Abstract
As high-performance computing resources have become increasingly available, new modes of computational processing and experimentation have become possible. This tutorial presents the Extreme-scale Model Exploration with Swift/T (EMEWS) framework for combining existing capabilities for model exploration approaches (e.g., model calibration, metaheuristics, data assimilation) and simulations (or any “black box” application code) with the Swift/T parallel scripting language to run scientific workflows on a variety of computing resources, from desktop to academic clusters to Top 500 level supercomputers. We will present a number of use-cases, starting with a simple agent-based model parameter sweep, and ending with a complex adaptive parameter space exploration workflow coordinating ensembles of distributed simulations. The use-cases are published on a public repository for interested parties to download and run on their own.
1. INTRODUCTION
Modern simulation-based application studies are campaigns consisting of large numbers of simulations with many possible variations. Simulations may be run with different parameters, possibly as part of an automated model parameter optimization, classification, or, more generally, model exploration (ME). Constructing the software to run such studies at the requisite computational scales is often unnecessarily time-consuming and the resulting software artifacts are typically difficult to generalize and package for other users.
In this tutorial, we present a solution for many of the challenges in running large-scale simulation studies. Our framework, Extreme-scale Model Exploration with Swift/T (EMEWS), uses the general-purpose parallel scripting language Swift (Armstrong et al. 2014) to generate highly concurrent simulation workflows. These workflows enable the integration of external ME algorithms to coordinate the running and evaluation of large numbers of simulations. The general-purpose nature of the programming model allows the user to supplement the workflows with additional analysis and post-processing as well.
Here, we focus on agent-based models (ABMs). Extracting knowledge from ABMs requires the use of approximate, heuristic ME methods involving large simulation ensembles. To improve the current state of the art it has been noted elsewhere that: “… there is a clear need to provide software frameworks for metaheuristics that promote software reuse and reduce developmental effort.” (Boussad, Lepagnot, and Siarry 2013) Our design goals are to ease software integration while providing scalability to the largest scale (petascale plus) supercomputers, running millions of ABMs, thousands at a time. Initial scaling studies of EMEWS have shown robust scalability (Ozik, Collier, and Wozniak 2015). The tools are also easy to install and run on an ordinary laptop, requiring only an MPI (Message Passing Interface) implementation, which can be easily obtained from common OS package repositories.
Figure 1 illustrates the main components of the EMEWS framework. The main user interface is the Swift script, a high-level program. The core novel contributions of EMEWS are shown in green, these allow the Swift script to access a running ME algorithm. This algorithm can be expressed in Python, R, C, C++, Fortran, Julia, Tcl, or any language supported by Swift/T. We provide a high-level queue-like interface with (currently) two implementations: EQ/Py and EQ/R (EMEWS Queues for Python and R). These allow the Swift script to obtain candidate model parameter inputs and return model outputs to the ME. The simulation models are distributed by the Swift/T runtime over a potentially large computer system, but smaller systems that run one model at a time are also supported. The simulations can be implemented as external applications called through the shell, or in-memory libraries accessed directly by Swift (for faster invocation).
Figure 1.
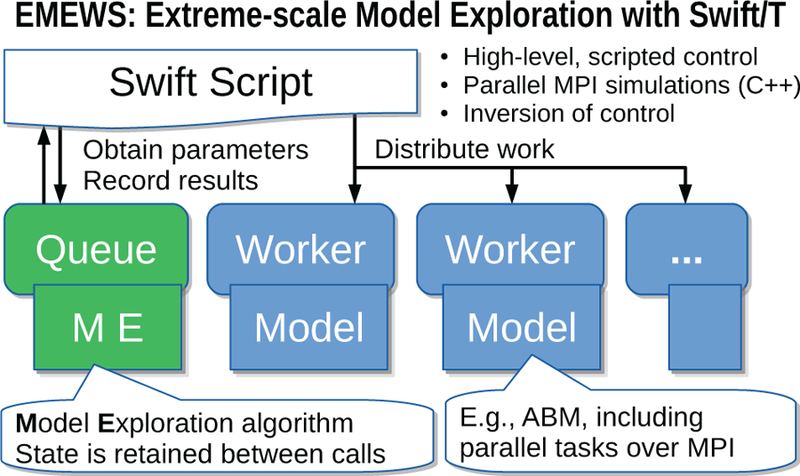
Overview of Extreme-scale Model Exploration with Swift/T (EMEWS) framework.
EMEWS thus offers the following contributions to the science and practice of simulation ME studies: 1) it offers the capability to run very large, highly concurrent ensembles of simulations of varying types; 2) it supports a wide class of model exploration algorithms, including those increasingly available to the community via Python and R libraries; and 3) it offers a software sustainability solution, in that simulation studies based around EMEWS can easily be compared and distributed.
Our tutorial will present these contributions in more detail. The examples use cases presented in this paper are generally runnable Swift (not pseudocode) and are published on a public repository (EMEWS Tutorial Website 2016).
The remainder of this paper is organized as follows. In Section 2, we describe related ABM software, ME libraries, and workflow toolkits. In Section 3, we describe the Swift programming model in detail with examples. In Section 4, we provide complete use cases in the ABM context. We summarize our contributions in Section 5.
2. RELATED WORK
2.1. Capabilities of existing ABM tools
Many ABM toolkits allow users to define a parameter space and then enable automated iteration through that space. The parameter space is defined in a toolkit-specific format specifying each parameter in terms of a range, and a step value, or as a list of elements. The toolkit takes this a priori determined parameter space as input and executes the required simulation runs. We briefly survey this and more advanced capabilities for model exploration in widely used ABM toolkits.
Repast Simphony (North et al. 2013) is the Java based toolkit of the Repast Suite. Given a parameter space as input, Repast Simphony’s batch run functionality can divide that space into discrete sets of parameter values and execute simulations over those discrete sets in parallel. The simulations can be run on a local machine, on remote machines accessible through secure shell (ssh), in the cloud (e.g., Amazon EC2) or on some combination of the three. Using an InstanceRunner interface, Repast Simphony models can be launched by other control applications such as a bash, PBS, or Swift scripts. For example, Ozik et al. (2014) describe how the InstanceRunner can be used with Swift to perform an adaptive parameter sweep using simulated annealing, and the InstanceRunner is used in use cases (§4.1) and (§4.2) described in this paper.
