SUMMARY
Microglia from different nervous system regions are molecularly and anatomically distinct, but whether they also have different functions is unknown. We combined lineage-tracing, single-cell transcriptomics and electrophysiology of the mouse retina and showed that adult retinal microglia shared a common developmental lineage and were long-lived but resided in two distinct niches. Microglia in these niches differed in their interleukin 34 dependency and functional contribution to visual information processing. During certain retinal degeneration models, microglia from both pools relocated to the subretinal space, an inducible disease-associated niche that was poorly accessible to monocyte-derived cells. This microglial transition involved transcriptional reprogramming of microglia, characterized by reduced expression of homeostatic checkpoint genes and upregulation of injury-responsive genes. This transition was associated with protection of the retinal pigment epithelium from damage caused by disease. Together, our data demonstrate that microglial function varies by retinal niche, thereby shedding light on the significance of microglia heterogeneity.
Keywords: microglia, microglial heterogeneity, IL-34, retinal degeneration, retinitis pigmentosa
Graphical Abstract

eTOC:
Whether microglia that reside in different central nervous system niches have distinct functions is unclear. O’Koren and Yu et al. now reveal key functional differences in microglia across two anatomically distinct locations in the retina during homeostasis. Furthermore, in degenerative conditions, microglia migrate to another location where they undergo transcriptional reprogramming and contribute to retinal cytoprotection.
INTRODUCTION
Microglia are highly specialized tissue macrophages that serve as immune sentinels of the central nervous system (CNS) parenchyma (Hanisch and Kettenmann, 2007; Kreutzberg, 1996; Ransohoff and Perry, 2009; Reyes et al., 2017), and are appreciated as key players in neural synapse remodeling during development and plasticity (Nimmerjahn et al., 2005; Paolicelli et al., 2011; Schafer et al., 2012; Squarzoni et al., 2014; Tremblay et al., 2010; Wake et al., 2009). Despite having such wide-ranging activities, microglia are typically thought of and studied as a single pool of cells. Supporting this concept, microglia are currently understood to share a common lineage from yolk sac (YS) erythromyeloid progenitors (Ginhoux et al., 2010; Gomez Perdiguero et al., 2015), and thought to be maintained throughout adulthood independently of circulating monocytes (Ajami et al., 2007; Mildner et al., 2007).
Nonetheless, the adaptability of microglia to local environments suggests that their characteristics and functionality are not uniform across the CNS. Indeed, their density, morphology, and marker expression vary by brain region, and even across layers of the same region, suggesting the possible existence of local variations in their function (de Haas et al., 2008; Jinno et al., 2007; Lawson et al., 1990; Vela et al., 1995). More evidence can be gleaned by the presence of regional variations in microglia dependency on interleukin-34 (IL-34), the alternate ligand of colony stimulating factor 1 (Csf1) receptor (Lin et al., 2008), which contributes to microglia maintenance in adult mouse brain (Greter et al., 2012; Wang et al., 2012). Recent data pointing to a microglia heterogeneity model are based on the existence of region-specific transcriptomic profiles of these cells in the brain (Grabert et al., 2016), and even between individual midbrain nuclei (De Biase et al., 2017), as well as possible differences when compared to the spinal cord (Matcovitch-Natan et al., 2016).
Together, these data raise a fundamental question of whether microglia have distinct functions in different CNS niches. However, it is not currently possible to disrupt microglia in a site-specific manner, and thus direct evidence for heterogeneous functions is lacking. This problem extends to microglia that occupy de novo disease niches arising in neurodegenerative conditions, (Itagaki et al., 1989; Keren-Shaul et al., 2017; Krasemann et al., 2017; Yamasaki et al., 2014), particularly as recruited monocyte-derived macrophages (mo-MFs), seen in certain disease settings, are difficult to distinguish from resident microglia (O'Koren et al., 2016).
We addressed these questions at the level of the retina, where microglia primarily reside in two distinct synaptic compartments of the neural parenchyma, termed the outer and inner plexiform layers (OPL and IPL, respectively). Our data showed that adult retinal microglia were YS-derived and long-lived, but only the IPL population was dependent on IL-34 in their maintenance. We further showed that IL-34-dependent microglia from this niche contributed to normal processing of cone-driven visual information. Despite these differences, microglia from both layers were able to relocate to the subretinal space (SRS) during retinal degeneration. Furthermore, subretinal microglia (srMG) led to the protection of retinal pigment epithelium (RPE) structural integrity. In contrast, under these conditions, monocyte-derived cells recruited to the retina were mostly restricted to the neural parenchyma. Therefore, our data demonstrate that retinal microglia function varies by niche, both in the steady state and in degenerative diseases.
RESULTS
Retinal Microglia Arise from Yolk Sac Precursors and Are Distinct from Other Ocular Macrophages.
We first interrogated the ontogeny and longevity of retinal microglia and compared it to the longevity of other macrophages in different eye structures (Figure 1A). Whereas the YS-origin of brain microglia has been well-established (Ginhoux et al., 2010), the origin of retinal microglia has not been examined. We addressed this in Runx1Mer-Cre-Mer; RosaR26R-eYFP embryos by pulsing with tamoxifen 4’OHT at E7.5. This approach will label Runx1 specifically in YS progenitors with eYFP. Retinal suspensions from progeny mice at 8 weeks were analyzed by flow cytometry. Brain microglia and age-matched Cre−/− littermates were analyzed as controls. We found that YFP+ microglia in retina were present to the same extent as brain (Figure 1B and 1C), indicating that retinal microglia are YS-derived.
Figure 1. Fate Mapping Reveals Ontogeny, Longevity and Phenotype of Retinal Microglia.
(A) Schematic showing the anatomy of the eye.
(B) Flow cytometry plots depict yolk-sac derived microglia in retina. Pregnant Runx1MCreM; RosaR26R-eYFP mice received tamoxifen 4’OHT at E7.5 days. Tissues were collected from 8 wk old progeny for flow cytometry.
(C) Dot plots (mean ± SEM) summarize the percentage of Runx1-YFP+ microglia shown in (B) from 2 independent experiments. Each dot represents one mouse (n = 4 to 6 per group).
(D) Representative images show distribution of short-lived (fGFP−) and long-lived (fGFP+) macrophages. Adult Cx3cr1Cre-ER; RosaR26R-fGFP mice received tamoxifen and collected 1 yr later. (outer and inner nuclei layer, ONL and INL; outer and inner plexiform layer, OPL and IPL; ganglion cell layer, GCL; ciliary body, CB; Optic N, nerve). Choroid layer boundary is delineated (white lines). Scale bars, 100 μm.
(E, F) Adult Cx3cr1Cre-ER; RosaP26R-fGFP mice received tamoxifen and tissues collected at 48 hrs (baseline) and 1 yr. Representative flow cytometry plots of macrophages at 1-yr post or control (E). Representative dot plots compare the percentage of fGFP+ macrophages between 48-hr and 1-yr (F) from 2 independent experiments. Grey areas (mean ± SD) show the percentage of control (n=9 eye tissues). Each dot represents one mouse (n = 6 per group).
(G, H) tSNE clustering of long- and short-lived macrophages (G) and tissue specificity of long-lived macrophages (H).
See also Figure S1.
We next investigated the longevity of retinal microglia, as they are a type of long-lived macrophages. Adult Cx3cr1Cre-ER; RosaR26R-fGFP mice (Goldmann et al., 2013; Parkhurst et al., 2013) were tamoxifen-pulsed at 6-10 weeks, and analyzed at 48 hours to ascertain baseline Cre-recombination and, again 1 year later to determine longevity (Figure S1A-D). We also evaluated optic nerve, choroid, iris and ciliary body and cornea for comparison. We found that in ocular CNS tissues that ~55% of total macrophages in optic nerve and 85% in retina were fGFP+ (Figure 1D-F). For non-CNS tissues, ~50% of total macrophages in iris and ciliary body were fGFP+, and only few were fGFP+ in cornea and choroid (Figure 1D-F), suggesting that these tissues were comprised of short-lived mo-MFs. Thus, we conclude that retinal microglia are long-lived, and other eye compartments also harbor long-lived macrophages.
We next compared the phenotype of retinal microglia to other long-lived macrophages in optic nerve and iris and ciliary body via multi-dimensional flow cytometry mapping. Analysis was performed at 1-year post tamoxifen of adult Cx3cr1Cre-ER; RosaR26R-fGFP mice. Markers included CD45, CD11b, CD11c, CD64, Ly6G, Ly6C and MHC class II. Total macrophages were grouped based on long-lived (fGFP+) vs. short-lived (fGFP−) status. We found that fGFP+ macrophages and their fGFP− counterparts clustered differently by t-Stochastic Neighbor Embedding (tSNE) (Figure 1G), and their phenotypes were significantly different (p≤0.001) mostly due to increased CD11c and MHC class II expression in short-lived macrophages (Figure S1E). We also found that fGFP+ macrophages from each individual compartment clustered differently (Figure 1H and S1F). These data indicate that both ontogeny and location affect the phenotype of long-lived macrophages.
Inner Plexiform Layer is an IL-34 Dependent Microglia Niche
Next, we looked for clues of retinal microglia heterogeneity in normal adult retina, particularly between IPL vs OPL pools. We examined the role of IL-34 in maintenance of retinal microglia. Levels of Il34 mRNA were compared relative to Csf1 in retina, using skin (including epidermis) as a positive control and lymph node as a negative control of IL-34 dependency. Similar to skin, Il34 levels in retina were greater than lymph node (Figure 2A), thus supporting a role of IL-34 in the retina. We quantified microglia in the retina of adult Il34LacZ/LacZ a Il34-deficient mouse strain, relative to Il34+/LacZ mice and found that Il34LacZ/LacZ mice had a 25.8% reduction in microglia number (Figure 2B). This reduction was observed in IPL, whereas no significant reduction was detectable in OPL (Figure 2C-E). Hence, IL-34 dependency is restricted to IPL microglia, thereby establishing niche-specific heterogeneity in microglia maintenance between OPL and IPL pools.
