Abstract
Background
There is a scarcity of data on pneumococcal serotypes carried by children in Ethiopia. We studied pneumococcal nasopharyngeal carriage rate, serotypes, and risk factors among children with community acquired pneumonia (CAP).
Methods
A prospective observational cohort study was performed in children with CAP, aged 0–15 years, in 2 pediatric emergency departments in Addis Ababa, Ethiopia. Nasopharyngeal swabs were cultured, and serotypes of Streptococcus pneumoniae were determined by sequencing the cpsB gene and by the Quellung reaction. Risk factors were analyzed by using binary logistic regression.
Results
Nasopharyngeal swabs were collected from 362 children with CAP. Pneumococcal carriage rate was 21.5% (78 of 362). The most common serotypes were 19A (27%), 16F (8.5%), and 6A (4.9%). In addition, 8.5% of the pneumococcal isolates were nontypeable. In bivariate analysis, children with a parent that smokes were more likely to carry pneumococci (crude odds ratio, 3.9; 95% confidence interval [CI], 1.2–12.3; P = .023) than those with parents that do not smoke. In multivariable analysis, living in a house with ≥2 rooms (adjusted odds ratio [AOR], 0.48; 95% CI, 0.28–0.82; P = .007) and vaccination with ≥2 doses of 10-valent pneumococcal conjugate vaccine (PCV10) (AOR, 0.37; 95% CI, 0.15–0.92; P = .033) were protective of pneumococcal carriage.
Conclusions
Five years after introduction of PCV10 in Ethiopia, the vaccine-related serotype 19A was predominant in the nasopharynx of children with CAP. Continued evaluation of the direct and indirect impact of PCV10 on pneumococcal serotype distribution in Ethiopia is warranted.
Keywords: community-acquired pneumonia, Ethiopia, nasopharyngeal pneumococcal carriage, PCV10
Community-acquired pneumonia (CAP) accounts for considerable morbidity and mortality among children in both developed and developing countries. A 2015 estimate indicated that, globally, there were 138 million (95% uncertainty interval [UI], 86–226) episodes of pneumonia in children aged <5 years, 0.9 million (95% UI, 0.8–1.1) of which led to deaths. Ethiopia was among the 5 countries in which, collectively, 49% of the global pneumonia deaths occurred [1].
In children <5 years, Streptococcus pneumoniae is the most common bacterial cause of pneumonia [2]. Introduction of 7-valent pneumococcal conjugate vaccine (PCV7) initially in the United States in 2000; and the further enhanced formulations, PCV10 and PCV13, in more than 129 countries around the world, has proven to be quite effective in significantly reducing the burden of pneumococcal diseases such as meningitis, pneumonia, and sepsis [3].
However, accurate assessment of the real impact of PCV introduction on pneumococcal pneumonia is difficult and is hampered by the difficulty in identifying the cause of lower respiratory tract infections [4]. One of the main reasons is that the site of infection (lung) is not easily accessible for sample collection. The preferred specimens for diagnosis of invasive disease are sterile site specimens such as blood and pleural fluid. Although organisms identified by blood culture are generally known to be indicative for the etiology of pneumonia, most pneumonia cases are not bacteremic [5].
Due to the difficulty of obtaining suitable samples, studies have used nasopharyngeal samples along with other samples to establish the etiology of CAP [6, 7]. Although pneumococcal etiology of pneumonia cannot be established with certainty by the mere presence of S pneumoniae in the nasopharynx during pneumonia, it is widely accepted that nasopharyngeal carriage of S pneumoniae precedes infection [8]. Pneumococcal carriage is also more likely during respiratory infections, and a pneumococcal disease is more likely to be caused by the serotype that is present in the nasopharynx [4, 6].
The PCV10 was introduced without a catch-up campaign in Ethiopia in 2011 as a 3-dose primary series at 6, 10, and 14 weeks of age and no booster dose (3p + 0) [9]. There is scarcity of data on pneumococcal serotypes carried by healthy children or children with respiratory infections in Ethiopia. A population-based study between 2006 and 2007 in Northern Ethiopia before and after mass azithromycin for trachoma remains the only study in which pre-PCV10 nasopharyngeal pneumococcal serotyping was performed. According to this study, serotypes 19F (9.15%), 3 (4.9%), 6A (4.6%), 6B (3.8%), and 23F (3.8%) were the most common serotypes [10]. The aim of this study was to determine the pneumococcal carriage rate, serotype distribution, and risk factors in children with CAP in Ethiopia.
METHODS
Study Setting
The study was carried out in pediatric emergency departments (PEDs) of Tikur Anbessa Specialized Hospital and the Yekatit 12 Hospital, Addis Ababa, Ethiopia.
