Abstract
Objectives:
To evaluate prognostic markers, prostate-specific antigen (PSA), prostate health index (PHI) and prostate volume indexed measures (PSAD and PHID) for predicting positive prostate cancer biopsies in magnetic resonance (MR) transrectal ultrasound fused versus non-fused transrectal ultrasonography biopsy.
Methods:
A retrospective cohort of 211 patients that had at least 1 suspected MR lesion, Prostate Imaging-Reporting and Data System ≥ 3 and subsequent biopsy (2015 to 2017). Clinical characteristics and prognostic biomarkers were evaluated as predictors of prostate cancer detection by type of biopsy guidance (fused versus non-fused).
Results:
One-hundred twenty-one patients had non-fused and 90 had fused biopsies. PHI and PHID had greater area under the receiver operating characteristics curve (AUC) in predicting positive biopsies than PSA or PSAD for both non-fused and fused biopsy. PHI 0.78 (95% CI 0.67 to 0.88) and PHID 0.82 (95% CI 0.73 to 0.91) had the greatest AUC for predicting biopsy results for non-fused and fused biopsies, respectively. Multiple-variable models did not improve model fit compared to single variables. Based on Youden’s index, a cut-off value of 45.9 for PHI in non-fused and 0.64 for PHID in fused biopsies would reduce the number of negative biopsies by 77.3% and 63.4%, respectively, but the percentage of missed clinically significant cancer biopsies would be 19% and 12%, respectively.
Conclusions:
Our findings demonstrate that the choice of prognostic biomarkers for predicting positive biopsies is a function of the biopsy guidance method. Volume indexed derivatives appear to have greater value when a MRI-US fused method is used.
Keywords: Prostate cancer, magnetic resonance imaging, prostate specific antigen, prostate health index, prostate biopsy
Introduction
Prostate cancer (PCa) is the second-most common cause of cancer-related death in men in the United States [1]. Screening relies primarily on measurement of serum prostate-specific antigen (PSA) levels [2, 3] and digital rectal examination (DRE), despite limited sensitivity and specificity [2]. PSA also can be elevated in benign prostatic hyperplasia (BPH) and prostatitis. Given the inverse relationship between prostate volume and the likelihood of a positive biopsy and more aggressive cancer [4], PSA density (PSA divided by prostate volume) has been studied as a PSA derivative for cancer. In addition to improved accuracy in predicting positive biopsy results compared with PSA [5–7], PSA density (PSAD) has shown a strong correlation with cancer aggressiveness [6, 8]. The prostate health index (PHI) incorporates the measurements of PSA, free PSA, and [–2] pro-PSA to develop a probability score for cancer risk. Studies have shown that PHI has increased accuracy in predicting cancer compared with PSA [9, 10]. There is an association of higher PHI values with more aggressive cancers [10, 11]. PSA density (PSAD) also has been shown to be a better predictor of prostate cancer than PSA, which has led to the evaluation of PHI density (PHID) for the diagnosis of prostate cancer. Two recent studies have found that PHID demonstrated the highest discrimination value for clinically-significant cancer [7, 12].
Needle biopsy is the gold standard for prostate cancer diagnosis. Traditionally, biopsies have been performed using a transrectal approach with ultrasound guidance (TRUS). Magnetic resonance-TRUS fusion biopsies can increase the positive biopsy rate compared to TRUS by targeting the suspected lesions identified on MR [13].
Prior studies that have examined patient characteristics, PSA and PSA derivatives have not controlled for the biopsy guidance method when determining the predictors of positive biopsy results [12, 14–19]. We hypothesized that the performance of PSA and PSA derivatives for predicting positive biopsies of multiparametric MRI (mpMRI) suspicious lesions (Prostate Imaging Reporting and Data System, PIRADS ≥ 3) vary as a function of the guidance method, whether MR-TRUS fusion (FUS) biopsy or TRUS biopsies without MR fusion (NFUS).
