Abstract
Objectives.
To characterize Staphylococcus aureus isolates recovered from hospitalized children and to determine the concordance between colonizing and invasive isolates.
Study design.
Children with culture-confirmed, community-onset, invasive S. aureus infections were enrolled in this prospective case series from a large children’s hospital over a 5-year period. Colonization isolates were obtained from the anterior nares, oropharynx, and inguinal folds and were compared with invasive isolates via repetitive-element, sequence-based PCR testing. Isolates with a ≥96% genetic match were characterized as concordant.
Results.
A total of 86 S. aureus isolates (44 invasive, 42 colonization) were collected from 44 children with invasive infections. 64% of invasive isolates were MSSA, and clinical isolates were genetically diverse. 59% of cases had a colonizing S. aureus isolate at the time of hospitalization. Of those who were colonized, at least one of their colonization isolates was indistinguishable from the infecting isolate in 88% of cases. Patients with invasive MSSA were significantly more likely to have a concordant MSSA colonization isolate present compared with patients with invasive MRSA (61% vs 38%, P < .05).
Conclusions.
Invasive MSSA infection was more common than MRSA infection in this pediatric cohort, and patients with MSSA infection were significantly more likely than those with MRSA infection to have concordant colonizing isolates across multiple anatomic sites. These findings warrant larger scale validation and may have important infection control and epidemiologic implications, as unlike MRSA, transmissibility of MSSA largely is ignored in healthcare settings.
Keywords: Staphylococcus aureus, MRSA, colonization, molecular epidemiology
Staphylococcus aureus is a major cause of invasive hospital-associated and community-associated bacterial infections globally. From 1999 to 2005, the number of hospitalizations related to S. aureus increased by 62%.1 In the same period, the percentage of methicillin-resistant Staphylococcus aureus (MRSA) infections in the lungs or blood leading to hospitalization more than tripled.1 However, since 2015, the rate of invasive MRSA infections in the United States has decreased, likely due, in part, to improved infection control practices.2
The burden of antimicrobial resistance in S. aureus extends well beyond methicillin resistance. The rates of resistance to clindamycin, a commonly used anti-staphylococcal agent, are increasing in both adults and children.3 Moreover, not all strains of S. aureus are equivalent in virulence. S. aureus produces numerous, diverse virulence factors that facilitate establishment and persistence of infection in humans. Although only a small number of MRSA clones dominated the community-associated MRSA (CA-MRSA) epidemic of the 2000s, S. aureus strains asymptomatically colonizing the nose of healthy individuals are much more genetically diverse.4,5 It is unclear whether this degree of genetic variability exists between colonizing and invasive methicillin-susceptible (MSSA) isolates that have emerged in the last decade. A primary goal of the current study was to describe the current molecular epidemiology of invasive S. aureus in a large pediatric medical center. This analysis included assessment for genes encoding Panton-Valentine leukocidin (PVL), a member of the bi-component leukocidin family of toxins produced by S. aureus that has been closely associated with the CA-MRSA epidemic strain;6 LukAB (also known as LukGH), the most recently described leukocidin that is critical for virulence in numerous animal models and is present in all clinical S. aureus isolates studied to date;7–9 and the accessory gene regulator (agr) locus, a global regulator of several S. aureus virulence factors.10
The second major goal of this study was to examine paired invasive and colonizing S. aureus isolates to determine the genotypic concordance between colonization and invasive isolates. Few data are available regarding genotypic concordance between invasive and colonizing MSSA (as opposed to studies of noninvasive MSSA clinical isolates or MRSA disease), or colonization characteristics across anatomic sites. Current infection control protocols at many institutions typically focus exclusively on isolating patients with MRSA infection, and suppressing or eradicating MRSA colonization, ignoring MSSA infection and colonization. Determining the prevalence of genotypically concordant MSSA colonization at various sites in patients with invasive infection may inform future infection control strategies.
Methods
All patients under 19 years of age who were admitted within a 5-year period (January 2013 to December 2017) to the Upstate Golisano Children’s Hospital (GCH) in Syracuse, New York with culture-confirmed, community-onset invasive S. aureus (CO-ISA) disease were eligible for the study. Invasive S. aureus disease was defined as recovery of S. aureus from a sterile site, including blood, bones, joint, lungs, and central nervous system. Cultures obtained within 48 hours of admission met criteria for community-onset disease. All protocols were reviewed and approved by the GCH Institutional Review Board. The sample size of the study represents all eligible subjects admitted during the study period.