Patient Recruitment and Data Collection
We carried out a prospective observational cohort study from September 1, 2016 to August 30, 2017 in the same group of patients from which blood samples were collected for studying bacteremic CAP (described previously in [11]). Children were enrolled in the study if they were between the ages 0 to 15 years, presented at the 2 PEDs with clinical or radiological CAP, and fulfilled the following criteria: (1) cough and/or dyspnea with tachypnea and or chest indrawing; and/or (2) radiological confirmation of pneumonia by the attending radiologist according to World Health Organization (WHO) guidelines [12]; (3) first symptoms appearing within the last 2 weeks; and (4) no intake of antibiotics that was ascertained by historical recall, and no hospital or healthcare admissions within 2 weeks before the current admission. Tachypnea was defined as a respiratory rate of ≥60 inhalations/minute in children <2 months, ≥50 inhalations/minute in children 2–12 months, ≥40 inhalations/minute in children ≥1–5 years, and ≥20 inhalations/minute in children >5 years [13, 14]. Cases with underlying comorbidity or immunodeficiency were excluded. Trained research nurses approached parents/guardians of children who fulfilled the inclusion criteria for CAP during the study period. A structured questionnaire was used to obtain sociodemographic and other relevant clinical data.
Laboratory Procedures
Sample Collection
Sample collection and storage was done based on the standard method for detection of upper respiratory carriage of S pneumoniae recommended by WHO [15]. Nasopharyngeal swabs were collected from children suspected with CAP, using flocked mini-tip rayon swabs (Copan, Brescia, Italy), inserted deep into the nasopharynx, rotated, and advanced until resistance was found. Swabs were immediately placed in a tube with 1 mL skimmed milk-tryptone-glucose-glycerol (STGG) media. The inoculated STGG tubes were initially stored at −20°C in the respective hospitals for a maximum of 1 week and transported to the Armauer Hansen Research Institute (AHRI) on ice packs. Upon arrival at AHRI, they were stored at −80°C for a maximum of 1 month until they were cultured.
Culture and Identification
A 100-μL aliquot of each sample was pipetted and plated onto blood agar (BA) and BA with 2.5 mg of gentamicin per liter (BA + G), and the sample was streaked out using a sterile loop. The BA and BA + G plates were incubated at 37°C in ~5% CO2 or in candle jar for 18–24 hours. Initial identification of S pneumoniae was made on the basis of colony morphology, optochin susceptibility, and bile solubility at AHRI. Further identification on all isolates by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and amplification of the autolysin (lytA) gene were then performed at the Laboratory Bacteriology Research (Ghent University, Belgium).
Deoxyribonucleic Acid Extraction and Amplification of lytA
Deoxyribonucleic acid (DNA) extraction from S pneumoniae isolates was carried out by alkaline lysis as described previously [16]. Amplification of a 101-base pair (bp) fragment of the lytA gene was carried out as previously described [17], using the primers lytA F (5’-ACGCAATCTAGCAGATGAAGCA-3’) and lytA R (5’-TCGTGCGTTTTAATTCCAGCT-3’). The cycling conditions were 95°C for 5 minutes, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C, with a final extension at 72°C for 7 minutes. The polymerase chain reaction (PCR) amplicons were analyzed with 1.0% agarose gel electrophoresis.
PCRSeqTyping
PCRSeqTyping was performed by amplifying and sequencing the cpsB gene (encoding for phosphotyrosine phosphatase) as previously described [18]. Primers used to amplify 1061 bp of the cpsB gene were forward primer cps1-FP (5’-GCAATGCCAGACAGTAACCTCTAT-3’) and 2 reverse primers, cps2-RP (5’-CCTGCCTGCAAGTCTTGATT-3’) and cps-2538-RP (5’-CTTTACCAACCTT TGTAATCCAT-3’). The reaction mixture contained 3 µL of alkaline DNA extract, 15 µL of 2× FastStart Master Mix (Roche, Basel, Switzerland), and 0.4 µM of each primer, made up to a final volume of 30 µL with DNase/RNase-free distilled water. The cycling conditions were an initial denaturation step at 94°C for 5 minutes, followed by 35 amplification cycles of 94°C for 30 seconds, 50°C for 30 seconds, and 72°C for 1 minute. The PCR amplicons were analyzed with 1.0% agarose gel electrophoresis.
Twenty microliters of the amplicons were sent for sequencing to GATC Biotech (Constance, Germany), and sequencing was performed using the Sanger sequencing technique. cpsB sequences were used to interrogate GenBank database (http://www.ncbi.nlm.nih.gov/blast). Serotype was assigned based on the serotype of the highest Basic Local Alignment Search Tool (BLAST) bit score identity of the cpsB nucleotide sequence query with the reference sequences from GenBank.
Serotyping
Serotyping was performed by the Quellung reaction [19] using pool and group antisera, obtained from the Statens Serum Institut (Copenhagen, Denmark), at the Belgian Pneumococcal Reference Laboratory, Universitair Ziekenhuis, Katholieke Universiteit Leuven, Leuven, Belgium.
Final serotype was assigned using a combination of Quellung and PCRSeqTyping results. Because Quellung is the currently available gold standard [15], for all isolates on which Quellung serotyping was performed, the result was used as a final serotype. For isolates that were correctly typed to serotype level using PCRSeqTyping, some were also typed using Quellung to validate the results.
Statistical Analysis
Data were initially entered in REDCap (Vanderbilt University, Nashville, TN), exported into Excel, and analyzed using PASW Statistics 20 software (SPSS Inc., Chicago, IL). Descriptive statistics were used to analyze sociodemographic and clinical characteristics. Continuous variables were expressed as median and interquartile range (IQR), whereas categorical variables were expressed as frequencies and percentages. To identify predictors of pneumococcal carriage, bivariate analysis using binary logistic regression was performed. Variables significant at P < .1 were then used in a multivariable model to identify independent associations. Variables in the multivariable model were considered to be significant when P < .05. Results from the logistic regression analyses are reported as crude odds ratios and adjusted odds ratios (AORs) with 95% confidence interval (CIs).