NetLogo (Wilensky 1999) has both GUI (BehaviorSpace) and command line based batch run capabilities. Stonedahl (2011) developed the Behavior Search tool which provides an easy to use interface for running an included set of heuristic model exploration techniques, e.g., simulated annealing, genetic algorithm, on NetLogo models. The MASON simulation library (Luke et al. 2005) offers a set of capabilities for creating ABMs. Its modularity allows MASON models to be called either via the command line or as libraries from Java-based programs (e.g., see ECJ in §2.2) for model exploration purposes. Repast (New BSD), NetLogo (GPL v2) and MASON (AFL v3) are all open source software.
AnyLogic is a proprietary multi-method simulation toolkit. AnyLogic comes with the ability to carry out Experiments, including optimization, calibration, and user-defined custom experiments using the AnyLogic engine Java API. (Note that many of the experiment capabilities are available only for AnyLogic Professional and University Researcher editions.)
None of the ABM toolkits on their own offer the capabilities or scope, in terms of flexible, simple integration of external model exploration tools and performance on massively parallel computing resources, that the EMEWS framework provides, but they can all be integrated into EMEWS.
2.2. Model exploration libraries and frameworks
Here we briefly survey existing model exploration (ME) libraries and frameworks. While most can be used as standalone ME tools, some of the these libraries can also be used as ME modules within the EMEWS framework. Most of the following software falls under the metaheuristics umbrella. For an overview of metaheuristics see Luke (2013), for reviews of more metaheuristics frameworks see Parejo et al. (2011) and for parallel metaheuristics frameworks see Alba, Luque, and Nesmachnow (2013).
OptTek offers proprietary tools for metaheuristic optimization capabilities and the ability to wrap custom objective functions. ECJ s an open source (AFL v3) research system for evolutionary computation, and can be used for developing evolutionary algorithms. ParadisEO, published under the CeCILL license, and MALLBA (Alba et al. 2007) published under a non-commercial license, are also frameworks for metaheuristics. SOF (Carillo et al. 2016) is a relative newcomer to the ME area, and is focused on NetLogo and MASON simulations, providing a generic simulator interface for other types of simulations. Dakota combines a number of optimization, design of experiment and uncertainty quantification libraries developed by Sandia National Laboratories (e.g., DDACE, HOPSPACK), in addition to other external libraries.
With the proliferation of freely accessible libraries published in Python and R, we present a very small subset of notable ones relevant to model exploration, some of which are utilized in use cases (§4.2) and (§4.3). In Python, the Distributed Evolutionary Algorithms in Python (DEAP) toolkit (Fortin et al. 2012) is a framework for developing evolutionary algorithms. The scikit-learn package provides a large number of machine learning methods and Theano and Lasagne enable deep learning, capabilities which are useful for surrogate/meta-modeling and active learning (Settles 2012) applications. On the R side, the EasyABC (Jabot, Faure, and Dumoulin 2013) and abc (Csillery, Francois, and Blum 2012) packages provide approximate Bayesian computation (ABC) capabilities that are increasingly being applied to ABM (Beaumont 2010, Hartig et al. 2011). For machine learning and other statistical applications, R includes packages such as caret (Kuhn 2008), randomForest (Liaw and Wiener 2002), and many others.
2.3. Workflow software: Large scale
The word “workflow” itself needs definition, as it can mean different things in different contexts. Here, we use a workflow to mean a system that: 1) conceptually breaks the computation into a number of discrete, coarse-grained steps, which are linked together via dataflow control; 2) allows the user to mingle software and data components of different types, running on different resources; 3) offers a very high-level programming interface, allowing the user to rapidly rearrange the workflow without modifying the lower-level components; and 4) automatically manages available concurrency by analyzing the dataflow control structure.
The Make system (Feldman 1979) is a primordial exemplar of the workflow system. Makefile recipes break the script into coarse sections (1), which call various programs through a standard (shell) interface on various data types (2), allowing the user to easily rewire the workflow process (3), and offers automatic concurrency (for GNU Make, since 1999 [Smith 2016]) (4).
Numerous approaches, languages, and tools exist to express and execute workflows in parallel and on distributed resources. Often these have tradeoffs in terms of the flexibility vs. familiarity of the expression model; the complexity and scalability of the parallelism; and the difficulty of execution on distributed and/or heterogeneous parallel resources. We briefly survey here several representative approaches, which address one or more parts of our definition above.
Language-neutral scripting tools:
MyCluster (Walker, Xu, and Chandar 2009) comprised extensions to the UNIX shell that allow a user to define a dataflow graph, including the concepts of fork, join, cycles, and key-value aggregation, but which execute on single parallel systems or clusters. These mechanisms, however, do not generalize to highly parallel and distributed environments.
Library-based approaches:
MapReduce (Dean and Ghemawat 2004) is a programming model and a runtime system to support the processing of large-scale datasets. Unlike Swift, which is tailored for composing workflows from application programs, like Repast Simphony or Repast HPC simulations, the MapReduce programming model is more oriented to using key-value pairs as input or output datasets.
Static DAG tools:
DAGMan (Thain, Tannenbaum, and Livny 2005) is a workflow engine that manages Condor jobs organized as directed acyclic graphs (DAGs) of explicit task precedence. It has no knowledge of data flow, and in a distributed environment it requires a higher-level, data-cognizant layer. Pegasus (Deelman et al. 2005) translates a workflow graph into location-specific DAGMan input, adding data staging, inter-site transfer, and data registration. These approaches lack the expressiveness of Swift’s functional programming model for the composition of adaptive ME workflows.
Language-based approaches:
Dryad (Isard et al. 2007) is an infrastructure for running data-parallel programs on a parallel or distributed system. Dryad graphs are explicitly developed by the programmer; Dryad appears to be used primarily for clusters and well-connected groups of clusters in single administrative domains and in Microsoft’s cloud. Dryad lacks the scalability that Swift provides for execution on supercomputers.
Visual workflow management environments:
Many workflow systems (e.g., Taverna (Oinn et al. 2004), Kepler (McPhillips et al. 2009), Galaxy (Goecks et al. 2010), and CloudSME (Taylor et al. )) are based on comparable data-driven computing models but lack Swift’s scalability, its simple generality for supporting arbitrary applications, and its provider-based architecture for broad platform support. Recent work has integrated Swift’s execution model into the Galaxy user interface model, to provide the best benefits of both workflow models (Maheshwari et al. 2013a, Maheshwari et al. 2013b).