Material and Methods
Study Design
This retrospective study (January 2015 to December 2017) was approved by the Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act (HIPAA). A waiver of informed consent was granted. Individual chart review was performed to determine the clinical history, laboratory results, results of previous MRIs and biopsies. The inclusion criteria were: patients with PHI test and 3T MR exam with at least one suspicious MR identified lesion with a PI-RADS score of ≥3 prior to biopsy. Excluded were patients with no identified lesion on mpMRI, PI-RADS scores <3 lesions, and lesions that did not fit our index lesion criteria. The index lesion was defined as the lesion with the highest PIRADS score (≥3) and largest size on apparent diffusion coefficient map images for peripheral zone lesions or T2-weighted images for transition zone lesions.
Multiparametric MR Imaging and Biopsy Protocol
MpMRI examinations were performed using 3T MR scanners with T2-weighted, axial dynamic contrast-enhanced (DCE), and diffusion-weighted imaging using a phased-array body coil without endorectal coil. All mpMRI studies were interpreted by expert genitourinary radiologists and assigned suspicion scores based on the standardized PI-RADS version 2 criteria scoring system [20].
Patients underwent transrectally biopsy either with FUS or NFUS based on the urologist’s preference. The images were segmented by dedicated genitourinary radiologists using the UroNav, Philips-Invivo platform. The mpMR images were postprocessed, and suspicious lesions were marked using DynaCAD (InVivo, Philips). The FUS biopsy was performed with the previously identified mpMRI lesions superimposed using the T2-weighted MR on the TRUS image. After the fusion, the probe/needle alignment is maneuvered free-hand with automated software guidance to the virtual MRI lesions for targeting. The lesions were sampled by an end-fire or side-fire TRUS probe. The systematic TRUS biopsy included at least 12 cores. When FUS biopsies were obtained, typically one to four cores were additionally obtained from each marked lesion.
Pathologic review was performed by two dedicated genitourinary subspecialty pathologists assigned the Gleason score (GS) ranging from 6–10. The highest Gleason score was recorded for each patient. Gleason scores were then divided into five grade groups (GG): GG1 equals GS6, GG2 equals GS7 (3+4=7), GG3 equals GS7 (4+3=7), GG4 equals GS8, GG5 equals GS9–10 [21]. Patients in group GG1 or above were considered positive cancers (PCa) biopsies and GG2 or greater as clinically significant cancer.
The baseline PSA and PHI values were obtained from the electronic medical record. PHI values can be calculated with the formula [−2]proPSA/fPSA x √PSA. Density values were calculated using the PSA or PHI value divided by the prostate volume determined from the MR by the prolate ellipsoid formula (height × width × length × π/6). The prostate width and length were measured on the axial T2-weighted image and height on the mid-sagittal T2-weighted image.
Statistical methods
The primary outcome was the determination of the association of patient characteristics, serum PSA and derivatives (PSAD, PHI and PHID) and their combinations with a PCa biopsy using FUS and NFUS methods. The univariable (unadjusted) association of patient characteristics between FUS and NFUS biopsy groups and between patients with a positive and negative biopsy were compared using the Mann-Whitney U test and a chi-squared statistic.
Multi-variable (adjusted) modeling of the association of patient characteristic and serum prognostic marker was performed using binary logistic regression. A base model included age, DRE and prior biopsy. Models were then created by the addition of either PSA or PSAD and then PHI or PHID, dependent upon whether the size adjusted prognostic marker had a greater area under the curve in univariable analysis, until a model including base + PSA(D) + PHI(D) was created. Multicollinearity of variables included in the logistic models was assessed by evaluating the tolerance (>0.1), variance inflation factor (VIF < 10), and the condition index (<30). The AUC for each model was compared to the prior model as well as the individual serum prognostic marker to determine the value of the multiparameter models.
