Abstract
Canine circovirus (CanineCV) was detected, together with canine parvovirus (CPV), in samples from an outbreak of fatal gastroenteritis in dogs in Argentina. We obtained the full-length genome of this recently discovered virus by overlapping PCR, designated strain UBA-Baires. Sequence analysis revealed a highly conserved genome but also showed several unique mutations in amino acids from the capsid protein that have not been previously reported. Phylogenetic analysis shows that this strain is more closely related to European strains than to viruses detected in North America or Asia. Although the pathogenic role of CanineCV in dogs is still unclear, this study highlights the importance of CanineCV as a coinfecting virus in disease development. To our knowledge, this is the first report of the involvement of CanineCV in severe clinical disease in dogs in South America. Our results expand our information on the geographical extent of this virus and contribute to the understanding of its role in disease.
Introduction
A newly described canine circovirus (CanineCV) was identified recently in serum from healthy dogs [1]. Since then, several countries have reported the molecular detection of CanineCV in cases associated with clinical disease, ranging from sudden death of puppies with bloody diarrhea to respiratory syndromes [2–6]. Lesions found include necrotizing vasculitis, lymphoid necrosis, granulomatous inflammation and necrosis of intestinal crypts and Peyer’s patches. Some characteristics of the disease observed in dogs (enteritis, pneumonia, lymphadenitis) were also present in pigs infected with porcine circovirus 2 (PCV-2), which was found to be genetically related [7].
A number of case-control studies have been published, most of them describing significant association between diarrhea and CanineCV detection [8–11]. The role of this new virus in pathogenesis is however difficult to assess since it is also found in feces from healthy dogs. Furthermore, the first description was done from serum samples from six healthy animals [1] and there have been recent reports of CanineCV in serum samples of dogs from Brazil and China [12, 13]. Additionally, coinfection with other viral pathogens, mainly canine parvovirus (CPV), but also canine distemper virus, canine coronavirus, and canine influenza virus is present in CanineCV positive dogs with clinical symptoms at rates between 50 to 100% [4, 6, 8–11]. Viral coinfection is also a main feature of PCV-2 infection [14, 15].
Circoviruses belong to the family Circoviridae, genus circovirus; they are small, non-enveloped virus with a circular single strand DNA genome of around 2000 bp [16]. The genome contains two main (and opposite) transcription units which encode two ORFs, a replicase associated protein (Rep) and the capsid protein (Cap). CanineCV sequences submitted to Genbank are quite similar differing slightly from those identified in wildlife. Nevertheless, the division of CanineCV into two genotypes (CanineCV-1 and Canine CV-2) based on the cap gene sequence has been proposed very recently [12].
In this work, we present, to our knowledge, the first report of a complete full-length CanineCV genome sequence from South America.
Materials and methods
Ethics statement
This study did not involve any animal experiments. Tissue samples were collected during necropsy of dead animals for laboratory analysis, with consent from the dog’s owner.
Animal description and sample collection
Severe disease outbreaks occurred in a mixed-breed colony in Buenos Aires between 2014 and 2015, in a cascade fashion. The age of affected dogs ranged from 50 days to one year, belonging to different breeds. The attending veterinarian described the situation as a highly contagious disease of very rapid onset and development, characterized by anorexia, vomiting and hemorrhagic diarrhea lasting from three to 7 days. In total, 18 puppies died, and the rest recovered within a week. All dogs had been properly vaccinated against CPV, canine distemper virus and infectious canine hepatitis virus. Fresh tissues obtained from pancreas, liver, kidney, lung, lymph nodes, spleen, large intestine and stomach belonging to three dead animals were submitted to the Virology Department of the Faculty of Veterinary Sciences at the University of Buenos Aires for testing. Samples belonging to surviving animals or to the other dead dogs were not available.