Figure 2. IL-34 Dependent Microglia Niche is Located at the IPL and These Microglia Functionally Contribute to Cone Bipolar Cell Output.
(A) Bar graphs (mean ± SEM) compare Il34 mRNA levels relative to Csf1 in skin (n=6), retina (n=6), and lymph node (n=4) from 2 independent experiments. GAPDH was used as a loading control. Level of Il34 in lymph node is set as 1.
(B) Dot plots (mean ± SEM) depict the percentage of microglia compared to total viable singlets in Il34 mutant mice analyzed by flow cytometry. Each dot represents one mouse.
(C, D) Representative images of microglia (IBA1, grey; DAPI, blue) from cross-sections (C) and retina flatmounts (D) compare the microglia distribution in OPL and IPL of Il34 mutant mice. Data were collected from 3 independent experiments. Scale bars, 100 μm.
(E) Dot plots (mean ± SEM) depict microglia density shown in (D) from 4 independent experiments. Each dot represents one mouse (n = 6 or 10 per group).
(F, G) Images show specific localization of Il34 promoter guided β-gal (green) in RGC layer (Brn3a, red; DAPI, blue). Scale bars, 50 μm.
(H) RNAScope of Il34 in Atoh7 mutant mice (Il34, yellow; DAPI, blue). Scale bar, 50 μm.
(I, J) ERG recordings of scotopic b-wave and photopic flicker response shown in (I) and (J), respectively. Scotopic b-waves are shown as rod driven and cone drive components (I). Data (Il34+/LacZ, n = 6; Il34LacZ/LacZ n = 10) were from 2 independent experiments.
See also Figure S2.
To determine the cellular source of IL-34, we examined LacZ expression driven by Il34 promoter in Il34+/LacZ mice, using Il34+/+ as controls. We detected β-gal only in BRN3A+ retinal ganglion cells (RGC) (Figure 2F and 2G). We also examined Il34 mRNA using RNAScope in RGC-deficient Atoh7−/− mice (Brown et al., 2001) and found that Il34 expression in RGC layer was greatly diminished compared to Atoh7+/− mice (Figure 2H). Some Il34 mRNA was also detected at the top of the IPL, where amacrine cells reside. Our results are consistent with mined data (Siegert et al., 2012) (Figure S2A). Together, our findings suggest that IL-34 is produced primarily by RGCs and contributes to microglia maintenance in the IPL.
IL-34 Dependent Microglia in the IPL Specifically Contribute to Visual Function
To address the functional contribution of the IL-34-dependent pool of microglia in the IPL, we used electroretinography (ERG) to assess light responses of IL-34 deficient mice. No changes were observed in dark-adapted (scotopic) a-wave (Figure S2B), a measure of photoreceptor light-excitation. However, scotopic b-wave responses showed a selective reduction at hi-light intensities (Figure 2I and S2C-E). Similar defect was observed for photopic b-waves (generated by cones under fully saturating rod responses, Figure S2F) and for photopic flicker ERGs (Figure 2J). This reduction did not appear to be related to neuronal Csf1r expression, as confocal microscopy from adult Csf1r-eGFP retinas of macrophage Fas-Induced apoptosis transgenic mice showed that GFP labeling only colocalized with IBA1+ cells (data not shown). Instead, because microglia loss in IL-34 deficient mice is IPL specific, our data suggest this phenotype likely originates from abnormal hyperpolarizing feedback regulation in the IPL (Diamond, 2017). However, no gross changes in synapse number that cone bipolar cells form were detected (Figure S2G-L), thereby pointing to a possible qualitative defect of these synapses. These findings suggest IL-34 dependent microglia pool in the IPL are qualitatively involved in feedback regulation at cone bipolar cell axons, thus revealing a niche-specific functional contribution of microglia.
Subretinal Space is a Microglia-Dominant Immune Niche in Models of Photoreceptor Degeneration
We next analyzed microglia responses to photoreceptor degeneration, which is associated with abnormal accumulation of phagocytes in the SRS (Calippe et al., 2017; Gupta et al., 2003; Ng and Streilein, 2001; Thanos, 1992). We first characterized microglia using a light damage (LD) model of photoreceptor degeneration (Noell et al., 1966). We induced LD in Cx3cr1YFP-Cre-ER/wt; RosaR26R-DTR/wt mice (Buch et al., 2005), which allows inducible tracking (or depletion) specifically of microglia. We bred these mice onto a CB6F1 background (i.e. Rpe65L450M/+) which confers LD susceptibility (Danciger et al., 2003). These CB6F1 :Cx3cr1YFP-Cre-ER/wt; RosaR26R-DTR/wt (i.e. F1-iDTR) mice were received tamoxifen at 6-10 weeks and subjected to LD at least 6 weeks later (Figure S3A). Retinas were immunostained with DTR antibody to label lineage-traced microglia (Figure S3B). In LD, for maximally damaged regions, we observed few remaining microglia in the neuroretina but an abundance in the SRS (Figure 3A). In contrast, mo-MFs were found in the neuroretina but not the SRS (Figure 3A, 3B and S3C), indicating distinct distributions. To determine if mo-MFs can fill the SRS in the absence of microglia, we performed the same experiment following microglia depletion by administering diphtheria toxin (DT) prior to LD induction (Figure S3A). We observed that mo-MFs were still not able to occupy the SRS (Figure 3A). Likewise, immunostaining of RPE flatmounts revealed that only microglia adhered to apical RPE (Figure 3C and 3D). These data suggest that the SRS is a microglia-dominant immune niche in the LD model, occupied by microglia migrating from both IPL and OPL pools.
Figure 3. The SRS is a Microglia-Dominant Immune Cell Niche in Models of Photoreceptor Degeneration.
(A) Representative images from cross sections (Cx3cr1YFP cells, green; DTR, red) show microglia in SRS of F1-iDTR mice in LD. See Fig.S3A for details. DAPI, blue. Scale bar,100 μm.
(B) Dot plots show the percentage of DTR+ cells among Cx3cr1+ cells in undamaged and damaged areas of F1-iDTR mice from 2 independent experiments. Each dot represents one mouse (n = 5).
(C) Representative images (YFP, green; DTR, red) from RPE flatmounts show the attachment of microglia onto the RPE in LD. Scale bar, 100 μm.
(D) Dot plots (mean ± SEM) depict microglia depletion efficiency in LD shown in (C). Each dot represents one mouse (n = 5 per group).
(E) Immunofluorescence images show SRS phagocytes (IBA1, magenta) in RhoP23H/wt retinas. DAPI, blue. Scale bar, 50 μm.
(F) RPE flatmounts show DTR+ microglia (YFP, green; DTR, red) on the RPE of P23H-iDTR mice at P30. Tissues were collected 3 wks post last tamoxifen. Scale bar, 100 μm.
(G) Images of RPE flatmounts show depletion of microglia (IBA1, grey) in female P23H-iDTR mice at P60. See Fig.S3A for details. Scale bar, 100 μm.
(H) Dot plots (mean ± SEM) depict microglia depletion efficiency shown in (G) from 3 independent experiments. Each dot represents one mouse (n = 3 or 6 per group).
See also Figure S3.
Because LD induces vigorous acute damage, we determined if microglia similarly occupy the SRS in the course of a more chronic retinal degeneration. We used RhoP23H/wt mice that have an autosomal dominant P23H knockin mutation in rhodopsin (Sakami et al., 2011), which reasonably phenocopies a major form of retinitis pigmentosa (Dryja et al., 1990). We found IBA1+ cells accumulate in the SRS of these mice with age (Figure 3E). Thus, we next generated RhoP23H/wt; Cx3cr1 YFP-Cre-ER/wt; RosaR26R-DTR/wt mice (i.e. P23H-iDTR) to ask if microglia occupy this niche. Microglia were labeled by tamoxifen pulse at P4 and P7 and assessed 3 weeks later. This analysis revealed that all the Cx3cr1+ phagocytes adhering to the RPE in P23H-iDTR mice were microglia (Figure 3F). We then depleted microglia in tamoxifen-pulsed P23H-iDTR mice by administering DT at P21 and P28 and examined RPE flatmounts at P60 (Figure S3A). Females were used because depletion efficiency was more consistent, possibly due to sexual dimorphic effects on microglia (Thion et al., 2018). Similar to LD, microglia depletion resulted in reduction in the number of RPE-adhered microglia (Figure 3G and 3H). Taken together, microglia are the only phagocytic cells able to efficiently occupy the SRS and adhere to the RPE in both LD and RhoP23H/wt models.
