Abstract
Fluorescent proteins have been extensively engineered and applied as optical indicators for chloride in a variety of biological contexts. Surprisingly, given the biodiversity of fluorescent proteins, a naturally occurring chloride sensor has not been reported to date. Here, we present the identification and spectroscopic characterization of the yellow fluorescent protein from the jellyfish Phialidium sp. (phiYFP), a rare example of a naturally occurring, excitation ratiometric, and turn-on fluorescent protein sensor for chloride. Our results show that chloride binding tunes the pKa of the chromophore Y66 and shifts the equilibrium from the fluorescent phenolate form to the weakly fluorescent phenol form. The latter likely undergoes excited state proton transfer to generate a turn-on fluorescence response that is pH dependent. Moreover, anion selectivity and mutagenesis in the chloride binding pocket provide additional evidence for the proposed chloride sensing mechanism. Given these properties, we anticipate that phiYFP, with further engineering, could be a new tool for imaging cellular chloride dynamics.
Graphical Abstract
Chloride is the most abundant, biologically relevant anion with intracellular concentrations ranging up to 70 mM in eukaryotic cell types.(1,2) The mobilization of chloride across extracellular and intracellular membranes is intimately linked to a variety of biological functions including cell volume, pH regulation, cell division, muscle contraction, and neuroexcitation.(3–5) Moreover, mutations or differential expression of universally expressed chloride channels can lead to chloride dysregulation in a wide range of diseases including cystic fibrosis, epilepsy, and chronic pain.(6–9) Our understanding of chloride in these contexts has been aided by small molecule and genetically encoded fluorescent biosensors that can provide both spatial and temporal resolution. Quinolinium and acridinium-based sensors are pH-independent and have been widely used to measure intracellular chloride, even though these sensors undergo collisional quenching and can be difficult to target to subcellular compartments.(3,10–12) However, Clensor, is a more recent advance that combines nucleic acids with a chloride-sensitive acridinium scaffold to afford a pH-independent, ratiometric chloride sensor for the quantification of chloride exclusively in acidic organelles.(13,14)
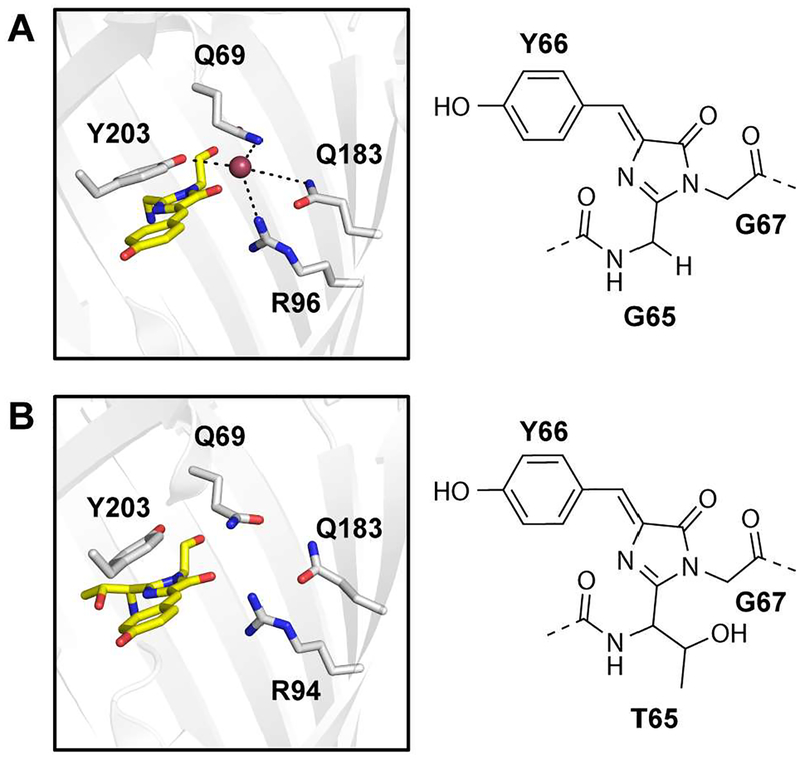
In parallel, genetically encoded biosensors based on yellow fluorescent protein H148Q (avYFP-H148Q), an engineered chloride-sensitive variant of green fluorescent protein (avGFP) from the jellyfish Aequorea victoria, have been developed.(15,16) Relative to avGFP, avYFP-H148Q has five amino acid substitutions: S65G, V68L, S72A, H148Q, and T203Y.(16) The latter π-stacks with the chromophore Y66 and is essential for chloride binding.(16) In addition to Y203, the chloride binding pocket consists of Q69, R96, and Q183 with hydrophobic residues lining the coordination sphere.(16) This has been confirmed in the crystal structure of avYFP-H148Q bound to iodide (PDB ID: 1F09, Figure 1A).(16) It is through these interactions that chloride tunes the pKa of the chromophore Y66 and shifts the equilibrium from the more fluorescent phenolate form to the weakly fluorescent phenol form, resulting in a turn-off fluorescence response. Even though avYFP-H148Q is a pH dependent, turn-off fluorescent sensor for chloride, it has been extensively engineered, along with avGFP, and fused to other fluorescent proteins for intensity or ratiometric-based imaging of cellular chloride.(2,10,17–23) Ratiometric sensors including Clomeleon, Cl-Sensor, and Super-Clomeleon provide a quantitative measure of chloride but are still pH dependent, thus requiring rigorous controls and calibrations.(2,10,17–20) Most recently, to account for these pH changes, LSSmClopHensor provides a dual readout of chloride and pH.(10,21–23)
Figure 1.