DNA extraction and PCR detection
The presence of CanineCV DNA was analyzed by PCR. Tissues were processed for DNA extraction using DNazol (Invitrogen). One hundred ng of DNA were used in a PCR reaction using 25 pmoles of each primer set (Table 1), 1X reaction buffer, 1 unit GoTaq (Promega), 200 μM dNTPs in a total volume of 50ul. Primers were designed according to conserved sequences of CanineCV deposited in Genbank using the Primer-BLAST program tool (NCBI-NIH). In a first approach, the primer set For genomic/Rev 533 was utilized to amplify a 533 bp fragment from the Rep gene for diagnostic purposes, after which positive samples were further tested with other primer sets in order to cover the entire genome for sequencing purposes (Table 1).
Table 1. List of primers used in this study.
| Primer sequence 5’-3’ | Name and location | Genomic position |
|---|---|---|
| ATGGCTCAAGCTCAGGTTG | For genomic | 1–20 |
| CTTGCGCGAGCTGCTCCTTA | For 456 | 456–478 |
| CCGCACAGAACCTCCACTTC | Rev 533 | 514–533 |
| AAT AAACAGCCT CGAAGGGG | Rev 1055 | 1036–1055 |
| GTTCCATAGCAACGGGGTCT | For 1734 | 1734–1753 |
| CATGCGCGTGCGGAGACATG | Rev 1910 | 1910–1929 |
| GTCGGAGGAGCGGAGGTGT | Rev genomic | 2045–2063 |
Samples were additionally tested for CPV. Two μL (10 ng) of extracted DNA were used in a PCR reaction that amplifies a 583 bp fragment of the VP2 gene using 555for and 555rev primers; this fragment includes a MboII restriction site identified by Buonavoglia et al. which allows the differentiation of CPV-2c strains from other CPV strains [17]. The resulting amplicon was digested with MboII to detect the presence of the CPV-2c strain´s characteristic substitution in VP2 gene (T to A at nt 4026) as described previously [17].
Full-genome sequencing of CanineCV strain
In order to determine the full-length genome of the CanineCV strain from Argentina, a positive sample was tested with the additional primer sets listed in Table 1 and sequenced twice. Resulting amplicons were eluted after agarose gel electrophoresis with Easy Pure Quick gel Extraction kit (TransGen Biotech) and submitted twice for sequencing to Unidad Genómica, Instituto de Biotecnología (INTA, Argentina) and to GenBiotech (Buenos Aires, Argentina), following the dideoxy-nucleotide chain termination method using the primers described above. The full-length genome was assembled using the Mafft software (EMBL-EBI). The complete sequence was named strain UBA-Baires and deposited in Genbank under accession number MK033608.
Sequence analysis
For phylogenetic analysis of the complete CanineCV genome sequence, we set up a matrix that included the 22 different complete CanineCV sequences deposited so far in GenBank. Details of selected sequences are shown in Table 2. This data set was organized using BioEdit software (v. 7.2.5) [18] and aligned with ClustalW (version 1.82)[19]. A maximum likelihood (ML) phylogenetic tree of these sequences was obtained using PhyML (version 3.1) [20]. For this analysis, we utilized the TN93+I+G model of nucleotide substitution (determined using Smart Model Selection) [21, 22] with sub-tree pruning and regrafting (SPR) tree improvement and 10 starting random trees. The robustness of individual nodes on the phylogeny was estimated using 1000 bootstrap replicates. Phylogenetic trees were visualized and edited with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Table 2. Full-length genome sequences of CanineCV selected for phylogenetic analysis.
