Abstract
N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated ion channels, which are critically involved in brain development, learning and memory, cognition, as well as a number of neurological diseases and disorders. Structural biology of NMDARs has been challenging due to technical difficulties associated with assembling a number of different membrane protein subunits. Here, we review historical X-ray crystallographic studies on isolated extracellular domains, which are still the most effective mean to delineate compound binding modes, as well as the most recent studies using electron cryo-microscopy (cryo-EM). A number of NMDAR structures accumulated over the past 15 years provide insights into the hetero-tetrameric assembly pattern, pharmacological specificities elicited by subtypes and alternative splicing, and potential patterns of conformational dynamics, however, many more important unanswered questions remain.
Keywords: N-methyl-D-aspartate receptors, multi-heteromeric membrane proteins, X-ray crystallography, cryo-EM, neuropharmacology
Introduction
The N-methyl-D-aspartate receptors (NMDARs) belong to the ionotropic glutamate receptor (iGluR) family and play critical roles in excitatory neurotransmission and synaptic plasticity. They are crucially involved in brain development as well as learning and memory, and their malfunctioning has been associated with a broad range of neurological diseases and disorders including depression, stroke, seizure, schizophrenia, and Alzheimer’s disease [1–3]. NMDARs primarily co-localize with other non-NMDAR iGluRs, including α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) and kainate receptors, at the post-synapse and contribute to excitatory postsynaptic potentials (EPSPs) upon binding of the neurotransmitter, glutamate. While the early phase and the amplitude of EPSPs are mainly dominated by the activities of non-NMDARs, the late decay phase is governed by the slow deactivation kinetics of NMDARs [4,5]. Opening of NMDARs results in a high calcium flux and requires both binding of neurotransmitters and a relief of a magnesium block by depolarization, thereby coincidently detecting information from the pre-synapse (glutamate release) and the post-synapse (depolarization) – the necessary functional features that initiate synaptic modifications for neuroplasticity according to the Hebbian theory [6].
Unlike non-NMDARs which can exist as homotetramers, NMDARs only function as heterotetramers composed of GluN1 subunits containing eight splice variants (1a-4a and 1b-4b) and GluN2A/B/C/D and/or GluN3A/B subunits. The GluN1 and GluN3 subunits bind glycine or D-serine, whereas the GluN2 subunits bind glutamate. Hence, for the most prevalent GluN1/GluN2 subtypes in the brain, binding of both glycine (or D-serine) and glutamate is required for channel opening whereas the GluN1/GluN3 subtype requires only glycine for the channel activation [7,8]. Like non-NMDAR subunits, every NMDAR subunit is composed of an amino-terminal domain (ATD), a ligand-binding domain (LBD), a transmembrane domain (TMD), and a carboxy-terminal domain (CTD). However, there is little or no primary sequence similarity in the ATDs and CTDs between NMDARs and non-NMDARs. Consistently, the ATDs and CTDs of NMDARs are involved in functions that are distinct from non-NMDARs. For example, the NMDAR ATDs allosterically regulate open probability and deactivation speed, and some also bind allosteric compounds that inhibit or potentiate the channel activities [9–14] while such role of the non-NMDARs ATDs has not been observed, thus far. The CTDs of the GluN2 NMDARs are substantially long (ranging from 396 to 626 residues for rat GluN2) compared to those of non-NMDARs (~80 residues). The GluN2 CTDs have been proposed to work as ‘hubs’ for scaffolding postsynaptic density proteins to mediate cellular signaling for neuronal plasticity [15]. The GluN1 CTDs (~50–100 residues) contain one to two calmodulin binding sites to govern a mode of inhibition called calcium-dependent inactivation, the cytoplasmic mechanism to prevent overshoot of calcium influx [16–18]. Although outside the scope of this review, it is worth mentioning that a complex structure between the peptide from a part of the C1 cassette of the GluN1 CTD and calmodulin has been reported previously [19]. This review will focus on recent structures, including intact NMDARs as well as isolated NMDAR extracellular domains, as others have extensively reviewed pharmacological aspects of NMDARs [9,10,20–22].
High-resolution crystallography of NMDAR extracellular domains
Historically, structural studies of NMDARs lagged behind those of non-NMDARs due to technical difficulties associated with their hetero-oligomeric nature as described above. That is, sample preparation involving recombinant expression and protein purification required optimization to allow and maintain proper assembly of hetero-oligomers. Prior to 2014, all available structures on the NMDAR were limited to the isolated extracellular domains solved by X-ray crystallography, i.e. the GluN1/2/3 LBDs [23–30] and GluN1/GluN2B ATDs [11,31,32]. Despite the availability of an intact homotetrameric structure of another iGluR, the AMPA receptor [33], it became clear from studies on the GluN1-GluN2B ATD heterodimers that the overall architecture of the intact NMDAR would be rather distinct [11]. Specifically, the ‘twisted’ bi-lobe architecture of the NMDAR ATDs compared to non-NMDAR ATDs [11,31] would make it sterically impossible for NMDARs to follow the arrangement of the homomeric AMPAR ATDs [34–36] and homo- or heteromeric kainate receptor ATDs [37–39]. The later structural studies on the intact NMDARs (Fig. 1a) [40,41] took full advantage of these NMDAR domain structures and also showed that the architectures and the patterns of subunit arrangement in these isolated domains are preserved in the intact receptors, therefore demonstrating the physiological relevance of the isolated domain structures (Fig. 1b, 1c). The key advantage for conducting studies on the isolated extracellular domains is their high-resolution capability (up to ~1.5 Å for the GluN1-GluN2A LBD heterodimers and ~2.5 Å for GluN1-GluN2B ATD heterodimers), which has been unattainable by single particle electron cryo-microscopy (cryo-EM) or X-ray crystallography of intact NMDARs. The high-resolution crystallographic studies on the isolated domains have been instrumental especially in unraveling precise chemical natures of bindings of agonists, antagonists, and some allosteric modulators [12,27,42–48]. The limitation of this fragmented approach, however, is the difficulty in studying compounds potentially binding at the inter-ATD-LBD or inter-LBD-TMD interfaces [49–53].
