Abstract
Rationale:
Truncation mutations in the MYBPC3 gene, encoding for cardiac myosin-binding protein C (MyBP-C), are the leading cause of hypertrophic cardiomyopathy (HCM). Whole heart, fiber and molecular studies demonstrate that MyBP-C is a potent modulator of cardiac contractility, but how these mutations contribute to HCM is unresolved.
Objectives:
To readdress whether MYBPC3 truncation mutations result in loss of MyBP-C content and/or the expression of truncated MyBP-C from the mutant allele and determine how these mutations effect myofilament sliding in human myocardium.
Methods and Results:
Septal wall tissue samples were obtained from HCM patients undergoing myectomy (n=18) and donor controls (n=8). The HCM samples contained 40% less MyBP-C and reduced levels of MyBP-C phosphorylation, when compared to the donor control samples using quantitative mass spectrometry. These differences occurred in the absence of changes in the stoichiometry of other myofilament proteins or production of truncated MyBP-C from the mutant MYBPC3 allele. The functional impact of MYBPC3 truncation mutations on myofilament sliding was determined using a total internal reflection microscopy (TIRFM) single particle assay. Myosin-thick filaments containing their native complement of MyBP-C, and actin-thin filaments decorated with the troponin/tropomyosin calcium regulatory proteins, were isolated from a subgroup of the HCM (n=4) and donor (n=5) heart samples. The maximal sliding velocity of native thin filaments was enhanced within the C-zones of the native thick filaments isolated from the HCM samples, when compared to velocity within the C-zones of thick filaments isolated from the donor samples. Analytical modeling demonstrated that the 40% reduction in MyBP-C content was sufficient to enhance the myofilament sliding velocity, as observed in the TIRFM assay.
Conclusions:
HCM-causing MYBPC3 truncation mutations result in a loss of MyBP-C content that enhances maximal myofilament sliding velocities, only where MyBP-C is localized within the C-zone. These findings support therapeutic rationale for restoring normal levels of MyBP-C and/or dampening maximal contractile velocities for the treatment of human HCM.
1. Introduction
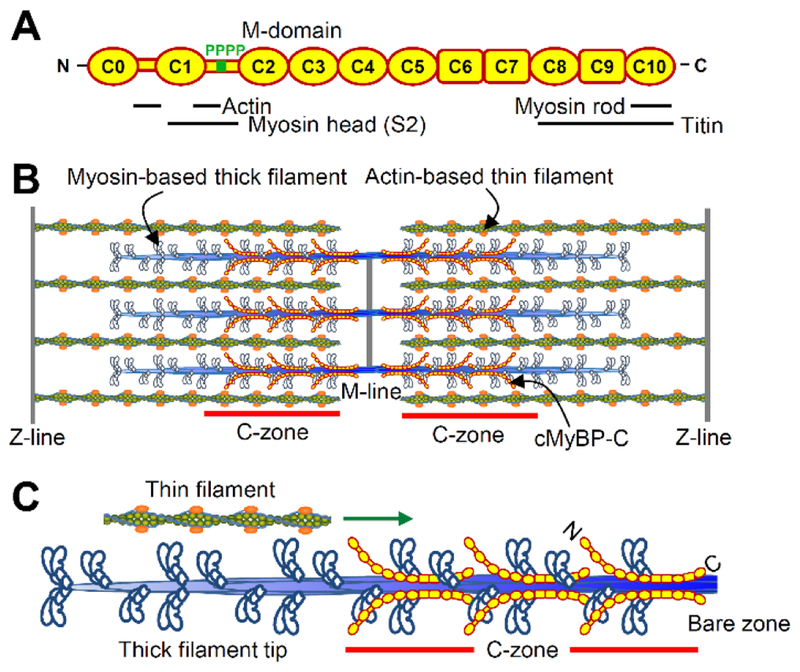
Heterozygous truncation mutations in the MYBPC3 gene are the leading cause of hypertrophic cardiomyopathy (HCM), affecting 1 in 500 individuals (1). The product of the MYBPC3 gene, cardiac myosin-binding protein C (MyBP-C), is a 140-kDa immunoglobulin protein superfamily member (Fig. 1A), and a key component of the sarcomere, being parallel arrays of myosin-based thick and actin-based thin filaments, necessary for myocardial contraction (Fig. 1B). Cardiac MyBP-C is comprised of eleven Ig- or Fn-like domains, a Pro-Ala linker between C0 and C1, and the M-domain, a flexible hinge between C1 and C2 that contains phosphorylatable serines. MyBP-C’s C-terminal domains tether MyBP-C to the myosin rod (2) while its N-terminal domains extend away from the thick filament backbone (3) and interact with actin and/or the myosin head (Fig. 1A). Studies have demonstrated that MyBP-C’s N-terminal domains are capable of initiating thin filament sliding at low calcium levels during the early phase of cardiac contraction and modulating maximal thin filament sliding velocities (4–7). While these mechanistic phenomena have been observed to occur within the C-zones of native mouse thick filaments (8, 9), similar studies have not been carried out with thick filaments isolated from human myocardium. Species dependent differences in MyBP-C’s N-terminal domain interactions (7, 10, 11) and heartrate, allow for the possibility that the molecular mechanisms of MyBP-C differ in mouse and human myocardium.
Fig. 1.
MyBP-C within the cardiac muscle sarcomere. (A) MyBP-C being comprised of eleven immunoglobulin- (oval) or fibronectin-like (square) domains, a Pro-Ala linker between C0 and C1, and a flexible hindge between C1 and C2 containing four potential phosphorylation sites (i.e. M-domain). Binding partner interactions shown below. (B) Interdigitating myosin-thick and actin-thin filaments form the sarcomere. MyBP-C is localized to the C-zones. (C) A ~250 nm thin filament sliding along one-half of a thick filament, as observed in the single molecule TIRFM assay.
While most human HCM-causing sarcomere gene mutations encode full-length proteins with single amino acid substitutions, ~90% of MYBPC3 mutations create a premature termination codon in one allele (12). These heterozygous MYBPC3 truncation mutations were expected to produce mutant MyBP-C that lacks the C-terminal domains necessary for localization within the thick filament C-zones (Fig. 1C). The majority of evidence suggests that HCM-causing MYBPC3 truncation mutations result in a reduction in MyBP-C content (13–15) and despite significant efforts, truncated MyBP-C has not been detected in human myocardium (13–17). Such studies have used staining for total protein content on acrylamide gels or western blotting with MyBP-C domain specific antibodies. To readdress whether human MYBPC3 truncation mutations result in a reduction in MyBP-C content and/or the production of truncated MyBP-C, interventricular septal wall myectomy samples were obtained from a cohort of genotyped HCM patients with MYBPC3 truncation mutations (n=18) and explanted donor controls (n=8). Quantitative mass spectrometry demonstrated that the HCM samples contained 40% less MyBP-C than the donor controls, with no detectable expression of truncated MyBP-C. To determine the impact of the MYBPC3 truncation mutations on actomyosin-based myofilament sliding, native myosin-thick filaments, containing endogenous levels of MyBP-C, and native actin-thin filaments, decorated with the calcium dependent troponin/tropomyosin regulatory proteins, were isolated from a subset of the human heart samples (n=9 samples). The sliding of short, ~250 nm, shards of the native thin filaments, was observed on native thick filaments in areas with only myosin (thick filament tips) or a mixture of myosin and MyBP-C (C-zones) (Fig. 1C). The experimental results combined with analytical modeling demonstrate that reduced MyBP-C content enhances myofilament contractile mechanics within the C-zone of thick filaments isolated from human HCM hearts with septal wall thickening.