Single-Cell RNA-Seq Uncovers the Distinct Transcriptome of subretinal Microglia in Photoreceptor Degeneration
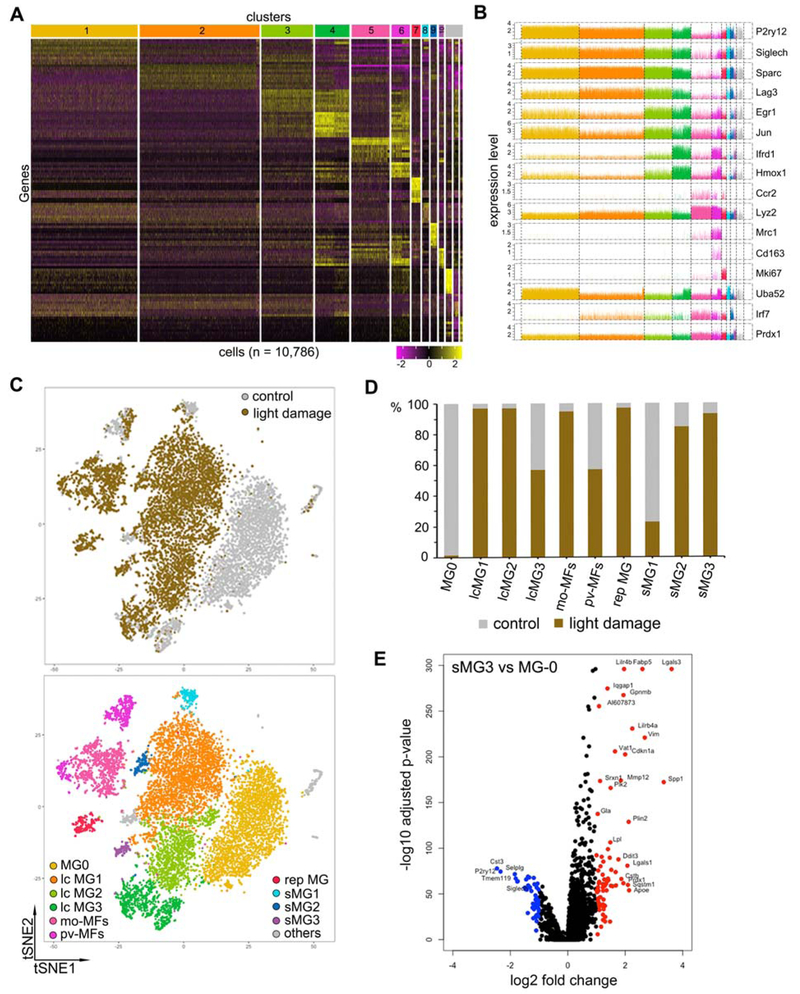
To gain insight into the transcriptional changes of srMG during photoreceptor degeneration, we applied single-cell RNA-seq (scRNA-seq) using the 10x Genomics platform (Figure S4A and S4B). Analysis was performed on FACS-sorted live Cx3cr1YFP+ cells from pooled neuroretinas of normal (n=5) and LD (n=8) mice. Using Seurat (Butler et al., 2018), total 10,786 cells passed quality control and unsupervised clustering produced >10 clusters (Figure 4A and Table S1). We observed 4 large clusters of microglia (lcMG) that showed high expression of P2ry12, Siglech, Sparc and other microglia markers (Bennett et al., 2016; Butovsky et al., 2014; Gautier et al., 2012), with corresponding low expression of myeloid cell markers, including Ccr2, Mrc1, and Lyz2 (Figure 4A-C and S4C). The MG0 cluster was comprised solely from normal retinas, whereas cells in other 3 large clusters came mostly from LD retinas (i.e. lcMG1 thru 3) with high expression of Lag3, Egr1, Ifrd1, and/or Hmox1 (Figure 4A-C and S4C). Small microglia clusters (sMG) included a replicating population (rep MG) with high Mki67 expression (Hughes et al., 2003; Zeiss and Johnson, 2004) (Figure 4A-C). Moreover, sMG1 was found in both normal and LD, whereas sMG2 and sMG3 were found almost exclusively in LD (Figure 4A-C). Clusters of mo-MFs and perivascular (pv) macrophages were enriched for myelomonocytic genes, including Ccr2, Mrc1, Lyz2, and Cd163 (Figure 4A-C and S4C). We excluded “other” clusters as they were a contaminant from photoreceptors or contained <2% of the cells.
Figure 4. Single-Cell RNA-Seq Uncovers A Distinct Microglia Type Associated with Retinal Degeneration.
(A) Heatmap of unsupervised clustering analysis featuring top 10 discriminative genes per cluster. Expression level is scaled based on z-score distribution. Data were collected from FACS-sorted live Cx3cr1YFP+ cells from pooled neuroretinas of normal (n=5) and LD (n=8) mice.
(B) Expression level of selected marker genes for clusters in (A) is shown.
(C) tSNE plots of scRNA-seq show unsupervised clusters. Top, control and LD; bottom, 10 major clusters.
(D) Bar graphs show sample components of each cluster in (C).
(E) Volcano plot shows the fold change of genes (log2 scale) and significance (−log 10 scale) between sMG3 and MG0. Upregulated genes, red; downregulated genes, blue. P-values adjusted based on Bonferroni correction.
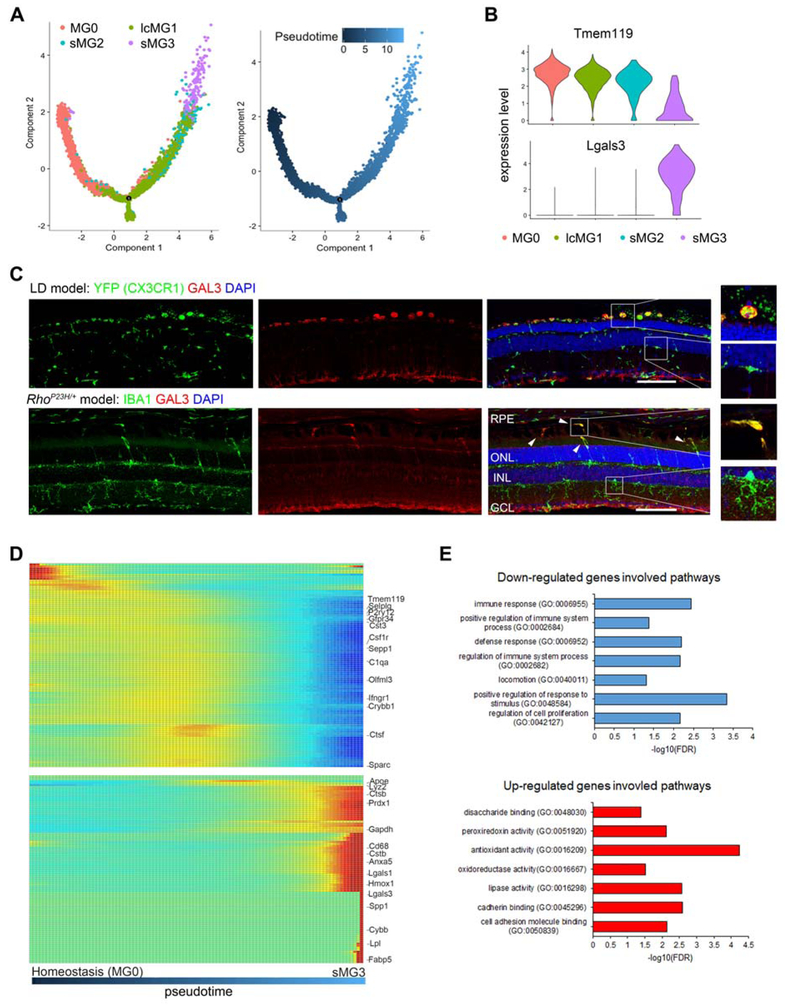
Next, we looked for the cluster corresponding to srMG in LD. We focused on small clusters since srMG are fewer relative to other populations in LD. While sMG2 and sMG3 were both candidates, only sMG3 had a large number of down-regulated homeostatic genes, including Tmem119, P2ry12, and Siglech (Figure 4D, S4D and S4E), a phenotype reported in other CNS diseases (Chiu et al., 2013; Holtman et al., 2015; Keren-Shaul et al., 2017; Krasemann et al., 2017; Wang et al., 2015a). Hence, sMG3 is particularly distinct from steady state microglia, although no microglia clusters from LD were enriched in MHC class II (Figure S4D). To help determine if sMG3 corresponds to srMG in LD, we performed a trajectory analysis with the top 1000 variably expressed genes (Qiu et al., 2017). By setting MG0 as the root state, we hypothesized that the cluster corresponding to srMG would be found at the final state of this trajectory. We excluded non-microglia clusters (mo-MFs and pv-MFs) and rep MG. While our resultant trajectory possessed two main branches, one of them contained sMG3 as the final state (Figure S5A-C). The same was true in subsequent trajectory on clusters of this branch only (Figure 5A and 5B). Hence, these data pointed to sMG3 as the corresponding population of srMG in LD.
Figure 5. Microglia that Occupy the SRS in Photoreceptor Degeneration Undergo Transcriptional Reprograming.
(A) Trajectory analysis suggests transition of different microglia subtypes (left) along pseudotime (right). Pseudotime of MG0 cluster is set as 0. Top 1000 variably expressed genes from clustering analysis are used.
(B) Violin plots depict expression changes of marker genes across clusters as indicated.
(C) Representative images show GAL3+ (red) srMG in LD (top: YFP, green) and RhoP23H (bottom: IBA1, green). Images were acquired in three independent experiments from 6 and 4 mice, respectively. See Table S2 for details. Scale bars, 100 μm.
(D) Heatmap illustrates dynamic transition from MG0 to sMG3 along pseudotime: top, downregulated genes; bottom, upregulated genes. Genes are clustered and ordered based on expression profile. Values beyond the scale range are set to minimum or maximum.
(E) Bar graphs show top ranked pathways by fold enrichment and significance (−log 10 scale) from GO-term pathway enrichment analysis. Top, downregulated genes (blue); bottom, upregulated (red). Top 100 downregulated and 100 upregulated genes in between sMG3 and MG0 are used for the analysis.
To validate this hypothesis, we immunostained LD retinas for proteins of enriched genes in sMG3, including Lgals3, Lpl, and Cd68. We found that srMG are positively labeled for Galectin-3 (GAL3), whereas Cx3cr1+ cells in the neuroretina were not (Figure 5C). RNAScope of Lgals3 corroborated this observation (Figure S5D). The same pattern was observed for LPL and Cd68 (Figure S5F), although LPL also labeled other retinal cells. Also consistent with the sMG3 signature, we observed P2RY12 immunolabeling of srMG was weak compared to robust labeling of Cx3cr1+ cells in neuroretina (Figure S5F). To determine if the sMG3 cluster might also be relevant in RhoP23H/wt mice, we immunostained for GAL3 and found that srMG were uniquely positive for this marker (Figure 5C and S5E). Also, weak P2RY12 immunolabeling was likewise noted for srMG, but not for those in neuroretina (Figure S5G). The srMG in RhoP23H/wt were also Cd68+, whereas Lpl was not (Figure S5G). The latter may reflect the slower nature of photoreceptor degeneration in the RhoP23H/wt mice. Thus, our data validate the sMG3 cluster as srMG in LD and suggests a phenotypic overlap with those srMG in RhoP23H/wt mice.