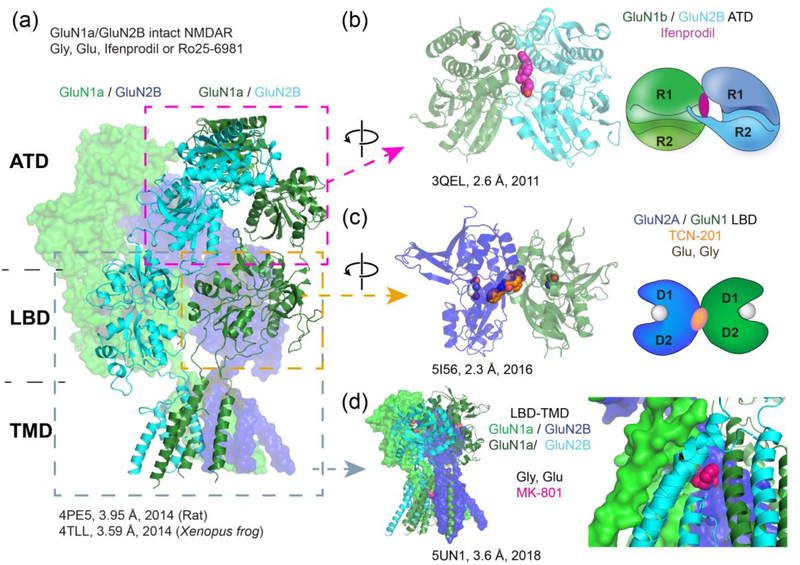
Figure 1. Crystallographic studies on the NMDAR.
(a) Crystal structures of the intact GluN1-GluN2B di-heteromeric NMDARs (without CTD) from rat and Xenopus frog showing a GluN1-GluN2B-GluN1-GluN2B arrangement (GluN1: green/light green, GluN2B: cyan/dark blue) and a domain-swap between the ATD layer and the LBD layer. (b-d) Fragmented approach to effectively monitor ligand binding sites at high resolution. (b) The crystal structure of the GluN1b-GluN2B ATD heterodimers shows the ifenprodil (pink) binding site at the GluN1b-GluN2B subunit interface. Binding of ifenprodil promotes the closure of the bi-lobed structure defined by the upper (R1) and lower (R2) lobes for the GluN2B ATD. (c) A number of crystal structures of NMDAR LBDs, including the ones of GluN1, GluN2A, and GluN3A, have been solved over the years with agonists, competitive antagonists, and allosteric modulators. Shown here is the crystal structure of the GluN1-GluN2A LBDs complexed to glycine (light gray), glutamate (light gray), and TCN-201 (orange). Glutamate and glycine bind at the cleft between the upper (D1) and lower (D2) lobes, whereas TCN-201 binds at the LBD dimer interface. (d) The crystal structure of the GluN1ΔATD – GluN2BΔATD NMDAR shows the binding site of the channel blocker MK-801 (red). Removing the ATDs changes the subunit arrangement in the LBD layer, which highlights the importance of ATDs for proper heterotetrameric subunit assembly. Resolutions of the electron-density maps (in Å) for the respective structures (with PDB code) are stated as in the Protein Data Bank.
Structural studies of intact NMDARs
The major shortcoming in the studies of isolated ATDs and LBDs is the lack of insights into inter-domain and inter-subunit interactions in the context of the heterotetrameric assembly. This is especially important in NMDARs because the ATDs regulate the ion channel activities [13,14] by interacting with the LBDs, a functional characteristic not present in non-NMDARs. However, how ATDs from different GluN2 subunits result in different channel open probabilities and how binding of allosteric modulators such as ifenprodil induce conformational changes in the ATD and result in allosteric inhibition of the GluN1-GluN2B ion channel at the TMD remained elusive. These fundamental questions drove structural biologists to obtain the first crystal structures of the intact GluN1-GluN2B NMDARs (from either rat or Xenopus frog), which were technically challenging [40,41]. For example, proper hetero-multimeric assembly of GluN1-GluN2B NMDARs required the usage of baculovirus-mediated gene transduction of mammalian cells (BacMam) [41,54] or baculovirus-mediated expression in insect cells using the Drosophila Hsp70 promoter instead of the conventional polyhedron promoter [40]. Availability of the high-resolution ATD [11] and LBD heterodimeric structures [24] facilitated obtaining sufficient phase information to result in electron density for the TMD from the diffraction data at around 4 Å. Improved resolution was achieved in both independent studies by introducing a disulfide crosslink between the two GluN2B subunits at the ATD, and by adding an allosteric inhibitor that binds at the ATD inter-subunit interface (ifenprodil or Ro25–6981), possibly suggesting a high degree of flexibility in the extracellular region. These intact tetrameric structures of the allosterically inhibited, closed channel with full or partial agonists bound have shown the overall assembly of alternating GluN1-GluN2B-GluN1-GluN2B subunits, the arrangement of “dimer of heterodimers” in the ATD and LBD layer, and a pseudo-fourfold symmetry in the TMD. Similar to AMPARs [33], domain swapping is observed in the extracellular region of the intact NMDARs, where the ATDs and LBDs change their respective heterodimer partners. Interestingly, structures of the desensitized GluK2 kainate receptor show a pinwheel arrangement for the LBD layer [55,56], different from any of the NMDAR or AMPAR structure to date. Considering the similarity between the GluK2 kainate receptors and the GluA2 AMPARs, a similar domain-swapping may also take place in non-desensitized states of the GluK2. The most pronounced difference between non-NMDARs and NMDARs is the inter-domain arrangement between the ATD and the LBD layers: the ATDs and LBDs in NMDARs have extensive inter-domain and inter-subunit interactions whereas the ATD-LBD interactions are minimal in non-NMDARs. Consequently, the NMDARs have a more compact ‘hot-air balloon’ architecture compared to the more extended ‘Y-shaped’ structure of AMPARs and kainate receptors.