The diagnostic utility of PSA and derivatives for predicting a positive biopsy was determined by constructing receiver operating characteristic curves (ROC). The area under the ROC curve (AUC) for each pair of prognostic markers was compared using the method of DeLong. The cut-off value for the model with the greatest AUC was calculated at the point of maximization of Youden’s J index and at sensitivities of 95 and 100 percent. The number of unnecessary biopsies was determined as the number of negative biopsies identified below the cut-off point of the prognostic marker out of total negative biopsies. The number of missed diagnoses was calculated as the number of negative biopsies identified above the cut-off point of the prognostic marker out of total positive biopsies.
The study sample was based on a convenience sample of all available patients that had serum PSA and derivatives, MR and biopsy performed from 2015 to 2017 to ensure that all patients had an MRI examination using a 3T scanner with similar protocols. Given that at least 80 subjects/lesions were included in each group, the study had 80% power to detect at least one predictor for each biopsy method with an odds ratio of at least 2 at an alpha of 0.05 using binary logistic regression.
Statistical analysis was performed using RStudio version 1.1.463 (Integrated Development for R. RStudio, Inc., Boston, MA; URL: http://www.rstudio.com/) and R version 3.5.2, release date 12/20/2018 (The R Foundation for Statistical Computing, Vienna, Austria). Sample size calculations were made using PASS 15, power analysis and sample size software (2017), NCSS, LLC, Kaysville UT, USA, ncss.com/software/pass.
Results
Two hundred and thirty-three patients had serum PSA and derivatives, MR examinations and a biopsy performed. The median (5th and 95th percentile) time between MR examination and biopsy was 29 (7 to 140) days. Twenty-two were excluded because no lesion was identified on MR or the highest PI-RADS lesion was less than 3.
FUS sampling provided a greater percentage of positive biopsies compared to NFUS sampling (54.4% to 27.2%, difference 27.2%, 95% CI of the difference 13% to 41%, P<0.001) (Table 1). The only difference in characteristics between FUS and NFUS methods was for DRE findings. Table 2 illustrates a comparison between PCa and negative biopsy groups for patient characteristics, PSA and derivatives, and MR variables. Univariable differences were found for prior biopsy, PI-RADS score, prostate volume, maximum lesion size, PHI score, PHID and PSAD.
Table 1:
Characteristics of patients receiving non-fused versus fused biopsies.
| Method of biopsy | P | ||
|---|---|---|---|
| Non-fused (n=121) | Fused (n=90) | ||
| Age (y) | 67 (59 to 70) | 65 (59 to 70) | 0.48 |
| Race n (%) | |||
| African American | 10 (8) | 9 (10) | 0.76 |
| White | 85 (71) | 65 (72) | |
| Hispanic | 5 (4) | 1 (1) | |
| Other | 15 (12) | 11 (12) | |
| Declined to answer | 6 (5) | 4 (5) | |
| Family history n (%) | |||
| No | 56 (46) | 46 (52) | 0.78 |
| Yes | 32 (27) | 22 (24) | |
| Unknown | 33 (27) | 22 (24) | |
| Digital rectal examination findings n (%) | |||
| Normal | 15 (12) | 39 (43) | <0.001 |
| Abnormal | 80 (66) | 34 (38) | |
| Not documented | 26 (22) | 17 (19) | |
| Prior biopsy n (%) | |||
| None | 57 (47) | 53 (59) | 0.10 |
| Negative | 64 (53) | 37 (41) | |
| PIRADS score | |||
| 3 | 57 (47) | 37 (41) | 0.49 |
| 4 | 50 (41) | 38 (42) | |
| 5 | 14 (12) | 15 (17) | |
| MR prostate volume (cm3) | 51 (34 to 77) | 49 (34 to 74) | 0.88 |
| Lesion maximum size (mm) | 10 (7 to 12) | 12 (7 to 15) | 0.19 |
| Location of lesion in prostate MR | |||
| Peripheral zone | 86 (72) | 58 (65) | 0.66 |
| Transition zone | 27 (22) | 21 (24) | |
| Peripheral plus transition zone | 4 (3) | 7 (8) | |
| Central zone | 4 (3) | 3 (3) | |
| Prostate Health Index (PHI) (Score) | 39.5 (29.4 to 54.8) | 42.5 (30.0 to 53.8) | 0.06 |
| PHI density | 0.77 (0.47 to 1.31) | 0.88 (0.53 to 1.38) | 0.16 |
| Prostate specific antigen (PSA) ng/mL | 5.6 (3.3 to 9.5) | 5.6 (4.0 to 9.4) | 0.22 |
| PSA density | 0.10 (0.07 to 0.17) | 0.11 (0.08 to 0.21) | 0.12 |
| Biopsy Gleason Grade Group n (%)a | |||
| GG1 | 12 (10) | 16 (18) | <0.001 |
| GG2 | 13 (10) | 22 (24) | |
| GG3 | 6 (5) | 3 (3) | |
| GG4 | 2 (2) | 6 (7) | |
| GG5 | 0 | 2 (2) | |
Data presented as n (%) of column or median (quartile).