Comparison of the crystal structures (left) and chromophores (right) for the (A) avYFP-H148Q (PDB ID: 1F09) bound to iodide (purple sphere) and (B) wild-type phiYFP from the jellyfish Phialidium sp. (PDB ID: 4HE4). Residues are shown as sticks in gray and labeled with the single letter amino acid code and residue number. The chromophores are shown as sticks in yellow, and the oxygen and nitrogen atoms are in red and blue, respectively.
Inspired by this body of work and given the diversity of fluorescent proteins in Nature, we were curious if a fluorescent protein chloride sensor with different properties could exist.(24,25) In this Communication, we report the identification and spectroscopic characterization of the yellow fluorescent protein from the jellyfish Phidlidium sp. (phiYFP), a previously unidentified turn-on fluorescent protein sensor for chloride that is pH-dependent but operates in an excitation ratiometric mode independently of a fused protein. Anion selectivity and mutagenesis in the chloride binding pocket suggests that the observed spectral properties arise from an excited-state proton transfer (ESPT) event.
Using the BLAST search algorithm, we determined that phiYFP (UniProt ID: Q6RYS7) is a naturally occurring homolog with the highest sequence identity (51%) to avYFP-H148Q (Figure S1). To our surprise, multiple sequence alignment revealed that phiYFP has the same set of amino acids, including Q69, R94, Q183, and Y203, at the positions corresponding to the chloride binding pocket in avYFP-H148Q (Figure S2). The similarity in the arrangement of these amino acids can be clearly seen in the phiYFP crystal structure that was previously solved without a halide (PDB ID: 4HE4, Figure 1B).(26) Compared to avYFP-H148Q, the chromophore in phiYFP is made up of T65, Y66, and G67 and can undergo conversion from the phenolate to the phenol form in a pH dependent manner (pKa = 6.6).(27) Based on these observations, we hypothesized that phiYFP could be a fluorescent chloride sensor with unique properties.
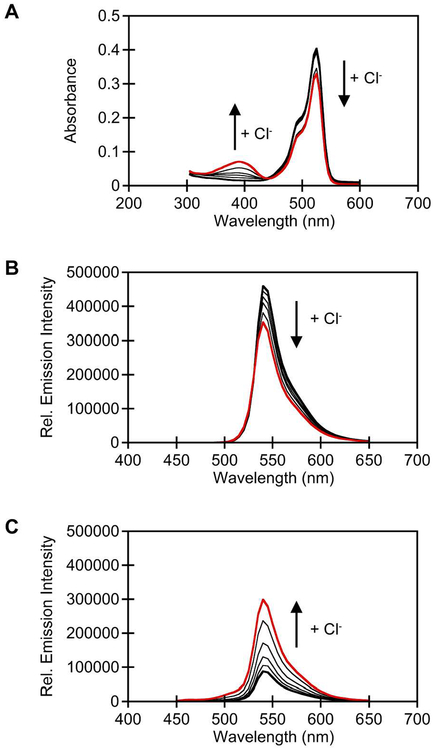
Here, we find that in the absence of chloride phiYFP has one major absorption band at 525 nm that corresponds to the phenolate form of the chromophore. The relative intensity of this absorption band does not change from pH 5 to 9, and we do not observe a pH dependent equilibrium with the phenol form of the chromophore as previously described (Figure S3A).(27) It is important to note that prior studies with phiYFP were carried out in buffers containing sodium chloride, giving rise to the differences we observed(27) However, upon lowering the pH to 4.5 or adding 400 mM chloride, the relative intensity of the absorption band decreases at 525 nm, a clear isosbestic point is observed at 450 nm, and a new absorption band appears at 400 nm corresponding to the phenol form of the chromophore (Figure S3B). In the presence of 400 mM chloride, the phiYFP chromophore pKa increases from 4.9 to 5.4, with the most observable absorbance changes occurring at pH 5 > 5.5 > 6 (Figure S3C). Given these results, we carried out chloride titrations at these pH values to show that the chromophore equilibrium is indeed dependent on the concentration of chloride like avYFP-H148Q (Figures 2A, S4A–S4C).(16)
Figure 2.