Technological breakthroughs in cryo-EM in recent years have allowed many large protein complexes to be solved at a rapid speed [57–59], including more NMDAR structures in distinct functional states. While the resolutions of cryo-EM structures are typically lower (~ 4.5–15 Å) than those of crystal structures, they offer insights into large domain movements, which can be gleaned by extensive 3D classification in single particle analysis. Cryo-EM studies by Zhu et al and Tajima et al captured conformations distinct from the crystal structures in the agonist/allosteric modulator-bound condition [40,41], thereby providing the first dynamic views of NMDARs and yielding structural insights into receptor activation, allosteric modulation, and antagonism [60,61]. The structure of the GluN1-GluN2B NMDAR in the presence of glycine and glutamate but in the absence of ATD-targeting allosteric inhibitors showed unique conformations in the extracellular domains that increases the distance between the GluN1-GluN2B ATD dimers, which is caused mainly by the opening of the GluN2B ATD bi-lobes compared to the ifenprodil-bound form. The majority of the captured 3D classes showed the ‘open’ GluN2B ATD bi-lobes and ‘closed’ LBD bi-lobes as observed in the previous agonist-bound crystal structures [11,24], and the TMD channel with the ‘closed’ gates, which could represent the ‘non-active’ pre-open or desensitized state [60,61]. One 3D class derived from ~16% of the NMDAR particles in the cryo-EM study by Tajima et al showed a conformation likely representing the ‘active’ state, but unfortunately, the TMDs are not well resolved in this particular 3D class and thus, limit mechanistic insights into gating [61] (Fig. 2a). Nevertheless, in this 3D class, in addition to the ‘opening’ of the GluN2B ATD bi-lobe, the inter-GluN1-GluN2B orientation is rotated by ~12°, which becomes translated to a rotation of the GluN1-GluN2B LBD dimers (Fig. 2a). This concerted movement between the ATD and the LBD increases the distances between the gating-ring residues that are proximal to the TMD channel by as much as ~20 Å compared to the other ‘non-active’ 3D classes, which may generate sufficient tensions in the LBD–TMD linker for ion channel gating [62,63]. The validity of naming this conformation ‘active’ was supported initially by the observation that trapping the ‘active’ GluN1-GluN2B subunit conformation at the ATD by chemical cross-linkers resulted in potentiation of agonist-induced currents as measured by electrophysiology [61]. Further support comes from recent work that trapped the ‘active’ conformation at the LBD layer by a disulfide bond, which also resulted in augmented activity reflected by a higher opening probability and mean open time [64]. Despite the significant advances represented by the above findings, the ‘active state’ structure with open gate and pore at the TMD is still required to obtain the complete mechanism of NMDAR activation.
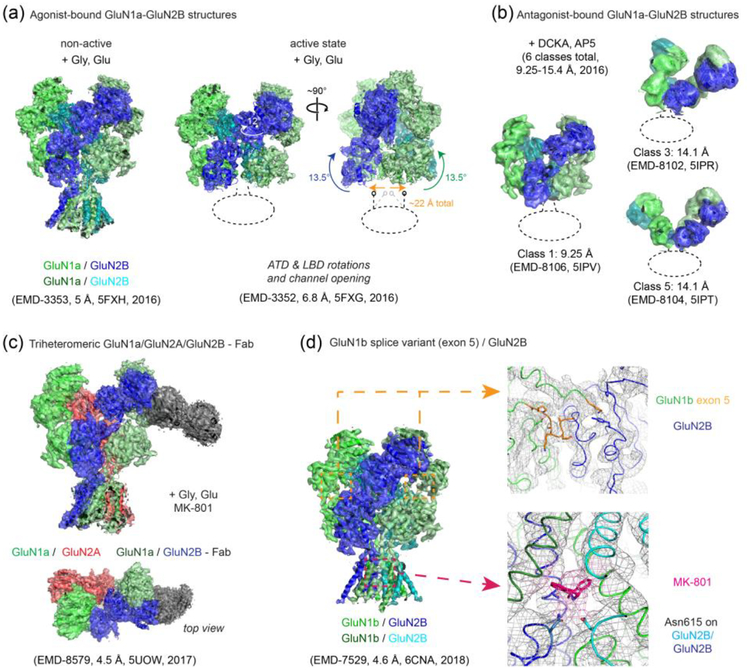
Figure 2. Single particle cryo-EM studies on the rat and Xenopus NMDARs.