Compositional programming:
Workflow systems can offer rich programming environments. Program Composition Notation and Strand (Foster 1996) emphasized coarse-grained, compositional programming to boost developer productivity when parallelizing execution of software components.
3. SWIFT/T: HIGH-PERFORMANCE WORKFLOW LANGUAGE
The Swift language allows the developer to rapidly implement workflows that satisfy the definition in (§2.3). The Swift/T implementation of Swift focuses on high-performance workflows to utilize TOP500 machines (Top500 2016). Such systems are characterized by high concurrency (tens of thousands of processor cores or more) and low-latency networks. Swift/T can utilize these systems well, producing over a billion tasks per second (Armstrong et al. 2014). As a workflow language, Swift composes existing software components into an overall workflow. The following sections describe Swift/T features relevant to EMEWS. For more details, see the Swift/T website.
3.1. Syntax
The Swift language uses C-like syntax and conventional data types such as int, float, and string. It also has typical control constructs such as if, for, and foreach. Swift code can be encapsulated into functions, which can be called recursively. As shown in Figure 2, Swift can perform typical arithmetic and string processing tasks quite naturally. Swift also has a file type, that allows dataflow processing on files.
Figure 2.

Sample Swift syntax.
3.2. External execution
Swift is primarily designed to call into external user code, such as simulations or analysis routines implemented in various languages. Like many other systems, Swift/T supports calls into the shell. However, this is not efficient at large scale, and so Swift/T also supports calls into native code libraries directly.
An example use of Swift for shell tasks is shown in Figure 3. This example demonstrates a fragment of a build system. The user defines two app functions, which compile and link a C language file. Swift app functions differ from other Swift functions in that they operate primarily on variables of type file.
Figure 3.

Swift used as a Makefile.
Other forms of external execution in Swift/T allow the user to call into native code (C/C++/Fortran) directly by constructing a package with SWIG. Such libraries can be assembled with dynamic or static linking; in the static case, the Swift script and the native code libraries are bundled into a single executable with minimal system dependencies for the most efficient loading on a large-scale machine.
3.3. Concurrency
The key purpose of Swift is to gain concurrency easily and correctly. This is accomplished in Swift through the use of dataflow instead of control flow. In Swift, there is no instruction pointer, execution is triggered wherever possible limited only by data availability. This results in an implicitly parallel programming model. Two modes of concurrency are shown in Figure 4, both based on the ready availability of i. Computing the ith Fibonacci number relies on two concurrent recursive calls, and iteration over an array of known values allows for parallel execution. Ordering can be forced with the statement1=>statement2 syntax, which creates an artificial data dependency.
Figure 4.

Swift concurrency modes.
3.4. Support for interpreted languages
Swift/T also provides high-level, easy to use interfaces for Python, R, Julia and Tcl, allowing the developer to pass a string of code into the language interpreter for execution (via its C or C++ interface). These interpreters are optionally linked to the Swift runtime when it is built. This allows the user to tightly integrate Swift logic with calls to the interpreters, as the interpreter does not have to be launched as a separate program for each call. This is a crucially significant performance benefit on very large scale supercomputers, enabling us to make millions of calls to the interpreter per second (Wozniak et al. 2015).
3.4.1. Python
Many users desire to access Python from the top level of the scientific workflow; and optionally call down from the interpreted level into native code, to gain high-performance operations for numerical methods or event-based simulation. A popular example of this model is Numpy, which provides a high-level interface for interaction, with high-performance, vendor-optimized BLAS, LAPACK, and/or ATLAS numerical libraries underneath. (Python, R, Julia and Tcl each support calling to native code in some way; Tcl was designed for this purpose.)
One use of Python from Swift/T is shown in Figure 5. In this example, a short module is defined in F.py which provides an addition function named f(). A call to this function from Swift/T is shown in python-f.swift lines 3–5. The string containing the python code is populated with the Pythonic % operator, which fills in values for x and y at the conversion specifiers %i. The Python function F.f() receives these values, adds them, and returns the result as a string. Swift/T receives the result in z and reports it with the Swift/T builtin trace() function. Thus, data can easily be passed to and from Python with Pythonic conventions; only stringification is required. To execute, the user simply sets PYTHONPATH so that the Python interpreter can find module F, and runs swift-t.
Figure 5.

Swift use of Python.
3.4.2. R
The R support in Swift/T is similar to the Python support. An example use case is shown in Figure 6. This script intends to run a collection of simulations in parallel, then send result values to R for statistical processing. The first section (lines 1–2) simply imports requisite Swift packages. The second section (lines 3–7) defines the external simulation program, which is implemented as a call to the bash shell random number generator, seeded with the simulation number i. The output goes to temporary file o. The third section (lines 8–12) calls the simulation a number of times, reading the output number from disk and storing it in the array results. The fourth section (lines 13–16) computes the mean of results via R. It joins the results into an R vector, constructed with the R function c(), uses the R function mean(), and returns the mean as a string mean that is printed by Swift.
Figure 6.
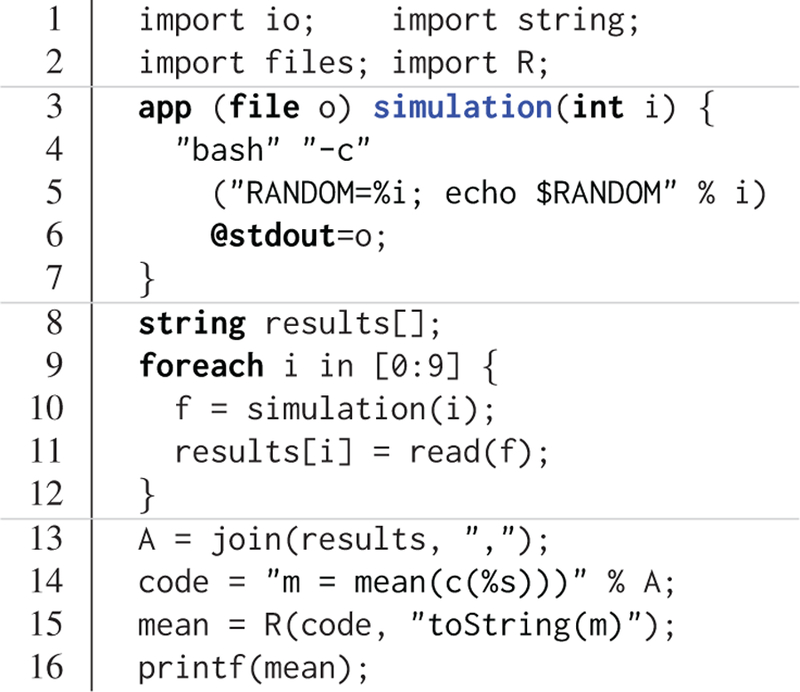
Swift use of R.