Ethical Approval
The study was approved by the AHRI/All Africa Leprosy Rehabilitation and Training Hospital Ethics Review Committee, Addis Ababa University Institutional Review Board, Yekatit 12 Medical College Ethics Committee, and the National Research Ethics Review Committee (No. 310/194/2017). Written informed consent was obtained from parents/guardians of all participants before collection of samples.
RESULTS
Characteristics of the Study Population
Among 5402 children aged 0–15 years admitted to the emergency departments of Black Lion and Yekatit 12 hospitals during the study period, 856 (15.8%) were admitted with CAP and 583 (68%) of them met the inclusion criteria. Among those eligible, 362 (62%) consented to participate in the study, 109 (18.7%) refused, and 112 (19.3%) were not approached for consent due to various reasons. Of the children enrolled in the study, 208 (57.5%) were boys (Table 1). The median age of the children was 9 months (IQR, 3–18 months), and most of the children (80.1%) were infants (28 days–1 year). The pneumococcal nasopharyngeal carriage rate was 21.5% (78 of 362). Although the number of children who had a parent that smokes was small, 12 (3.3%), the pneumococcal carriage rate was the highest (50%) in these children. A total of 340 (93.9%) of the children have received at least 2 doses of PCV10.
Table 1.
Association Between Sociodemographic and Clinical Risk Factors and Pneumococcal Carriage Among Children Aged 0–15 Years With CAP
| All Children (n = 362) | Colonized With Pneumococci (n = 78) | Not Colonized With Pneumococci (n = 284) | Bivariate Analysis | Multivariable Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| Variable | Category | No. (%) | No. (%) | No. (%) | COR (95% CI) | P a | AOR (95% CI) | P a | |
| Gender | Male | 208 (57.5) | 36 (17.3) | 172 (82.7) | Ref | Ref | |||
| Female | 154 (42.5) | 42 (27.3) | 112 (72.7) | 1.79 (1.08–2.97) | .024 | 1.72 (1.03–2.89) | .040 | ||
| Age | <28 days | 3 (0.8) | 0 (0.0) | 3 (100) | (0.00) | .999 | - | - | |
| 28 days–1 year | 290 (80.1) | 63 (21.7) | 227 (78.3) | 1.94 (0.24–16.08) | .538 | - | - | ||
| 2 years–5 years | 61 (16.9) | 14 (23.0) | 47 (77.0) | 2.08 (0.24–18.42) | .509 | - | - | ||
| ≥6 years | 8 (2.2) | 1 (12.5) | 7 (87.5) | Ref | |||||
| Premature birth | No | 297 (82) | 63 (21.2) | 234 (78.8) | Ref | ||||
| Yes | 65 (18) | 15 (23.1) | 50 (76.9) | 1.11 (0.59–2.11) | .74 | - | - | ||
| Birth weight | <2.5 kg | 40 (11) | 9 (22.5) | 31 (77.5) | Ref | ||||
| ≥2.5 kg | 322 (89) | 69 (21.4) | 253 (78.6) | 0.94 (0.43–2.07) | .876 | - | - | ||
| Breastfeeding | No | 138 (38.1) | 24 (17.4) | 114 (82.6) | Ref | ||||
| Yes | 224 (61.9) | 54 (24.1) | 170 (75.9) | 1.51 (0.88–2.58) | .133 | - | - | ||
| Siblings aged <5 years | No | 294 (81.2) | 66 (22.4) | 228 (77.6) | Ref | ||||
| Yes | 68 (18.8) | 12 (17.6) | 56 (82.4) | 0.74 (0.38–1.46) | .387 | - | - | ||
| Day care/preschool attendance | No | 326 (90.1) | 67 (20.6) | 259 (79.4) | Ref | ||||
| Yes | 36 (9.9) | 11 (30.6) | 25 (69.4) | 1.70 (0.79–3.63) | .17 | - | - | ||
| No. of household members | ≤3 | 115 (31.8) | 25 (21.7) | 90 (78.3) | Ref | ||||
| 4–6 | 220 (60.8) | 47 (21.4) | 173 (78.6) | 0.98 (0.56–1.69) | .937 | - | - | ||
| ≥7 | 27 (7.5) | 6 (22.2) | 21 (77.8) | 1.03 (0.37–2.82) | .956 | - | - | ||
| No. of rooms in the house | 1 | 105 (29) | 33 (31.4) | 72 (68.6) | Ref | ||||
| ≥2 | 257 (71) | 45 (17.5) | 212 (82.5) | 0.46 (0.28–0.78) | .004 | 0.48 (0.28–0.82) | .007 | ||
| Parental smoking | No | 350 (96.7) | 72 (20.6) | 278 (79.4) | Ref | ||||
| Yes | 12 (3.3) | 6 (50.0) | 6 (50.0) | 3.86 (1.21–12.33) | .023 | 3.0 (0.90–9.97) | .073 | ||