(a) Multiple conformational states of the rat GluN1-GluN2B NMDARs bound to the agonists, glycine and glutamate. The cryo-EM density maps and the molecular models for the GluN1 subunits are colored green and light green whereas those for the GluN2B subunits are colored cyan and dark blue. Transitioning from the non-active state to the active state requires ‘opening’ of the bi-lobed structure of the GluN2B ATD and inter-subunit reorientation of the GluN1 and GluN2B ATDs by ~12°, which translates to rotation movement of the GluN1-GluN2B LBD heterodimers by ~13° and onto the gate of the TMD channel (arrows show movement from “non-active 2” state to “active” state [[61]]). Unresolved TMDs are marked with dotted ellipses/lines. (b) Binding of the antagonists, DCKA and AP5, leads to large movements in the extracellular domains as shown by selected 3D classes of the Xenopus GluN1a-GluN2B NMDARs with different conformations and the disrupted pattern of the “dimer of dimers” assembly. The color code is as in panel (a). (c) The overall assembly pattern of the tri-heteromeric Xenopus GluN1a-GluN2A-GluN2B NMDARs is similar to that of the di-heteromeric GluN1-GluN2B NMDARs in that the GluN1-GluN2-GluN1-GluN2 subunit arrangement is conserved. Binding of anti-GluN2B Fab (dark gray) facilitated single particle analysis by distinguishing the GluN2B subunit (dark blue) from the GluN2A subunit (red). (d) The cryo-EM structure of the GluN1b-GluN2B NMDAR splicing variant containing exon 5 in the GluN1b subunit shows that the exon 5-encoded motif (orange) is located at the ATDLBD interface and interacts with both the GluN1b LBD and GluN2B LBD to strengthen the hetero-dimeric interaction. This structure also showed cryo-EM density for a channel blocker, MK-801, and interacting residues such as GluN2B Asn615 (shown as sticks) in the TMD channel. Similar density for MK-801 was also observed in the cryo-EM structure of the tri-heteromeric Xenopus GluN1a-GluN2A-GluN2B NMDARs. Resolutions of the EM maps (in Å) for any given class (with EMD code) are stated as in the EM Data Bank, which were determined by the current gold-standard Fourier Shell Correlation (FSC) cutoff at 0.143.
Cryo-EM structures at 9–15 Å resolution of the GluN1-GluN2B NMDAR prepared in the presence of competitive antagonists 5,7-dichlorokynurenic acid (DCKA) and D-2-amino-5-phosphonovalerate (D-AP5) showed 3D classes with robust separation of the extracellular region compared to the agonist-bound form [60]. Based on the cryo-EM density and rigid-body model fitting, the study concluded that the gating-ring in those antagonist-bound NMDAR structures is ruptured in a way that the “dimer of dimers” arrangement is no longer maintained to mediate signal transduction from the LBDs to the TMD. Similar to the ‘active’ state [61], the TMDs were not resolved in those structures, thus, limiting the precise insights into how competitive antagonism works at the level of the ion channel. Furthermore, whether the structures of the antagonist-bound state resemble that of the resting state (i.e. no ligands) remains to be investigated. The observation in the cryo-EM study was further validated by double-electron-electron-resonance (DEER) experiments, which measures the distance between spin labelled residues to estimate inter-domain and inter-lobe distances [60]. Generally, it is important to carefully validate observed conformational changes in cryo-EM structures through a number of ways including functional assays, biochemical cross-linking, and DEER experiments, since a plethora of experimental factors in protein sample preparation as well as the vitrification process of cryo-EM sample preparation (e.g., shearing forces in vitreous ice, detergents, types of grids) can often lead to dissociation and deformation of protein assembly. For all of the observed conformational alterations associated with activation, competitive antagonism, and allosteric inhibition, it would be interesting to examine them by alternative methods such as luminescence resonance energy transfer (LRET) [65–68] and single-molecule fluorescence resonance energy transfer (smFRET) [69–72]. The key advantage of those methods is that the experiments can be conducted at room temperature, whereas DEER experiments are typically performed at cryogenic temperatures (~80 K) [73] and hence might not validate physiological relevance convincingly. Furthermore, probing conformational alteration using computational approaches [74,75] may provide an insightful path to understand the mechanistic details. The major challenge in this area of research is to correlate a number of modal patterns of activation observed by electrophysiology to the protein conformational landscape [76–80].
Addressing more specific questions
With the completion of de novo structures of the intact NMDARs and observation of fundamental conformational movement, the structural biology of NMDARs is at the stage where more specific questions can be pursued. Examples are structures of tri-heterotetrameric NMDARs composed of GluN1, GluN2A, and GluN2B [81], splice variants [82], and channel blocker binding [82,83].
In addition to di-heteromeric NMDARs, tri-heteromeric NMDARs containing three distinct subunits are present in the adult brain [84]. The first identified tri-heteromeric NMDARs was the GluN1-GluN2A-GluN2B combination [85]. Later studies demonstrated that the tri-heteromeric NMDARs are present in the synapses to mediate synaptic transmission and neuroplasticity [86–88]. Curiously, the GluN1-GluN2A-GluN2B tri-heteromeric NMDAR has functional properties, which are characteristic of both GluN1-GluN2A and GluN1-GluN2B di-heteromeric NMDARs but are more highly weighted toward the GluN1-GluN2A NMDAR [86,89,90]. The recent cryo-EM structure of the GluN1-GluN2A-GluN2B NMDARs from Xenopus laevis provided a possible answer to this phenomenon [81]. Technical difficulties included assembly of the GluN1-GluN2AGluN2B tri-heteromeric NMDAR proteins in a recombinant expression system and distinction of GluN2A and GluN2B during the single particle analysis due to the high structural similarity between the GluN2A and GluN2B subunits, which is reflected by their high sequence identity/similarity (71%/83% for Xenopus GluN2A/B without the CTD). By labeling the GluN2B subunit with an anti-GluN2B Fab fragment, Lü et al could however elegantly distinguish the subunits in the intact tri-heteromer [81] (Fig. 2c). The structure showed more extensive interactions between the GluN2A ATD and the LBD than between the GluN2B ATD and the LBD, possibly explaining why the pharmacological properties of these asymmetric tri-heterotetramers lean towards the di-heterotetrameric GluN1-GluN2A such as nano-molar zinc inhibition potency and a faster glutamate deactivation time course [86,90,91]. The most recent cryo-EM structures of the di-heteromeric GluN1-GluN2A NMDAR [92] now allow the field to make more detailed comparisons with the tri-heteromeric NMDARs and will help provide further explanation to the asymmetric pharmacological properties described above.