3.5. Features for large-scale computation
Swift/T has multiple other features to support the needs of workflow applications, including support for locations and MPI tasks. The underlying call from Swift to external code (via the shell, a script, or native code) is called a leaf function as its execution is opaque to Swift. These features are accessed by Swift/T annotations that are applied to the leaf function invocation. A generic annotation takes the form
type result = @key=value f(params);
where the key and value denote the annotation type.
3.5.1. Task locations
Task locations allow the developer to specify the location of task execution in the system. Locations are optional; Swift/T defaults to placing the next task in a location determined by the load balancer. Locations can be used to direct computation to part of the system for multiple reasons (Duro et al. 2016). In a data-intensive application, tasks can be sent to the location containing the data they desire to process. In a workflow with resident tasks (Ozik, Collier, and Wozniak 2015), certain processes retain state from task to task, and can be queried by sending a task to that process.
A location object L in Swift/T is a data structure containing an MPI rank R and optionally other location-aware scheduling constraint information. The MPI rank is the target of the location, it is simply a rank integer in the overall Swift/T run, representing a single process. The code in Figure 7 shows the Swift steps for looking up a hostname, constructing a location object, and sending a task there.
Figure 7.

Use of location in Swift.
3.5.2. Parallel tasks
Swift/T offers the ability to construct workflows containing large numbers of MPI tasks that run on variably-sized communicators (Wozniak et al. 2013). These tasks obtain a communicator of a programmatically-determined size, specified with the @par annotation, and (optionally) destroy it on completion. Swift/T obtains the given number of worker processes and constructs these communicators using the MPI_Comm_create_group() function in MPI 3. Parallel tasks can be configured to run on contiguous processes, or on any available processes. The @par annotation can be combined with other annotations such as @location.
4. USE CASES
4.1. Simple Workflows with ABM
For a first demonstration ABM use case, we begin with an example of a Swift/T parallel parameter sweep to explore the parameter space of a model. A second example will build on this, illustrating how to construct a Swift/T scripted workflow that begins with multiple model runs being performed in parallel and concludes with some post-run analysis on the model outputs, performed in R. (The full Use Case One [UC1] project can be found at the tutorial website [EMEWS Tutorial Website 2016] where, in addition to the Repast Simphony examples for use cases §4.1 and §4.2, we also provide NetLogo and MASON examples.) The example model used here is an adaptation of the JZombies demonstration model distributed with Repast Simphony (Collier and North 2015). (The fictional Zombies versus Humans model is intended to illustrate that Swift/T and Repast Simphony are domain agnostic.) The model has two kinds of agents, Zombies and Humans. Zombies chase the Humans, seeking to infect them, while Humans attempt to evade Zombies. When a Zombie is close enough to a Human, that Human is infected and becomes a Zombie. During a typical run all the Humans will eventually become Zombies.
These agents are located in a two dimensional continuous space where each agent has a x and y coordinate expressed as a floating point number (and in a corresponding discrete grid with integer coordinates). Movement is performed in the continuous space and translated into discrete grid coordinates. The grid is used for neighborhood queries (e.g. given a Zombie’s location, where are the nearest Humans). The model records the grid coordinate of each agent as well as a count of each agent type (Zombie or Human) at each time step and writes this data to two files. The initial number of Zombies and Humans is specified by model input parameters zombie_count and human_count, and the distance a Zombie or Human can move at each time step is specified by the parameters zombie_step_size and human_step_size.
In order for Swift/T to call an external application such as the Zombies model, the application must be wrapped in a leaf function (§3.2). The Zombies model is written in Java which is not easily called via Tcl and thus an app function is the best choice for integrating the model into a Swift script. Repast Simphony provides command line compatible functionality, via an InstanceRunner (§2.1) class, for passing a set of parameters to a model and performing a single headless run of the model using those parameters. We have wrapped that command line invocation of Repast Simphony in a bash script repast.sh (EMEWS Tutorial Website 2016) that eases command line usage.
The Swift app function that calls Repast Simphony is shown in the top of Figure 9. Prior to the actual function definition, the environment variable T_PROJECT_ROOT is accessed. This variable is used to define the project’s top level directory, relative to which other directories (e.g., the directory that contains the Zombies model) are defined. On line 2, the app function definition begins. The function returns two files, one for standard output and one for standard error. The arguments are those required to run repast.sh: the name of the script, the current run number, the directory where the model run output should be written, and the model’s input scenario directory. The body of the function calls the bash interpreter passing it the name of the script file to execute and the other function arguments as well as the project root directory. @stdout=out and @stderr=err redirect stdout and stderr to the files out and err. It should be easy to see how any model or application that can be run from the command line and wrapped in a bash script can be called from Swift in this way.
Figure 9.
Zombies model parameter sweep.