Gleason Grade Group (GGx) based on 2014 International Society of Urological Pathology Consensus Conference.
Table 2:
Characteristics of patient with and without prostate cancer diagnosis.
| Non-cancer (n=129) | Cancer (n=82) | P | |
|---|---|---|---|
| Age (y) | 65 (60 to 69) | 68 (61 to 72) | 0.14 |
| Race n (%) | |||
| African American | 10 (8) | 9 (11) | 0.54 |
| White | 89 (69) | 61 (74) | |
| Hispanic | 5 (4) | 1 (1) | |
| Other | 18 (14) | 8 (10) | |
| Declined to answer | 7 (5) | 3 (4) | |
| Family history n (%) | |||
| No | 65 (50) | 37 (45) | 0.73 |
| Yes | 31 (24) | 23 (28) | |
| Unknown | 33 (26) | 22 (37) | |
| Digital rectal examination findings n(%) | |||
| Normal | 30 (23) | 24 (29) | 0.20 |
| Abnormal | 76 (59) | 38 (46) | |
| Not documented | 23 (18) | 20 (25) | |
| Prior biopsy n (%) | |||
| None | 58 (45) | 52 (63) | 0.01 |
| Negative | 71 (55) | 30 (37) | |
| PIRADS score | |||
| 3 | 68 (53) | 26 (32) | <0.001 |
| 4 | 54 (42) | 34 (41) | |
| 5 | 7 (5) | 22 (27) | |
| MR prostate volume (cm3) | 53 (40 to 83) | 41 (30 to 56) | <0.001 |
| Lesion maximum size (mm) | 10 (7 to 13) | 11 (8 to 16) | 0.03 |
| Location of lesion in prostate on MR | |||
| Peripheral | 85 (66) | 59 (72) | 0.21 |
| Transition | 34 (26) | 14 (17) | |
| Peripheral plus Transition | 4 (4) | 7 (8) | |
| Central | 5 (4) | 2 (3) | |
| Prostate Health Index (PHI) (Score) | 36.5 (28.3 to 44.9) | 49.3 (38.5 to 77.2) | <0.001 |
| PHI density | 0.61 (0.41 to 1.03) | 1.26 (0.76 to 1.96) | <0.001 |
| Prostate specific antigen (PSA) ng/mL | 5.6 (3.5 to 8.6) | 5.6 (3.9 to 11.7) | 0.15 |
| PSA density (ng·ml−1/cm3) | 0.09 (0.06 to 0.14) | 0.15 (0.08 to 0.25) | <0.001 |
Data presented as median (quartiles) or n (%). Cancer diagnosis defined as GG1 or greater 1 based on 2014 International Society of Urological Pathology Consensus Conference.