Another important feature of NMDARs is their differential pH-sensitivities controlled by alternative splicing [93]. It has been known that inclusion of the 21 amino acids encoded by exon 5 in GluN1 (in GluN1–1b to GluN1–4b splice variants) reduces potency of proton inhibition. A recent cryo-EM structure of the GluN1b-GluN2B receptor at 4.6 Å showed that the exon 5-encoded motif changes the local structure and protrudes from the GluN1 ATD core to form unique interactions with the GluN1 and GluN2B LBDs, thereby strengthening the heterodimeric GluN1-GluN2B LBD interaction [82] (Fig. 2d). This cryo-EM structure along with the structure-based experiments linked the exon 5 paradigm with the previous finding that the pH-sensor may lie at the GluN1-GluN2 LBD interface [94].
Improved cryo-EM density maps or electron density maps from X-ray crystallography for the TMD have now even enabled the field to locate the binding sites of small molecule channel blockers, such as MK-801 [82,83]. The cryo-EM study showed clear cryo-EM density of MK-801, which interacts closely with the residue Asn615 of the pore-lining M3 helices of rat GluN2B [81,82]. These findings were corroborated by X-ray crystallography as it generally remains the method of choice when probing binding of small molecules. In this case, removing the ATD helped in obtaining crystals with improved resolution and showed electron densities for the channel blocker MK-801 (Fig. 1d) [83]. While the LBD arrangement in these crystals seems different from the intact NMDARs containing the ATD, the TMD channel is assembled correctly, thus, the crystal structure serves as a valid tool to assess binding of channel blockers.
Conclusions
With the general structural framework of the NMDAR set and steadily improving resolution in cryo-EM studies, more detailed questions surrounding NMDAR subtypes as well as functional states and some compound binding patterns can be probed. However, at this point, X-ray crystallography remains to be the superior method for obtaining detailed compound binding mechanisms that are useful for therapeutic interventions. Combining all those structures with spectroscopic and computational experiments and correlating the outcomes with functional readouts from electrophysiology is the desired approach to complete the sophisticated mechanistic scheme of NMDARs.
Highlights.
The X-ray crystallographic structures of the intact NMDARs showed unique domain and subunit assembly patterns explaining the distinct functions from the non-NMDARs.
Implementation of single particle cryo-electron microscopy revealed multiple conformations of the intact NMDARs that are linked to functions.
X-ray crystallography on isolated domains of NMDARs remains the best method to observe compound binding modes at high-resolution.
Despite significant advances over the last decade, sophisticated questions such as modal gating of NMDARs have not been well explained by the structural biology at this point.
Acknowledgement
This work was supported by grants from the National Institutes of Health (MH085926 and GM105730), Robertson funds at Cold Spring Harbor Laboratory, Austin’s purpose, and Stanley Institute of cognitive genomics (all to H.F.). J.X.W. was supported by a PhD fellowship of the Boehringer Ingelheim Fonds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
| Karakas E, Furukawa H: Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 2014, 344:992–997. Lee C-H, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E: NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 2014, 511:191–197. |
** First structures of assembled heterotetrameric NMDARs via X-ray crystallographic studies for rat and frog NMDARs, showing alternating subunit arrangement and domain swap between the ATD layer and the LBD layer. |
| Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H: Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 2016, 10.1038/nature17679 | ** Cryo-EM structures of three major different agonist-bound classes suggest concerted movements of the ATD and the LBD are required to activate the NMDAR ion channel. The structure-based functional experiments validated the structural observation. |
| Lu W, Du J, Goehring A, Gouaux E: Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 2017, 355:eaal3729 | ** First structure of the triheteromeric NMDAR using antibody fragments to distinguish highly similar subunits. |
| Regan MC, Grant T, McDaniel MJ, Karakas E, Zhang J, Traynelis SF, Grigorieff N, Furukawa H: Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron 2018, 98:521–529 e523. | ** Cryo-EM structure of GluN1b-GluN2B shows the structural organization of the exon 5- encoded motif at the ATD-LBD interface and a channel blocker MK-801 in the TMD. |
| Karakas E, Simorowski N, Furukawa H: Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 2011, 475:249–253. | * Crystal structure of Xenopus GluN1 ATD and rat GluN2B ATD heterodimer shows that the ifenprodil binding site resides at the GluN1- GluN2B subunit interface. This study shows that the pattern of inter-GluN1-GluN2B interactions at ATD is distinct from those of non-NMDARs. |
| Gielen M, Retchless BS, Mony L, Johnson JW, Paoletti P: Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 2009, 459:703–707 Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF: Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 2009, 29:1204512058. |
* The subunit identity of ATD and the linker between ATD and LBD (ATD-LBD linker) influences channel open probability of the NMDARs, as shown by swapping the ATD and the ATD-LBD linker between GluN2A, GluN2B and GluN2D subunits. Similarly, authors use GluN2A-GluN2D chimeras to show that the ATD not only influences channel open probability, but also mean open duration and deactivation time courses by extensive patch-clamp experiments. |
| Sobolevsky AI, Rosconi MP, Gouaux E: X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 2009, 462:745–756. | * First crystal structure of the intact AMPAR whose overall fold is distinct and less compact compared to the NMDAR. |