We next used our validated trajectory to study transcriptional changes of srMG in photoreceptor degeneration. The top 100 upregulated genes from MG0 (i.e. steady state) and sMG3 (i.e. srMG) clusters were plotted over pseudotime (Figure 5D). Our analysis revealed that gene expression kinetics across pseudotime was a dynamic process. Some genes were induced temporarily during intermediary states of the trajectory, such as Apoe, Ppiaf4, Lsp1, and Alox5ap, whereas others were upregulated very late in the trajectory, such as Lgals3, Cd63, Fabp5, Lpl, Cybb, Mmp12 and Spp1. Importantly, the latter was accompanied by downregulation of homeostatic microglia genes, such as Tmem119, Selplg, P2ry12, Hexb, Gpr34 and Cx3cr1.
We also performed a Gene Ontology Enrichment Analysis of the top altered genes in sMG3 relative to MG0 and found that down-regulated genes included immune response pathways, whereas enriched genes included cell recognition and adhesion pathways (Figure 5E). The latter agrees with our finding that srMG adhere to the RPE. Moreover, enriched pathways such as lipid metabolism and antioxidant pathway genes in sMG3 (Figure 5E), may point to the possibility that srMG are protective in this setting. In sum, our data suggest that srMG undergo a dynamic reprogramming that may result in a protective phenotype.
Subretinal Microglia Contribute to Retinal Pigment Epithelial Protection in Photoreceptor Degeneration
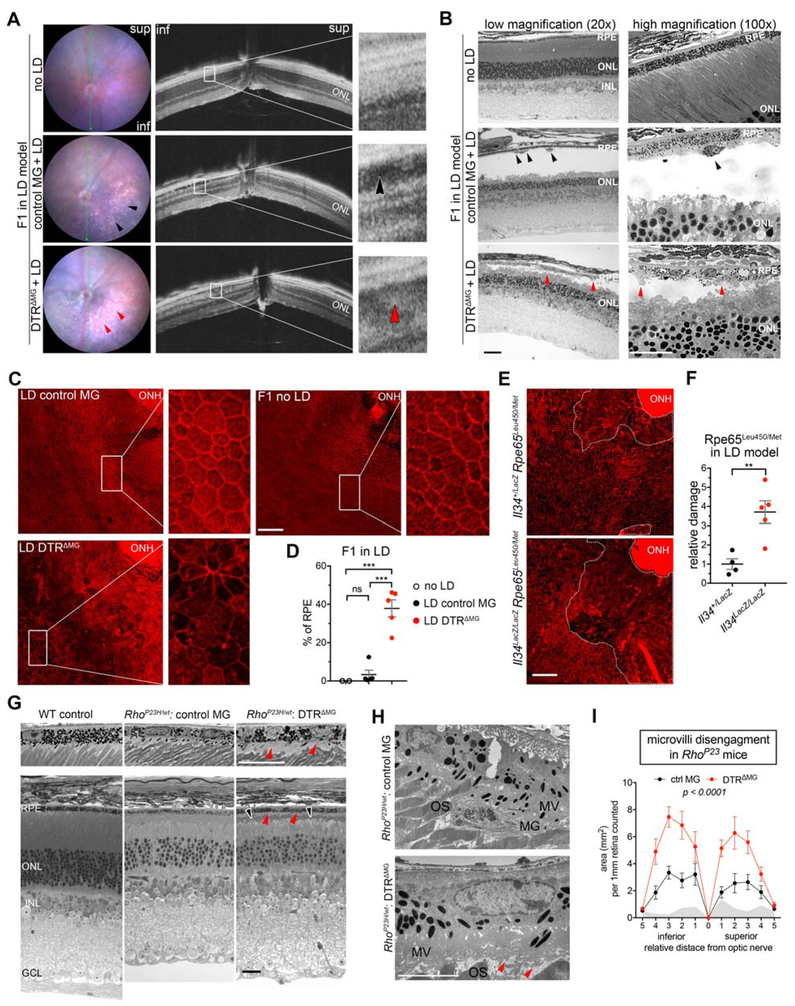
Final experiments addressed the functional role of srMG in retinal degeneration. We took advantage of F1-iDTR mice to conditionally deplete microglia in LD. Two control lines used here either had the same genotype but did not receive DT or were DTR-negative littermates that did receive DT. Retinas were examined in live mice using fundoscopy and optical coherence tomography (OCT). Histological analysis was performed on plastic embedded retinal cross-sections and phalloidin-stained RPE flatmounts.
Our results showed that LD most severely affected the inferior retina, adjacent to optic nerve head (Figure 6A). There, fundoscopy of control showed punctate legions that were absent in the DTRΔMG fundus (Figure 6A), which corresponded histologically with SRS phagocytes (Figure 6A and 6B). This finding is consistent with (Luhmann et al., 2009), though our conditional depletion approach indicates these are microglia. OCT scans revealed hypo-reflectivity at outer segment regions of control, whereas the same location in DTRΔMG mice displayed hyper-reflective scattering (Figure 6A and 6B). The latter corresponded histologically to outer segment debris (Figure 6A and 6B), suggesting a role for microglia in their removal. We also observed massive amounts of pyknotic TUNEL+ nuclei in the ONL of DTRΔMG mice (Figure 6B and S6A), indicating a role for microglia in swift clearance of photoreceptor somata debris as well.
Figure 6. SrMG Functionally Contribute to Protection of RPE Structural Integrity in Photoreceptor Degeneration.
(A) In vivo retinal imaging of live CBF1 mice (left: fundus microscopy; right: OCT) shows morphological changes in LD. Black and red arrow indicate hypo- and hyper-reflectivity of outer retina, respectively.
(B) Histology reveals accumulation of subretinal debris in LD described in (A). Scale bar, 100 μm (left) and 50 μm (right). Black arrows indicate SRS phagocytes; red arrows indicate photoreceptor debris.
(C) Representative images of RPE flatmounts show F-actin (phalloidin, red) change of the RPE in LD. ONH, optical nerve head. Scale bar, 100 μm.
(D) Dot plots (mean ± SEM) depict the percentage of dysmorphological RPE in (C) from 3 independent experiments. Each dot represents one mouse.
(E) Images of RPE flat mounts show loss of distinguishable RPE around the ONH of Il34 deficient mice (Rpe65Leu450/Met450) upon LD. Dashed circles indicate the damaged area. Scale bar, 100 μm.
(F) Dot plots (mean ± SEM) depict the relative fold change of damaged area in (E). Each dot represents one mouse (n = 4 or 5 per group).
(G) Histological images shows outer segment shrinking in DTRΔMG retina of RhoP23H/wt mice at P60. Two magnifications are shown. Scale bars, 100 μm. Black arrows indicate shortened outer segments, and red arrows indicate RPE microvilli disengagement from outer segments.
(H) Images of TEM show disorganized apical microvilli of RPE in RhoP23H/wt: DTRΔMG retina as described in (E). MG, microglia; MV, microvilli; OS, outer segments. Scale bar, 5 μm.
(I) The plots (mean ± SEM) quantify the area between outer segment tips and apical side of the RPE cell body described in (E) from 3 independent experiments. Grey areas show one WT control. Control MG, n = 8; DTRΔMG, n = 10.
See also Figure S6.
Next, we analyzed phalloidin stained RPE. We did not observe any obvious F-actin alterations between control and naïve mice. However, the RPE of DTRΔMG showed loss of apical F-actin and dysmorphic lateral F-actin (Figure 6C, 6D and S6B). This is indicative of pathological changes to apical microvilli and general RPE stress, thus implicating srMG in the protection of RPE during LD. Our findings earlier that both IL-34 dependent and independent microglia pools migrate to the SRS in LD led us to next address if the IL-34 dependent pool contributes to RPE protection. We crossed CB6F1 back to C57BL/6J background for 9 generations and generated Il34 mutant mice with Rpe65L450M/+ to confer LD susceptibility. After challenge with LD, RPE flatmounts of these mice were stained with phalloidin and examined by confocal microscopy. Though OCT showed Il34LacZ/LacZ and Il34+/LacZ mice experienced less ONL thinning (data not shown) relative to CB6F1 mice, there was a marked difference in RPE changes based on Il34 genotype. RPE damage was significantly greater in Il34LacZlacZ relative to hemizygous controls. In addition to dysmorphology, these nullizygous mice had larger areas that showed loss of distinguishable RPE (Figure 6E, 6F and S6C). Moreover, lower numbers of microglia in steady state Il34LacZ/LacZ retina was conserved in the LD setting (Figure S6D). Our findings indicate that srMG protect RPE and that IL-34 dependent pool of microglia contributes to this protection.
We next addressed if there is a similar protective role of srMG in RhoP23H/wt mice. We induced conditional depletion of microglia in P23H-iDTR female mice, as described above. Controls were the same genotype that did not receive DT or were DTR-negative RhoP23H/wt littermates that received DT. Unlike the LD model, we were not able to find obvious differences by fundoscopy (Figure S6E) nor in phalloidin-stained RPE (data not shown), which could be explained by the less acute nature of this degeneration. Nonetheless, we observed existence of shortened and disorganized outer segments (Figure 6G), suggesting augmented defects to photoreceptor morphology. The ONL counts were slightly greater in RhoP23H/wt DTRΔMG mice relative to control RhoP23H/wt mice, which was again associated with increased number of pyknotic and TUNEL+ nuclei (Figure S6F-J). Thus, similar to the LD setting, depletion of microglia leads to the accumulation of dead photoreceptor somata.