Patients with invasive S. aureus disease were identified through a daily laboratory report to capture all eligible patients. S. aureus isolates were obtained directly from the clinical Microbiology Laboratory. In addition, swabs for culture were collected from the anterior nares, the oropharynx, and inguinal folds of each subject admitted with CO-ISA infection within 7 days of the sterile-site culture collection. Specific clinical information including age, sex, discharge diagnosis, intensive care unit stay, and recent antibiotic use was recorded. Routine antibiotic susceptibility testing was performed according to the standards of the Clinical and Laboratory Standards Institute.
Isolates were stored in sterile skim milk at −70°C until subculture for molecular analysis was performed. For molecular analysis, genomic DNA was extracted as previously described,4 using the QIAGEN MoBio DNA extraction kit. DNA was quantified using the NanoDrop 1000 (ThermoFisher) to confirm a concentration of 25–50 micrograms/mL. Purified genomic DNA was used as the template for PCR detection of genes encoding PVL, LukAB, arginine catabolic mobile element (ACME), staphylococcal cassette chromosome mec (SCCmec), and the accessory gene regulator (agr) locus type. Previously identified S. aureus strains containing or lacking the genes of interest, or of a particular SCCmec or agr locus type, were used as controls. Assignment of SCCmec type was determined using the multiplex strategy as previously reported.11 Repetitive element, sequence-based PCR (Diversilab System; Biomerieux, Durham, North Carolina) was used to determine genetic relatedness and assign clonal type based on pulsed-field gel electrophoresis (PFGE) USA types for S. aureus. The threshold for categorizing two isolates as concordant was a ≥96% genetic match.
Statistical analysis was performed using Prism6 by GraphPad. Significance of associations was analyzed using the Fisher exact test, and differences with p value < .05 were deemed significant.
Results
Characteristics of the patient population
In total, 143 cases of invasive S. aureus disease were identified during the study period; 44 parents/caregivers consented for their children to participate in the study within the timeframe required for collection of swab samples to determine colonization. Twenty-six children (59%) had at least one colonization strain detected. The median patient age was 10 years; 35% of subjects were female and 65% were male. The most common infections identified were bacteremia, pneumonia, osteomyelitis, and septic arthritis. Hospital length of stay varied widely among patients, with five patients remaining in the hospital three days or less, sixteen between 4–9 days, eight between 10–13 days, and fourteen greater than 14 days (Table I).
Table 1:
Clinical characteristics and concordance of invasive and colonization S. aureus isolates
| Patient ID | Age (years) | Duration of Hospitalization (days) | Invasive Isolate, PFGE Type1 | Infection Type2 | Number of colonization sites3 | Invasive concordance with colonization? |
|---|---|---|---|---|---|---|
| 10 | 0.8 | 40 | MRSA, USA300 | Pneumonia | 0 | N/A |
| 11 | 3.1 | 15 | MRSA, USA300 | SSTI | 0 | N/A |
| 15 | 0.1 | 37 | MRSA, USA200 | Pneumonia | 0 | N/A |