2. Methods (See also: online supplement)
2.1. Human heart tissue procurement
Myocardial tissue was obtained from the interventricular septum of subjects with HCM at the time of surgical myectomy to treat left ventricular outflow tract obstruction (n=18). HCM was diagnosed on the basis of standard criteria and in the absence of any cause for secondary hypertrophy (18). Genetic testing was performed following Clinical Laboratory Improvement Amendments in certified laboratories using a standard 18 HCM gene panel. Interventricular septal tissue was obtained from gift of life donor hearts at time of explant (n=8). Donor hearts were perfused with cardioplegia solution before removal. All tissues were snap-frozen in liquid nitrogen immediately after excision. Patient demographic and clinical data were recorded at the time of tissue collection. This study had the approval of the University of Michigan Institutional Review Board and subjects gave informed consent.
2.2. Mouse heart tissue
Apical tissue was collected from a well characterized mouse knock-in model harboring a homozygous mutation in MYBPC3 that caused C-terminal truncation and a wild-type (WT) mouse heart. Both mice were of the same age (3 months), sex (male) and strain (FVB) to minimize biological variability. The animals were euthanized in a carbon dioxide (CO2) chamber by slow flow of CO2 (10–30% of chamber volume per minute) followed by continued exposure for 15–30 minutes after breathing had stopped. The hearts were excised and snap-frozen in liquid nitrogen. All animal protocols were approved by the Institutional Animal Care and Use Committee at University of Cincinnati and were in accordance with the guidelines listed in the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health.
2.3. Quantitative liquid chromatography mass spectrometry
Individual pieces of intact heart muscle (1–2 mg), thick filament preparations and aliquots of C0C3 MyBP-C fragment, were solubilized in RapiGest SF Surfactant (Waters Corporation). The samples were reduced, alkylated and digested with trypsin (Promega). The resultant peptides were separated by ultra-high pressure liquid chromatography (LC) and analyzed using a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (MS). Peptide LC peak areas within each sample were identified and quantified using the Proteome Discoverer 2.2 (PD 2.2) software package with the Minora Feature Detector (Thermo Fisher Scientific) enabled. Peptide LC peak areas were normalized to account for differences in the amount of sample digested and loaded onto the LC column (19, 20). Normalization was achieved by dividing each peptide LC peak area by the average peptide LC peak area of the top 15 peptides shared between the α- (MYH6) and β-myosin heavy chain (MYH7) isoforms. These were the only sarcomeric myosin heavy chain isoforms expressed in the hearts. Data normalized using peptide LC peak areas from titin (TTN) did not differ. The relative abundance of proteins between sample groups were determined using a pairwise ratio analysis of the abundances of the top (maximum 15) ionizing peptides produced from the tryptic digestion of each protein. Relative molar abundances of individual proteins and protein isoforms were estimated from the average abundance of the top (maximum 3) ionizing tryptic peptides originating from the cleavage of each protein or protein isoform. This approach partly accounts for differences in peptide ionization efficiencies and allows for determination of molar protein abundances (20). Detailed methods for each analysis are described in the results and online supplement.
2.4. Protein preparation
Myosin-thick filaments, actin-thin filaments and monomeric myosin were isolated from the human heart samples. A human, truncated MyBP-C N-terminal protein fragment comprised of the C0 to C3 domains (Fig. 1A) was bacterially-expressed. Detailed methodology is described in the online supplement.
2.5. Fluorescence image acquisition
All filament sliding assays were performed at room temperature (22 ± 1°C) using a Nikon Eclipse Ti-U inverted microscope equipped with a Plan Apo 100X 1.49 N.A. oil emersion lens for through-the-objective total internal reflection microscopy (TIRFM) or epifluorescence microscopy as described (8, 9). Briefly, either a 532-nm diode-pumped, solid-state, 50-mW laser (Lasever Inc.) or Lumen 200-W metal arc lamp (Prior Scientific) were used for excitation. Images were collected using an intensified high-resolution XR/Turbo-Z, 10-bit digital camera (Stanford Photonics).
2.6. Quantification of native thin filament motion on native thick filaments
The movement of short, ~250 nm, native actin-thin filament shards (Fig. 1C), isolated from multiple pieces of a single donor heart were quantified on myosin thick-filaments isolated from 4 HCM (Table S1, Samples A-C and K) and 4 donor heart samples by TIRFM. The single particle imaging techniques were previously applied to myofilaments isolated from mouse hearts (8, 9). Briefly, the thick filaments from each human sample were non-specifically adhered to the surface of a glass flow-cell. The thin filaments were fluorescently-labeled with TRITC-phalloidin, observed landing on the thick filaments, and being translocated toward their bare zones using TIRFM (Fig. 1C). Rapid imaging and Gaussian fitting of the fluorescence allowed for high temporal (8.3 ms) and spatial (±20 nm) resolution of thin filament motion (8, 9). The average fraction of thin filaments moving (±SEM) on thick filament and thin filament sliding velocities (±SEM) at pCa 4 were determined from individual experiments. Two independent thick filament isolations and experiments were performed for each of the 4 HCM (Table S1, Samples A-C and K) and donor heart samples over the full range of calcium concentrations as described in the online supplement. The data were fitted with sigmoidal dose-response curves with variable slopes and statistical significance was determined by the sum-of-squares F test.
2.7. Modeling of thin filament motion on thick filaments
An analytic model accounting for the thick and thin filament spatial geometries in the assay (8) was implemented to predict the effect of reduced MyBP-C content on thin filament sliding velocity along the thick filaments (Fig. 1C), as described in the online supplement.
2.8. Quantification of thin filament motion on monomeric myosin
The movement of actin-thin filaments isolated from the same 4 HCM (Table S1, Samples A-C and K) and 4 donor heart samples used for the thick filament experiments were quantified on a surface of depolymerized monomeric myosin by epifluorescence as described in the online supplement. For each experiment, the myosin was isolated from multiple pieces of a single donor heart as described in the online supplement. The average fraction of thin filaments moving (±SEM) and thin filament sliding velocities (±SEM) were determined from the motion on the depolymerized myosin. Four independent thin filament isolations and experiments were performed for each of the 4 samples in the HCM and donor groups. The data were fitted with sigmoidal dose-response curves with variable slopes and statistical significance was determined by the sum-of-squares F test.
3. Results
3.1. Patient demographic, clinical, and genetic data
Group summaries of demographic and clinical data for the HCM and donor subjects are shown in (Table 1). Individual MYBPC3 truncating mutations are provided (Table S1). No subjects were related. Age and gender were not significantly different between the control and HCM groups. Septal wall thickness and ejection fractions were higher (P<0.05) in the HCM group when compared to the control group, as expected based on the severity of the disease and need for septal myectomy.
Table 1.