| Cooking fuel | Firewood | Yes | 58 (16) | 12 (14.7) | 46 (79.3) | 0.94 (0.47–1.88) | .862 | - | - |
| Charcoal | Yes | 219 (60.5) | 52 (23.7) | 167 (76.3) | 0.71 (0.42–1.21) | .209 | - | - | |
| Electricity | Yes | 209 (57.7) | 42 (20.1) | 167 (79.9) | 0.82 (0.49–1.35) | .433 | - | - | |
| Gas | Yes | 22 (6.1) | 6 (27.3) | 16 (72.7) | 1.39 (0.53–3.69) | .502 | - | - | |
| URTI in the last 3 months | No | 313 (86.5) | 64 (20.4) | 249 (79.6) | Ref | ||||
| Yes | 49 (13.5) | 14 (28.6) | 35 (71.4) | 1.56 (0.79–3.06) | .201 | - | - | ||
| LRTI in the last 3 months | No | 306 (84.5) | 67 (21.9) | 239 (78.1) | Ref | ||||
| Yes | 56 (15.5) | 11 (19.6) | 45 (80.4) | 0.87 (0.43–1.78) | .706 | - | - | ||
| Hospital admission in the last 3 months | No | 312 (86.2) | 64 (20.5) | 248 (79.5) | Ref | ||||
| Yes | 50 (13.8) | 14 (28.0) | 36 (72.0) | 1.51 (0.77–2.96) | .234 | - | - | ||
| Vaccination with PCV10 | ≤1 dose | 22 (6.1) | 9 (40.9) | 13 (59.1) | Ref | ||||
| ≥2 doses | 340 (93.9) | 69 (20.3) | 271 (79.7) | 0.37 (0.15–0.89) | .028 | 0.37 (0.15–0.92) | .033 | ||
| Nutritional statusb | Well nourished | 278 (76.8) | 56 (20.1) | 222 (79.9) | Ref | ||||
| Moderately malnourished | 36 (9.9) | 9 (25) | 27 (75) | 1.32 (0.59–2.97) | .5 | - | - | ||
| Severely malnourished | 48 (13.3) | 13 (27.9) | 35 (72.9) | 1.47 (0.73–2.97) | .279 | - | - |
Abbreviations: AOR, adjusted odds ratio; CAP, community-acquired pneumonia; CI, confidence interval; COR, crude odds ratio; LRTI, lower respiratory tract infection; PCV, pneumococcal conjugate vaccine; Ref, reference; URTI, upper respiratory tract infection.
a P value of binary logistic regression analysis. Values in bold: P < .05.
bWorld Health Organization child growth standard of weight for age z score was used to determine nutritional status: well nourished, z-score ≥ −2.0; moderately malnourished, −3.0 < z-score < −2.0; severely malnourished, z-score < −3.0.
Predictors of Pneumococcal Carriage
In bivariate analysis, children with a parent that smokes were more 3.9 times more likely to carry pneumococci (95% CI, 1.2–12.3; P = .023) than those with parents that do not smoke (Table 1). According to multivariable analysis, female sex was positively associated with pneumococcal carriage, whereas living in a house with ≥2 rooms and vaccination with ≥2 doses of PCV10 were negatively associated with pneumococcal carriage (Table 1). Girls were 1.7 times more likely to carry pneumococci than boys (95% CI, 1.03–2.89; P = .040). However, the CI for the mean carriage rate of boys and girls were overlapping (boys, 17% [95% CI, 12–22]; girls, 27% [95% CI, 20–34]). Children living in a house with 2 or more rooms were less likely to carry pneumococci compared with those living in a house with only 1 room (AOR, 0.48; 95% CI, 0.28–0.82; P = 0.007). Children who received at least 2 doses of PCV10 were also less likely to carry pneumococci than those who were unvaccinated or received only 1 dose (AOR, 0.37; 95% CI, 0.15–0.92; P = .033).
Pneumococcal Serotypes and Vaccine Coverage
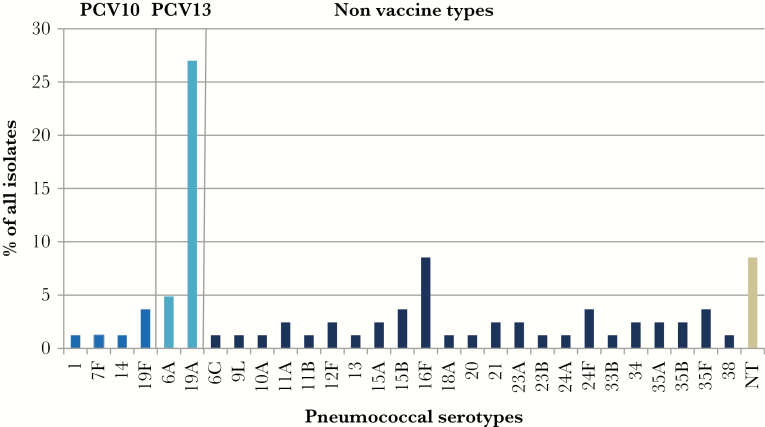
Of the 362 cultures from nasopharyngeal swabs, 78 (21.5%) yielded pneumococci. A total of 29 different serotypes plus nontypeable pneumococci were identified. The most common serotypes colonizing the nasopharynx of children with CAP were 19A (27%), 16F (8.5%), and 6A (4.9%) (Figure 1). In addition, 8.5% of the pneumococcal isolates were nontypeable. Most of the serotypes were non-PCV10 serotypes (95.1%). Vaccine-related serotypes (the additional 3 serotypes included in PCV13) constituted 31.9% of all serotypes. The PCV13 would increase the potential coverage from 4.9% to 39.2%.
