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–w303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li L, Kim E, Yuan H, et al. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–19709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Garrenton LS, Stefan CJ, McMurray MA, et al. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci, USA. 2010;107:11805–111810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gallego O, Betts MJ, Gvozdenovic‐Jeremic J, et al. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol Syst Biol. 2010;6:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vonkova I, Saliba AE, Deghou S, et al. Lipid cooperativity as a general membrane-recruitment principle for PH domains. Cell Rep. 2015;12:1519–1530. [DOI] [PubMed] [Google Scholar]
- [61].Morales-Johansson H, Jenoe P, Cooke FT, et al. Negative regulation of phosphatidylinositol 4,5-bisphosphate levels by the INP51-associated proteins TAX4 and IRS4. J Biol Chem. 2004;279:39604–39610. [DOI] [PubMed] [Google Scholar]
- [62].Schmidt O, Weyer Y, Fink MJ, et al. Regulation of Rab5 isoforms by transcriptional and post-transcriptional mechanisms in yeast. FEBS Lett. 2017;591:2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Barr FA. Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol. 2013;202:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 2014;6:a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. Embo J. 1994;13:1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Toshima JY, Nishinoaki S, Sato Y, et al. Bifurcation of the endocytic pathway into Rab5-dependent and -independent transport to the vacuole. Nat Commun. 2014;5:3498. [DOI] [PubMed] [Google Scholar]
- [67].Gerrard SR, Bryant NJ, Stevens TH. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Peplowska K, Markgraf DF, Ostrowicz CW, et al. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–750. [DOI] [PubMed] [Google Scholar]
- [69].Cabrera M, Arlt H, Epp N, et al. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J Biol Chem. 2013;288:5166–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Buvelot Frei S, Rahl PB, Nussbaum M, et al. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10 and Ypt11. Mol Cell Biol. 2006;26:7299–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chavrier P, Parton RG, Hauri HP, et al. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. [DOI] [PubMed] [Google Scholar]
- [72].Singer-Kruger B, Stenmark H, Zerial M. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J Cell Sci. 1995;108:3509–3521. [DOI] [PubMed] [Google Scholar]
- [73].Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. [DOI] [PubMed] [Google Scholar]
- [74].Lachmann J, Barr FA, Ungermann C. The Msb3/Gyp3 GAP controls the activity of the Rab GTPases Vps21 and Ypt7 at endosomes and vacuoles. Mol Biol Cell. 2012;23:2516–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346:561–576. [PMC free article] [PubMed] [Google Scholar]
- [76].Agarwala AK. How to explain the AKT phosphorylation of downstream targets in the wake of recent findings. Proc Natl Acad Sci USA. 2018;115:E6099–E6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lučić I, Rathinaswamy MK, Truebestein L, et al. Conformational sampling of membranes by Akt controls its activation and inactivation. Proc Natl Acad Sci USA. 2018;115:E3940–E3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ebner M, Sinkovics B, Szczygieł M, et al. Localization of mTORC2 activity inside cells. J Cell Biol. 2017;216:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Jethwa N, Chung GH, Lete MG, et al. Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J Cell Sci. 2015;128:3456–3465. [DOI] [PubMed] [Google Scholar]
- [80].Goto-Silva L, McShane MP, Salinas S, et al. Retrograde transport of Akt by a neuronal Rab5-APPL1 endosome. Sci Rep. 2019;9:2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kovalski JR, Bhaduri A, Zehnder AM, et al. The functional proximal proteome of oncogenic Ras includes mTORC2. Mol Cell. 2019;73:830–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Esters H, Alexandrov K, Constantinescu AT, et al. High-resolution crystal structure of S. cerevisiae Ypt51(DeltaC15)-GppNHp, a small GTP-binding protein involved in regulation of endocytosis. J Mol Biol. 2000;298:111–121. [DOI] [PubMed] [Google Scholar]
- [84].Yu Y, Li S, Xu X, et al. Structural basis for the unique biological function of small GTPase RHEB. J Biol Chem. 2005;280:17093–17100. [DOI] [PubMed] [Google Scholar]