Patient-specific demographic and clinical data.
| Patient group | Age | Gender (% male) | Septal thickness (mm) | LV ejection fraction (%) | LV outflow tract gradient (mmHg) |
|---|---|---|---|---|---|
| HCM (n=18) | 41 ± 3 | 44 | 25 ± 3* | 73 ± 2* | 76 ± 9 |
| Donor (n=8) | 52 ± 6 | 38 | 10 ± 0.6 | 65 ± 2 | N/A |
P<0.05 for difference between HCM and donor groups.
3.2. Heterozygous MYBPC3 truncation mutations result in a reduction in MyBP-C content in human HCM myocardium
An average of 11,768 ± 238 tryptic peptides were identified and quantified in each of the human heart samples. The summed abundance of the peptides coming from the thick filament proteins, thin filament proteins, and titin (Fig. 2A) accounted for 48.0 ± 1.1% and 45.0 ± 1.0% of the total summed peptide abundances in the donor and HCM heart sample groups, respectively. The similarity between HCM and donor groups demonstrated that the sarcomeric protein content was proportional to the hypertrophy. Therefore, the increase in septal wall thickness was not merely due to fibrosis.
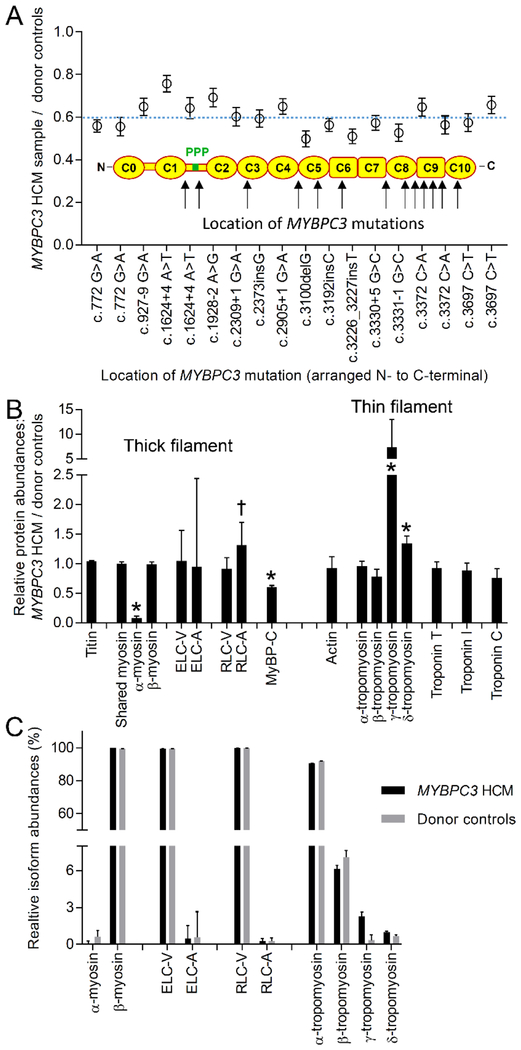
Fig. 2.
Abundances of sarcomeric contractile proteins in HCM and donor control samples. (A) Relative abundance of MyBP-C in the individual HCM samples when compared to the donor control group by pairwise analysis using the top 15 ionizing peptides in the MS data. Blue dashed line indicates the group mean. (B) Relative abundances of sarcomeric proteins in the HCM samples when compared to the donor control group by pairwise analysis from the top (maximum 15) ionizing peptides. *P<0.01; †P<0.05. (C) Relative protein isoform abundances calculated from the averages of the top (maximum 3) ionizing peptides for each protein isoform in the HCM and donor sample groups.
The donor heart samples contained 11.4 ± 0.4 moles of peptides shared between α- and β-myosin heavy chain isoforms for each mole of MyBP-C, which corresponds to 5.7 molecules of myosin heavy chain for each molecule of MyBP-C. This stoichiometry was similar to that observed in mouse thick filaments (8) and expectations from structural studies (21). The relative abundance of MyBP-C (P<0.01) was reduced by 40% in the HCM sample group when compared to the control group (Fig. 2A) using a pairwise ratio analysis of the top 15 ionizing MyBP-C tryptic peptides in each sample. The magnitude of the reduction in MyBP-C content (24–50% reduction) did not correlate with the location of the MYBPC3 mutation in the individual HCM samples (Fig. 2A). This demonstrated that all of the truncation mutations examined reduced MyBP-C content to similar levels despite there being differences in the location of the truncation within the MYBPC3 gene.
3.3. MYBPC3 truncation mutations have minimal impact on the expression of other thick and thin filament proteins
The relative expression of the other thick and thin filament proteins in the HCM and donor sample groups (Fig. 2B) and the molar abundance of protein isoforms (Fig. 2C) were quantified by MS. The lack of difference in the relative abundances of titin, shared myosin heavy chains and actin between the HCM and donor groups demonstrated that there were no overall differences in the titin, thick and thin filament stoichiometry (Fig. 2B). Changes in the relative abundances of α-myosin heavy chain (MYH6; P<0.01), atrial myosin regulatory light chain (MYL7; P<0.05), γ-tropomyosin (TPM3; P<0.01) and δ-tropomyosin (TPM4; P<0.01) protein isoforms were observed between sample groups (Fig. 2A). Such changes were consistent with reports from end-stage failing human heart samples (22–24). However, unlike MyBP-C, which expresses a single isoform in cardiac muscle, α-myosin heavy chain, atrial myosin regulatory light chain, γ-tropomyosin and δ-tropomyosin are co-expressed with other isoforms. The overall abundances of these specific isoforms were <2.5% that of the major isoform for each protein in the HCM and donor groups (Fig. 2C) when quantified using the top (maximum 3) ionizing tryptic peptides from each isoform (20). Therefore, although the changes in relative abundance of the isoforms were significant between groups (Fig. 2B), the overall abundances (Fig. 2C) suggested that the relative changes would have minimal impact on the composition of the thick and thin filaments in either group.
To validate the mass spectrometry-based approach, the same analytical routine was applied to triplicate samples from the apex of a mouse heart with a well-characterized homozygous MYBPC3 truncation mutation (25) and a wild-type control (Fig. S1). MyBP-C was nearly ablated in the MYBPC3 mutant mouse (0.01% of wild-type). Differences (P<0.01) were observed in the abundances of both α- and β-myosin heavy chain isoforms and skeletal myosin regulatory light chain isoform (MYL1) in the MYBPC3 mutant mouse samples when compared to the wild-type mouse samples (Fig. S1A). Whereas the adult human heart expresses 99% β-myosin heavy chain (Fig. 2C) the wild-type mouse heart expresses 99% α-myosin heavy chain (Fig. S1B). The significant changes in the relative abundances of α- and β-myosin heavy chain isoforms in the mouse samples (Fig. S1A) were indicative of a shift in expression from 99% α-myosin heavy chain to 39% α- and 61% β-myosin heavy chains in the MYBPC3 mutant heart samples (Fig. S1B). These changes in expression were similar to findings with standard gel-based methodologies (25, 26).