Our full script performs a simple parameter sweep using the app function to run the model. The parameters to sweep are defined in a file where each row of the file contains a parameter set for an individual run. The script will read these parameter sets and launch as many parallel runs as possible for a given process configuration, passing each run a parameter set. The Swift script is shown in Figure 9. The script (EMEWS Tutorial Website 2016) uses two additional functions that have been elided to save space. The first, cp_message_center, calls the unix cp command to copy a Repast Simphony logging configuration file into the current working directory. The second, make_dir, calls the Unix mkdir command to create a specified directory. Script execution begins in line 8, calling the cp_message_center app function. In the absence of any data flow dependency, Swift statements will execute in parallel whenever possible. However, in our case, the logging file must be in place before a Zombie model run begins. The => symbol enforces the required sequential execution: the code on its left-hand side must complete execution before the code on the right-hand side begins execution. Lines 11 and 12 parse command line arguments to the Swift script itself. The first of these is the name of the unrolled-parameter-file that contains the parameter sets that will be passed as input to the Zombies model. Each line of the file contains a parameter set, that is, the random_seed, zombie_count, human_count, zombie_step_size and human_step_size for a single model run. The parameter set is passed as a single string (e.g. random_seed = 14344, zombie_count = 10 …) to the Zombies model where it is parsed into the individual parameters. The scenario argument specifies the name of the Repast Simphony scenario file for the Zombies model. This defaults to “scenario.rs” if the argument is not given. In line 13 the built-in Swift file_lines function is used to read the upf file into an array of strings where each line of the file is an element in the array. The foreach loop that begins on line 14 executes its loop iterations in parallel. In this way, the number of model runs that can be performed in parallel is limited only by hardware resources.
The variable s is set to an array element (that is, a single parameter set represented as a string) while the variable i is the index of that array element. Lines 15 and 16 create an instance directory into which each model run can write its output. The => symbol is again used to ensure that the directory is created before the actual model run that uses that directory is performed in line 20. Lines 17 and 18 create file objects into which the standard out and standard error streams are redirected by the repast function (Figure 9). The Repast Simphony command line runs allows for the parameter input to be passed in as a file and so in line 19 the parameter string s is written to a upf.txt file in the instance directory. Lastly, in line 20, the app function, repast, that performs the Zombie model run is called with the required arguments.
In this script we have seen how to run multiple instances of the Zombies model in parallel, each with a different set of parameters. Our next example builds on this by adding some post-run analysis that explores the effect of simulated step size on the final number of humans. This analysis will be performed in R and executed within the Swift workflow. We present this in two parts. The first describes the changes to the foreach loop to gather the output and the second briefly describes how that output is analyzed to determine the “best” parameter combination.
This example assumes an unrolled-parameter-file where we vary zombie_step_size and human_step_size. For each run of the model, that is, for each combination of parameters, the model records a count of each agent type at each time step in an output file. As before the script will iterate through the “upf” file performing as many runs as possible in parallel. However an additional step that reads each output file and determines the parameter combination or combinations that resulted in the most humans surviving at time step 150 has been added. The relevant parts of the new script (EMEWS Tutorial Website 2016) are shown in Figure 10. Here the repast call is now followed by the execution of an R script (lines 2–3, a Swift multiple-line string literal) that retrieves the final number of humans from the output file. The R script reads the CSV file produced by a model run into a data frame, accesses the last row of that data frame, and then the value of the human_count column in that row. The count_humans string variable holds a template of the R script where the instance directory (line 3) in which the output file (counts.csv) is written can be replaced with an actual instance directory. Line 11 performs this substitution with the directory for the current run. The resulting R code string is evaluated in line 12 using the Swift R() function. In this case, the res variable in the R script (line 3) contains the number of surviving humans, and the second argument in the R call in line 15 returns that value as a string. This string is then placed in the results array at the ith index.
Figure 10.

Zombies sweep with human count.
An additional workflow step in which R is used to determine the indices of the maximum values in the results array can be seen in the full script (EMEWS Tutorial Website 2016). Given that the value in results[i] is produced from the parameter combination in upf_lines[i], the index of the maximum value or values in the array is the index of the “best” parameter combination or combinations. Swift code is used to iterate through the array of best indices as determined by R and write the corresponding best parameters to a file.
4.2. Workflow control with Python-based external algorithms
Due to the highly non-linear relationship between ABM input parameters and model outputs, as well as feedback loops and emergent behaviors, large-parameter spaces of realistic ABMs cannot generally be explored via brute force methods, such as full-factorial experiments, space-filling sampling techniques, or any other a priori determined sampling schemes. This is where adaptive, heuristics-based approaches are useful and this is the focus of the next two use cases.
In (Ozik, Collier, and Wozniak 2015) we describe an inversion of control (IoC) approach enabled by resident Python tasks in Swift/T and simple queue-based interfaces for passing parameters and simulation results, where a metaheuristic method (GA) developed with DEAP (Fortin et al. 2012) is used to control a large workflow. Our second use case shows how this is done with the EQ/Py extension. The benefit of using external libraries directly is threefold. First, there is no need to port the logic of a model exploration method into Swift/T, thereby removing the (possibly prohibitive) effort overhead and the possibility for translation errors. Second, the latest methods from the many available model exploration toolkits (e.g., those in §2.2) can be easily compared with each other for utility and performance. Third, the external libraries are not aware of their existence within the EMEWS framework, so methods developed without massively parallel computing resources in mind can be nonetheless utilized in such settings.
In this use case we continue with the Repast Simphony JZombies demonstration model. For resident tasks, which retain state, the location of a worker is used so that the algorithm state can be repeatedly accessed. The EQ/Py extension (EMEWS Tutorial Website 2016) provides an interface for interacting with Python-based resident tasks at specific locations. Figure 11 shows how EQ/Py is used in the current example. We import the extension in line 1. The deap function is defined to take the arguments py_rank (a unique rank), iters (the number of GA iterations), trials (the number of stochastic variations per parameter combination, or individual), pop (the number of individuals in the GA population), and seed (the random seed to use for the GA). A location ME is generated from ME_rank in line 4. This location is passed to the EQPy_init_package call, along with a package name (deap_ga), which loads the Python file named deap_ga.py (found by setting the appropriate PYTHONPATH environment variable), initializes input and output queues, and starts the run function in the deap_ga.py file, before returning.
Figure 11.

Setting up a DEAP call from Swift.
At this point the resident task is available to interact with through the EQPy_get() and EQPy_put() calls, which get string values from and put string values into the resident task OUT and IN queues, respectively. The first call to EQPy_get() (line 8) is made in order to push initialization parameters to the resident task via EQPy_put(ME, algo_params) (line 9). Then the doDEAP() function, to be discussed next, is called and, when it finishes executing, EQPy_stop() is called to shut down the resident task.