| 29 | 5.7 | 8 | MRSA, USA300 | MSKI | 0 | N/A |
| 32 | 4.0 | 35 | MRSA, NT | Pneumonia | 0 | N/A |
| 34 | 10.2 | 9 | MRSA, USA300 | MSKI | 0 | N/A |
| 41 | 0.6 | 33 | MRSA, USA300 | MSKI | 0 | N/A |
| 43 | 10.1 | Not available | MRSA, USA200 | MSKI | 0 | N/A |
| 16 | 9.4 | 6 | MRSA, USA300 | MSKI | 1 (N) | No |
| 25 | 9.3 | 10 | MRSA, USA1000 | MSKI | 1 (T) | No |
| 28 | 17.6 | 21 | MRSA, USA100 | CLABSI | 1 (T) | Yes (T) |
| 33 | 17.7 | 13 | MRSA, USA100 | MSKI | 1 (T) | Yes (T) |
| 37 | 12.8 | 7 | MRSA, USA300 | MSKI | 1 (T) | Yes (T) |
| 45 | 14.0 | 10 | MRSA, USA300 | Pneumonia | 1 (T) | Yes (T) |
| 38 | 2.2 | 102 | MRSA, USA300 | SSTI | 3 (G, N, T) | Yes (G) |
| 30 | 1.1 | 31 | MRSA, NT | Pneumonia | 2 (G, N) | Yes (G, N) |
| 4 | 1.4 | 19 | MSSA, USA400 | CLABSI | 0 | N/A |
| 9 | 16.8 | 10 | MSSA, USA300 | Peritonitis | 0 | N/A |
| 12 | 3.3 | 23 | MSSA, USA900 | VPS | 0 | N/A |
| 23 | 0.9 | 2 | MSSA, USA300 | CLABSI | 0 | N/A |
| 24 | 17.5 | 6 | MSSA, USA200 | MSKI | 0 | N/A |
| 27 | 17.0 | 13 | MSSA, USA600 | SSTI | 0 | N/A |
| 31 | 13.9 | 5 | MSSA, NT | MSKI | 0 | N/A |
| 36 | 6.0 | 11 | MSSA, USA700 | VPS | 0 | N/A |
| 40 | 13.8 | 30 | MSSA, NT | MSKI | 0 | N/A |
| 44 | 15.0 | 7 | MSSA, USA300 | Bacteremia | 0 | N/A |
| 8 | 0.3 | 15 | MSSA, USA300 | UTI | 1 (G) | Yes (G) |
| 1 | 13.7 | 6 | MSSA, USA300 | MSKI | 1 (N) | No |
| 2 | 10.5 | 5 | MSSA, NT | MSKI | 1 (N) | Yes (N) |
| 7 | 12.7 | 8 | MSSA, NT | CLABSI | 1 (N) | Yes (N) |
| 14 | 5.2 | 10 | MSSA, USA300 | RPA | 1 (N) | Yes (N) |
| 18 | 8 | 3 | MSSA, NT | CLABSI | 1 (N) | Yes (N) |
| 19 | 14.9 | 7 | MSSA, USA800 | MSKI | 1 (N) | Yes (N) |
| 20 | 16.6 | 5 | MSSA, USA800 | MSKI | 2 (N, T) | Yes (N, T) |
| 39 | 11.1 | 3 | MSSA, USA200 | MSKI | 2 (N, T) | Yes (N, T) |
| 13 | 12.9 | 4 | MSSA, USA200 | CLABSI | 2 (G, N) | Yes (N) |
| 6 | 16.2 | 17 | MSSA, USA 100 | Endocarditis | 2 (G, N) | Yes (G) |
| 21 | 14.5 | 8 | MSSA, USA800 | Bacteremia | 2 (G, N) | Yes (N) |
| 22 | 11.4 | 4 | MSSA, USA200 | MSKI | 2 (G, N) | Yes (G, N) |
| 35 | 9.5 | 2 | MSSA, USA200 | MSKI | 2 (G, N) | Yes (G, N) |
| 42 | 17.4 | 6 | MSSA, USA300 | Bacteremia | 2 (G, N) | Yes (G, N) |
| 5 | 12.1 | 12 | MSSA, USA300 | MSKI | 2 (N, T) | Yes (N, T) |
| 17 | 0.01 | 33 | MSSA, USA200 | MSKI | 3 (G, N, T) | Yes (G) |
| 3 | 5.3 | 3 | MSSA, NT | SSTI | 3 (G, N, T) | Yes (G, N, T) |
NT = non-typeable strain
MSKI = musculoskeletal infection, CLABSI = Central line-associated bloodstream infection, VPS = Ventriculoperitoneal shunt, SSTI = Skin and soft tissue infection, UTI = Urinary Tract Infection, RPA = Retropharyngeal Abscess
N=colonization isolate obtained from the nares, G = colonization isolate obtained from the inguinal folds and T = colonization isolate obtained from oropharynx
Molecular Characteristics of Clinical Isolates
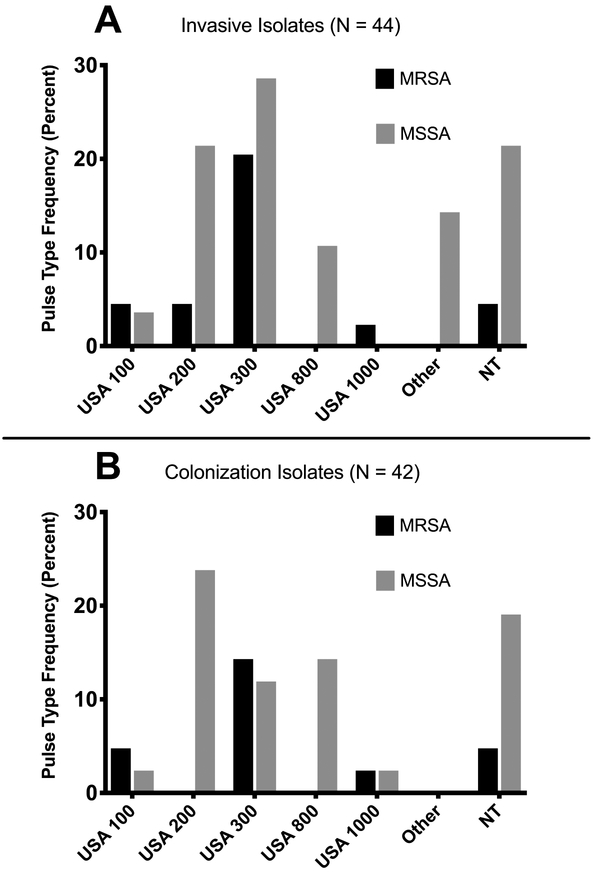
Overall, 86 S. aureus isolates were collected from 44 children (44 invasive isolates and 42 colonization isolates, Table 1). 64% (28/44) of invasive S. aureus isolates were MSSA. The most frequent clonotype of the total 86 invasive and colonizing S. aureus isolates was USA300, representing 34% (29) of all isolates (41% of invasive isolates and 26% of colonization isolates, Figure). Overall, 20% (17) of isolates were classified as USA200, 11% (9) USA800, and 7% (6) USA100; 21% (18) were non-typeable. Of the MRSA isolates, 56% of isolates were classified as USA300 (9/16 invasive disease and 6/11 colonization), and 15% were classified as USA100. MSSA isolates were more diverse, with USA200 most frequently encountered (13/59, 22% of isolates), followed by USA 300 (12/59 (20%)).