Figure 1.
Nasopharyngeal carriage of pneumococcal serotypes in children with community acquired pneumonia. NT, nontypeable; PCV, pneumococcal conjugate vaccine.
PCRSeqTyping
Of the 78 isolates that were subjected to PCRSeqTyping, amplification of the cpsB gene (1061 bp) was possible for 72 (92.3%) isolates. After sequence analysis, 30 (41.7%) of the 72 isolates were identified at the serotype level, 11 (15.3%) were identified at the serogroup level, and 31 (43.1%) were identified at subtype level (Table 2). Isolates that were identified at subtype level were assigned into homologous groups and amplified using group-specific primers as previously indicated [18] and sequenced. Of 11 isolates that were initially identified to subtype level and amplified using group-specific primers, only 1 was resolved to serotype level.
Table 2.
Quellung Serotyping and PCRSeqTyping Results of Pneumococcal Isolates
| Serotype (na) | PCRSeqTyping (na) | Quellung (na) |
|---|---|---|
| 1 | 1/14/16F/19F | 1 |
| 6A (4) | 6A/6B/6C (3) | 6A (3) |
| 6A | NT | |
| 6C | 6A/6B/6C | 6C |
| 9L | 9I/9N/13/20 | 9L |
| 10A | 10A | NT |
| 11A (2) | 11A/11D/18F (2)c | 11A (2) |
| 11B | 11B/11Cb | 11B |
| 12F (2) | 12A/12B/12F/44/46 (2)b | 12F (2) |
| 13 | 13/20b | 13 |
| 14 | 1/7/14/16F/19F/ 1 | 14 |
| 15A (2) | 15A/15F (2) | 15A (2) |
| 15B (3) | 15B/15C/19B/23A/24F(3) | 15B (3) |
| 16F (7) | 16F/19F (5) | 16F (5) |
| 16F (2) | NT (2) | |
| 18A | 11B/11C/18F/23B | 18A |
| 19A (22) | 19A (6) | 19A (6) |
| 19A (12) | NT (12) | |
| 4/10A/10B/17F | 19A | |
| 1/7/14/16F/19A/19F/40 | 19A | |
| 4, 19A/19V/33A/33B/33C/33F | 19A | |
| 19F (3) | 19A/19F | 19F |
| 1/14/16F/19A/19F | 19F | |
| 19F | NT | |
| 20 | 9N/9I/13/20 | 20 |
| 21 (2) | 21 | 21 |
| 21 | NT | |
| 23A (2) | 23A | 23A |
| 23A | NT | |
| 24A | 24A | 24A |
| 24F (3) | 19B/24F, 23A (2) | 24F (2) |
| 24F | 24F | |
| 33B | 6A/6B/17A/33B/33D/34 | 33B |
| 34 (2) | 17A/34 | 34 |
| 34 | NT | |
| 35A (2) | 17F/35A/35B/35C/35F (2)b | 35A (2) |
| 35B (2) | 35A/35B/35C/35F (2)b | 35B (2) |
| 35F (3) | 35A/35B/35C/35F (2) b | 35F (2) |
| 35F, 47Fb | 35F | |
| 38 | Negatived | 38 |
| Nontypeable (7) | 10C/10F | Negative |
| 17A/34 | Negative | |
| Negatived (5) | Negative (5) |
Abbreviations: NT, not tested; PCR, polymerase chain reaction.
aNumber of isolates if more than 1.
bIsolates that were initially identified to subtype level and amplified using group-specific primers.
cIsolate that was resolved from subtype to serotype level in the second step of PCRSeqTyping.
d cpsB-PCR negative. All other isolates were both cpsB and lytA PCR positive.
Quellung Serotyping
A total of 51 isolates—6 cpsB negative, 31 identified to subtype level, 5 identified to serogroup level, and 9 identified to serotype level (as positive controls)—were serotyped using the Quellung reaction. Five of the 6 cpsB-negative isolates were also Quellung negative, whereas 1 was serotype 38 (Table 2).
Of the 31 isolates that were identified to subtype level by PCRSeqTyping, 29 were correctly serotyped according to the Quellung reaction, whereas 2 were Quellung negative. All 6 isolates that were identified to serogroup level by PCRSeqTyping were correctly serotyped according to Quellung reaction.
Final serotype was assigned using a combination of Quellung and PCRSeqTyping results. For all isolates on which Quellung serotyping was performed (51), the result was used as a final serotype. For the remaining isolates (27), which were typed to serotype level using PCRSeqTyping, the result was used as a final serotype. This was done taking into consideration that there was a 100% concordance between PCRSeqTyping and Quellung for the 9 isolates (positive controls), which were similarly typed to serotype level by PCRSeqTyping.
DISCUSSION
Nasopharyngeal carriage of S pneumoniae, besides being a source of pneumococcal transmission between people, is also known to precede infection [20], and carriage of vaccine-type (VT) pneumococci has been reported to be associated with pneumonia [21]. We studied pneumococcal nasopharyngeal carriage rate, serotypes, and associated risk factors in children with CAP.