| Dolino DM, Chatterjee S, MacLean DM, Flatebo C, Bishop LDC, Shaikh SA, Landes CF, Jayaraman V: The structure- energy landscape of NMDA receptor gating. Nat Chem Biol 2017, 13:1232–1238. | * Single-molecular FRET studies probe different conformational states of the channel gate region of full-length GluN1/GluN2A receptors with several ligands. |
References
- 1.Paoletti P, Bellone C, Zhou Q: NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature reviews. Neuroscience 2013, 14:383–400. [DOI] [PubMed] [Google Scholar]
- 2.du Bois TM, Huang X-F: Early brain development disruption from NMDA receptor hypofunction: Relevance to schizophrenia. Brain Research Reviews 2007, 53:260–270. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Sheng M: NMDA receptors in nervous system diseases. Neuropharmacology 2013, 74:69–75. [DOI] [PubMed] [Google Scholar]
- 4.Lester RA, Jahr CE: NMDA channel behavior depends on agonist affinity. J Neurosci 1992, 12:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lester RA, Clements JD, Westbrook GL, Jahr CE: Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature 1990, 346:565–567. [DOI] [PubMed] [Google Scholar]
- 6.Hebb DO: The Organization of Behavior: New York: Wiley & Sons; 1949. [Google Scholar]
- 7.Cummings KA, Popescu GK: Protons Potentiate GluN1/GluN3A Currents by Attenuating Their Desensitisation. Sci Rep 2016, 6:23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cul J, Tu S, Sevarino KA, Nakanishi N, et al. : Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature 2002, 415:793–798. [DOI] [PubMed] [Google Scholar]
- 9.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R: Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews 2010, 62:405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu S, Paoletti P: Allosteric modulators of NMDA receptors: multiple sites and mechanisms. Current opinion in pharmacology 2015, 20:14–23. [DOI] [PubMed] [Google Scholar]
- 11.Karakas E, Simorowski N, Furukawa H: Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature 2011, 475:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H: Molecular Basis for Subtype Specificity and High-Affinity Zinc Inhibition in the GluN1-GluN2A NMDA Receptor Amino-Terminal Domain. Neuron 2016, 10.1016/j.neuron.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gielen M, Retchless BS, Mony L, Johnson JW, Paoletti P: Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature 2009, 459:703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF: Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci 2009, 29:12045–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG: Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nature neuroscience 2000, 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 16.Ehlers MD, Zhang S, Bernhadt JP, Huganir RL: Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell 1996, 84:745–755. [DOI] [PubMed] [Google Scholar]
- 17.Ehlers MD, Fung ET, O’Brien RJ, Huganir RL: Splice variant-specific interaction of the NMDA receptor subunit NR1 with neuronal intermediate filaments. J Neurosci 1998, 18:720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iacobucci GJ, Popescu GK: Resident Calmodulin Primes NMDA Receptors for Ca(2+)-Dependent Inactivation. Biophys J 2017, 113:2236–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA: The NMDA Receptor NR1 C1 Region Bound to Calmodulin: Structural Insights into Functional Differences between Homologous Domains. Structure 2007, 15:1603–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karakas E, Regan MC, Furukawa H: Emerging structural insights into the function of ionotropic glutamate receptors. Trends in biochemical sciences 2015, 40:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Regan MC, Romero-Hernandez A, Furukawa H: A structural biology perspective on NMDA receptor pharmacology and function. Current Opinion in Structural Biology 2015, 33:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen KB, Yi F, Perszyk RE, Furukawa H, Wollmuth LP, Gibb AJ, Traynelis SF: Structure, function, and allosteric modulation of NMDA receptors. J Gen Physiol 2018, 150:1081–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa H, Gouaux E: Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. The EMBO journal 2003, 22:2873–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa H, Singh SK, Mancusso R, Gouaux E: Subunit arrangement and function in NMDA receptors. Nature 2005, 438:185–192. [DOI] [PubMed] [Google Scholar]
- 25.Inanobe A, Furukawa H, Gouaux E: Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 2005, 47:71–84. [DOI] [PubMed] [Google Scholar]
- 26.Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jorgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF: Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Molecular pharmacology 2013, 84:114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vance KM, Simorowski N, Traynelis SF, Furukawa H: Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nature communications 2011, 2:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvist T, Greenwood JR, Hansen KB, Traynelis SF, Bräuner-Osborne H: Structure-based discovery of antagonists for GluN3-containing N-methyl-d-aspartate receptors. Neuropharmacology 2013, 75:324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML: Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J 2008, 27:2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Belcher J, Berger AJ, Mayer ML, Lau AY: Conformational analysis of NMDA receptor GluN1, GluN2, and GluN3 ligand-binding domains reveals subtype-specific characteristics. Structure 2013, 21:1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karakas E, Simorowski N, Furukawa H: Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. The EMBO journal 2009, 28:3910–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T: Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience 2011, 31:3565–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobolevsky AI, Rosconi MP, Gouaux E: X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 2009, 462:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E: Crystal structure and association behaviour of the GluR2 amino-terminal domain. The EMBO journal 2009, 28:1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukumaran M, Rossmann M, Shrivastava I, Dutta A, Bahar I, Greger IH: Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J 2011, 30:972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR: Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol 2009, 392:1125–1132. [DOI] [PubMed] [Google Scholar]