Spectroscopic characterization of wild-type phiYFP (5 μM) at pH 5.5. (A) UV-visible response to chloride. Emission response to chloride (B) with λex = 480 nm and (C) with λex = 400 nm. Arrow direction corresponds to increasing chloride concentrations. All spectra were acquired in 50 mM MES buffer, pH 5.5 in the presence of 0 (bold), 25, 50, 100, 200, and 400 mM chloride (red). The average of three technical replicates with standard error of the mean is reported (Figure S4).
Next, we evaluated the fluorescence properties of phiYFP as a function of chloride at pH 5.5, which is higher than the chromophore pKa in the presence of chloride. With λex = 480 nm, apo phiYFP has a single emission peak at 540 nm (Φ = 0.44) (Figures 2B, S4H). This emission maximum corresponds to the excited state of the phenolate form of the chromophore.(27) Addition of 400 mM chloride quenches this emission by 23% (Φ = 0.49) (Figure 2B, S4H). Interestingly, with λex= 400 nm, apo phiYFP has a single emission centered at 540 nm (Φ = 0.02, Figures 2C, S4E), and the addition of chloride triggers a 3.5-fold (Φ = 0.06) increase in fluorescence intensity with no shift in the emission maximum, making phiYFP excitation ratiometric (Figures 2C, S4E, Table S1). Similar results were also obtained under conditions of constant ionic strength (Figure S7). The phenol form of the chromophore is fluorescent at lower pH values, but its emission maximum is at 510 nm (Figure S4D). As such, we speculate that like avGFP, phiYFP can undergo ESPT from the chromophore Y66 to a proton acceptor in the chloride binding pocket to generate the fluorescent phenolate form of the chromophore.(28–30) The turn-on emission response is unique to phiYFP and is not observed with avYFP-H148Q (Figures S8).
Given the turn-on fluorescence response and larger dynamic range with λex = 400 nm, we further characterized the chloride binding properties of phiYFP at this excitation wavelength. Even though it is known that phiYFP is a weak dimer in solution, Hill plot analysis indicates that chloride binding is not cooperative (ηH = 0.90 ± 0.07, Figure S5E).(27) The apparent Kd for chloride binding to phiYFP was calculated using a single site binding model and is 384 ± 46 mM (Figure S5B, Table S1).