We also observed continuous and diffuse structural inclusions along the apical RPE in RhoP23H/wt DTRΔMG mice (Figure 6G). By ultrastructural analysis via transmission electron microscope, this phenotype was explained by the disengagement of the distal end of photoreceptor outer segments from RPE microvilli from the distal end of photoreceptor outer segments, which normally interdigitate this photoreceptor compartment. In contrast, RPE microvilli interdigitation was better-preserved in control RhoP23H/wt mice, with microglia found surrounded by RPE microvilli (Figure 6H). Quantification of the area between the outer segment tips and RPE cell body underscored the prevalence of microvilli disengagement in RhoP23H/wt DTRΔMG mice (Figure 6I). Of note, this disengagement trended greater in the inferior retina, which degenerate most rapidly in RhoP23H/wt (Figure 6I and S6J), indicating that this phenotype correlates with the severity of ongoing pathology. These data suggest that srMG protect RPE-photoreceptor interactions, thereby prolonging the structural integrity of the outer retina in RhoP23H/wt mice.
DISCUSSION
Despite emerging data that microglia could be phenotypically heterogeneous (De Biase et al., 2017; Grabert et al., 2016), direct evidence of functional heterogeneity has been lacking. We now demonstrate that microglia do, in fact, have unique functional specializations at different locations of the retina.
We showed that ~85% of macrophages in the retina were long-lived, which are mostly microglia except for a small fraction of perivascular macrophages (Dick et al., 1995; O'Koren et al., 2016). This pattern is typical for neural tissues (Prinz et al., 2017), including the optic nerve where we demonstrated that the fraction of long-lived macrophages was > 50%. This was also the case for the iris and ciliary body, which are similarly separated by the blood-tissue barriers. In contrast, neither choroid nor cornea harbored a significant population of long-lived macrophages. Multidimensional flow cytometry mapping revealed that, whereas the profiles of all long-lived macrophages differ from their short-lived counterparts, long-lived cells residing within different ocular compartments themselves have unique signatures. Hence, both ontogeny and location influence phenotype of long-lived macrophages.
In this study, we focused primarily on characterizing the heterogeneity of retinal microglia. Although commonly derived from YS progenitors and long-lived, these cells occupied two distinct niches, the OPL and IPL. Only IPL microglia were dependent on IL-34, a cytokine we show was expressed predominantly by RGCs. Moreover, IL-34 deficient mice had reduced ERG responses in the cone-driven channel of vision. Our data suggest that IL-34 dependent microglia may assist secondary feedback inputs that bipolar cells receive in the IPL known to shape signal amplitude and kinetics. Indeed, only ERG responses specifically produced by bipolar cells (b-waves) were affected in the absence of IL-34. Our result complements a previous report that ablating all retinal microglia causes a reduction of both a- and b-wave responses, indicative of defects at both photoreceptor and bipolar cell levels (Wang et al., 2016). Collectively these findings suggest that each sub-population of microglia contribute to different steps of visual processing. Whether IL-34 dependency is only important for microglia maintenance or is also directly involved in shaping their functional contributions in the context of visual processing requires further investigation.
Our second major finding is that microglia from both niches migrated to the SRS in photoreceptor degeneration, where they adhered to apical RPE. In contrast, mo-MFs under the same conditions, occupied the neuroretina vacated by microglia, and neither migrated into the SRS nor adhered to the RPE in any significant number. We previously documented this pattern in LD (O'Koren et al., 2016; Reyes et al., 2017), which was subsequently recapitulated in the NaIO3 induced RPE injury model (Ma et al., 2017). We now extend our finding to the RhoP23H/wt model of retinitis pigmentosa. Moreover, even when we depleted microglia, thereby eliminating any competition for SRS, mo-MFs were still unable to take up this location during LD. Therefore, in the models and timepoints analyzed, the SRS serves as a microglia-dominant immune niche.
Our data further suggest that emergence of srMG under pathological conditions is accompanied by their stepwise transcriptional reprogramming. Trajectory analysis revealed this involves downregulation of homeostatic genes, like Tmem119, P2ry12, Siglech, Hexb, Selplg, Sparc, and Sall1, and upregulation of neurodegeneration-linked genes, like Lgals3, Lpl, Cd68, Fabp5, Lilr4b, Apoe, Trem2, Cstb, and Sqstm1. Polymorphisms in several of the latter genes (e.g. Apoe, Sqstm1, and Lpl) have been linked with retina degenerative diseases (Blanton et al., 1991; Hoffman et al., 1989; McKay et al., 2011; Scheetz et al., 2016; Wang et al., 2015b). Likewise, some genes have been implicated in brain and spinal cord neurodegenerations (Chiu et al., 2013; Fritsche et al., 2016; Holtman et al., 2015; Keren-Shaul et al., 2017; Krasemann et al., 2017).
Finally, our data suggest that srMG protect RPE from disease-associated damage. We showed that these microglia, including the IL-34 dependent pool, prevented morphological RPE damage in LD. Similarly in RhoP23H/wt mice, srMG promoted interdigitation of RPE microvilli with photoreceptor outer segments, a key interaction required for vision. An example of microvilli disengagement was recently described in BEST1 mutation affecting RPE in canines (Guziewicz et al., 2018). Microglia also accumulate in the SRS of aged mice (Ma et al., 2017), but whether these cells are likewise protective in this context requires further investigation. Together, our results represent a significant break from conventional thinking, which holds that phagocytes in the SRS in retinal degeneration only contribute to ongoing pathology (Calippe et al., 2017; Karlstetter et al., 2015; Reyes et al., 2017). The potential impact of these findings is that it opens doors to consider neuroprotective features of srMG as a potential therapeutic target for vision preservation.
In summary, we showed that microglia in the adult retina have niche-dependent functional specializations, both in health and disease. Normally, microglia occupied two plexiform layers where their maintenance and ability to support interneuronal communications differed by IL-34 dependency. However, in the disease settings examined, both pools underwent a massive migration to the SRS where they adhered to the apical RPE and protected the structural integrity of the outer retina and RPE. The challenge of future studies is to define how each of these major functions is fulfilled on the mechanistic level.
STAR★Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel R. Saban (daniel.saban@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J, BALB/cJ, Cx3cr1tm1.1(cre)Jung, Cx3cr1YFP-Cre-ER/YFP-Cre-ER and the inducible DTR mouse line, Rosa26R26R-DTR/R26R-DTR mice were purchased from Jackson Laboratories. Rosa26R26R-fGFP/R26R-fGFP mice were kindly provided by Dr. Brigid L.M. Hogan (Duke University). Il34LacZ/LacZ Aoth7−/− and RhoP23H/P23H mice were generated as described previously (Brown et al., 2001; Greter et al., 2012; Sakami et al., 2011). CB6F1: Cx3cr1Cre-ER/wt; R26R26R-DTR/wt (F1-iDTR), F1 progeny of BALB/cJ and Cx3cr1Cre-ER; R26R26R-DTR mice of C57BL/6J background, were generated for light damage model. CB6F1: Cx3cr1Cre-ER/wt and CB6F1 mice were also used as available. CB6F1 mice were also backcrossed with C57BL/6J background mice for 9 generations, during which Il34LacZ transgene was introduced to generate Il34LacZ; Rpe65Leu450/Met450 mice for light damage. RhoP23H mice were crossed with Cx3cr1Cre-ER; R26R26R-DTR mice to generate RhoP23H/wt; Cx3cr1Cre-ER/wt; R26R26R-DTR/wt (P23H-iDTR) mice for inherited retinal degeneration model. If not otherwise stated, mice used were mixed-gender and 6-12 weeks of age. All mice herein did not carry rd8 mutation and were bred and housed at a barrier-free and specific-pathogen-free facility at Duke University School of Medicine (Durham, NC). All procedures were approved by the Institutional Animal Care and Use Committee at Duke University, and the procedures were carried out in accordance with the approved guidelines. Runx1Cre-ER/wt and Rosa26R26R-eYFP/R26R-eYFP mice were generated and used previously as described (Ginhoux et al., 2010). These mice were bred and housed in the Biomedical Resource Centre, Singapore. All experiments and procedures done on these mice were approved by the Institutional Animal Care and User Committee (IACUC) of A*STAR (Biopolis, Singapore) (Authorization No.: IACUC 151071) in accordance with the guidelines of the Agri-Food and Veterinary Authority (AVA) and National Advisory Committee for Laboratory Animal Research (NACLAR) of Singapore.
METHOD DETAILS
Tamoxifen Treatment
Tamoxifen (Sigma-Aldrich) was dissolved in corn oil to a stock concentration of 20 mg/ml. 75 mg/kg of tamoxifen was intraperitoneally injected twice with one day in between injections. Unless otherwise stated, mice were 6 to 10 weeks of age when given tamoxifen. For the study of yolk sac derived retinal microglia, tamoxifen and progesterone were dissolved in corn oil and to a final stock concentration of 7.5 mg/ml and 3.75 mg/ml progesterone. A single dose of 60 mg/kg tamoxifen and 30 mg/kg of progesterone were injected intraperitoneally into pregnant mice at 7.5 days post-conception.
Tissue Harvesting
Both brain and eye tissues from adults were harvested from freshly euthanized mice. If required, intravenous perfusion with anti-CD45 was performed prior to euthanasia to label intravascular MACROPHAGES. Brains were cut into small pieces, incubated in HBSS containing 10% fetal bovine serum and Collagenase type IV (Sigma) for one hour and then passed through a 19G syringe to obtain a homogeneous cell suspension. Eyes were gently enucleated. Corneas and lens were removed to expose the retina under a dissection microscope. The retina was gently separated from the RPE/choroid using forceps, and tissues including retina, iris and ciliary body, optic nerve, the RPE/choroid/sclera were collected. Eye tissues were further digested in 1.5 mg/ml collagenase A and 0.4 mg/ml DNase I (Roche) for 1 hour at 37 °C with agitation. Single cell suspensions were generated by passing through 70 μm filters. Blood was collected via cardiac puncture. Cells were then treated with red blood cell lysis buffer (Sigma-Aldrich) and thoroughly washed and immunolabeled for flow cytometry.