- 37.Kumar J, Mayer ML: Crystal structures of the glutamate receptor ion channel GluK3 and GluK5 amino-terminal domains. J Mol Biol 2010, 404:680–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar J, Schuck P, Jin R, Mayer ML: The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol 2009, 16:631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar J, Schuck P, Mayer ML: Structure and assembly mechanism for heteromeric kainate receptors. Neuron 2011, 71:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karakas E, Furukawa H: Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 2014, 344:992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee C-H, Lü W, Michel JC, Goehring A, Du J, Song X, Gouaux E: NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 2014, 511:191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H: Structural insights into competitive antagonism in NMDA receptors. Neuron 2014, 81:366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackos DH, Lupardus PJ, Grand T, Chen Y, Wang T-M, Reynen P, Gustafson A, Wallweber HJA, Volgraf M, Sellers BD, et al. : Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function. Neuron 2016, 89:983–999. [DOI] [PubMed] [Google Scholar]
- 44.Volgraf M, Sellers BD, Jiang Y, Wu G, Ly CQ, Villemure E, Pastor RM, Yuen PW, Lu A, Luo X, et al. : Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design. J Med Chem 2016, 59:2760–2779. [DOI] [PubMed] [Google Scholar]
- 45.Romero-Hernandez A, Furukawa H: Novel Mode of Antagonist Binding in NMDA Receptors Revealed by the Crystal Structure of the GluN1-GluN2A Ligand-Binding Domain Complexed to NVP-AAM077. Mol Pharmacol 2017, 92:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lind GE, Mou T-C, Tamborini L, Pomper MG, De Micheli C, Conti P, Pinto A, Hansen KB: Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits. Proceedings of the National Academy of Sciences 2017, 114:E6942–E6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi F, Mou T-C, Dorsett KN, Volkmann RA, Menniti FS, Sprang SR, Hansen KB: Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron 2016, 91:1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stroebel D, Buhl DL, Knafels JD, Chanda PK, Green M, Sciabola S, Mony L, Paoletti P, Pandit J: A Novel Binding Mode Reveals Two Distinct Classes of NMDA Receptor GluN2B-selective Antagonists. Mol Pharmacol 2016, 89:541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khatri A, Burger PB, Swanger SA, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF: Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Molecular pharmacology 2014, 86:548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen KB, Traynelis SF: Structural and mechanistic determinants of a novel site for noncompetitive inhibition of GluN2D-containing NMDA receptors. J Neurosci 2011, 31:3650–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perszyk R, Katzman BM, Kusumoto H, Kell SA, Epplin MP, Tahirovic YA, Moore RL, Menaldino D, Burger P, Liotta DC, et al. : An NMDAR positive and negative allosteric modulator series share a binding site and are interconverted by methyl groups. 2018:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaiser TM, Kell SA, Kusumoto H, Shaulsky G, Bhattacharya S, Epplin MP, Strong KL, Miller EJ, Cox BD, Menaldino DS, et al. : The Bioactive Protein-Ligand Conformation of GluN2C-Selective Positive Allosteric Modulators Bound to the NMDA Receptor. Mol Pharmacol 2018, 93:141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yelshanskaya MV, Singh AK, Sampson JM, Narangoda C, Kurnikova M, Sobolevsky AI: Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs. Neuron 2016, 91:1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC: BacMam System for High-Level Expression of Recombinant Soluble and Membrane Glycoproteins for Structural Studies. Protein expression and purification 2008, 62:160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S: Structural mechanism of glutamate receptor activation and desensitization. Nature 2014, 514:328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyerson JR, Chittori S, Merk A, Rao P, Han TH, Serpe M, Mayer ML, Subramaniam S: Structural basis of kainate subtype glutamate receptor desensitization. Nature 2016, 537:567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henderson R: From Electron Crystallography to Single Particle CryoEM (Nobel Lecture). Angew Chem Int Ed Engl 2018, 57:10804–10825. [DOI] [PubMed] [Google Scholar]
- 58.Frank J: Single-Particle Reconstruction of Biological Molecules-Story in a Sample (Nobel Lecture). Angew Chem Int Ed Engl 2018, 57:10826–10841. [DOI] [PubMed] [Google Scholar]
- 59.Campbell MG, Cheng A, Brilot AF, Moeller A, Lyumkis D, Veesler D, Pan J, Harrison SC, Potter CS, Carragher B, et al. : Movies of ice-embedded particles enhance resolution in electron cryo-microscopy. Structure 2012, 20:1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu S, Stein RA, Yoshioka C, Lee C-H, Goehring A, McHaourab HS, Gouaux E: Mechanism of NMDA Receptor Inhibition and Activation. Cell 2016, 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N, Furukawa H: Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 2016, 10.1038/nature17679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP: Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nature neuroscience 2014, 17:914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Twomey EC, Sobolevsky AI: Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry 2018, 57:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esmenjaud JB, Stroebel D, Chan K, Grand T, David M, Wollmuth LP, Taly A, Paoletti P: An inter-dimer allosteric switch controls NMDA receptor activity. Embo j 2018, 10.15252/embj.201899894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sirrieh RE, MacLean DM, Jayaraman V: Subtype-dependent N-methyl-D-aspartate receptor amino-terminal domain