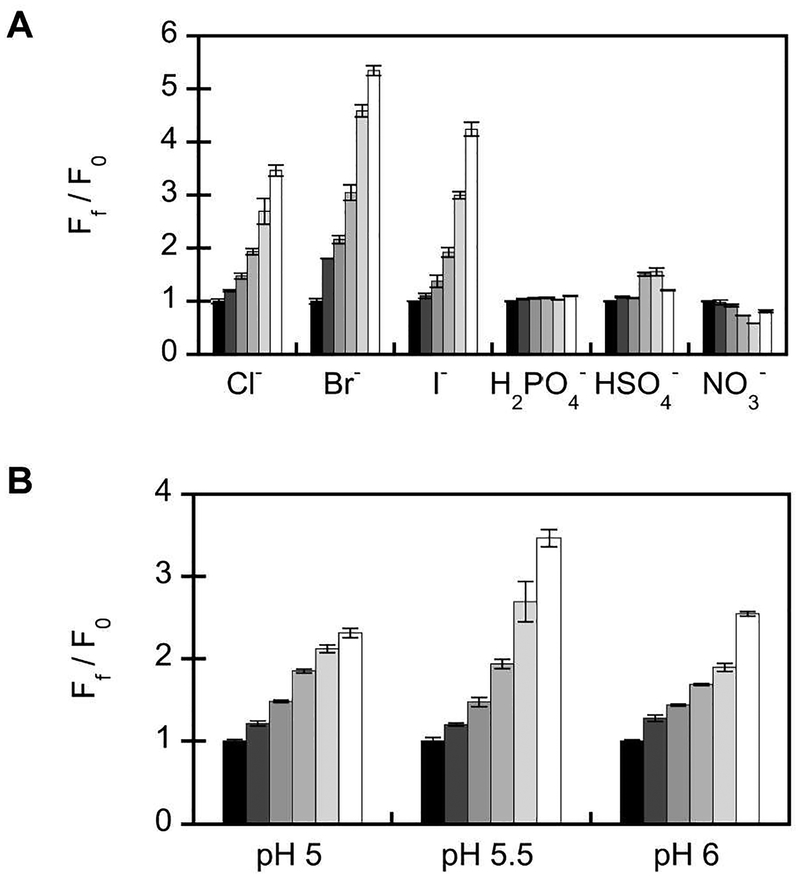
The fluorescence response of phiYFP to other halides and oxyanions (λex = 400 nm) at pH 5.5 is shown in Figure 3 (Table S1). As expected, phiYFP binds to bromide (5.4-fold turn-on, Kd = 106 ± 5 mM) and iodide (4.2-fold turn-on, Kd = 50 mM ± 2 mM) (Figures S9–S12). With λex = 480 nm, the emission is quenched in the presence of both halides (Figures S9–S12). No change is observed, irrespective of the excitation, in the presence of phosphate and sulfate, which equilibrate towards dihydrogen phosphate and hydrogen sulfate at pH 5.5 (Figures S13–S15). However, the fluorescence is quenched by 19% in the presence of nitrate even though the chromophore equilibrium is shifted to the phenol form as seen in the absorbance spectra (Kd = 1 9 mM ± 4 mM, Figures S16B, S17B). Similar quenching occurs with λex = 480 nm (Figure S16H). This suggests that nitrate binding could prevent ESPT from the phenol form of the chromophore to a proton acceptor in the chloride binding pocket, as speculated above. Of note, an emission ratiometric response from 540 nm to 510 nm is observed in the presence of iodide and nitrate with λex = 400 nm (Figures S11E, S16E). The anion selectivity of phiYFP is similar to that observed with previously reported avYFP and avGFP-based chloride sensors.(10,16,21)
Figure 3.
Anion selectivity and pH profile of wild-type phiYFP (5 μM). (A) Emission response to 0, (F0, black bar), 25, 50, 100, 200, and 400 mM (white bar) chloride, bromide, iodide, dihydrogen phosphate, hydrogen sulfate, and nitrate. Spectra were acquired in 50 mM MES, pH 5.5 with λex = 400 nm. (B) Emission response to 0, (F0, black bar), 25, 50, 100, 200, and 400 mM (white bar) chloride. Spectra were acquired in 50 mM citrate, pH 5 (black bars), 50 mM MES, pH 5.5, and 50 mM MES, pH 6 with λex = 400 nm. The average of three technical replicates with standard error of the mean is reported. The sodium salt was used for all of the anions tested.
This turn-on emission response is not only a function of the chloride concentration but also of pH (Figure 3B, Table S1). At pH 5 with λex = 480 nm, phiYFP has a single emission at 540 nm that is quenched by 73% with 400 mM chloride. However, with λex = 400 nm, phiYFP has an emission ratiometric response where the emission shifts from 540 nm to 510 nm with chloride (2.3-fold turn-on, Kd = 290 mM ± 44 mM, Figures S4E, S5A). Since the pH tested was lower than the chromophore pKa with chloride, ESPT would not be favored to the same extent giving rise to the phenol emission at 510 nm. At pH 6 in the presence of chloride, the emission intensity at 540 nm does not significantly change (p > 0.5) with λex = 480 nm but increases by 2.5-fold with λex = 400 nm (Kd = 306 ± 38 mM, Figure S4F, S5C). A similar pH dependence is observed for the other anions tested (Figures S9–S17). The differences observed in the emission responses are likely due to how both anion binding and the protonation state of ionizable residues affect the chromophore equilibrium at each pH.