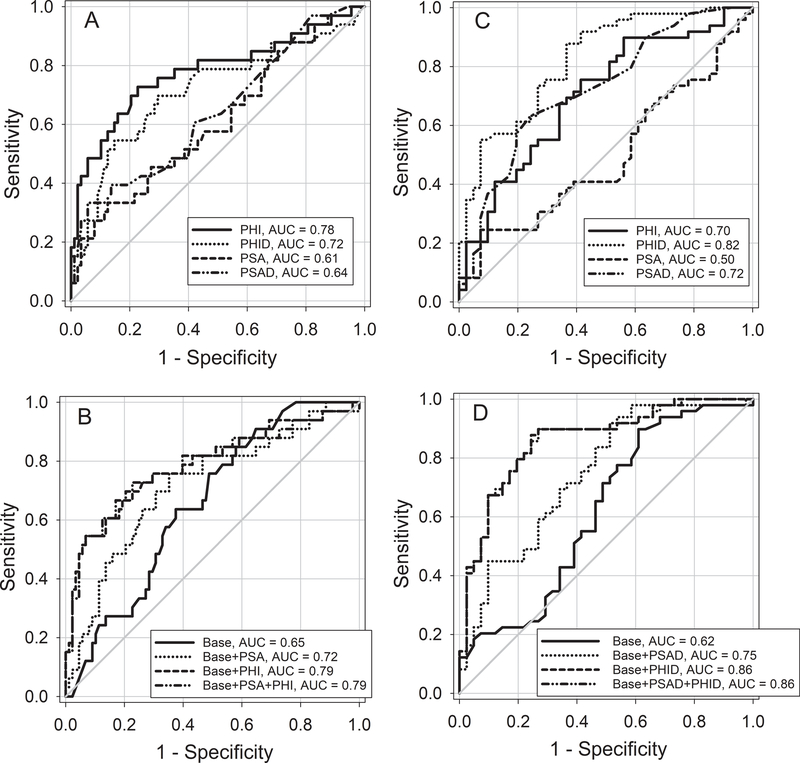
In NFUS patients, PHI had the greatest AUC for the detection of a positive biopsy (Figure 1, panel A) and was greater than PSA, difference 0.17 (95% CI of the difference 0.05 to 0.29, P=0.006), PSAD, difference 0.13 (95% CI of the difference 0.02 to 0.25, P=0.02), but not PHID, difference 0.06 (95% CI of the difference −0.04 to 0.16, P=0.05). Models developed from clinical characteristics and serum PSA and derivatives did not increase the AUC compared to the PHI (Figure 1, panel B). In FUS patients, PHID had the greatest AUC for the detection of a positive biopsy (Figure 1, panel C), and was greater than PSA, difference 0.32 (95% CI of the difference 0.17 to 0.48, P<0.001), PSAD, difference 0.09 (95% CI of the difference 0.01 to 0.18, P=0.03), and PHI, difference 0.12 (95% CI of the difference 0.02 to 0.22, P=0.02). Models developed from clinical characteristics and serum PSAD and derivatives did not increased the AUC compared to the PHID (Figure 1, panel D).
Figure 1.
Receiver-operating analysis (ROC) curves for patients with prostate biopsy. Panel A: Univariable ROC curves for PSA, PSAD, PHI and PHID with prostate biopsy using non-fused (NFUS) sampling. AUC’s and 95% CI are PSA 0.61 (0.49 to 0.72), PSAD 0.64 (0.53 to 0.75), PHI 0.78 (0.67 to 0.88), and PHID 0.72 (0.60 to 0.83).
Panel B: Multi-variable ROC curves for base model (age, DRE and prior biopsy) alone or with the addition of PSA, PHI and PSA plus PHI for NFUS sampling. AUC’s and 95% CI are Base 0.65 (0.55 to 0.75), Base + PSA 0.72 (0.68 to 0.82), Base + PHI 0.79 (0.68 to 0.89), Base + PSA + PHI 0.79 (0.68 to 0.89).
Panel C: Univariable ROC curves for PSA, PSAD, PHI and PHID with prostate biopsy sampled using fused (FUS) sampling. AUC’s and 95% CI are PSA 0.50 (0.38 to 0.62), PSAD 0.72 (0.62 to 0.83), PHI 0.70 (0.59 to 0.81), and PHID 0.82 (0.73 to 0.91).