| GenBank Accession Number | Origin | Year | Host | Virus ID | Reference |
|---|---|---|---|---|---|
| MK033608 | Argentina | 2018 | dog | UBA-Baires/2018/ARG | This study |
| JQ821392 | USA | 2012 | dog | 214/2012/USA | Kapoor et al., 2012 |
| KC241982 | USA | 2013 | dog | UCD1/2013/USA | Li et al., 2013 |
| KC241983 | USA | 2013 | dog | UCD3/2013/USA | Li et al., 2013 |
| KC241984 | USA | 2013 | dog | UCD2/2013/USA | Li et al., 2013 |
| KF887949 | Germany | 2014 | dog | Ha13/2014/GER | Halami et al., 2014 (unpublished) |
| KJ530972 | Italy | 2014 | dog | Bari/2014/ITA | Decaro et al., 2014 |
| KT283604 | Germany | 2015 | dog | FUBerlin/2015/GER | Plog et al., 2015 (unpublished) |
| KT946839 | China | 2015 | dog | JZ98/2015/CHI | Zheng et al., 2015 (unpublished) |
| KT734812 | Italy | 2016 | wolf | CB6293/2016/ITA | Zaccaria et al., 2016 |
| KT734813 | Italy | 2016 | dog | AZ2972/2016/ITA | Zaccaria et al., 2016 |
| KT734814 | Italy | 2016 | wolf | TE4016/2016/ITA | Zaccaria et al., 2016 |
| KT734815 | Italy | 2016 | wolf | AZ4133/2016/ITA | Zaccaria et al., 2016 |
| KT734816 | Italy | 2016 | badger | AZ4438/2016/ITA | Zaccaria et al., 2016 |
| KT734817 | Italy | 2016 | dog | AZ5212/2016/ITA | Zaccaria et al., 2016 |
| KT734819 | Italy | 2016 | wolf | AZ5586/2016/ITA | Zaccaria et al., 2016 |
| KT734820 | Italy | 2016 | wolf | AZ663/2016/ITA | Zaccaria et al., 2016 |
| KT734821 | Italy | 2016 | dog | TE6685/2016/ITA | Zaccaria et al., 2016 |
| KT734822 | Italy | 2016 | wolf | TE7482/2016/ITA | Zaccaria et al., 2016 |
| KT734823 | Italy | 2016 | dog | PE8575/2016/ITA | Zaccaria et al., 2016 |
| MG266899 | China | 2017 | dog | CD17/2017/CHI | Yan et al., 2017 (unpublished) |
| MF797786 | China | 2018 | dog | XF16/2018/CHI | Je et al., 2018 (unpublished) |
| MF457592 | USA | 2018 | dog | OH19098/2018/USA | Wang et al., 2018 (unpublished) |
Results
Detection of CanineCV
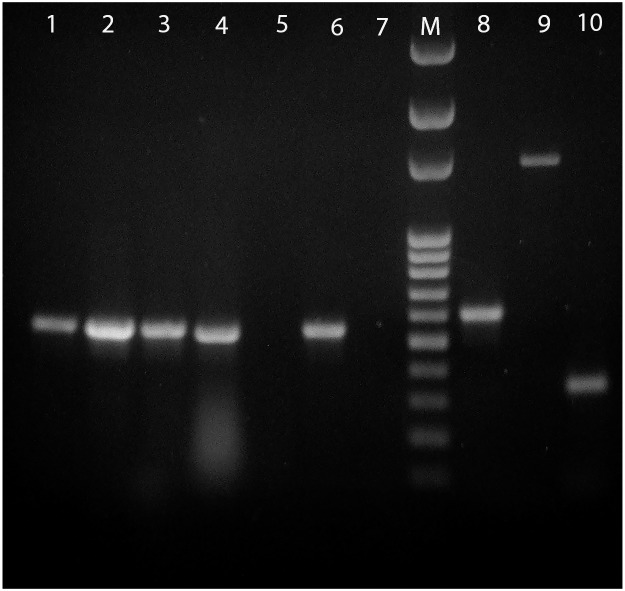
Pooled tissue samples from two of the three animals tested positive for CanineCV with the primer set For genomic/Rev 533. Then, individual organs were further screened with the same primer set finding that all tissues (pancreas, liver, kidney, lung, lymph nodes, spleen and large intestine), except for the stomach, were positive as well. DNA from one of these tissues (lymph node) was randomly selected for further analysis with the remaining primer sets in order to assemble the entire CanineCV genome sequence, which was concluded to be of 2063 bp (Fig 1). PCR analysis detected the presence of CPV in all samples, except for the stomach (Fig 2A). Moreover, all generated amplicons were digested with MboII, indicating the presence of the glutamine at VP2 aa 426 characteristic of CPV-2c strains [17] (Fig 2B).