With spectroscopic data showing that phiYFP is a turn-on fluorescent sensor for chloride, we turned our attention to validate the putative chloride binding pocket. It is quite likely that chloride binds near the chromophore and due to electrostatic repulsion, shifts the chromophore equilibrium to the neutral phenol form. Based on the crystal structure avYFP-H148Q bound to iodide, we identified Q69 as a starting point for mutagenesis in phiYFP. Of the three amino acids in avYFP-H148Q that interact with iodide, Q69 is the closest (3.2 Å, Figure 1A).(16) Moreover, mutations at this position in avYFP variants can reduce chloride affinity and do not significantly interfere with chromophore maturation, which we deemed useful for a plate reader screening assay with phiYFP.(18,31) Site-saturation mutagenesis of phiYFP was carried out at Q69, and we identified two variants, Q69L and Q69H, with reduced chloride affinity (Figures 4A, 4B). Similar to wild-type phiYFP at low pH, the chromophore in both variants can undergo conversion from the phenolate form to the phenol form (Figures S18A, S19A). In the presence of 400 mM chloride, the chromophore pKa of both Q69L and Q69H increases from 4.7 to 5.2 (Figures S18C, S19C).
Figure 4.
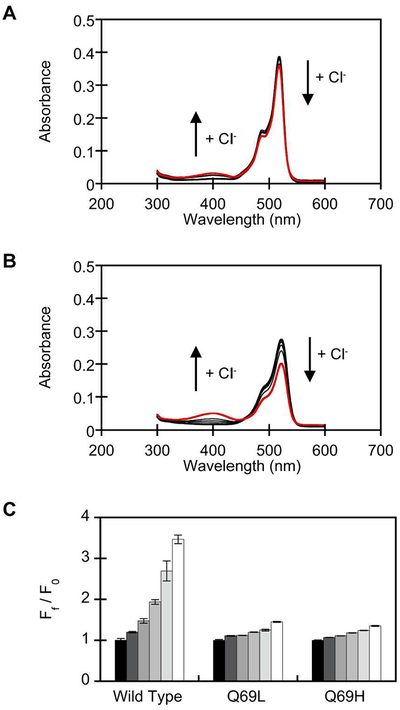
Spectroscopic characterization of phiYFP Q69L (4.4 μM) and Q69H (4.4 μM) at pH 5.5. UV-visible response of (A) Q69L and (B) Q69H to 0, 25, 50, 100, 200, and 400 mM chloride (red). Arrows correspond to increasing chloride concentrations. (C) Emission response wild-type phiYFP and variants to 0 (F0, black bar), 25, 50, 100, 200, and 400 mM (white bar) chloride. Spectra were acquired in 50 mM MES, pH 5.5 with λex = 400 nm. The average of three technical replicates with standard error of the mean is reported.
Like wild-type phiYFP, apo Q69L has a single emission peak at 525 nm with λex = 400 nm and λex = 480 nm, and apo Q69H has a single emission peak at 535 nm with λex = 400 nm and λex = 480 nm (Figures S20, S21). These emission maxima correspond to the phenolate form of the chromophore, indicating that ESPT is not affected in the absence of chloride. In the presence of chloride, Q69L shows a 1.5-fold increase with λex = 400 nm and no significant change with λex = 480 nm (p > 0.05, Figures 4C, S20). For Q69H, the fluorescence response increases by 1.7-fold with λex = 400 nm, and the fluorescence response is quenched by 36% with λex = 480 nm (Figures 4C, S21). The emission response to chloride for Q69L and Q69H at both excitation wavelengths is still significantly less than wild-type phiYFP (p < 0.001). The apparent Kd values for Q69L and Q69H, could not be determined due to weak binding or collisional quenching because there is a linear correlation between the fluorescence intensity and chloride concentrations (Figures S20E, S21E). At this position, the mutation from glutamine to leucine removes a hydrogen bond donor and histidine, depending on its orientation and protonation state, can alter the hydrogen bonding network in the chloride binding pocket, thus interfering with the formation of a strong coordination complex.