3.4. Truncated MyBP-C is not expressed from the mutant MYBPC3 allele in HCM hearts at detectable levels
The genetic sites of truncation within the 18 HCM samples had a broad distribution (amino acid 257 to 1232) that would produce truncated MyBP-C with masses ranging from 22.5 to 135.3 kDa (Table S1). All of the truncated MyBP-C products expected from the mutant MYBPC3 allele would lack portions of the C-terminal domain that tether MyBP-C to the thick filament backbone within the C-zone (Fig. 1). A subset of the truncated MyBP-C expressed from the mutant MYBPC3 alleles would produce unique C-terminal peptides when digested with trypsin during the sample preparation for LCMS. To determine whether these unique peptides were present in the digested HCM samples, a custom database was generated that contained the unique peptide sequences for the truncated MyBP-C products. The LCMS data files were searched against the database using SEQUEST and manually searched using the exact peptide masses and theoretical spectra. None of the unique C-terminal peptides were found in the analyses.
A second, quantitative mass-balance approach that did not require the identification of the unique peptides was also used to determine whether truncated MyBP-C is expressed in the human HCM heart samples. A pairwise peptide abundance ratio analyses was carried out using 76 MyBP-C peptides identified in each of the HCM and donor heart samples. These 76 peptides covered 68% of MyBP-C’s sequence. The abundance of each tryptic peptide LC peak area in the HCM samples was divided by the median abundance of the same peptide LC peak area in the donor sample group. The resultant values for each peptide were then used to determine a mean N- to C-terminal peptide ratio for each HCM sample. These ratios were generated from the abundances of tryptic peptides N- (10 to 71 peptides) and C-terminal (3 to 64 peptides) to the genetic sites of truncation in each sample (Table S1). Neither the N- to C-terminal ratios for the HCM group mean (Fig. 3A) (1.00 ± 0.05; ±SEM) nor individual HCM samples (Fig S2A; ±SD) differed from 1. The large standard error of mean for the individual measurements (Fig. S2A) was due to the necessary inclusion of peptides with low ionization efficiencies (Figs. S2B and S2C) and variance in digestion efficiencies for single samples. This variance was minimized by averaging the individual ratios for the 18 samples in the HCM group (Fig. 3A).
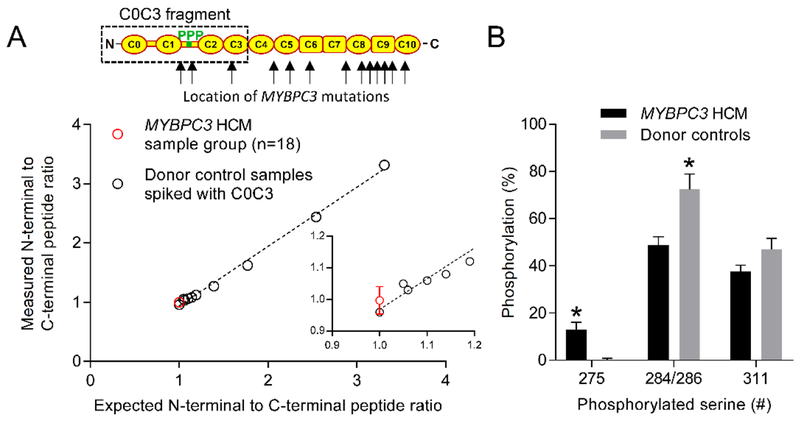
Fig. 3.
Quantification of the abundance of truncated MyBP-C and phosphorylation in the human heart samples. (A) Red data point (±SEM) is the grouped HCM N- to C-terminal MyBP-C peptide ratio determined from peptides N- and C-terminal to the first amino acid altered by each MYBPC3 mutation in the HCM samples. Black data points are N- to C-terminal peptide ratios generated from donor controls samples spiked with increasing concentrations of a bacterially expressed human C0C3 MyBP-C protein fragment. (B) Percent phosphorylation determined for serines 275, 284/286 and 311 in the HCM and donor samples. *P<0.01
To validate this approach and determine the limit of detection empirically, the same analytic routine was applied to tryptic peptides from the digestion of a donor heart sample that was spiked with aliquots of tryptic peptides from the digestion of a bacterially expressed human C0C3 MyBP-C fragment. A linear increase was observed in the N- to C-terminal ratio (Fig. 3A). The root mean square error being 0.059 was indicative of a <10% limit of detection (27). This demonstrated that >90% of the tryptic peptides found in the HCM samples were generated from the digestion of full-length MyBP-C. The expression of the truncated MyBP-C in the HCM samples at levels below the detection limit of the assay would be <6% that of full-length MyBP-C in the donor heart samples, because the HCM samples only contain 60% full-length MyBP-C (Fig. 2A).
3.5. Phosphorylation of MyBP-C serine 284/286 was reduced in the human HCM heart samples
Thick filament samples were solubilized in RapiGest SF Surfactant (Waters Corporation), reduced, alkylated, digested with trypsin, and analyzed by LCMS. Multiple peptides containing phosphate on serine 275, 284, 286 or 311 being: 273RTSpLAGGGR281, 283ISpDSHEDTGILDFSSLLK300, 282RISpDSHEDTGILDFSSLLK300,282RISDSpHEDTGILDFSSLLK300 and 307TPRDSpKLEAPAEEDVWEILR326, where p is phosphate, were identified in MS/MS fragmentation spectra. All four of these sites were within the M-domain and no other phosphorylation sites were identified in other regions of MyBP-C. Phosphate could be assigned to serine 284 and 286 in the MS/MS fragmentation spectra (Fig. S3), as previously reported in failing hearts (28). However, manual interrogation of the MS/MS spectra suggested that the fragmentation pattern with phosphate on serine 284 (Fig. S3A) was nearly identical to that with phosphate on serine 286 (Fig. S3B). Therefore, the position of phosphate was not precisely localized in the analyses and possibly only exists on serine 284 as previously reported for kinase-treated bacterially expressed fragments (29). Another phosphorylation site, being serine 304 has also been observed within the M-domain of mouse MyBP-C (8) and kinase-treated bacterially expressed fragments by mass spectrometry (29). Four phosphorylation sites have been observed in human HCM heart samples by phosphate affinity SDS-PAGE (30). However, the short 303DSpFR306 or 302RDSpFR306 tryptic phosphopeptides containing serine 304 were not detected in any of the analyses.
The abundance of phosphorylation at each site was determined by mass-balance from the loss in abundance of the non-phosphorylated peptides in each sample, as previously described (8). Ratios were generated from peptide LC peak areas of the corresponding non-phosphorylated peptides in the human heart and bacterially expressed human C0C3 MyBP-C samples. Noting that the bacterially expressed C0C3 samples were fully dephosphorylated. Phosphorylation of serine 275 was increased while serine 284/286 was decreased in the HCM samples when compared to the control group (Fig. 3B). The decrease in phosphorylation of serine 284/286 was consistent with previous studies (14, 30).