Figure: Genetic diversity of S. aureus colonization and invasive disease isolates obtained in this study, by pulsed-field gel electrophoresis (PFGE) type*, stratified by Invasive and Colonization isolates.
1A: Each bar represents the relative percentage of a given PFGE type among all invasive disease isolates (n=45). Although USA300 was the most common pulsotype among both MRSA and MSSA populations (~20% of each), isolates were diverse, with no clone representing the majority of isolates.
1B: Each bar represents the relative percentage of a given PFGE type among all colonization isolates (n=42). Colonization isolates were quite diverse, with no pulsotype representing more than a quarter of the group.
*NT = non-typeable strain (not assignable to any PFGE type)
Molecular characteristics of invasive and colonizing isolates are provided in Table 2. Of 27 MRSA isolates, 18 (67%) contained the ACME locus and 20 (74%) were PVL-positive. 22 (82%) were mec Class B, and 82% possessed agr type I. Of 59 MSSA isolates, 8 (14%) were PVL positive, and 19 (32%) possessed agr type I. Two of the MSSA isolates were ACME-positive. All isolates (both MRSA and MSSA) contained the lukAB gene.
Table 2:
Prevalence of purported virulence genes of all isolates collected from 44 children hospitalized with invasive S. aureus infection, 2013 through 2017.*
| Molecular Characteristics | All N=86 | MRSA N=27 | MSSA N=59 | All Invasive N=44 | All Colonization N=42 | Invasive MRSA N=16 | Invasive MSSA N=28 | Colonization MRSA N=11 | Colonization MSSA N=31 |
|---|---|---|---|---|---|---|---|---|---|
| agr Type I | 47.7 | 81.5 | 32.2 | 56.8 | 38.0 | 87.5 | 42.9 | 81.8 | 25.8 |
| agr Type II | 24.4 | 11.1 | 30.5 | 20.4 | 28.6 | 6.3 | 25.0 | 18.2 | 29.1 |
| agr Type III | 27.9 | 3.7 | 39.0 | 22.7 | 33.0 | 6.3 | 32.1 | 0 | 45.1 |
| pvl Presence | 38.3 | 74.1 | 13.6 | 38.6 | 26.2 | 81.2 | 17.9 | 72.7 | 9.6 |
| lukAB Presence | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| acme Presence | 23.3 | 66.7 | 3.4 | 27.3 | 19.0 | 68.8 | 7.1 | 72.7 | 0 |
| SCCmec IVB | 22.1 | 81.5 | N/A | 29.5 | 21.4 | 87.5 | N/A | 81.8 | N/A |
| SCCmec IIA | 3.5 | 11.1 | N/A | 2.3 | 4.8 | 6.3 | N/A | 18.2 | N/A |
| SCCmec V/VIIC | 1.2 | 3.7 | N/A | 2.3 | 0 | 6.3 | N/A | 0 | N/A |
All values in %
Genetic Relatedness between Colonization and Invasive S. aureus Isolates
In 18/44 patients (41%), S. aureus was not isolated from one or more colonization sites. For the 26 (59%) patients colonized with S. aureus at the time of hospitalization, at least one colonization isolate was concordant with the corresponding invasive isolate in 23/26 cases (88%, Table 1). Concordance between disease isolates and nasal colonization isolates (56%) was higher than for the inguinal folds (36%) or oropharynx (28%, p<0.05). Colonization with a concordant strain was significantly more common for children with MSSA infections than those with MRSA infection (17/28 [61%] vs 6/16 [38%]; p < .05).