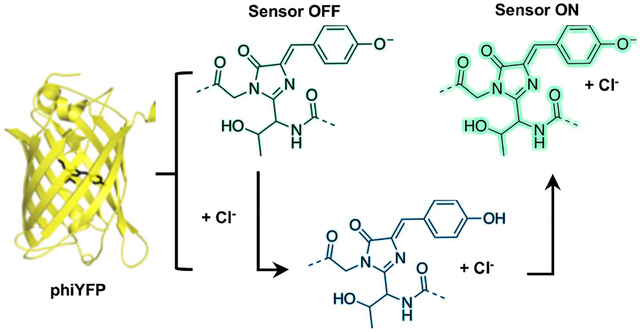
To close, here we have identified and characterized phiYFP from the jellyfish Phialidium sp. as a naturally occurring, excitation ratiometric, and turn-on fluorescent sensor for chloride. Our data shows that chloride binding in the ground state tunes the pKa of the chromophore Y66 and shifts the equilibrium from the phenolate to phenol form, which likely undergoes ESPT resulting in a turn-on fluorescence response (Figure S22). Not only does our study demonstrate that naturally occurring fluorescent proteins can have new properties, but also how these can be starting points to potentially create functional tools. Future efforts will focus on structural characterization and detailed study of the proposed ESPT mechanism(32) Moreover, we anticipate that upon shifting the operational pH and improving chloride affinity of phiYFP, we can take advantage of its excitation ratiometric properties for live cell imaging applications.
Supplementary Material
ACKNOWLEDGMENT
We thank Professor Alex Lippert, Professor Gabriele Meloni, and the Dodani Lab for helpful discussions.
Funding Sources
This work was supported by startup funds from The University of Texas at Dallas, the Welch Foundation (AT-1918–20170325), and the National Institutes of Health (1R35GM128923–01) to S.C.D.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website.
Experimental methods and supporting figures (PDF)
The authors declare no competing financial interests.
REFERENCES
- (1).Reuter D, Zierold K, Schröder WH, Frings S (1998) A depolarizing chloride current contributes to chemoelectrical transduction in olfactory sensory neurons in situ, J. Neurosci 18, 6623–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bregestovski P, Waseem T, and Mukhtarov M (2009) Genetically encoded optical sensors for monitoring of intracellular chloride and chloride-selective channel activity, Front. Mol. Neurosci 2, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Verkman AS (1990) Development and biological applications of chloride-sensitive fluorescent indicators, Am. J. Physiol 259, C375–C388. [DOI] [PubMed] [Google Scholar]
- (4).Suzuki M, Morita T, and Iwamoto T (2006) Diversity of Cl–channels, Cell. Mol. Life Sci 63, 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Puljak L, and Kilic G (2006) Emerging roles of chloride channels in human diseases, Biochim. Biophys. Acta 1762, 404–413. [DOI] [PubMed] [Google Scholar]
- (6).Verkman AS, and Galietta L (2009) Chloride channels as drug targets, Nat. Rev 8, 153–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Planells-Cases R, and Jentsch TJ. (2009) Chloride channelopathies, Biochim. Biophys. Acta 1792, 173–189. [DOI] [PubMed] [Google Scholar]
- (8).Gagnon M, Bergeron MJ, Lavertu G, Castonguay A, Tripathy S, Bonin RP, Perez-Sanchez J, Coull JA, and De Koninck Y (2013) Chloride extrusion enhancers as novel therapeutics for neurological diseases, Nat. Med 19, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Peretti M, Angelini M, Savalli N, Florio T, Yuspa SH, and Mazzanti M (2015) Chloride channels in cancer: Focus on chloride intracellular channel 1 and 4 (CLIC1 and CLIC4) proteins in tumor development and as novel therapeutic targets, Biochim. Biophys. Acta 1848, 2523–2531. [DOI] [PubMed] [Google Scholar]
- (10).Arosio D, and Ratto GM (2014) Twenty years of fluorescence imaging of intracellular chloride, Front. Cell. Neurosci 8, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Geddes CD (2001) Optical halide sensing using fluorescence quenching: theory, simulations and applications-a review, Meas. Sci. Technol 12, R53–R88. [Google Scholar]
- (12).Ashton TD, Jolliffe KA, and Pfeffer FM (2015) Luminescent probes for the bioimaging of small anionic species in vitro and in vivo, Chem. Soc. Rev 44, 4547–4595. [DOI] [PubMed] [Google Scholar]
- (13).Saha S, Prakash V, Haider S, Chakraborty K, and Krishnan Y (2014) A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells, Nat. Nanotechnol 10, 645–652. [DOI] [PubMed] [Google Scholar]