In this study, pneumococcal nasopharyngeal carriage rate was 21.5%. Our findings were lower than results from a similar study in Mozambique (45.1%) [21], which was performed 1 to 3 years after introduction of PCV10 in the country. However, our results were higher than results from a similar study in China (17.3%) [22] performed before the introduction of PCV in the country using nasopharyngeal aspirate cultures.
The most plausible reason for the low carriage rate in our study might be previous antibiotic exposure. Although we tried to excluded patients exposed to antibiotics within 2 weeks of sampling, this was done using historic recall by parents or guardians. Studies have indicated that antibiotic exposure reduces yield of S pneumoniae from nasopharyngeal culture by approximately 30% [21, 23]. In addition, in children with pneumonia and with high rate of exposure to antibiotics, compared with culture, PCR has been able to provide a 45% enhancement in pneumococcal yield [21]. Therefore, our results might have been affected by possible antibiotic exposure before swabbing, which was further compounded by the use of culture instead of PCR.
We identified 29 different serotypes plus nontypeable pneumococci. Carriage rate of PCV10 serotypes was 4.9%. The residual VT carriage in our study is lower than results obtained in Kenya [24] and Mozambique [21]. In Kenya, 5 years after introduction of PCV10 with 3p + 0 schedule with a catch-up vaccination, the residual VT colonization was 6% in children less than 5 years of age and 7% in those aged 5–14 years [24]. In Mozambique, 1–3 years after the introduction of PCV10 with 3p + 0 schedule, residual VT colonization in children with pneumonia was 18.6% and pneumonia was associated with VT pneumococcal colonization [21]. Using more mature programs, countries such as the United States and the United Kingdom have been able to achieve near elimination of VT carriage [25, 26], which has led to the consideration of lower dose (2 + 1 and 1 + 1) regimens [27]. Although our findings indicate a residual carriage of VT pneumococci that is lower than other countries using the same vaccination program, the number of VT isolates in our study is low. Therefore, it is important to perform large-scale carriage studies to assess both the direct and indirect effects of PCV10 in Ethiopia.
Our findings also indicate that serotype 19A is the predominant serotype (27% of all isolates) followed by 16F (8.5% of all isolates), nontypeables (8.5% of all isolates), and 6A (4.9% of all isolates). In the pre-PCV10 study by Keenan et al [10] in Northern Ethiopia, serotype 19F (9.15%) was the predominant serotype, whereas serotype 19A constituted only 2.1% of all isolates. In another study on the impact of PCV10 on nasopharyngeal carriage of pneumococci in healthy children in Addis Ababa, Ethiopia, serotype 6A (5.0%) was the most common serotype [28]. One notable finding is that the percentage of serotype 6A remained similar in the pre-PCV10 study in Northern Ethiopia (4.6%), in the previous study in Addis Ababa (5.0%), and in our study (5.1%). A similar trend in the carriage of serotype 19A and 6A has been reported in children <5 years in Kenya whereby carriage of serotype 19A increased, whereas there was no effect on serotype 6A carriage 5 years after introduction of PCV10 in the country [24]. Likewise, in Mozambique, 6 months to 2 years after introduction of PCV10 in the country, colonization prevalence for the 3 additional serotypes included in PCV13 (3, 6A, and 19A) increased significantly (12.4% to 20.7%; P = .009), which was mainly driven by an increase in the prevalence of 19A [24].
Contrary to these findings, however, in a clinical trial in Finland that assessed the effectiveness of PCV10 using vaccine schedules of 3 primary doses with a booster (3p + 1) or 2 primary doses with a booster (2p + 1), a trend towards decreased carriage of serotype 19A was observed [29]. Likewise, in Brazil, 3 years after introduction of PCV10, which is given in a 3p + 1 schedule, although carriage of vaccine-related serotypes increased (10.8%–21.0%; P < .0001), it was driven primarily by a rise in serotype 6C (1.8%–11.2%; P < .0001), and carriage of serotypes 6A and 19A did not significantly change [30]. Therefore, it is important to continuously monitor the distribution of serotypes both in carriage and disease in the post-PCV10 era in Ethiopia.
We performed molecular serotyping using PCRSeqTyping [18] and performed the Quellung reaction [19] for all isolates that could not be typed to serotype level using PCRSeqTyping and 9 isolates that were typed to serotype level by PCRSeqTyping. Although the Quellung method remains the gold standard for pneumococcal serotyping, it is laborious, expensive, and requires technical expertise and experience. Availability of cpsB sequences of the 90 pneumococcal serotypes [31] has paved the way for the development of Sequetyping [32], a method based on amplification of the cpsB gene with 1 pair of primers and sequencing. This was further developed into PCRSeqTyping [18]. In our study, only 38.5% of the isolates were identified to serotype level, whereas 6 (7.7%) were cpsB negative in the first step and only 1 additional isolate was resolved from subtype to serotype level during the second step. In the study by Nagaraj et al [18], 64.8% of the 91 pneumococcal reference strains and 89.3% of the 28 clinical isolates were identified to serotype level during the first step, and all of the strains/isolates were identified to the serotype level during the second step. Our results also indicated that of the 6 cpsB-negative isolates, 5 were also Quellung negative, whereas 1 was serotype 38. Serotype 38 was one of the serotypes predicted in silico to be nonamplifiable [32].