Flow Cytometry Staining and Acquisition
This method and the antibody panel used for immunophenotyping eye MACROPHAGES was previously established (O'Koren et al., 2016), including antibody titrations and fluorescence minus one (FMO) controls. Briefly, single cell suspensions of tissues were transferred into PBS for staining with Aqua Live/Dead viability dye (Thermo Fisher Scientific) for 30 minutes and then washed with PBS. Cells were incubated in the blocking solution containing 5% normal mouse serum, 5% normal rat serum, and 1% Fc block (eBiosciences) for 10 min and subsequently stained with a combination of fluorophore-conjugated primary antibodies against mouse CD45, Ly6C, Ly6G, CD64, CD11b, CD11c (Biolegend), F4/80 (eBiosciences) and MHC class II (BD Biosciences), at room temperature for 20 minutes. After staining, cells were washed and fixed with 0.4% paraformaldehyde in PBS. Data was acquired with BD Fortessa flow cytometer using BD FACSDiva software (BD Biosciences). Raw flow cytometry data was analyzed using FlowJo software (FlowJo LLC).
Labeling of intravascular CD45 cells
This procedure was previously described. Briefly, mice were anesthetized by i.p. injections of ketamine/xylazine (120 and 20 mg/kg, respectively). Once anesthetized mice were injected retroorbitally with 3.0 μg of biotin-labeled anti-CD45 (Biolegend, San Diego, CA). Mice were euthanized 5 minutes post injection to ensure labeling of blood cells.
Histology
For cryo-histology, dissected eye tissues were fixed in 2% paraformaldehyde in PBS for 3 hours at room temperature and then washed with PBS. Tissues were successively dehydrated with 15% sucrose, 30% sucrose before embedded in OCT and frozen on dry ice. Frozen sections (15-25 μm) or RPE flat mounts were immunostained with antibodies against GFP/YFP (Invitrogen), IBA1, β-galactosidase, DTR, galectin-3, LPL, CD68, CD11b. Cell nuclei were counterstained with DAPI. Images were acquired with a Leica SP5 confocal microscopy, a Nikon A1 confocal microscopy or Nikon C1/C2 confocal microscopy. Maximum projections of z-stacks with 0.5 to 1 μm interval step were shown in a constant tissue volume.
For plastic-histology and TEM histology, euthanized mice were immediately fixed via trans-cardial perfusion with 0.1% cacodylate buffer (pH = 7.2) containing 2% paraformaldehyde and 2% glutaraldehyde. The eye tissues were post-fixed in the same fixative for at least 24 hours and processed in a solution of 2% osmium tetraoxide in 0.1% cacodylate buffer, following by dehydration with gradient ethanol from 50% to 100% and propylene oxide, and infiltration of propylene oxide: epoxy 812 compound with the ratio of 1:1 overnight under the vacuum. Samples were further processed with pure epoxy 812 compound the next day and embedded in fresh epoxy 812 compound resins at 65°C overnight. Semi-thin cross sections (0.5 μm) for plastic histology across the block were stained with 1% methylene blue. Images were acquired using Olympus Ckx31 inverted microscope with Lumenera’s INFINITY1-2 COMS digital camera. Ultra-thin cross sections for TEM were cut at 65nm −75nm thickness by using a Leica EM CU7 and contrast stained with 2% uranyl acetate, 4% lead citrate solution. Images of ultrathin sections were acquired using a JEM-1400 TEM (JEOL) using an ORIUS (1000) CCD 35mm port camera.
RNA Isolation and Quantitative Real-Time PCR
Total RNA was isolated by RNeasy Micro Kit (Qiagen) and reverse transcribed using Superscript III Kit (Invitrogen) according to the manufactory protocol. Real-time PCR was performed using pre-formulated probes for indicated transcripts and Universal PCR Mastermix (Applied Biosystems). The results were analyzed by the comparative threshold cycle method and normalized by GAPDH as an internal control.
RNAScope In Situ Hybridization
Eyes from Aoth7+/−, Aoth7−/− and light damaged CBF1 adult mice were fixed with 4% paraformaldehyde in PBS for 24 hours at room temperature and then washed twice with PBS. Retinas isolated were cryoprotected with 30% sucrose and 0.02% sodium azide in PBS and embedded in Tissue Freezing Medium (General Data; Cincinnati, OH). Retinas were sectioned at 16 microns and placed onto Superfrost Plus slides. For RNAScope labeling, tissues were pre-treated according to the manufacturer’s instructions for fixed frozen tissue (Advanced Cell Diagnostics, Newark, CA). Probes were hybridized and amplified using the RNAScope Fluorescent Multiplex Kit according to manufacturer’s instructions. Slides were counterstained with DAPI and imaged on a Nikon A1 confocal system. Z-stack images with one μm step size were acquired using a 60X objective (Plan Apochromat, NA 1.40) with oil immersion. Images were imported into FIJI image J, and single slices of the z-stacks were chosen for presentation.
In Vivo Visual Function Analysis by Electroretinography (ERG)
Electroretinography was performed as previously described(Toomey et al., 2015). Briefly, mice were dark-adapted for at least 4 hours, pupils dilated with 0.5% (wt/vol) tropicamide and 1.25% (wt/vol) phenylephrine and anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg). Scotopic and photopic electroretinogram (ERG) responses, were recorded using an Espion E2 system (Diagnosys LLC, Lowell, MA), at increasing flash intensities (scotopic: 2.5 × 10−5, 5 × 10−5, 5 × 10−4, 5 × 10−3, 0.05, 0.5, 5, 50, and 500 cd·s/m2; photopic: 5, 50, and 500 cd·s/m2 with a background light intensity of 25.5 cd·s/m2). Recordings consisted of single flash presentations, repeated 0–15 times to verify the response reliability and improve the signal-to-noise ratio, if required. Low-pass frequency filtering of 50 Hz was applied to remove oscillatory potentials and noise.
Optical Coherence Tomography (OCT) and Fundus Imaging
Mouse eyes were topically dilated with one drop of a 1:1 mixture of 1% tropicamide and 10% phenylephrine sulphate. Mice were anesthetized via intraperitonially injection with ketamine (90-100mg/kg) and xylazine (5-10mg/kg) The corneas were kept moist with regular application of GenTeal lubricant eye gel. Eyes were imaged using the Micron IV retinal imaging system (Phoenix Research Labs, CA, USA). Images were analyzed by software provided by vendor.
Conditional Genetic Depletion of Microglia
BALB/c and RhoP23H/P23H mice were crossed with Cx3cr1CrB-ER/wt; R26DTR/wt to generate tamoxifen-F1-iDTR and P23H-iDTR for the light damage and inherited retinal degeneration models, respectively. After tamoxifen treatment, two doses of 0.5 μg of diphtheria toxin (DT, Sigma-Aldrich) were injected intraperitoneally with one day in between injections before harvest for light damage. For P23H-iDTR mice, mice were injected with DT twice at 21- and 28-days post-partum and harvested for experiment four weeks after receiving the last DT injection at P60. The same genotype littermates that did not receive DT or DTR-negative littermates that received DT were used as controls for both experiments.
Light Damage
Light damage of CB6F1/J or Il34LacZ Rpe65Leu450/Met450 mice was induced as previously described (O'Koren et al., 2016). Briefly, mice were adapted in darkness for eight hours, and eyes were dilated with 1% atropine sulfate (Bausch & Lomb, Tamp, FL) and 10% phenylephrine hydrochloride (Paragon BioTeck, INC., Portland, OR). Mice were then placed in a ventilated reflective container that contained available food and hydrogel. A cool white-light LED light source (Fancierstudio, San Francisco, CA) was placed above the container with 10,000 lux output adjusted using a luxometer. After four hours of light challenge, the mice were returned to the housing facility with normal lighting and bred for additional five days before experiments.
Single Cell Isolation and Sorting
Retinas were isolated from LD or control CB6F1/J male mice at eight months of age. Retinas from 5 to 8 mice per group were pooled and digested to generate single cell suspensions as described above. Cells were stained with PE before FACS sorting. Live Cx3cr1YFP+ cells were sorted by FACS, and ~6,000 to 8,000 cells of each sample were collected.
Single Cell RNA-seq Library Preparation and Sequencing (Drop-seq)
The samples were prepared at the core facility of DMPI Molecular Genomics (Duke) for the 10x Genomics single cell RNA-seq library, and then sequenced using HiSeq4000 with a read length of 150bp by GCB Sequencing and Genomic Technologies (Duke) and analyzed using Cell Ranger pipelines.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of microglia in OPL and IPL
Retina flat mounts were stained with IBA1 antibody. Images were acquired by scanning between INL to ONL for microglia in OPL and between GCL to INL for microglia in IPL. Maximum projections were used for quantification. The density of microglia was calculated by averaging at least three images of each mouse.
Analysis of ERG Data
ERG data analysis and fitting were performed as described (Herrmann et al., 2010) using GraphPad Prism 7.0 software. For scotopic b-waves, rod-driven responses to low light stimuli were shown separate from cone-additive responses to high intensity light stimuli, as previously described. Briefly, data points from the a-wave and b-wave stimulus-response curves were fitted to by equation 1 (Herrmann et al., 2010) using the least-square fitting procedure. For all electroretinographic data, two-way ANOVA with Bonferroni’s post hoc test was performed. P-value < 0.05 was considered as a statistically significant difference.