- (14).Chakraborty K, Leung K, and Krishnan Y (2017) High lumenal chloride in the lysosome is critical for lysosome function, Elife. 6,1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jayaraman S, Haggie P, Wachter RM; Remington SJ, and Verkman AS (2000) Mechanism and cellular applications of a green fluorescent protein–based halide sensor, J. Biol. Chem 275, 6047–6050. [DOI] [PubMed] [Google Scholar]
- (16).Wachter RM, Yarbrough D, Kallio K and Remington SJ (2000) Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein, J. Mol. Biol 301, 157–171. [DOI] [PubMed] [Google Scholar]
- (17).Kuner T, and Augustine GJ (2000) A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons, Neuron. 27, 447–459. [DOI] [PubMed] [Google Scholar]
- (18).Grimley JS, Li L, Wang W, Wen L, Beese LS, Hellinga HW, and Augustine GJ. (2013) Visualization of synaptic inhibition with an optogenetic sensor developed by cell-free protein engineering automation, J. Neurosci, 33, 16297–16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Markova O, Mukhtarov M, Real E, Jacob Y, and Bregestovski P (2008) Genetically encoded chloride indicator with improved sensitivity, J. Neurosci. Methods 170, 67–76. [DOI] [PubMed] [Google Scholar]
- (20).Zhong S, Navaratnam D, and Santos-Sacchi J (2014) A genetically-encoded YFP sensor with enhanced chloride sensitivity, photostability and reduced pH interference demonstrates augmented transmembrane chloride movement by Gerbil Prestin (SLC26a5), PLoS One 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Arosio D, Garau G, Ricci F, Marchetti L, Bizzarri R, Nifosì R, and Beltram F (2007) Spectroscopic and structural study of proton and halide ion cooperative binding to GFP, Biophys. J 93, 232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Arosio D, Ricci F, Marchetti L, Gualdani R, Albertazzi L, and Beltram F (2010) Simultaneous intracellular chloride and pH measurements using a GFP-based sensor, Nat. Methods 7, 516–518. [DOI] [PubMed] [Google Scholar]
- (23).Paredes JM, Idilli AI, Mariotti L, Losi G, Arslanbaeva LR, Sato SS, Artoni P, Szczurkowska J, Cancedda L, Ratto GM, Carmignoto G, Arosio D (2016) Synchronous bioimaging of intracellular pH and chloride based on LSS fluorescent protein, ACS Chem. Biol 11, 1652–1660. [DOI] [PubMed] [Google Scholar]
- (24).Labas YA, Gurskaya NG, Yanushevich YG, Fradkov AF, Lukyanov KA, Lukyanov SA, and Matz MV (2002) Diversity and evolution of the green fluorescent protein family, Proc. Natl. Acad. Sci. USA 99,4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV (2008) Diversity and evolution of coral fluorescent proteins, PLoS One 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pletneva NV, Pletnev VZ, Souslova E, Chudakov DM, Lukyanov S, Martynov VI, Arhipova S, Artemyev I, Wlodawer A, Dauter Z, and Pletnev S (2013) Yellow fluorescent protein phiYFPv (Phialidium): structure and structure-based mutagenesis, Acta Crystallogr., Sect. D: Biol. Crystallogr 69,1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Pakhomov AA, and Martynov VI (2011) Probing the structural determinants of yellow fluorescence of a protein from Phialidium sp., Biochem. Biophys. Res. Commun 407, 230–235. [DOI] [PubMed] [Google Scholar]
- (28).Chattora M, King BA, Bublitz GU, and Boxer SG (1996) Ultra-fast excited state dynamics in green fluorescent protein: multiple states and proton transfer, Proc. Natl. Acad. Sci. USA 93, 8362–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tonge PJ, and Meech SR (2009) Excited state dynamics in the green fluorescent protein, J. Photochem. Photobiol. A 205, 1–11. [Google Scholar]
- (30).Dhamija S, Thakur B, Guptasarma P, and De AK (2018) Probing the excited state dynamics of Venus: origin of dual-emission in fluorescent proteins, Faraday Discuss. 207, 39–54. [DOI] [PubMed] [Google Scholar]
- (31).Griesbeck O, Baird GS, Campbell RE, Zacharias DA, and Tsien RY (2001) Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications, J. Biol. Chem 276, 29188–29194. [DOI] [PubMed] [Google Scholar]
- (32).Tang L, Wang Y, Zhu L, Kallio K, Remington SJ, and Fang C (2018) Photoinduced proton transfer inside an engineered green fluorescent protein: a stepwise-concerted-hybrid reaction, Phys. Chem. Chem. Phys, 20, 12517–12526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.