The cpsB gene is known to have extensive interstrain variation due to natural transformation [31, 33, 34], to which regional differences might also further contribute to the variation. However, it is unclear why in our study PCRSeqTyping failed to resolve the serotype to the serotype level in so many isolates. Further studies from different regions are required to validate PCRSeqTyping.
Besides identifying pneumococcal serotypes, we also identified risk factors for pneumococcal carriage in Ethiopian children with CAP. In bivariate analysis, parental smoking was a risk factor associated with pneumococcal carriage. Children with a parent that smokes were 3.9 times more likely to carry pneumococci than those with parents that do not smoke. A similar finding has been reported in a study from Israel [35].
In multivariable analysis, living in a house with 2 or more rooms and vaccination with 2 or more doses of PCV10 were protective of pneumococcal carriage. Although results of the multivariable analysis indicate that girls were more likely to carry pneumococci than boys, the mean carriage rates have overlapping CIs, indicating that there is weak to no evidence of pneumococcal carriage difference by gender. Children living in a house with 2 or more rooms were less likely to carry pneumococci compared with those living in house with only 1 room, which is in agreement with a previous study from Gondar, Ethiopia [36]. Children who have received 2 or more doses of PCV10 were also less likely to carry pneumococci compared with those who have received only 1 dose or less. Similar to our findings, in a population-based study that assessed the direct effect of PCV10 on pneumococcal carriage in children in Brazil, pneumococcal carriage was significantly lower in children who received 2 or more doses of PCV10 compared with those who were either unvaccinated or received only 1dose [37].
Our study had some limitations. Previous antibiotic usage was recorded using historic recall, and we were not able to sample urine to detect recent antibiotic usage. Besides this, due to the involvement of many nurses in sample collection, personal differences in correctly swabbing the nasopharynx may have also contributed to the low level of pneumococcal carriage. Therefore, the pneumococcal carriage rate we reported could be an underestimation.
CONCLUSIONS
Five years after the introduction of PCV10-based vaccination against pneumococci in Ethiopia, non-PCV10 serotypes were the most common serotypes carried in the nasopharynx of children with CAP. Overall, serotype 19A, which is a PCV13 serotype, was predominant. In multivariate analysis, living in a house with 2 or more rooms and receiving 2 or more doses of PCV10 were protective of pneumococcal colonization.
Acknowledgments
We express our sincere gratitude to all of the children and guardians who participated in the study. We also thank the physicians and nurses who participated in clinical diagnosis and sample collection and the Armauer Hansen Research Institute (AHRI) data management team for data entry.
Disclaimer. The funders had no role in study design, data collection, analysis, interpretation and writing the manuscript. The views expressed here are those of the authors and do not necessarily represent the views of the Belgian Development Cooperation or AHRI.
Financial support. This research was supported by a PhD Scholarship for A. A. N. from the Belgian Development Cooperation through VLIR-UOS. Additional funding was received from AHRI.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. McAllister DA, Liu L, Shi T, et al. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob Health 2019; 7:e47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rudan I, O’Brien KL, Nair H, et al. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health 2013; 3:010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alicino C, Paganino C, Orsi A, et al. The impact of 10-valent and 13-valent pneumococcal conjugate vaccines on hospitalization for pneumonia in children: a systematic review and meta-analysis. Vaccine 2017; 35:5776–85. [DOI] [PubMed] [Google Scholar]
- 4. Chappuy H, Keitel K, Gehri M, et al. Nasopharyngeal carriage of individual Streptococcus pneumoniae serotypes during pediatric radiologically confirmed community acquired pneumonia following PCV7 introduction in Switzerland. BMC Infect Dis 2013; 13:357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hammitt LL, Murdoch DR, Scott JA, et al. Specimen collection for the diagnosis of pediatric pneumonia. Clin Infect Dis 2012; 54(Suppl 2):S132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bénet T, Sylla M, Messaoudi M, et al. Etiology and factors associated with pneumonia in children under 5 years of age in Mali: a prospective case-control study. PLoS One 2015; 10:e0145447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holter JC, Müller F, Bjørang O, et al. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis 2015; 15:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simell B, Auranen K, Käyhty H, et al. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 2012; 11:841–55. [DOI] [PubMed] [Google Scholar]
- 9. Ministry of Health Federal Republic of Ethiopia. Introducing Pneumococcal Conjugate Vaccine in Ethiopia: Training Manual for Health Workers. Ethiopia: Addis Ababa; 2011. [Google Scholar]
- 10. Keenan JD, Sahlu I, McGee L, et al. Nasopharyngeal pneumococcal serotypes before and after mass azithromycin distributions for trachoma. J Pediatric Infect Dis Soc 2016; 5:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Negash AA, Asrat D, Abebe W, et al. Bacteremic community-acquired pneumonia in Ethiopian children: etiology, antibiotic resistance, risk factors, and clinical outcome. Open Forum Infect Dis 2019; 6:ofz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cherian T, Mulholland EK, Carlin JB, et al. Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ 2005; 83:353–9. [PMC free article] [PubMed] [Google Scholar]