Fig 1. Detection of CanineCV.
Gel electrophoresis of PCR reactions for CanineCV with different tissues and primers. Lanes 1–7 set For genomic/Rev 533. Lane 1: spleen, 2: mesenteric lymph node, 3: liver/lung, 4: pancreas, 5: stomach, 6: large intestine, 7: negative control. Line 8: For 456/Rev 1055, Line 9: For 456/Rev 1910, Line 10: For 1734/Rev genomic. M: molecular weight marker 100 bp DNA Ladder (Solis BioDyne).
Fig 2. Detection of CPV.
Gel electrophoresis of (A) PCR reactions for CPV. Lane 1: positive control, 2: negative control, 3: Mesenteric lymph node, 4: Spleen, 5: Lung, 6: Stomach, 7: Liver, 8: Large intestine, M: Molecular weight marker 100 bp DNA Ladder (Solis BioDyne). (B) MboII Restriction Fragment Length Polymorphism of Large intestine amplicon. M: Molecular weight marker 100 bp Ladder, U: Undigested Large intestine amplicon, D: MboII digested Large intestine amplicon.
Genome characterization
Sequence analysis revealed strong similarities to those of other previously described CanineCV. The complete genome of strain UBA-Baires was 2063 nt in length with a GC content of 51.5%. Genome characteristics include two putative ORFs synthesized from both DNA strands and two intergenic regions. These ORFs encode a putative viral replicase (Rep) and a capsid protein (Cap), which are 303 and 270 aa long, respectively. The 5’ intergenic region, located between the start codons and encompassing 135 nt, contains a conserved stem-loop structure and a 9 nt sequence (TAGTATTAC) for initiation of rolling cycle replication. The overall nt identity of strain UBA-Baires ranged from 98% (strain Bari/411-13) to 85% (isolate UCD3-478). When compared to other CanineCV, both Rep and Cap ORFs were, as expected, highly conserved including sequences related to rolling circle replication (FTINN, TPHLQ and CSK) and dNTP binding (GCGKS) in the non-structural gene (Rep). However, four aa changes within the Cap gene and one within the Rep gene were found, which have not been previously reported for other CanineCV sequences being unique to strain UBA-Baires (Table 3). These mutations were confirmed after two independent sequencing runs directly from PCRs amplicons. No mutations were observed in the intergenic regions.
Table 3. Amino acid substitutions in the replicase and capsid proteins unique to CanineCV strain UBA-Baires.