Synapse Quantification
For quantification of synapses, images were acquired using a Nikon A1 confocal system. Z-stacks (106 μm × 106 μm × 0.33 μm step size; 9-15 steps/z-stack) were acquired using a 60X objective (Plan Apochromat, NA 1.40) with oil immersion and 2X optical zoom. Three independent fields of view (FOV) were used for analysis of ON and OFF sublaminae for each animal. All FOV were sampled from a similar retinal eccentricity. Images were deconvolved using Nikon Elements Extended Resolution plugin, then imported into ImageJ. Minor brightness/contrast adjustments were made similarly across all images. Maximum intensity projections of three consecutive 0.33 μm z-sections (for a total z-depth of 1 μm) were used for synapse analysis using the Puncta Analyzer plugin for ImageJ (Ippolito and Eroglu, 2010). 3-5 maximum intensity projections were averaged to obtain the number of colocalized puncta per field (dots on graphs). The border between OFF and ON sublaminae was defined based on histologic landmarks (Koh et al., 2018). Published protocols were followed to count synapses based on the colocalization of presynaptic and postsynaptic markers (Ippolito and Eroglu, 2010; Koh et al., 2018). In brief, the projected images were divided into individual channels (presynaptic and postsynaptic markers); background was subtracted for each channel (rolling ball radius = 50); thresholds were adjusted manually to detect individual puncta while minimizing background; and puncta smaller than 4 pixels were filtered out. For OPL synapses: PNA/mGluR6 colocalization was counted by manual examination of z-stacks. Since the number of z slices varied somewhat across stacks used for the analysis, the mean PNA+ cone pedicles reported are normalized to a field depth of 1 μm. For all conditions, N=2 animals per genotype, with 3 FOV per animal. Single slices of the z-stacks were chosen for presentation of representative images. Standard error was calculated from 6 FOV from 2 animals per genotype.
Quantification in LD Model
Images for quantification in LD model were acquired by a confocal microscopy with by constant volume z-scans, and the maximum projections were used for analysis. To compare the microglia % in damaged and undamaged areas of OPL and IPL under LD setting, retina cross sections of F1-iDTR mice were stained with YFP and DTR. The number of DTR+ and Cx3cr1+ cells in OPL and IPL was manually counted between damaged and undamaged areas of each mouse. The sum of DTR+ cells and the sum of ratio Cx3cr1+ cells in each mouse were used to calculate the ratio between DTR+ and Cx3cr1+ cells, and the mean ratio of five mice were shown.
To quantify the microglia depletion efficiency in LD model, RPE flatmounts of F1-iDTR mice treated with DT or not were stained with YFP. The number of microglia was manually counted. The area size per field was measured using Image J, and four fields per eye were analyzed. The depletion efficiency was shown as the mean frequency of microglia per one mm2 area counted.
To quantify the % of the dysmorphic RPE in LD model, RPE flatmounts with microglia depleted or not were stained with phalloidin for F-actin cytoskeleton. RPE cells that exhibited either altered lateral or lost apical F-actin morphology were considered as dysmorphology. Five random regions per field and four fields per eye were assessed. The numbers of abnormal and total RPE cells were counted in each field, and the mean of RPE dysmorphology % in four fields was calculated for each mouse.
Quantification in Il34LacZ/LacZ Rpe65Leu450/Met450 Mice Upon LD
Phalloidin staining was performed as in CBF1 LD model described above. Four images around the optic disc were acquired using 20x objective lens. Total areas and the damaged areas with no distinguishable RPE morphology were measured. The % of damaged RPE areas was calculated for each mouse and normalized to the one of Il34+/LacZ Rpe65Leu450/Met450 mice.
Quantification in RhoP23H Models
Due to inconsistent depletion efficiency in male RhoP23H mice, only female mice were included in the quantification of RhoP23H depletion study. To quantify the microglia depletion efficiency in RhoP23H model, RPE flatmounts of RhoP23H: iDTR mice treated with DT or not were stained with IBA1. Images were acquired by a confocal microscopy with by constant volume z-scans. The number of microglia was manually counted in maximum projection. The area size per field was measured using Image J, and four fields per eye were analyzed. The depletion efficiency was shown as the mean frequency of microglia per one mm2 area counted.
To quantify the microvilli disengagement area between the outer segment tips and the RPE cell body in RhoP23H mice, ten images (five images from the inferior and five from the superior) of a semi-thin retina cross section across the optic nerve were acquired using Olympus microscopy with 40x lens for each mouse. The brightness and contrast of images were adjusted to highlight the disengagement areas, and the same settings were applied to all images analyzed. Microvilli disengagement areas and lengths of retina counted per image were selected and measured using Image J. Data were shown as the mean area of subretinal space per one mm retina counted. Moreover, the frequency of pyknotic nuclei in OPL was also counted in these images. The sum of pyknotic nuclei number in the entire cross section was calculated for each mouse, and the mean frequency of pyknotic nuclei in OPL was shown.
TUNEL Staining for Detection of In Situ Cell Death
This assay was performed using in situ cell death detection kit (Roche) according to manufacturer’s instruction. Briefly, cross sections of light damage model and RhoP23H mice with or without microglia depletion were stained for TUNEL and DAPI. At least three images of each animal were acquired and analyzed. TUNEL positive cells in the ONL were quantified relative to the area in RhoP23H mice.
Single Cell Data Processing
Single cell data from Cell Ranger were further analyzed using Seurat version 2.0 (Butler et al., 2018). These data initially contain 27998 genes of 6821 cells from light damage retina and 4003 cells from normal control. Genes that were expressed by more than three cells and all the cells that expressed more than 200 genes were kept for analysis. “FilterCells” function was used to conservatively remove the effect of doublet cells in Drop-seq, and the thresholds were determined based on mitochondrial gene % from normal control. Totally, 16256 genes and 10786 cells were used for downstream analysis.
A Combined Graph-based Clustering of Single Cell Data
Clustering analysis was conducted using Seurat version 2.0. After normalizing data using “LogNormalize” method with the scale factor of 10,000, finding variably expressed genes (mean.function = ExpMean, dispersion.function = LogVMR) with 1,209 genes identified, scaling and regressing unwanted variation, principle component analysis (PCA) was performed to reduce dimensionality of our dataset with top 50 PC calculated. Based on p-values from JackStrawPlot, top 40 PCs (p-value < 0.01) were used to find clusters. 11 different clusters were identified including one cluster of photoreceptors contamination. Only the clusters, the sizes of which are larger than 2% of cells, were considered for downstream analysis. Heatmap of single cell clusters was conducted using DoHeatMap function with top 10 markers for each cluster tSNE plots were applied using top 40 PC as input to visualize the structure of data in two dimensions.
Identifying Markers for Each Cluster
Positively differentially expressed genes (biomarkers) of each cluster were identified using FindAllMarkers function with default LRT test in Seurat package. To compare the upregulated and downregulated genes between two clusters, the fold change of gene expression and adjusted p-values based on Bonferroni correction using all genes in the dataset were calculated with FindMarkers function. Markers across all clusters were shown in bar graphs and violin plots (probability distributions across clusters).
Trajectory Analysis
To understand the transition between different microglia subtypes in response to light damage, trajectory analysis was conducted using monocle version 2.2.0 (Qiu et al., 2017). The analysis was performed on all microglia clusters (without rep MG) in clustering analysis: MG0, lc MG1, lc MG2, lc MG3, sMG1, sMG2 and sMG3. As monocle allows only one root state, MG0 was used as the initial microglia and the undamaged proportion of lc MG3 was excluded for the analysis. CellDataSet Object was created using the data from clustering analysis. To construct single cell trajectory, an unsupervised gene list was chosen including top 1000 variably expressed genes in clustering analysis. After reducing dimension of the data with DDRTree, cells were ordered into pseudotime. Furthermore, a branch of the whole pseudotime trajectory was extracted, and a separate trajectory was constructed on the following microglia clusters: MG0, lc MG1, sMG2, and sMG3. The heatmap of MG0, lc MG1, sMG2, and sMG3 clusters along pseudotime trajectory was also generated using top 100 up-regulated and 100 down-regulated genes of sMG3 compared with MG0.
GO Pathway Enrichment Analysis
GO pathway enrichment analysis were performed using PANTHER Classification System (http://pantherdb.org/). Top 100 up-regulated and 100 down-regulated genes of sMG3 compared with MG0 were used.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data normality and homogeneity of variance were assessed using Kolmogorov-Smirnov test and Levene’s test, respectively. Log transformation was conducted to fit the test assumptions when necessary. Two group comparisons were analyzed using unpaired Student’s two-tailed t-test. Comparisons of three groups and/or multiple factor comparisons were analyzed using ANOVA with Bonferroni’s post hoc test. To assess significance, a p < 0.05 was considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Data quantification were performed blindly. All sample sizes and p-values were indicated in graphs and figure legends. tSNE plots of short- and long-lived macrophages were generated using Cytobank. (https://www.cytobank.org). Graphs of single cell RNA-seq data were generated using R studio, and all other statistical graphs were generated using GraphPad Prism 7 software.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is TBD.
Supplementary Material
Highlights:
Retina has 2 microglia pools differing by niche and IL-34 dependency.
In homeostasis, IL-34 dependent microglia contribute to neuronal function.
In degeneration, both pools relocate toward the retinal pigmented epithelium (RPE).
This transition reprograms microglia and is associated with RPE protection.