3.6. Maximal myofilament sliding velocities are enhanced within the C-zone of native thick filaments isolated from HCM heart samples
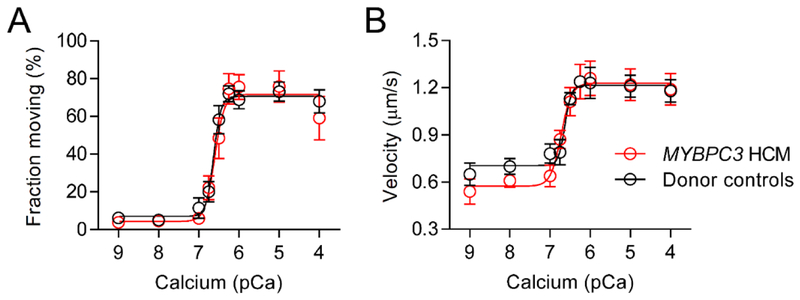
To determine the impact of MYBPC3 truncation mutations on actomyosin function at the myofilament level, native myosin-thick filaments were mechanochemically isolated from 4 HCM (Table S1, Samples A-C and K) and 4 donor human heart samples. Neither the average MyBP-C content in the 4 HCM samples (63 ± 5%) nor the myosin heavy chain to MyBP-C molar ratio in the 4 donor samples (11.3 ± 0.7) differed from the group means (60 ± 2% and 11.4 ± 0.4). Native actin-thin filaments were isolated from pieces of a single donor heart sample, fluorescently-labeled with TRTC-phalloidin and fragmented to produced ~250 nm shards (8). The short native thin filaments were observed sliding on the thick filaments (Fig. 1C) with TIRFM as previously described for mouse filaments (8, 9). At high calcium levels (pCa 4) the average number of thin filaments observed sliding on thick filament isolated from HCM (28 ± 5) and donor (29 ± 6) hearts in each experiment, did not differ. In both groups, the fraction of thin filaments moving decreased sigmoidally in a calcium dependent manner (Fig. 4A). This demonstrated that calcium tightly regulated myosin’s access to actin, as expected for actin filaments decorated with the troponin and tropomyosin calcium regulatory proteins (31). The overall calcium sensitivity (pCa50) was slightly decreased (P<0.05) on thick filaments isolated from the HCM heart samples when compared to those from donor control samples (Fig. 4A).
Fig. 4.
Thin filament motion on thick filaments isolated from HCM (n=4) and donor (n=4) heart samples. (A) Fraction of thin filaments moving on thick filament isolated from the donor (black; pCa50=6.6±0.1) and HCM heart samples (red; pCa50=6.3±0.1) versus calcium concentration. The pCa50 differed, P<0.05. (B and C) Displacement versus time traces for thin filaments sliding on thick filament isolated from (B) donor and (C) HCM heart samples at pCa 4 demonstrating 2 phases of velocity (n=15 filaments). (D) Average ± SEM velocities of thin filaments sliding on the tips and within the C-zones of thick filaments isolated from donor (black, n=50 filaments) and HCM (red, n=57 filaments) heart samples. Thick filament tip and C-zone velocities predicted from an analytical model using donor heart data with a theoretical 40% reduction in MyBP-C content. *P<0.01
At maximal calcium levels (pCa 4), the majority of the thin filament motion on the thick filaments isolated from the donor (86%, Fig. 4B) and HCM samples (71%, Fig. 4C) was characterized by two distinct phases of velocity. In both sample groups, the second phase of velocity was slower, indicative of motion within the thick filament Czone (Fig. 1C), as demonstrated using mouse thick filaments (8, 9). While the velocity on the tips of the thick filaments (Fig. 1C) isolated from the donor and HCM heart samples was similar between groups, the velocity within the C-zone was enhanced (P<0.01) on thick filaments isolated from the HCM samples when compared to those isolated from donor samples (Fig. 4D). This demonstrated that the functional impact of the MYBPC3 truncation mutations in the HCM heart samples was localized to the thick filament C-zone.
To determine whether the differences in myofilament function in the HCM and donor heart samples were restricted to the thick filament C-zone, native thin filaments were isolated from pieces of the same 4 HCM (Table S1, Samples A-C and K) and 4 donor heart samples. The native thin filaments were fluorescently-labeled with TRITC-phalloidin and observed sliding on surface coated with randomly oriented monomeric myosin molecules isolated from pieces of a single donor heart. Both the fraction of filaments moving (Fig. 5A) and their sliding velocities (Fig. 5B) increased sigmoidally with calcium concentration. Neither calcium sensitivity (pCa50), the fraction of filaments moving, nor velocity over the range of calcium concentrations tested differed between groups. This demonstrated that the native thin filaments isolated from the HCM and donor hearts were functionally equivalent in the myofilament sliding assay.
Fig. 5.
Motion of thin filaments isolated from HCM (n=4) and donor (n=4) heart samples on monomeric myosin isolated from a single donor heart sample. (A) Fraction of thin filaments moving and (B) sliding velocities versus calcium concentration.
3.7. Reduction in MyBP-C content enhances myofilament sliding velocities within the C-zone
An analytic model was implemented to determine whether a 40% reduction in MyBP-C content was sufficient to enhance myofilament sliding velocities within the C-zone. The model accounted for geometrically constrained perturbations in the mechanical interactions between the 250 nm thin filaments and ensembles of myosin and MyBP-C molecules that interact with the thin filament as it slides along the thick filament (Fig. S4A) (8). The model recapitulated the two phases of velocity (2.1 μm/s, fast, 0.9 μm/s, slow) observed on the thick filaments isolated from the donor heart samples (Fig. S4B). Reduction of MyBP-C content by 40% in the model enhanced the second phase of velocity (Figs. S4B and S4C, 0.9 to 1.4 μm/s) to the speed observed in the C-zone of thick filaments isolated from the HCM heart samples (Fig. 4D, 1.4 μm/s). This supported that the 40% reduction in MyBP-C content in the thick filaments isolated from the HCM heart samples was sufficient to enhance maximal myofilament sliding velocities within the C-zone.
4. Discussion
Mechanistic studies using mouse model systems have enriched our understanding of the structure and function of MyBP-C, but why heterozygous MYBPC3 truncation mutations result in human HCM remains unresolved. Species-dependent differences in MyBP-C’s domain specific interactions (7, 10, 11) and rates of actomyosin contraction, raise the possibility that MyBP-C’s function may differ in mouse and human myocardium. Despite such differences, our findings suggest that MyBP-C plays similar mechanistic roles in the modulation of myofilament sliding in mice and humans.
A quantitative mass spectrometry based approach was used to demonstrate that HCM causing heterozygous truncation mutations in the MYBPC3 gene result in a 40% reduction in MyBP-C content (Fig. 2A) in adult diseased myocardium. This was consistent with reports from other groups for similar cohorts of patients (13–15) but differed from a report by Helms et al. (17) using western blot analysis. This reduction in MyBP-C content appears to be restricted to samples with MYBPC3 truncation mutations rather than a product of adverse hypertrophic remodeling (13, 15, 17). The lack of significant difference in MyBP-C content in a similar cohort of HCM and donor heart samples reported by Helms et al. (17) may have arose from the use of GAPDH for normalization or from noise associated with western blot analysis. Major strengths of the MS approach are the lack of protein fractionation in the MS preparation and the use of peptides shared between the α- and β-myosin heavy chain isoforms for normalization. The approach ensures that all of the MyBP-C expressed within the heart samples is detected in the analyses, regardless of localization, and its content is determined relative to that of myosin, to which MyBP-C is strongly bound in the sarcomere (Fig. 1). The narrow, 24–50%, distribution in reduction of MyBP-C content in each of the HCM samples regardless of position of the MYBPC3 mutation (Fig. 2A), suggests these findings can be extrapolated to other MYBPC3 truncation mutations that cause HCM. It should be noted that mice with heterozygous MYBPC3 mutations demonstrate minimal, or no loss in MyBP-C content or the development of cardiac hypertrophy (32–34), unless challenged with transverse aortic constriction (34). Due to the lack of access to cardiac tissue from healthy individuals carrying similar MYBPC3 mutations, it is unknown whether carriers have reduced MyBP-C content or the reduction occurs with the progression of HCM.