| Protein name | Nt position | Nt substitution | Protein residue position | strain UBA-Baires | CanineCV strains |
|---|---|---|---|---|---|
| Replicase | 580 | C to G/A | 194 | Gln | Glu / Lys |
| Capsid | 1499 | T to A/G | 144 | Cys | Ser / Gly |
| 1522–1523 | CA to AC | 136 | Gln | Thr | |
| 1645 | T to A | 95 | Phe | Tyr | |
| 1694 | A to G | 79 | Thr | Ala |
Phylogeny
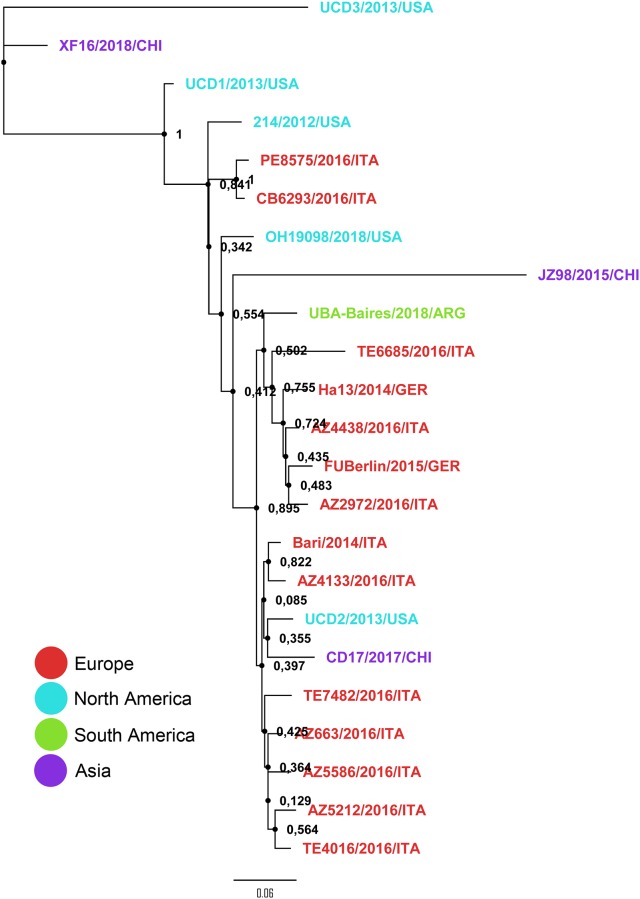
Phylogenetic evaluation of the genetic relationship with other CanineCV was performed using whole genome sequences published so far (Table 2). Among the CanineCV sequences, the strain from the present study clustered with viruses circulating recently in animals from Europe, mainly Germany and Italy, with strong bootstrap support, and was divergent from most strains from the USA and China (Fig 3).
Fig 3. Maximum likelihood tree of full-length sequences of CanineCV strains.
The sequences are colored based on their continental origin. Details of data set are shown in Table 2.
Discussion
Kapoor et al. reported for the first time the detection of CanineCV in the serum of healthy dogs in the USA [1]. Over the last five years, CanineCV has been detected in Italy, Germany, China, Thailand, Taiwan and Brazil [3–6, 8, 12, 13]. In the present study, we report an outbreak of a fatal disease associated with the presence of CanineCV in Argentina.
Although virus isolation was not attempted since it was proved unsuccessful in previous reports [3], the full-length genome of strain UBA-Baires was determined, revealing close similarity to European viruses. We found only one unique mutation in the rep gene, but surprisingly, four unique aa substitutions in the Cap protein, after two independent experiments using different sequencing facilities. Very recently, nine major variable regions have been proposed to exist within the cap gene of CanineCV [12]. Only one of the four mutations present in the cap gene of strain UBA-Baires is encompassed at these proposed locations (residue 95), suggesting that, until sequences from more extent geographical areas are analyzed, these variable regions need to be taken cautiously. Point mutations in the cap gene have been described for PCV2 and are a potential source of virus evolution, especially due to selective pressure caused by vaccination [23]. In the case of CanineCV, where vaccination has not been used, the mechanisms behind these aa substitutions are unclear; virus variability under natural infection has been shown to be very complex for PCV-2 [24]. In addition, the relevance of these changes, if any, is difficult to be evaluated at this moment when it is still under investigation the role of CanineCV as a sole cause of disease [25].