Figure 12 shows the main DEAP workflow loop, a general pattern for interacting with resident tasks. Unlike the foreach loop, which parallelizes the contents of its loop, the Swift for loop iterates in a sequential fashion, only guided by dataflow considerations. The for loop continues until the EQPy_get() call receives a message “FINAL”, at which point EQPy_get() is called again to retrieve the final results and doDEAP() exits the loop and returns (lines 12–15). Otherwise, the next set of parameters is obtained by splitting (line 17) the string variable retrieved on line 10. The contents of the pop array are individual parameter combinations, also referred to as individuals of a GA population. Each individual is then sent to a summary objective function obj which creates trials stochastic variations of the individual, evaluates their objective function (the number of Humans remaining, the count_humans R code from Figure 10) and returns the average value, (not shown here, full script [EMEWS Tutorial Website 2016] on tutorial website). Lines 25–29 transform the summary objective results for each individual into a string representation that can be evaluated within the Python resident task, and this value is sent to it via EQPy_put() (line 30).
Figure 12.
The main DEAP workflow loop.
The EQ/Py extension makes two functions, IN_get and OUT_put, available for the Python resident task and these can be used to pass candidate parameters to and get results from any Swift/T workflow. These functions are the complements to the EQPy_get() and EQPy_put() functions on the Swift/T side. The DEAP framework provides flexibility in defining custom components for its GA algorithms and we take advantage of this by overriding the map() function used to pass candidate parameters for evaluation to our custom evaluator with toolbox.register(“map”, queue map). The queue_map function executes calls to OUT_put and IN_get. In this way the Python resident task is unaware of being a component in an EMEWS workflow. The full Python resident task code (deap ga.py) along with the full DEAP use case can be found in the Use Case Two (UC2) project (EMEWS Tutorial Website 2016).
4.3. Calling a distributed MPI-based Model
In this use case, we will show how to integrate a multi-process distributed native code model written in C++ into a Swift/T workflow. The model is a variant of the Java Zombies model, written in C++ and using MPI and the Repast HPC toolkit (Collier and North 2012) to distribute the model across multiple processes. The complete two dimensional continuous space and grid span processes and each individual process holds some subsection of the continuous space and grid. The Zombies and Humans behave as before but may cross process boundaries into another subsection of the continuous space and grid as they move about the complete space. The HPC Zombies source, a Makefile, the various files required to integrate it with Swift/T, and the Swift scripts can be found in the Use Case Three (UC3) project (EMEWS Tutorial Website 2016). There, the HPC Zombie model runs are driven by an active learning (Settles 2012) algorithm using EQ/R, the R counter-part to EQ/Py described above (§4.2).
In contrast to the previous two examples the MPI-based HPC Zombies model is compiled as a shared library that exposes a Swift/T Tcl interface (Wozniak et al. 2015). Swift/T runs on Tcl and thus wrapping the library in Tcl provides tighter integration than an app function, but is also necessary in the case of multi-process distributed models that use MPI. Such models when run as standalone applications initialize an MPI Communicator of the correct size within which the model can be distributed and run. Since the HPC Zombies model uses MPI, as do all Repast HPC based models, it must be treated as an MPI library and passed an MPI communicator of the correct size when run from Swift/T (§3.5.2).
The first step in integrating the HPC Zombies model with Swift/T is to compile it as library, converting main() into a function that runs the model. The next step is to make that function callable from Tcl via a SWIG created binding. SWIG (Beazley 1996) is a software tool that generates the ‘glue code’ required for some target language, such as Tcl, to call C or C++ code. The SWIG tool processes an interface file and produces the ‘glue-code’ binding as a source code file. In this case, the C++ code we want to call from Tcl and ultimately from Swift is the Zombies model function: std::string zombies model run(MPI_Comm comm, const std::string& config, const std::string& parameters). The function takes the MPI communicator in which the model runs, the filename of a Repast HPC config file, and the parameters for the current run. When called, it starts a model run using these arguments. The SWIG interface (EMEWS Tutorial Website 2016) file is run through SWIG and the resulting source code is compiled with the HPC Zombies model library code. The result is a zombie_model_run function that is callable from Tcl. The Makefile target, ./src/zombies model wrapper.cpp in the Makefile (EMEWS Tutorial Website 2016) template is an example of this process.
The next step is to create the Swift bindings for the library function. The Swift bindings define how the zombie_model_run function will be called from Swift. The Swift code is shown in Figure 13. The function is annotated with @par (§3.5) allowing it to be called as a parallel function. The @dispatch=WORKER (§3.5) directs the function to run on a worker node. The function itself returns a string that contains the number of humans and zombies at each time step. For arguments, the function takes the config file file name and a string containing the parameters for the run. The model will parse the individual parameters from this params string. The final 3 parts of the function definition are the Tcl package name, the required package version, and the Tcl function to call in the package. With this code included in our Swift script (either directly or through an import), we can then run the HPC Zombies model with a call like:
string output = @par=4 zombies_model_run(config file, params);
Figure 13.
Zombies model Swift interface.
Our Swift binding references a zombies_model Tcl package and a zombies_model_tcl function in that package. The final step in integrating the HPC Zombies model with Swift is to create this package and the function. A Tcl package is defined by its pkgIndex.tcl file that specifies the libraries that need to be loaded as part of the package and the Tcl code that is in the package. Tcl has a built-in function ::pkg::create that can be used to create a pkgIndex.tcl given a package name, version, the name of the library to load, and the Tcl code file name. The HPC Zombies example uses some simple Tcl code to call this function as part of a Makefile to create the package (c.f. the zombies_tcl_lib Makefile target and the make-package.tcl script [EMEWS Tutorial Website 2016]).
The code in the zombies_model Tcl package that our Swift function calls is shown in Figure 14. For parallel tasks, Swift/T currently requires two Tcl procedures, a dataflow interface (line 1 f.) and a body (line 7 f.) that calls our zombies_model_run function. The rule command (line 2) registers dataflow execution: when all input IDs ins are available, zombies_model_body will be released to the load balancer and executed on an available subset of workers. The args are Swift-specific task settings, including the @par parallel settings, and are not accessed by the user. The body function retrieves the input parameter values from Swift by applying the provided retrieve_string function on the IDs, obtaining the configuration file name and the sample model parameters. The MPI subcommunicator for the parallel task and the current MPI rank in that communicator are accessed using functions by Swift (lines 11–12). The communicator comm will be of the size specified by the @par annotation. Our HPC Zombies library interface is called (line 15) and returns a string containing the model output that is stored in z_value. Only one process need store the output z in Swift memory; we use rank 0 (lines 16–17). (This interface / body pattern can easily be adapted for any MPI library, adjusting for any differences in the wrapped library function arguments and additional input/output parameters.) With the various bindings having been created, the HPC Zombies model can now be called from a Swift script.