conformations and modulation by spermine. The Journal of biological chemistry 2015, 290:12812–12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sirrieh RE, MacLean DM, Jayaraman V: Amino-terminal domain tetramer organization and structural effects of zinc binding in the N-methyl-D-aspartate (NMDA) receptor. The Journal of biological chemistry 2013, 288:22555–22564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rambhadran A, Gonzalez J, Jayaraman V: Subunit arrangement in N-methyl-D-aspartate (NMDA) receptors. J Biol Chem 2010, 285:15296–15301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rambhadran A, Gonzalez J, Jayaraman V: Conformational changes at the agonist binding domain of the N-methyl-D-aspartic acid receptor. J Biol Chem 2011, 286:16953–16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dolino DM, Chatterjee S, MacLean DM, Flatebo C, Bishop LDC, Shaikh SA, Landes CF, Jayaraman V: The structure-energy landscape of NMDA receptor gating. Nat Chem Biol 2017, 13:1232–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sasmal DK, Lu HP: Single-molecule patch-clamp FRET microscopy studies of NMDA receptor ion channel dynamics in living cells: revealing the multiple conformational states associated with a channel at its electrical off state. J Am Chem Soc 2014, 136:12998–13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dolino DM, Cooper D, Ramaswamy S, Jaurich H, Landes CF, Jayaraman V: Structural Dynamics of the Glycine-binding Domain of the N-Methyl-d-Aspartate Receptor. The Journal of biological chemistry 2015, 290:797–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacLean DM, Durham RJ, Jayaraman V: Mapping the Conformational Landscape of Glutamate Receptors Using Single Molecule FRET. Trends Neurosci 2018, 10.1016/j.tins.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Vera IMS, Blackburn ME, Galiano L, Fanucci GE: Pulsed EPR Distance Measurements in Soluble Proteins by Site-directed Spin-labeling (SDSL). Current protocols in protein science / editorial board, John E. Coligan … [et al.] 2013, 74:Unit-17.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dai J, Zhou H-X: Reduced Curvature of Ligand-Binding Domain Free-Energy Surface Underlies Partial Agonism at NMDA Receptors. Structure 2015, 23:228–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu A, Lau AY: Glutamate and Glycine Binding to the NMDA Receptor. Structure 2018, 26:1035–1043.e1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iacobucci GJ, Popescu GK: Kinetic models for activation and modulation of NMDA receptor subtypes. Curr Opin Physiol 2018, 2:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Iacobucci GJ, Popescu GK: NMDA receptors: linking physiological output to biophysical operation. Nat Rev Neurosci 2017, 18:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cummings KA, Popescu GK: Glycine-dependent activation of NMDA receptors. J Gen Physiol 2015, 145:513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borschel WF, Myers JM, Kasperek EM, Smith TP, Graziane NM, Nowak LM, Popescu GK: Gating reaction mechanism of neuronal NMDA receptors. J Neurophysiol 2012, 108:3105–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amico-Ruvio SA, Popescu GK: Stationary gating of GluN1/GluN2B receptors in intact membrane patches. Biophys J 2010, 98:1160–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lü W, Du J, Goehring A, Gouaux E: Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation. Science 2017, 355:eaal3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Regan MC, Grant T, McDaniel MJ, Karakas E, Zhang J, Traynelis SF, Grigorieff N, Furukawa H: Structural Mechanism of Functional Modulation by Gene Splicing in NMDA Receptors. Neuron 2018, 98:521–529 e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song X, Jensen MØ, Jogini V, Stein RA, Lee C-H, McHaourab HS, Shaw DE, Gouaux E: Mechanism of NMDA receptor channel block by MK-801 and memantine. Nature 2018, 556:515–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stroebel D, Casado M, Paoletti P: Triheteromeric NMDA receptors: from structure to synaptic physiology. Curr Opin Physiol 2018, 2:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY: Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 1994, 368:144–147. [DOI] [PubMed] [Google Scholar]
- 86.Sun W, Hansen KB, Jahr CE: Allosteric Interactions between NMDA Receptor Subunits Shape the Developmental Shift in Channel Properties. Neuron 2017, 94:58–64 e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Delaney AJ, Sedlak PL, Autuori E, Power JM, Sah P: Synaptic NMDA receptors in basolateral amygdala principal neurons are triheteromeric proteins: physiological role of GluN2B subunits. Journal of Neurophysiology 2013, 109:1391–1402. [DOI] [PubMed] [Google Scholar]
- 88.Foster KA, McLaughlin N, Edbauer D, Phillips M, Bolton A, Constantine-Paton M, Sheng M: Distinct Roles of NR2A and NR2B Cytoplasmic Tails in Long-Term Potentiation. Journal of Neuroscience 2010, 30:2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatton CJ, Paoletti P: Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron 2005, 46:261–274. [DOI] [PubMed] [Google Scholar]
- 90.Hansen Kasper B, Ogden Kevin K, Yuan H, Traynelis Stephen F: Distinct Functional and Pharmacological Properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA Receptors. Neuron 2014, 81:1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stroebel D, Carvalho S, Grand T, Zhu S, Paoletti P: Controlling NMDA receptor subunit composition using ectopic retention signals. J Neurosci 2014, 34:16630–16636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jalali-Yazdi F, Chowdhury S, Yoshioka C, Gouaux E: Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor. Cell 2018, 175:1520–1532.e1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Traynelis SF, Hartley M, Heinemann SF: Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science 1995, 268:873–876. [DOI] [PubMed] [Google Scholar]
- 94.Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P: Structural Rearrangements of NR1/NR2A NMDA Receptors during Allosteric Inhibition. Neuron 2008, 57:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]