The 40% reduction in MyBP-C content observed in the HCM hearts occurred without detectable expression of truncated MyBP-C from the mutant MYBPC3 gene. The lack of detectable truncated MyBP-C suggests that either no protein is synthesized from the mutant MYBPC3 allele, perhaps through nonsense mediated mRNA decay, or alternatively, the protein is synthesized but susceptible to rapid degradation. Based on the limit of detection in the MS assay (Fig. 3A), if truncated MyBP-C were expressed in the HCM myocardium, the concentration would be <0.4 μM, assuming 80 μM myosin within cardiac muscle tissue (35) and 11.4 to 1 ratio of myosin to MyBP-C. High levelsof expression of truncated MyBP-C fragments have profound effects on cardiac myocyte contractility (36) and cardiac structure and function (37). However, infusion of 1 μM exogenous C0C2 MyBP-C fragment into skinned human ventricular myocytes had no effect on calcium sensitization or maximal force generation at 2.3 μm sarcomere lengths (36). Low, ~7% or ~0.5 μM, expression of a 29 kDa MyBP-C N-terminal fragment in transgenic mouse hearts had no pathological effect on cardiac morphology, structure or function (37). These results collectively suggest the small amount of truncated MyBP-C, possibly expressed in the HCM hearts at levels below the detection limit of the MS assay, would have minimal to no effect on human cardiac structure or function.
To determine the impact of the reduction in MyBP-C content on actomyosin-based myofilament sliding, native myosin-thick filaments containing MyBP-C, and native actin-thin filaments containing troponin/tropomyosin, were isolated from the a subset of the human heart samples. Sliding of these filaments was examined in a single particle biophysical assay. The MyBP-C content in the 4 HCM samples was reduced (63 ± 5%) to a level similar to the group mean (60 ± 2%). Replicate myofilament isolations and biophysical measurements over the entire range of calcium were performed on each sample to account for analytical variability. The ~40% reduction in MyBP-C content in the thick filaments from the HCM heart samples resulted in a slight (P < 0.05) decrease in calcium sensitivity for the fraction of thin filaments sliding when compared to those isolated from donor controls (Fig. 4A). MyBP-C’s ability to enhance calcium sensitivity within the C-zones of mouse thick filaments was previously demonstrated to result from the binding of MyBP-C’s N-terminal domains to the actin thin filament, resulting in the displacement of tropomyosin at submaximal calcium levels (9). In vitro studies demonstrate that human MyBP-C’s N-terminal domains are also capable of displacing tropomyosin to sensitize the thin filament to calcium, albeit utilizing different MyBP-C N-terminal domains (7, 11). Thus, the link between the ~40% loss of MyBP-C content in the HCM hearts and decrease in calcium sensitivity (Fig. 4A) suggests that MyBP-C likely plays similar roles in mouse and human myocardium, aiding in thin filament activation at low calcium levels.
Mouse MyBP-C’s ability to enhance calcium sensitivity and the displacement of tropomyosin from the blocked position on the actin filament under physiological conditions were shown to be increased by MyBP-C dephosphorylation (9, 38, 39). The human HCM samples were dephosphorylated on serine 284/286. Thus, MyBP-C dephosphorylation was anticipated to increase the effective concentration of MyBP-C in the thick filaments isolated from the HCM samples. While dephosphorylation of serine 284/286 may have partially restored MyBP-C’s activating potential at the lowest calcium levels (pCa 7 and 6.75) under pathological conditions, the 40% loss in MyBP-C content in the HCM samples still resulted in an overall loss in calcium sensitivity (Fig. 4A).
Previous studies have demonstrated significant increases in calcium sensitivity of force production for skinned fibers isolated from a similar cohort of HCM heart samples when compared to fibers from donor samples (14) and in engineered heart tissues with MyBP-C truncation mutations (40). It is important to note that the myofilament-sliding assay measures different biophysical mechanics than those probed in the force assays (41). The myofilament assay probes mechanochemical motion generation from a small number of myosin molecular motors and the velocity reaches a maximum at physiological calcium concentrations (9). Force in the skinned fiber assays (14) and engineered heart tissues (40) only begin to rise at a calcium concentration (pCa 6 or greater) that produces ~75% of the maximal myofilament sliding velocity. Unlike velocity, force continues to rise at higher calcium concentrations due to an increasing number of myosin cross-bridges attaching to the thin filament (41). Moreover, a reduction in MyBP-C content in a similar cohort of human samples with MYBPC3 truncation has been demonstrated to reduce the fraction of myosin molecules in the super-relaxed state (15). Disruption of this super-relaxed state may increase the effective concentration of myosin molecules within the thick filament that are available for binding to actin. This may enhance the calcium sensitivity of force production in the muscle fibers lacking MyBP-C. Such biophysical phenomena are not detected in the myofilament sliding assay used in the current study.
At maximal calcium levels (pCa 4) the thin filament displacement trajectories were characterized by two distinct phases of velocity (Fig. 4B and C) as demonstrated for mouse thick filaments (8, 9). The average velocity within the C-zones of the HCM thick filaments was enhanced when compared to that in the C-zones of the donor thick filaments (Fig. 4D). Analytical modeling demonstrated that a 40% reduction in MyBP-C content within the C-zone of the thick filaments was sufficient to enhance the thin filament sliding velocity to that measured in the C-zone of the thick filaments isolated from the HCM heart samples (Fig. 4D). Previous studies have shown that calcium ablates the ability of phosphorylation to tune mouse MyBP-C’s function (39), so the effect of dephosphorylation of MyBP-C serine 284/286 in the HCM hearts was expected to have no effect on this measurement. The strong agreement between the measured and predicted C-zone sliding velocities (Fig. 4D) suggested that calcium would have a similar effect on human MyBP-C, because the change in velocity was solely dependent on MyBP-C content. It was not surprising that the thin filament sliding velocities on the tips of the thick filaments isolated from the HCM and donor samples did not differ (Fig. 4D) because there were no other significant changes in the overall composition of the thick filaments (Fig. 2C). Although post-translational modifications of thin filament proteins such as phosphorylation were not quantified in the MS assay, neither calcium sensitivity nor the maximal sliding velocity differed for thin filaments sliding on monomeric myosin in the absence of MyBP-C (Fig. 5). To our knowledge, no other reports have demonstrated differences in the function of native thin filaments isolated from similar cohorts of human heart samples. The similarity in the native thin filament structure and function suggest that the MYBPC3 mutations do not have significant secondary effects on thin filament remodeling.