ACKNOWLEDGEMENTS
This study was funded by: Bright Focus MDR (D.R.S), R01EY021798 (D.R.S), P30EY005722 (Duke Eye Center), R01EY022959 (V.Y.A), R01EY02161 (C.B.R), Bright Focus MDR (M.K), Research to Prevent Blindness (Duke Eye Center). F.G is an EMBO YIP awardee and is supported by SIgN core funding and as well as a Singapore NRF2016NRF-NRFI001-02. We would like to acknowledge the assistance of the Duke Molecular Physiology Institute Molecular Genomics core for the generation of data for the manuscript. We thank S. Finkelstein and E. Lobanova for provision of mice, P. Saloupis for help with OCT, and Y. Hao for TEM (Duke Eye Center). We thank S. Koh (Eroglu lab, Duke) for help with synapse quantification and B. Hogan (Duke) for gifting R26-fGFP mice.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ajami B, Bennett JL, Krieger C, Tetzlaff W, and Rossi FM (2007). Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 10, 1538–1543. [DOI] [PubMed] [Google Scholar]
- Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113, E1738–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton SH, Heckenlively JR, Cottingham AW, Friedman J, Sadler LA, Wagner M, Friedman LH, and Daiger SP (1991). Linkage mapping of autosomal dominant retinitis pigmentosa (RP1) to the pericentric region of human chromosome 8. Genomics 11, 857–869. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, and Glaser T (2001). Math5 is required for retinal ganglion cell and optic nerve formation. Development 128, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, and Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2, 419–426. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. (2014). Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calippe B, Augustin S, Beguier F, Charles-Messance H, Poupel L, Conart JB, Hu SJ, Lavalette S, Fauvet A, Rayes J, et al. (2017). Complement Factor H Inhibits CD47-Mediated Resolution of Inflammation. Immunity 46, 261–272. [DOI] [PubMed] [Google Scholar]
- Chiu IM, Morimoto ET, Goodarzi H, Liao JT, O'Keeffe S, Phatnani HP, Muratet M, Carroll MC, Levy S, Tavazoie S, et al. (2013). A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep 4, 385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danciger M, Lyon J, Worrill D, LaVail MM, and Yang H (2003). A strong and highly significant QTL on chromosome 6 that protects the mouse from age-related retinal degeneration. Invest Ophthalmol Vis Sci 44, 2442–2449. [DOI] [PubMed] [Google Scholar]
- De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, et al. (2017). Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron 95, 341–356 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas AH, Boddeke HW, and Biber K (2008). Region-specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia 56, 888–894. [DOI] [PubMed] [Google Scholar]
- Diamond JS (2017). Inhibitory Interneurons in the Retina: Types, Circuitry, and Function. Annu Rev Vis Sci 3, 1–24. [DOI] [PubMed] [Google Scholar]
- Dick AD, Ford AL, Forrester JV, and Sedgwick JD (1995). Flow cytometric identification of a minority population of MHC class II positive cells in the normal rat retina distinct from CD45lowCD11b/c+CD4low parenchymal microglia. Br J Ophthalmol 79, 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, Sandberg MA, and Berson EL (1990). A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature 343, 364–366. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, et al. (2016). A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 48, 134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, et al. (2013). A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16, 1618–1626. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, and Rodewald HR (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, and McColl BW (2016). Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat Neurosci 19, 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, Kundig TM, Frei K, Ginhoux F, Merad M, and Becher B (2012). Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity 37, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Brown KE, and Milam AH (2003). Activated microglia in human retinitis pigmentosa, late-onset retinal degeneration, and age-related macular degeneration. Exp Eye Res 76, 463–471. [DOI] [PubMed] [Google Scholar]
- Guziewicz KE, Cideciyan AV, Beltran WA, Komaromy AM, Dufour VL, Swider M, Iwabe S, Sumaroka A, Kendrick BT, Ruthel G, et al. (2018). BEST1 gene therapy corrects a diffuse retina-wide microdetachment modulated by light exposure. Proc Natl Acad Sci U S A 115, E2839–E2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch UK, and Kettenmann H (2007). Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 10, 1387–1394. [DOI] [PubMed] [Google Scholar]
- Herrmann R, Lobanova ES, Hammond T, Kessler C, Burns ME, Frishman LJ, and Arshavsky VY (2010). Phosducin regulates transmission at the photoreceptor-to-ON-bipolar cell synapse. J Neurosci 30, 3239–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AL, Makowka L, Cramer DV, Cai X, Banner B, Pascualone A, Hoffmann S, and Starzl TE (1989). Induction of stable chimerism and elimination of graft-versus-host disease by depletion of T lymphocytes from bone marrow using immunomagnetic beads. Surgery 106, 354–363. [PMC free article] [PubMed] [Google Scholar]
- Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, Wes PD, Moller T, Orre M, Kamphuis W, et al. (2015). Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun 3, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes EH, Schlichtenbrede FC, Murphy CC, Sarra GM, Luthert PJ, Ali RR, and Dick AD (2003). Generation of activated sialoadhesin-positive microglia during retinal degeneration. Invest Ophthalmol Vis Sci 44, 2229–2234. [DOI] [PubMed] [Google Scholar]
- Ippolito DM, and Eroglu C (2010). Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itagaki S, McGeer PL, Akiyama H, Zhu S, and Selkoe D (1989). Relationship of microglia and astrocytes to amyloid deposits of Alzheimer disease. J Neuroimmunol 24, 173–182. [DOI] [PubMed] [Google Scholar]
- Jinno S, Fleischer F, Eckel S, Schmidt V, and Kosaka T (2007). Spatial arrangement of microglia in the mouse hippocampus: a stereological study in comparison with astrocytes. Glia 55, 1334–1347. [DOI] [PubMed] [Google Scholar]
- Karlstetter M, Scholz R, Rutar M, Wong WT, Provis JM, and Langmann T (2015). Retinal microglia: just bystander or target for therapy? Prog Retin Eye Res 45, 30–57. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell 169, 1276–1290 e1217. [DOI] [PubMed] [Google Scholar]
- Koh S, Chen WJ, Dejneka NS, Harris IR, Lu B, Girman S, Saylor J, Wang S, and Eroglu C (2018). Subretinal Human Umbilical Tissue-Derived Cell Transplantation Preserves Retinal Synaptic Connectivity and Attenuates Muller Glial Reactivity. J Neurosci 38, 2923–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O'Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581 e569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzberg GW (1996). Microglia: a sensor for pathological events in the CNS. Trends Neurosci 19, 312–318. [DOI] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P, and Gordon S (1990). Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170. [DOI] [PubMed] [Google Scholar]
- Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, et al. (2008). Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 320, 807–811. [DOI] [PubMed] [Google Scholar]
- Luhmann UF, Robbie S, Munro PM, Barker SE, Duran Y, Luong V, Fitzke FW, Bainbridge JW, Ali RR, and MacLaren RE (2009). The drusenlike phenotype in aging Ccl2-knockout mice is caused by an accelerated accumulation of swollen autofluorescent subretinal macrophages. Invest Ophthalmol Vis Sci 50, 5934–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Zhang Y, Gao C, Fariss RN, Tam J, and Wong WT (2017). Monocyte infiltration and proliferation reestablish myeloid cell homeostasis in the mouse retina following retinal pigment epithelial cell injury. Sci Rep 7, 8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. [DOI] [PubMed] [Google Scholar]
- McKay GJ, Patterson CC, Chakravarthy U, Dasari S, Klaver CC, Vingerling JR, Ho L, de Jong PT, Fletcher AE, Young IS, et al. (2011). Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat 32, 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Bruck W, Priller J, and Prinz M (2007). Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci 10, 1544–1553. [DOI] [PubMed] [Google Scholar]
- Ng TF, and Streilein JW (2001). Light-induced migration of retinal microglia into the subretinal space. Invest Ophthalmol Vis Sci 42, 3301–3310. [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, and Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- Noell WK, Walker VS, Kang BS, and Berman S (1966). Retinal damage by light in rats. Invest Ophthalmol 5, 450–473. [PubMed] [Google Scholar]
- O'Koren EG, Mathew R, and Saban DR (2016). Fate mapping reveals that microglia and recruited monocyte-derived macrophages are definitively distinguishable by phenotype in the retina. Sci Rep 6, 20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, Hempstead BL, Littman DR, and Gan WB (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M, Erny D, and Hagemeyer N (2017). Ontogeny and homeostasis of CNS myeloid cells. Nat Immunol 18, 385–392. [DOI] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff RM, and Perry VH (2009). Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol 27, 119–145. [DOI] [PubMed] [Google Scholar]
- Reyes NJ, O'Koren EG, and Saban DR (2017). New insights into mononuclear phagocyte biology from the visual system. Nat Rev Immunol 17, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, and Palczewski K (2011). Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem 286, 10551–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheetz TE, Roos BR, Solivan-Timpe F, Miller K, DeLuca AP, Stone EM, Kwon YH, Alward WL, Wang K, and Fingert JH (2016). SQSTM1 Mutations and Glaucoma. PLoS One 11, e0156001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Cabuy E, Scherf BG, Kohler H, Panda S, Le YZ, Fehling HJ, Gaidatzis D, Stadler MB, and Roska B (2012). Transcriptional code and disease map for adult retinal cell types. Nat Neurosci 15, 487–495, S481–482. [DOI] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, and Garel S (2014). Microglia modulate wiring of the embryonic forebrain. Cell Rep 8, 1271–1279. [DOI] [PubMed] [Google Scholar]
- Thanos S (1992). Sick photoreceptors attract activated microglia from the ganglion cell layer: a model to study the inflammatory cascades in rats with inherited retinal dystrophy. Brain Res 588, 21–28. [DOI] [PubMed] [Google Scholar]
- Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, et al. (2018). Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell 172, 500–516 e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey CB, Kelly U, Saban DR, and Bowes Rickman C (2015). Regulation of age-related macular degeneration-like pathology by complement factor H. Proc Natl Acad Sci U S A 112, E3040–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, and Majewska AK (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela JM, Dalmau I, Gonzalez B, and Castellano B (1995). Morphology and distribution of microglial cells in the young and adult mouse cerebellum. J Comp Neurol 361, 602–616. [DOI] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, and Nabekura J (2009). Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J Neurosci 29, 3974–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao L, Zhang J, Fariss RN, Ma W, Kretschmer F, Wang M, Qian HH, Badea TC, Diamond JS, et al. (2016). Requirement for Microglia for the Maintenance of Synaptic Function and Integrity in the Mature Retina. J Neurosci 36, 2827–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. (2015a). TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160, 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, Barrow AD, Diamond MS, and Colonna M (2012). IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 13, 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Han Y, Zhang R, Qin L, Wang MX, and Ma L (2015b). CETP/LPL/LIPC gene polymorphisms and susceptibility to age-related macular degeneration. Sci Rep 5, 15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki R, Lu H, Butovsky O, Ohno N, Rietsch AM, Cialic R, Wu PM, Doykan CE, Lin J, Cotleur AC, et al. (2014). Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med 211, 1533–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiss CJ, and Johnson EA (2004). Proliferation of microglia, but not photoreceptors, in the outer nuclear layer of the rd-1 mouse. Invest Ophthalmol Vis Sci 45, 971–976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is TBD.