Panel D: Multi-variable curves for base model (age, DRE and prior biopsy) alone or with the addition of PSAD, PHID and PSAD plus PHID for FUS sampling. AUC’s and 95% CI are Base 0.62 (0.50 to 0.74), Base + PSAD 0.75 (0.64 to 0.85), Base + PHID 0.86 (0.78 to 0.94) and Base + PSAD + PHID 0.86 (0.78 to 0.94).
Binomial prediction values for PHI in NFUS and PHID in FUS biopsies at cutoff values for PCa shown in table 3. In NFUS patients, a PHI cut-off value of 45.9 would reduce the number of unnecessary biopsies by 68 of 88 (77.3%) but would result in missed PCa diagnosis in 9 of 33 (27.3%) patients. The number of missed GG2 or greater biopsies would be 4 of 21 (19.0%). At a sensitivity of 95% and 100%, PHI cut-off values of 24.7 and 21.0 would reduce unnecessary biopsies by 17 (19.3%) and 4 (4.5%) of 88, and the number of missed GG2 or greater biopsies would be 1 (4.8%) and 0 (0%) of 21, respectively. In FUS patients a PHID lcut-off value of 0.64 would reduce the number of unnecessary biopsies by 26 of 41 (63.4%) but would result in missed PCa in 6 of 49 (12%) patients. The number of missed GG2 or greater biopsies would be 3 of 33 (9%). At a sensitivity of 95% and 100%, PHID cut-off values of 0.52 and 0.32 would reduce unnecessary biopsies by 19 (46.3%) and 6 (14.6%) of 41, and the number of missed GG2 or greater biopsies would be 1 (3.0%) and 0 (0%) of 33, respectively.
Table 3:
Binary test results based on cut-off determination for PHI and PHID in NFUS and FUS prostate biopsies.
| Non-Fused Biopsies | |||
|---|---|---|---|
| Cut-off based on | Youden J index | Sensitivity = 95% | Sensitivity = 100% |
| PHI cut-off value | 45.9 | 24.7 | 21.0 |
| PHI Test positive/biopsy positive | 24 (GG1=7, ≥GG2=17) | 31 (GG1=11, ≥GG2=20) | 33 (GG1=12, ≥GG2=21) |
| PHI Test negative/biopsy positive | 9 (GG1=5, ≥GG2=4) | 2 (GG1=1, ≥GG2=1) | 0 |
| PHI Test positive/biopsy negative | 20 | 71 | 84 |
| PHI Test negative/biopsy negative | 68 | 17 | 4 |
| Sensitivity (%) | 73 (54 to 87) | 94 (80 to 99) | 100 (89 to 100) |
| Specificity (%) | 77 (67 to 86) | 19 (12 to 29) | 4 (1 to 11) |
| Positive Predictive Value (%) | 54 (42 to 74) | 30 (22 to 41) | 28 (20 to 37) |
| Negative Predictive Value (%) | 88 (77 to 93) | 89 (67 to 98) | 100 (39 to 100) |
| Fused Biopsies | |||
| PHID cut-off value | 0.64 | 0.52 | 0.32 |
| PHID Test positive/biopsy positive | 43 (GG1=13, ≥GG2=30) | 47 (GG1=15, ≥GG2=32) | 49 (GG1=16, ≥GG2=33) |
| PHID Test negative/biopsy positive | 6 (GG1=3, ≥GG2=3) | 2 (GG1=1, ≥GG2=1) | 0 |
| PHID Test positive/biopsy negative | 15 | 22 | 35 |
| PHID Test negative/biopsy negative | 26 | 19 | 6 |
| Sensitivity (%) | 88 (75 to 95) | 96 (86 to 99) | 100 (93 to 100) |
| Specificity (%) | 63 (47 to 78) | 46 (31 to 63) | 15 (5 to 29) |
| Positive Predictive Value (%) | 74 (61 to 85) | 68 (56 to 79) | 58 (47 to 69) |
| Negative Predictive Value (%) | 81 (63 to 93) | 90 (70 to 99) | 100 (54 to 100) |
Data presented as counts or estimate (95% CI).