In summary, MYBPC3 truncation mutations reduce MyBP-C content and enhance maximal thin filament sliding velocity within the thick filament C-zone. The effect of MyBP-C cannot be compensated by MyBP-C dephosphorylation to increase its molecular potency. These findings are consistent with studies showing that other HCM-associated sarcomere gene mutations enhance contractility (42, 43). These data support a rationale for developing therapies directed toward restoring normal levels of MyBP-C (44–46), dampening contractile velocities (47) or enhancing MyBP-C’s activity in the presence of high levels of calcium.
Supplementary Material
Highlights.
MYBPC3 truncation mutations are the leading cause of hypertrophic cardiomyopathy (HCM) but the effects of these mutations on MyBP-C content and myofilament contractility are unresolved.
Hearts from HCM patients with heterozygous MYBPC3 truncation mutations had reduced MyBP-C content and truncated MyBP-C was not expressed from the mutant MYBPC3 allele at detectable levels.
Reduced MyBP-C content in the HCM hearts enhanced the sliding velocity of native thin filaments on native thick filaments isolated from HCM hearts, only where MyBP-C is localized within the thick filament C-zones.
These data support therapeutic rationale for restoring normal levels of MyBP-C content and/or dampening maximal contractile velocities for the treatment of human HCM.
Acknowledgements
We thank Dr. D.M. Warshaw, Molecular Physiology and Biophysics Department at the University of Vermont (UVM), and G. Kennedy, Instrumentation and Modeling Facility (UVM), for providing us with fluorescence microcopy capabilities; S. Beck Previs, Molecular Physiology and Biophysics Department (UVM), for training J.S. on protein isolation and solution preparation; S. Beck Previs and J. Gulick, Cincinnati Children’s Hospital for clone generation and assistance with expression of the MyBP-C C0C3 fragment. M.J.P. and T.S.O. were supported by NIH grant R00 HL124041; J.S. and M.J.P. were supported by an American Physiological Society Undergraduate Summer Research Fellowship and Vermont Genetics Network Summer Research Fellowship; S.S. was supported by NIH Grants R01 HL130356, R01 HL105826, R01 AR06727 and R56 HL139680; S.M.D. was supported by AHA Grant in Aid, Children’s Cardiomyopathy Foundation, the Taubman Medical Research Institute, and the Cardiovascular Center Inaugural Grant Fund at the University of Michigan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors have no competing interests to declare.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE (1995) Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the cardia study. Coronary artery risk development in (young) adults. Circulation 92:785–789 [DOI] [PubMed] [Google Scholar]
- 2.Flashman E, Watkins H, Redwood C (2007) Localization of the binding site of the c-terminal domain of cardiac myosin-binding protein-c on the myosin rod. Biochem J 401:97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luther PK, Winkler H, Taylor K, Zoghbi ME, Craig R, Padrón R, Squire JM, Liu J. (2011) Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc Natl Acad Sci U S A. 108:11423–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris SP, Rostkova E, Gautel M, Moss RL. (2004) Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ Res 95:930–936 [DOI] [PubMed] [Google Scholar]
- 5.Razumova MV, Shaffer JF, Tu AY, Flint GV, Regnier M, Harris SP (2006) Effects of the n-terminal domains of myosin binding protein-c in an in vitro motility assay: Evidence for long-lived cross-bridges. J Biol Chem 281:35846–35854 [DOI] [PubMed] [Google Scholar]
- 6.Kampourakis T, Yan Z, Gautel M, Sun YB, Irving M (2014) Myosin binding protein-c activates thin filaments and inhibits thick filaments in heart muscle cells. Proc Natl Acad Sci U S A 111:18763–18768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mun JY, Previs MJ, Yu HY, Gulick J, Tobacman LS, Beck Previs S, Robbins J, Warshaw DM, Craig R (2014) Myosin-binding protein c displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc Natl Acad Sci U S A 111:2170–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM (2012) Molecular mechanics of cardiac myosin-binding protein c in native thick filaments. Science 337:1215–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Previs MJ, Prosser BL, Mun JY, Previs SB, Gulick J, Lee K, Robbins J, Craig R, Lederer WJ, Warshaw DM (2015) Myosin-binding protein c corrects an intrinsic inhomogeneity in cardiac excitation-contraction coupling. Science Advances 1: e1400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaffer JF, Wong P, Bezold KL, Harris SP (2010) Functional differences between the N-terminal domains of mouse and human myosin binding protein-C. J Biomed Biotechnol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris SP, Belknap B, Van Sciver RE, White HD, Galkin VE (2016) C0 and C1 N-terminal Ig domains of myosin binding protein C exert different effects on thin filament activation. Proc Natl Acad Sci U S A 113:1558–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL (2015) Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet Med 17:880–888 [DOI] [PubMed] [Google Scholar]
- 13.Marston S, Copeland O, Jacques A, Livesey K, Tsang V, McKenna WJ, Jalilzadeh S, Carballo S, Redwood C, Watkins H (2009) Evidence from human myectomy samples that mybpc3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ Res 105:219–222 [DOI] [PubMed] [Google Scholar]
- 14.van Dijk SJ, Dooijes D, dos Remedios C, Michels M, Lamers JM, Winegrad S, Schlossarek S, Carrier L, ten Cate FJ, Stienen GJ, van der Velden J (2009) Cardiac myosin-binding protein c mutations and hypertrophic cardiomyopathy: Haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 119:1473–1483 [DOI] [PubMed] [Google Scholar]
- 15.McNamara JW, Li A, Lal S, Bos JM, Harris SP, van der Velden J, Ackerman MJ, Cooke R, Dos Remedios CG. (2017) MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy. PLoS One 12:e0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marston S, Copeland O, Gehmlich K, Schlossarek S, Carrier L (2012) How do MYBPC3 mutations cause hypertrophic cardiomyopathy? J Muscle Res Cell Motil 33:75–80 [DOI] [PubMed] [Google Scholar]
- 17.Helms AS, Davis FM, Coleman D, Bartolone SN, Glazier AA, Pagani F, Yob JM, Sadayappan S, Pedersen E, Lyons R, Westfall MV, Jones R, Russell MW, Day SM (2014) Sarcomere mutation-specific expression patterns in human hypertrophic cardiomyopathy. Circ Cardiovascular genet 7:434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gersh BJ, Maron BJ, Bonow RO, Dearani JA, Fifer MA, Link MS, Naidu SS, Nishimura RA, Ommen SR, Rakowski H, Seidman CE, Towbin JA, Udelson JE, Yancy CW, American College of Cardiology Foundation/American Heart Association Task Force on Practice G, American Association for Thoracic S, American Society of E, American Society of Nuclear C, Heart Failure Society of A, Heart Rhythm S, Society for Cardiovascular A, Interventions, Society of Thoracic S (2011) ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: A reportof the American College of Cardiology Foundation/American Heart Association task force on practice guidelines. Circulation 124:e783–831 [DOI] [PubMed] [Google Scholar]