Discussion
In our study analyzing genetic relatedness between invasive and colonizing strains of S. aureus, we found that invasive MSSA was significantly more likely to be associated with concordant colonization compared with invasive MRSA, and patients with invasive S. aureus infections were significantly more likely to have a genetically matched nasal colonization strain than concordant throat or inguinal skin colonization. Furthermore, we observed substantial genetic diversity among both MRSA and MSSA invasive isolates, reflecting increased genetic diversity across invasive S. aureus infections than was seen during the height of the CA-MRSA epidemic in the US. The prevalence of the PVL in invasive MSSA strains has remained low whereas it remained high in invasive MRSA strains, underscoring an uncertain role of PVL in S. aureus pathogenesis.
In this series of hospitalized children with culture-proven, invasive S. aureus infection, MRSA accounted for 30% of isolates, consistent with recent national data.12 Of the MRSA isolates, USA300 remained the most common clonotype, accounting for 62% of MRSA isolates; however, this is a departure from our 2009 findings in which >80% of invasive isolates were classified as USA3004, suggesting that invasive MRSA disease is becoming more genetically diverse. In addition, USA300 was infrequently identified as a colonization isolate in our study, and MSSA isolates represented ~70% of the isolates across both colonization and invasive sites.
Previous studies have reported a concordance between colonization strains of S. aureus and strains causing noninvasive disease, such as skin and soft tissue infections. Evidence is conflicting, however, regarding the most likely site of concordance between noninvasive disease isolates and colonization isolates. In 1 study of deep skin abscesses, inguinal isolates were most likely to be concordant,13 and a similar study found that the throat was the most frequently concordant site of colonization.14 Even less is known about concordance between colonization isolates and invasive disease, particularly regarding invasive MSSA infection. This is an important distinction as the relative frequency of MSSA infections has increased (compared with MRSA) over the last decade.4 We observed a high degree of concordance between MSSA invasive isolates and colonization isolates, particularly those isolates from the anterior nares, similar to what has been previously demonstrated for MRSA15. This has important infection control implications, as nasal decolonization procedures are currently used in many hospitals for patients colonized with MRSA but not with MSSA.16
Our data indicate a low prevalence of the leukotoxin PVL in invasive MSSA isolates (only 16% of isolates harbored the pvl locus, compared with 80% of invasive MRSA isolates), consistent with previous reports.17 Although PVL has been well-described as an epidemiologic marker for community-onset MRSA infection, its role in pathogenesis is unclear, as invasive MSSA isolates typically lack this toxin.18 Of note, the lukAB gene, which encodes another bi-component leukocidin with potential significance in invasive human disease, was present in all clinical isolates in this study. LukAB is known to be produced in the setting of invasive human disease9 and may play an important role in virulence in the setting of human infection.
Our study has limitations. The study included children admitted with culture-proven, invasive S. aureus infection in whom we were able to obtain colonization swabs. As a result, the sample size is limited and precluded a comprehensive analysis for multiple covariates. A larger, multicenter study would be needed to assess subtle epidemiologic differences. Sites chosen for sampling were nares, oropharynx, and inguinal folds; other relevant sites, such as axilla and rectal areas, were not tested. Colonizing sites or density of colonization may differ for MSSA and MRSA. An additional limitation is delay in sampling specified mucocutaneous sites until after beginning parenteral antibiotic therapy in most cases. In addition, all patients were enrolled from a single U.S. medical center and the results may not be generalizable to a wider population.