In the case of PCV2, coinfection with other pathogens has been associated with different clinical outcomes [14]. Special attention was given to coinfection with porcine parvovirus and the syndrome could be reproduced experimentally [26, 27]. Similarly, CanineCV has been found in most cases accompanying other viral pathogens. In hemorrhagic enteritis, it is often associated to CPV, but also canine distemper virus and canine coronavirus with coinfection rates of 100% [4, 6], 70–80% [8, 9, 11] and 50% [10]. Only one report so far has found CanineCV as a primary agent [3]. Case-control studies carried out in different population of dogs suggest a high association between diarrhea and CanineCV, but CanineCV is still found in feces and serum from healthy dogs [8–11]. Nevertheless, one study showed that mortality rate was higher when CanineCV was found together with CPV [8]. Interestingly, in dogs with respiratory symptoms from Thailand, CanineCV was detected together with respiratory pathogens (canine influenza virus, canine parainfluenza virus, and canine respiratory coronavirus)[5]. In our case, all organs tested except for stomach, were also positive for CPV. Unfortunately, no histopathology was performed on these cases in order to confirm the presence of some of the lesions observed in previously described cases involving CanineCV, in particular vasculitis [25]. Surprisingly, in our case, sick animals had been vaccinated against CPV. This phenomenon was also described in a breeding colony in the USA where two severe outbreaks of hemorrhagic gastroenteritis occurred among animals vaccinated against CPV [4]. Again, the role of CanineCV in the context of vaccine efficacy against co-infecting viruses is still unclear and needs to be carefully analyzed. ISH studies carried out with clinical samples show that CanineCV infects mainly lymphoid tissues and cells [2, 5] suggesting that Infection with circovirus could cause immunosuppression and allow infection with CPV even in the case of vaccinated animals. Nevertheless, further experimental infections are necessary to elucidate the pathogenic involvement of CanineCV in disease pathogenesis.
To our knowledge, this is the first report of a complete genome sequence of CanineCV associated with disease from Latin America. A very recent paper published by Weber and col. characterizing the dog’s virome found CanineCV in the sera from seven healthy dogs from Brazil [13]. Phylogenetic analysis using a partial sequence from Rep revealed that the Brazilian virus seemed to be related to strains from China and the USA, among others. The tree topology is somehow different form the one we present in this study based on the full-length genome, in which the Argentinian strain grouped closer to European viruses. Hopefully, additional sequences from South America will be soon deposited in GenBank allowing a more thorough analysis of the epidemiology and evolution of this emerging virus and will aid in the comprehension of its role in disease.
Data Availability
All relevant data are within the manuscript. Virus sequence has been deposited in Genbank under accession number MK033608 as stated in the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Kapoor A, Dubovi EJ, Henriquez-Rivera JA, Lipkin WI. Complete genome sequence of the first canine circovirus. J Virol. 2012;86(12):7018 10.1128/JVI.00791-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, et al. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis. 2013;19(4):534–41. 10.3201/eid1904.121390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decaro N, Martella V, Desario C, Lanave G, Circella E, Cavalli A, et al. Genomic characterization of a circovirus associated with fatal hemorrhagic enteritis in dog, Italy. PLoS One. 2014;9(8):e105909 10.1371/journal.pone.0105909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thaiwong T, Wise AG, Maes RK, Mullaney T, Kiupel M. Canine Circovirus 1 (CaCV-1) and Canine Parvovirus 2 (CPV-2): Recurrent Dual Infections in a Papillon Breeding Colony. Vet Pathol. 2016;53(6):1204–9. 10.1177/0300985816646430 [DOI] [PubMed] [Google Scholar]