- 19.Previs MJ, VanBuren P, Begin KJ, Vigoreaux JO, LeWinter MM, Matthews DE (2008) Quantification of protein phosphorylation by liquid chromatography-mass spectrometry. Anal Chem 80:5864–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ (2006) Simultaneous qualitative and quantitative analysis of the escherichia coli proteome: A sweet tale. Mol Cell Proteomics 5:589–607 [DOI] [PubMed] [Google Scholar]
- 21.Luther PK, Bennett PM, Knupp C, Craig R, Padron R, Harris SP, Patel J, Moss RL (2008) Understanding the organization and role of myosin binding protein c in normal striated muscle by comparison with MyBP-C knockout cardiac muscle. J Mol Biol 384:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyata S, Minobe W, Bristow MR, Leinwand LA (2000) Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res 86:386–390 [DOI] [PubMed] [Google Scholar]
- 23.Morano I, Hadicke K, Haase H, Bohm M, Erdmann E, Schaub MC (1997) Changes in essential myosin light chain isoform expression provide a molecular basis for isometric force regulation in the failing human heart. J Mol Cell Cardiol 29:1177–1187 [DOI] [PubMed] [Google Scholar]
- 24.Marston SB, Copeland O, Messer AE, MacNamara E, Nowak K, Zampronio CG, Ward DG (2013) Tropomyosin isoform expression and phosphorylation in the human heart in health and disease. J Muscle Res Cell Motil 34:189–197 [DOI] [PubMed] [Google Scholar]
- 25.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, Turnbull DH, Georgakopoulos D, Kass D, Bond M, Niimura H, Schoen FJ, Conner D, Fischman DA, Seidman CE, Seidman JG (1999) Dilated cardiomyopathy in homozygous myosin-binding protein-c mutant mice. J Clin Invest 104:1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadayappan S, Gulick J, Osinska H, Martin LA, Hahn HS, Dorn GW 2nd, Klevitsky R, Seidman CE, Seidman JG, Robbins J (2005) Cardiac myosin-binding protein-c phosphorylation and cardiac function. Circ Res 97:1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernal E (2014) Limit of Detection and Limit of Quantification Determination in Gas Chromatography. Advances in Gas Chromatography: 57–81. Available online: https://www.intechopen.com/books/advances-in-gas-chromatography/limit-of-detection-and-limit-of-quantification-determination-in-gas-chromatography [Google Scholar]
- 28.Kooij V, Holewinski RJ, Murphy AM, Van Eyk JE (2013) Characterization of the cardiac myosin binding protein-c phosphoproteome in healthy and failing human hearts. J Mol Cell Cardiol 60:116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia W, Shaffer JF, Harris SP, Leary JA (2010) Identification of novel protein kinase a phosphorylation sites in the m-domain of human and murine cardiac myosin binding protein-c using mass spectrometry analysis. J Proteome Res 9:1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copeland O, Sadayappan S, Messer AE, Steinen GJ, van der Velden J, Marston SB (2010) Analysis of cardiac myosin binding protein-c phosphorylation in human heart muscle. J Mol Cell Cardiol 49:1003–1011 [DOI] [PubMed] [Google Scholar]
- 31.Homsher E, Kim B, Bobkova A, Tobacman LS (1996) Calcium regulation of thin filament movement in an in vitro. Biophys J. 70:1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrier L, Knöll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J Jr, Schwartz K, Chien KR (2004) Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 63:293–304 [DOI] [PubMed] [Google Scholar]
- 33.Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, Greaser ML, Powers PA, Moss RL (2002) Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 90:594–601 [DOI] [PubMed] [Google Scholar]
- 34.Barefield D, Kumar M, Gorham J, Seidman JG, Seidman CE, de Tombe PP, Sadayappan S (2015) Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice. J Mol Cell Cardiol. 79:234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balogh J, Merisckay M, Li Z, Paulin D, Arner A. (2002) Hearts from mice lacking desmin have a myopathy with impaired active force generation and unaltered wall compliance. Cardiovasc Res. 53:439–450. [DOI] [PubMed] [Google Scholar]
- 36.Herron TJ, Rostkova E, Kunst G, Chaturvedi R, Gautel M, Kentish JC (2006) Activation of myocardial contraction by the n-terminal domains of myosin binding protein-c. Circ Res 98:1290–1298 [DOI] [PubMed] [Google Scholar]
- 37.Razzaque MA, Gupta M, Osinska H, Gulick J, Blaxall BC, Robbins J (2013) An endogenously produced fragment of cardiac myosin-binding protein C is pathogenic and can lead to heart failure. Circ Res. 113:553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaffer JF, Kensler RW, Harris SP (2009) The myosin-binding protein c motif binds to f-actin in a phosphorylation-sensitive manner. J Biol Chem 284:12318–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Previs MJ, Mun JY, Michalek AJ, Previs SB, Gulick J, Robbins J, Warshaw DM, Craig R (2016) Phosphorylation and calcium antagonistically tune myosin-binding protein c’s structure and function. Proc Natl Acad Sci U S A 113:3239–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wijnker PJ, Friedrich FW, Dutsch A, Reischmann S, Eder A, Mannhardt I, Mearini G, Eschenhagen T, van der Velden J, Carrier L. (2016) Comparison of the effects of a truncating and a missense MYBPC3 mutation on contractile parameters of engineered heart tissue. J Mol Cell Cardiol 97;82–92 [DOI] [PubMed] [Google Scholar]
- 41.Homsher E, Wang F, Sellers J (1993) Factors affecting filament velocity in in vitro motility assays and their relation to unloaded shortening velocity in muscle fibers. Adv Exp Med Biol 332:279–289; [DOI] [PubMed] [Google Scholar]
- 42.Moore JR, Leinwand L, Warshaw DM (2012) Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res 111:375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spudich JA (2014) Hypertrophic and dilated cardiomyopathy: Four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys J 106:1236–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gedicke-Hornung C, Behrens-Gawlik V, Reischmann S, Geertz B, Stimpel D, Weinberger F, Schlossarek S, Precigout G, Braren I, Eschenhagen T, Mearini G, Lorain S, Voit T, Dreyfus PA, Garcia L, Carrier L (2013) Rescue of cardiomyopathy through u7snrna-mediated exon skipping in mybpc3-targeted knock-in mice. EMBO molecular medicine 5:1128–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mearini G, Stimpel D, Geertz B, Weinberger F, Kramer E, Schlossarek S, Mourot-Filiatre J, Stoehr A, Dutsch A, Wijnker PJ, Braren I, Katus HA, Muller OJ, Voit T, Eschenhagen T, Carrier L (2014) Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nature Communications 5:5515. [DOI] [PubMed] [Google Scholar]
- 46.Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, Koski A, Ji D, Hayama T, Ahmed R, Darby H, Van Dyken C, Li Y, Kang E, Park AR, Kim D, Kim ST, Gong J, Gu Y, Xu X, Battaglia D, Krieg SA, Lee DM, Wu DH, Wolf DP, Heitner SB, Belmonte JCI, Amato P, Kim JS, Kaul S, Mitalipov S (2017) Correction of a pathogenic gene mutation in human embryos. Nature 548:413–419 [DOI] [PubMed] [Google Scholar]
- 47.Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG, Seidman CE (2016) A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 351:617–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N (2007) Statistical significance of quantitative PCR. BMC bioinformatics 8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiverick KT, Thomas LL, Alpert NR (1975) Purification of cardiac myosin. Application to hypertrophied myocardium. Biochim Biophys Acta. 393:124–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.