The epidemiology of invasive, pediatric S. aureus infections in the U.S. continues to evolve, with a rising role of MSSA. We observed that children with invasive MSSA infections were more likely to have a corresponding colonization isolate, a finding that warrants validation on a larger, multicenter scale so that molecular epidemiologic findings can optimally inform infection control practices. Large-scale validation would also determine whether the detection of specific S. aureus isolates from colonizing sites can reliably predict the presence of S. aureus from a sterile site, specifically in patients for whom antibiotic therapy prior to sampling of a sterile site may reduce culture positivity.19
Acknowledgments
I.T. is supported by National Institute of Allergy and Infectious Diseases (NIAID) (1K23AI113150), serves as an investigator on studies funded by GlaxoSmithKline, and has served on scientific advisory boards for Horizon Pharma. J.S. is supported by the Upstate Golisano Children’s Hospital/Children’s Miracle Network and has served as a consultant for Pfizer. C.C. has joint research or grants with Pfizer, GSK, and Merck and has served on scientific advisory boards for Theravance Pharmaceuticals, GSK Vaccines, and Astellas Pharmaceuticals. The other authors declare no conflicts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis 2007;13:1840–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 2013;173:1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutter DE, Milburn E, Chukwuma U, Dzialowy N, Maranich AM, Hospenthal DR. Changing Susceptibility of Staphylococcus aureus in a US Pediatric Population. Pediatrics 2016;137. [DOI] [PubMed] [Google Scholar]
- 4.Thomsen I, McKenna BD, Saye EJ, Jimenez N, Edwards KM, Creech CB. Molecular distinctions exist between community-associated methicillin-resistant Staphylococcus aureus colonization and disease-associated isolates in children. Pediatr Infect Dis J 2011;30:418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suryadevara M, Clark AE, Wolk DM, Carman A, Rosenbaum PF, Shaw J. Molecular Characterization of Invasive Staphylococcus aureus Infection in Central New York Children: Importance of Two Clonal Groups and Inconsistent Presence of Selected Virulence Determinants. J Pediatric Infect Dis Soc 2013;2:30–39. [DOI] [PubMed] [Google Scholar]
- 6.Spaan AN, van Strijp JAG, Torres VJ. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat Rev Microbiol 2017;15:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, et al. Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol Microbiol 2011;79:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DuMont AL, Yoong P, Day CJ, Alonzo F 3rd, McDonald WH, Jennings MP, et al. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proceedings of the National Academy of Sciences of the United States of America 2013;110:10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomsen IP, Dumont AL, James DB, Yoong P, Saville BR, Soper N, et al. Children with invasive Staphylococcus aureus disease exhibit a potently neutralizing antibody response to the cytotoxin LukAB. Infection and immunity 2014;82:1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le KY, Otto M. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol 2015;6:1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2002;46:2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010;23:616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albrecht VS, Limbago BM, Moran GJ, Krishnadasan A, Gorwitz RJ, McDougal LK, et al. Staphylococcus aureus Colonization and Strain Type at Various Body Sites among Patients with a Closed Abscess and Uninfected Controls at U.S. Emergency Departments. J Clin Microbiol 2015;53:3478–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar N, David MZ, Boyle-Vavra S, Sieth J, Daum RS. High Staphylococcus aureus colonization prevalence among patients with skin and soft tissue infections and controls in an urban emergency department. J Clin Microbiol 2015;53:810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 2001;344:11–16. [DOI] [PubMed] [Google Scholar]
- 16.Mollema FP, Richardus JH, Behrendt M, Vaessen N, Lodder W, Hendriks W, et al. Transmission of methicillin-resistant Staphylococcus aureus to household contacts. J Clin Microbiol 2010;48:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shore AC, Tecklenborg SC, Brennan GI, Ehricht R, Monecke S, Coleman DC. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol 2014;52:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis 2006;194:1761–1770. [DOI] [PubMed] [Google Scholar]
- 19.Harris AD, Furuno JP, Roghmann MC, Johnson JK, Conway LJ, Venezia RA, et al. Targeted surveillance of methicillin-resistant Staphylococcus aureus and its potential use to guide empiric antibiotic therapy. Antimicrob Agents Chemother 2010;54:3143–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]