Figure 14.

Swift/T Tcl interface functions for HPC Zombies.
5. SUMMARY
In this tutorial paper, we have described a reusable, extensible software framework (EMEWS) for executing ME studies involving large simulation ensembles on massively parallel resources. The framework allows a great deal of flexibility in defining both the simulation model code and the ME algorithm. We demonstrated ABMs based on single-process Java and MPI-enabled C++, and the simple integration of external algorithms from R and Python. The overall workflow is controlled by a general-purpose script that can easily be extended by the user.
The key contributions of our framework are in scalability, variety of algorithms supported, and software sustainability. The underlying runtime system has previously been demonstrated to run at very large scale on Cray and IBM supercomputers. Our novel resident task plugin model allows 3rd party algorithms to be rapidly integrated. The use of such tools allows ME studies to be more easily packaged, distributed, and compared, which offers significant software sustainability benefit to the simulation community.
Figure 8.

Swift parallel tasks.
ACKNOWLEDGMENTS
This material is based upon work supported by the U.S. Department of Energy, Office of Science, under contract DE-AC02–06CH11357; by NSF awards ACI 1148443, BCS 1114851, DEB 1516428; and by NIH awards R01GM115839, R01AG047869, R01DA039934. This research used resources of the Argonne Leadership Computing Facility, which is a DOE Office of Science User Facility. This work was completed in part with resources provided by the Research Computing Center and the Beagle system at the University of Chicago.
AUTHOR BIOGRAPHIES
JONATHAN OZIK, Ph.D., is a Computational Scientist at Argonne National Laboratory and a Senior Fellow at the Computation Institute at the University of Chicago. His research focuses on agent-based modeling and large-scale model exploration. His email address is jozik@anl.gov.
NICHOLSON COLLIER, Ph.D., is a Software Engineer at Argonne National Laboratory and a staff member at the Computation Institute at the University of Chicago. His research focuses on agent-based and distributed agent-based modeling. His email address is ncollier@anl.gov.
JUSTIN M. WOZNIAK, Ph.D., is a Computer Scientist at Argonne National Laboratory and a Fellow at the Computation Institute at the University of Chicago. His research focuses on programming languages for large-scale computing. He received his Ph.D. at the University of Notre Dame. His email address is wozniak@mcs.anl.gov.
CARMINE SPAGNUOLO is a Ph.D. candidate in Computer Science at the University of Salerno and a Guest Graduate at Argonne National Laboratory. His research interests are in parallel algorithms, distributed systems, and agent-based simulations. His email address is cspagnuolo@unisa.it.
REFERENCES
- Alba E, Luque G, Garcia-Nieto J, Ordonez G, and Leguizamon G. 2007, April “MALLBA: A Software Library to Design Efficient Optimisation Algorithms”. International Journal of Innovative Computing Applications 1 (1): 74–85. [Google Scholar]
- Alba E, Luque G, and Nesmachnow S. 2013, January “Parallel Metaheuristics: Recent Advances and New Trends”. International Transactions in Operational Research 20 (1): 1–48. [Google Scholar]
- Armstrong TG, Wozniak JM, Wilde M, and Foster IT. 2014. “Compiler Techniques for Massively Scalable Implicit Task Parallelism”. In SC14: Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. [Google Scholar]
- Beaumont MA 2010. “Approximate Bayesian Computation in Evolution and Ecology”. Annual Review of Ecology, Evolution, and Systematics 41 (1): 379–406. [Google Scholar]
- Beazley DM 1996. “SWIG: An Easy to Use Tool for Integrating Scripting Languages with C and C++”. In Proceedings of the Fourth Annual USENIX Tcl/Tk Workshop. [Google Scholar]
- Boussad I, Lepagnot J, and Siarry P. 2013, July “A Survey on Optimization Metaheuristics”. Information Sciences 237:82–117. [Google Scholar]
- Carillo M, Cordasco G, Serrapica F, Scarano V, Spagnuolo C, and Szufel P. 2016, February “SOF: Zero Configuration Simulation Optimization Framework on the Cloud”. In 2016 24th Euromicro International Conference on Parallel, Distributed, and Network-Based Processing (PDP), 341–344. [Google Scholar]
- Collier N, and North M. 2012. “Parallel Agent-Based Simulation with Repast for High Performance Computing”. Simulation (90): 425–437. [Google Scholar]
- Collier N and North M 2015. “Repast Java Getting Started” Accessed Jul. 21, 2016 http://repast.sf.net/docs/RepastJavaGettingStarted.pdf.
- Csillery K, Francois O, and Blum MGB. 2012. “ABC: An R Package for Approximate Bayesian Computation (ABC)”. Methods in Ecology and Evolution 3 (3): 475–479. [DOI] [PubMed] [Google Scholar]
- Dean J, and Ghemawat S. 2004. “MapReduce: Simplified Data Processing on Large Clusters”. In Proceedings on Operating Systems Design and Implementation. [Google Scholar]
- Deelman E, Singh G, Su M-H, Blythe J, Gila Y, Kesselman C, Mehta G, Vahi K, Berriman GB, Good J, Laity A, Jacob JC, and Katz DS. 2005. “Pegasus: A Framework for Mapping Complex Scientific Workflows onto Distributed Systems”. Scientific Programming 13 (3): 219–237. [Google Scholar]
- Duro FR, Blas JG, Isaila F, Wozniak JM, Carretero J, and Ross R. 2016. “Flexible Data-aware Scheduling for Workflows over an In-memory Object Store”. In Proceedings of the ACM International Symposium on Cluster, Cloud and Grid Computing. [Google Scholar]
- EMEWS Tutorial Website 2016. “EMEWS Tutorial Website” Accessed Jul. 21, 2016 https://bitbucket.org/jozik/wintersim2016_adv_tut/src.