- 13. World Health Organization. Pocket Book of Hospital Care for Children: Guidelines for the Management of Common Illnesses With Limited Resources. Geneva: World Health Organization; 2013. [PubMed] [Google Scholar]
- 14. Long SS, Pickering LK, Prober CG.. Principles and Practice of Pediatric Infectious Diseases. London: Elsevier Inc.; 2012. [Google Scholar]
- 15. Satzke C, Turner P, Virolainen-Julkunen A, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013; 32:165–79. [DOI] [PubMed] [Google Scholar]
- 16. Vaneechoutte M, Claeys G, Steyaert S, et al. Isolation of Moraxella canis from an ulcerated metastatic lymph node. J Clin Microbiol 2000; 38:3870–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verhelst R, Kaijalainen T, De Baere T, et al. Comparison of five genotypic techniques for identification of optochin-resistant pneumococcus-like isolates. J Clin Microbiol 2003; 41:3521–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagaraj G, Ganaie F, Govindan V, Ravikumar KL. Development of PCRSeqTyping—a novel molecular assay for typing of Streptococcus pneumoniae. Pneumonia 2017:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sørensen UB. Typing of pneumococci by using 12 pooled antisera. J Clin Microbiol 1993; 31:2097–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 2004; 4:144–54. [DOI] [PubMed] [Google Scholar]
- 21. Adebanjo T, Lessa FC, Mucavele H, et al. Pneumococcal carriage and serotype distribution among children with and without pneumonia in Mozambique, 2014-2016. PLoS One 2018; 13:e0199363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu YY, Xie XH, Ren L, et al. Epidemiological characteristics of nasopharyngeal Streptococcus pneumoniae strains among children with pneumonia in Chongqing, China. Sci Rep 2019; 9:3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Driscoll AJ, Deloria Knoll M, Hammitt LL, et al. The effect of antibiotic exposure and specimen volume on the detection of bacterial pathogens in children with pneumonia. Clin Infect Dis 2017; 64:368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammitt L, Etyang AO, Morpeth SC, et al. Impact of 10-valent pneumococcal conjugate vaccine on invasive pneumococcal disease and nasopharyngeal carriage in Kenya. Lancet 2019. April 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Southern J, Andrews N, Sandu P, et al. Pneumococcal carriage in children and their household contacts six years after introduction of the 13-valent pneumococcal conjugate vaccine in England. PLoS One 2018; 13:e0195799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loughlin AM, Hsu K, Silverio AL, et al. Direct and indirect effects of PCV13 on nasopharyngeal carriage of PCV13 unique pneumococcal serotypes in Massachusetts’ children. Pediatr Infect Dis J 2014; 33:504–10. [DOI] [PubMed] [Google Scholar]
- 27. Goldblatt D, Southern J, Andrews NJ, et al. Pneumococcal conjugate vaccine 13 delivered as one primary and one booster dose (1 + 1) compared with two primary doses and a booster (2 + 1) in UK infants: a multicentre, parallel group randomised controlled trial. Lancet Infect Dis 2018; 18:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sime WT, Aseffa A, Woldeamanuel Y, Brovall S, Morfeldt E, Henriques-Normark B. Serotype and molecular diversity of nasopharyngeal Streptococcus pneumoniae isolates from children before and after vaccination with the ten-valent pneumococcal conjugate vaccine (PCV10) in Ethiopia. BMC Infect Dis 2019; 19:409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vesikari T, Forsten A, Seppä I, et al. Effectiveness of the 10-valent pneumococcal nontypeable Haemophilus influenzae protein D-conjugated vaccine (PHiD-CV) against carriage and acute otitis media-A double-blind randomized clinical trial in Finland. J Pediatric Infect Dis Soc 2016; 5:237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brandileone MC, Zanella RC, Almeida SC, et al. Effect of 10-valent pneumococcal conjugate vaccine on nasopharyngeal carriage of Streptococcus pneumoniae and Haemophilus influenzae among children in São Paulo, Brazil. Vaccine 2016; 34:5604–11. [DOI] [PubMed] [Google Scholar]
- 31. Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2006; 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leung MH, Bryson K, Freystatter K, et al. Sequetyping: serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J Clin Microbiol 2012; 50:2419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salter SJ, Hinds J, Gould KA, et al. Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. Microbiology 2012; 158:1560–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wen Z, Liu Y, Qu F, Zhang J. Allelic variation of the capsule promoter diversifies encapsulation and virulence in Streptococcus pneumoniae. Sci Rep 2016; 6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greenberg D, Givon-Lavi N, Broides A, et al. The contribution of smoking and exposure to tobacco smoke to Streptococcus pneumoniae and Haemophilus influenzae carriage in children and their mothers. Clin Infect Dis 2006; 42:897–903. [DOI] [PubMed] [Google Scholar]
- 36. Assefa A, Gelaw B, Shiferaw Y, Tigabu Z. Nasopharyngeal carriage and antimicrobial susceptibility pattern of Streptococcus pneumoniae among pediatric outpatients at Gondar University Hospital, North West Ethiopia. Pediatr Neonatol 2013; 54:315–21. [DOI] [PubMed] [Google Scholar]
- 37. Andrade AL, Ternes YM, Vieira MA, et al. Direct effect of 10-valent conjugate pneumococcal vaccination on pneumococcal carriage in children Brazil. PLoS One 2014; 9:e98128. [DOI] [PMC free article] [PubMed] [Google Scholar]