- 5.Piewbang C, Jo WK, Puff C, van der Vries E, Kesdangsakonwut S, Rungsipipat A, et al. Novel canine circovirus strains from Thailand: Evidence for genetic recombination. Sci Rep. 2018;8(1):7524 10.1038/s41598-018-25936-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaccaria G, Malatesta D, Scipioni G, Di Felice E, Campolo M, Casaccia C, et al. Circovirus in domestic and wild carnivores: An important opportunistic agent? Virology. 2016;490:69–74. 10.1016/j.virol.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 7.Allan G, Krakowka S, Ellis J, Charreyre C. Discovery and evolving history of two genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res. 2012;164(1–2):4–9. 10.1016/j.virusres.2011.09.013 [DOI] [PubMed] [Google Scholar]
- 8.Anderson A, Hartmann K, Leutenegger CM, Proksch AL, Mueller RS, Unterer S. Role of canine circovirus in dogs with acute haemorrhagic diarrhoea. Vet Rec. 2017;180(22):542 10.1136/vr.103926 [DOI] [PubMed] [Google Scholar]
- 9.Dowgier G, Lorusso E, Decaro N, Desario C, Mari V, Lucente MS, et al. A molecular survey for selected viral enteropathogens revealed a limited role of Canine circovirus in the development of canine acute gastroenteritis. Vet Microbiol. 2017;204:54–8. 10.1016/j.vetmic.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentil M, Gruber AD, Muller E. [Prevalence of Dog circovirus in healthy and diarrhoeic dogs]. Tierarztl Prax Ausg K Kleintiere Heimtiere. 2017;45(2):89–94. 10.15654/TPK-160701 [DOI] [PubMed] [Google Scholar]
- 11.Hsu HS, Lin TH, Wu HY, Lin LS, Chung CS, Chiou MT, et al. High detection rate of dog circovirus in diarrheal dogs. BMC Vet Res. 2016;12(1):116 10.1186/s12917-016-0722-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun W, Zhang H, Zheng M, Cao H, Lu H, Zhao G, et al. The detection of canine circovirus in Guangxi, China. Virus Res. 2019;259:85–9. 10.1016/j.virusres.2018.10.021 [DOI] [PubMed] [Google Scholar]
- 13.Weber MN, Cibulski SP, Olegario JC, da Silva MS, Puhl DE, Mosena ACS, et al. Characterization of dog serum virome from Northeastern Brazil. Virology. 2018;525:192–9. 10.1016/j.virol.2018.09.023 [DOI] [PubMed] [Google Scholar]
- 14.Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012;164(1–2):20–32. 10.1016/j.virusres.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segales J, Kekarainen T, Cortey M. The natural history of porcine circovirus type 2: from an inoffensive virus to a devastating swine disease? Vet Microbiol. 2013;165(1–2):13–20. 10.1016/j.vetmic.2012.12.033 [DOI] [PubMed] [Google Scholar]
- 16.Breitbart M, Delwart E, Rosario K, Segales J, Varsani A, Ictv Report C. ICTV Virus Taxonomy Profile: Circoviridae. J Gen Virol. 2017;98(8):1997–8. 10.1099/jgv.0.000871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, et al. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82(Pt 12):3021–5. 10.1099/0022-1317-82-12-3021 [DOI] [PubMed] [Google Scholar]
- 18.Hall T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41: 95–98. [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 21.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 22.Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34(9):2422–4. 10.1093/molbev/msx149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzo G, Tucciarone CM, Cecchinato M, Drigo M. Porcine circovirus type 2 (PCV2) evolution before and after the vaccination introduction: A large scale epidemiological study. Sci Rep. 2016;6:39458 10.1038/srep39458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correa-Fiz F, Franzo G, Llorens A, Segales J, Kekarainen T. Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies. Sci Rep. 2018;8(1):15469 10.1038/s41598-018-33849-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesavento PA, Murphy BG. Common and emerging infectious diseases in the animal shelter. Vet Pathol. 2014;51(2):478–91. 10.1177/0300985813511129 [DOI] [PubMed] [Google Scholar]
- 26.Allan GM, Kennedy S, McNeilly F, Foster JC, Ellis JA, Krakowka SJ, et al. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J Comp Pathol. 1999;121(1):1–11. 10.1053/jcpa.1998.0295 [DOI] [PubMed] [Google Scholar]
- 27.Ellis JA, Bratanich A, Clark EG, Allan G, Meehan B, Haines DM, et al. Coinfection by porcine circoviruses and porcine parvovirus in pigs with naturally acquired postweaning multisystemic wasting syndrome. J Vet Diagn Invest. 2000;12(1):21–7. 10.1177/104063870001200104 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript. Virus sequence has been deposited in Genbank under accession number MK033608 as stated in the manuscript.