- Feldman SI 1979. “Make - A Program for Maintaining Computer Programs”. Software - Practice and Experience 9. [Google Scholar]
- Fortin F-A, Rainville F-MD, Gardner M-A, Parizeau M, and Gagn C. 2012, July “DEAP: Evolutionary Algorithms Made Easy”. Journal of Machine Learning Research 13:2171–2175. [Google Scholar]
- Foster I 1996. “Compositional Parallel Programming Languages”. Transactions on Programming Languages and Systems 18 (4). [Google Scholar]
- Goecks J, Nekrutenko A, and Taylor J. 2010. “Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences”. Genome Biology 11 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig F, Calabrese JM, Reineking B, Wiegand T, and Huth A. 2011, August “Statistical Inference for Stochastic Simulation Models Theory and Application”. Ecology Letters 14 (8): 816–827. [DOI] [PubMed] [Google Scholar]
- Isard M, Budiu M, Yu Y, Birrell A, and Fetterly D. 2007. “Dryad: Distributed Data-parallel Programs from Sequential Building Blocks”. In Proceedings of EuroSys 2007. [Google Scholar]
- Jabot F, Faure T, and Dumoulin N. 2013, July “EasyABC: Performing Efficient Approximate Bayesian Computation Sampling Schemes Using R”. Methods in Ecology and Evolution 4 (7): 684–687. [Google Scholar]
- Kuhn M 2008. “Building Predictive Models in R Using the Caret Package”. Journal of Statistical Software 28 (1): 1–26.27774042 [Google Scholar]
- Liaw A, and Wiener M. 2002. “Classification and Regression by RandomForest”. R News 2 (3): 18–22. [Google Scholar]
- Luke S 2013, June Essentials of Metaheuristics (Second Edition). Lulu. [Google Scholar]
- Luke S, Cioffi-Revilla C, Panait L, Sullivan K, and Balan G. 2005, July “MASON: A Multiagent Simulation Environment”. Simulation 81 (7): 517–527. [Google Scholar]
- Maheshwari K, Rodriguez A, Kelly D, Madduri R, Wozniak JM, Wilde M, and Foster IT. 2013a. “Enabling Multi-task Computation on Galaxy-based Gateways using Swift”. In Proceedings of the Science Gateway Institute Workshop. [Google Scholar]
- Maheshwari K, Rodriguez A, Kelly D, Madduri R, Wozniak JM, Wilde M, and Foster IT. 2013b. “Extending the Galaxy Portal with Parallel and Distributed Execution Capability”. In Proceedings of the Fourth International Workshop on Data Intensive Computing in the Clouds 2013. [Google Scholar]
- McPhillips T, Bowers S, Zinn D, and Ludäscher B. 2009. “Scientific Workflow Design for Mere Mortals”. Future Generation Computing Systems 25 (5). [Google Scholar]
- North MJ, Collier NT, Ozik J, Tatara ER, Macal CM, Bragen M, and Sydelko P. 2013, March “Complex Adaptive Systems Modeling with Repast Simphony”. Complex Adaptive Systems Modeling 1 (1): 3. [Google Scholar]
- Oinn T, Addis M, Ferris J, Marvin D, Greenwood M, Carver T, Pocock M, Wipat A, and Li P. 2004. “Taverna: A Tool for the Composition and Enactment of Bioinformatics Workflows”. Bioinformatics 20 (17). [DOI] [PubMed] [Google Scholar]
- Ozik J, Collier NT, and Wozniak JM. 2015. “Many Resident Task Computing in Support of Dynamic Ensemble Computations”. In 8th Workshop on Many-Task Computing on Clouds, Grids, and Supercomputers Proceedings Austin, Texas. [Google Scholar]
- Ozik J, Wilde M, Collier N, and Macal CM. 2014, January “Adaptive Simulation with Repast Simphony and Swift”. In Euro-Par 2014: Parallel Processing Workshops, edited by Lopes L and others, Number 8805 in Lecture Notes in Computer Science, 418–429. Springer International Publishing. [Google Scholar]
- Parejo JA, Ruiz-Corts A, Lozano S, and Fernandez P. 2011. “Metaheuristic Optimization Frameworks: A Survey and Benchmarking”. Soft Computing 16 (3): 527–561. [Google Scholar]
- Settles B 2012, June “Active Learning”. Synthesis Lectures on Artificial Intelligence and Machine Learning 6 (1): 1–114. [Google Scholar]
- Smith Paul D.. [Jul. 20, 2016]; “GNU Make Jobserver Implementation”. 2016 Accessed. http://make.mad-scientist.net/papers/jobserver-implementation.
- Stonedahl FJ 2011. Genetic Algorithms for the Exploration of Parameter Spaces in Agent-based Models Ph. D. thesis, Northwestern University, Evanston, IL, USA: AAI3489404. [Google Scholar]
- Taylor SJE, Anagnostou A, Kiss T, Terstyanszky G, Kacsuk P, and Fantini N. “A Tutorial on Cloud Computing for Agent-based Modeling & Simulation with Repast”. In Proceedings of the 2014 Winter Simulation Conference. Piscataway, New Jersey: Institute of Electrical and Electronics Engineers, Inc. [Google Scholar]
- Thain D, Tannenbaum T, and Livny M. 2005. “Distributed Computing in Practice: The Condor Experience”. Concurrency and Computation: Practice and Experience 17 (2–4). [Google Scholar]
- Top500. [Jul. 20, 2016]; “Top 500 web site”. 2016 Accessed. http://www.top500.org.
- Walker E, Xu W, and Chandar V. 2009. “Composing and Executing Parallel Data-flow Graphs with Shell Pipes”. In Workshop on Workflows in Support of Large-Scale Science at SC’09
- Wilensky Uri 1999. “NetLogo” Accessed Jul. 21, 2016 http://ccl.northwestern.edu/netlogo/ Center for Connected Learning and Computer-Based Modeling, Northwestern University; Evanston, IL. [Google Scholar]
- Wozniak JM, Armstrong TG, Maheshwari KC, Katz DS, Wilde M, and Foster IT. 2015. “Interlanguage Parallel Scripting for Distributed-memory Scientific Computing”. In WORKS ‘15: Proceedings of the 10th Workshop in Support of Large-Scale Science. [Google Scholar]
- Wozniak JM, Peterka T, Armstrong TG, Dinan J, Lusk EL, Wilde M, and Foster IT. 2013. “Dataflow Coordination of Data-parallel Tasks via MPI 3.0”. In Proceedings of Recent Advances in Message Passing Interface (EuroMPI). [Google Scholar]