Discussion
The most important finding from our study was that PHI outperformed PSA as a diagnostic biomarker when either FUS or NFUS biopsy methods were used but that volume indexed PSAD and PHID were significantly better to use when targeted biopsies were performed. No combination of predictor variables increased the AUC’s of the NFUS or FUS biopsy samples compared to the PHI or PHID, respectively. In NFUS biopsy patients we found that in order to obtain high sensitivity (≥ 95%) for detection of GG2 or greater PCa the reduction in unnecessary biopsies would be only 19.3% and 4.5%, respectively. Whereas, in FUS biopsies 98% and 100% of GG2 PCa would be detected with a reduction in unnecessary biopsies of 46.3% and 14.6%, respectively. These findings have clinical importance since a reduction of 25% in the number of biopsies has been suggested to be a reasonable clinical goal for a serum biomarker when the number of missed diagnoses is ≥ 2% [7].
Our study also demonstrated that FUS biopsies yielded more positive PCa than NFUS biopsies. This corroborates the findings from previous studies in which MR-US fusion targeted biopsy had higher detection rates than other biopsy methods [22–27]. Regardless of method, a negative prior biopsy was more prevalent in the non-cancer group, while naïve-biopsy patients were more prevalent in the cancer group. Higher PI-RADS scores and lower volumes were also found in patients with positive biopsy. Maximum lesion size was slightly larger in cancer patients. Significant differences between the groups was found in PHI, PHID and PSAD values, with higher values found in patients with cancer. PHID had the greatest AUC for the detection of a positive biopsy.
Prior studies have examined the predictive value of PSA, PHI and volume-indexed values of the PSA and PHI and biopsy results. In African-American patients with abnormal DRE and suspicion of cancer (age and PSA), Tosoian et al found that PHI outperformed PSA alone, was associated with high grade prostate cancer and provided complementary information to MRI [18]. Not all subjects in their study had mpMRI scans or a targeted biopsy. In another study by the same group, in patients with a normal DRE, PHID had the highest discriminative ability for the diagnosis of PCa [7]. The authors did not report if all subjects had mpMRI suspicious (PI-RADS ≥ 3) lesions or the biopsy method. They estimated that at a PHID cut-off value of 0.43 unnecessary biopsies would be reduced by 38% while failing to detect 2% of cancers. In patients with a prior negative biopsy and normal DRE, Druskin et al reported that PHID outperformed PHI and other PSA derivatives in the diagnosis of clinically significant cancer [12]. In Druskin’s study, the diagnostic performance for PHID was improved by adding age, prior negative biopsy status and PI-RADS score. The method of biopsy was not specified, and the volume was measured using TRUS. Similar to our results, they found a threshold of 0.44 for PHID could avoid 35.3% biopsies but would miss 7.7% of clinically significant cancers. Friedl et al reported an overall detection rate of 55% of cancers when using targeted biopsy in patients with PI-RADS 3–5 with a prior negative biopsy [14]. In their study the optimal cutoff values for PHI and PHID were 59 and 0.79, respectively, yielding sensitivities of 69% and 84% and specificities of 82% and 62%, while avoiding 82% and 62% of unnecessary biopsies and failing to detect 31% and 16% of all cancers, respectively.
Currently, no consensus, has been reached in regard to an optimal cutoff value/threshold for PHI, PHID or PSAD, likely because of the heterogeneity of the studies performed. The National Comprehensive Cancer Network 2016 guidelines reports that for patients with multiple adverse factors should be shifted into the next highest risk group, but that a PSAD lower than 0.15 places them in the very low risk group [28]; however, using this cutoff we found that 22 of 138 (16%) of patients had clinically significant (GG2 or greater) biopsy results.
The results of our study should be interpreted only in the context of its limitations. It is a single center retrospective study and includes a relatively small number of cases limiting the ability to detect increased sensitivity in the multi-variable models. We included both naïve and repeat biopsies and the accuracy and cutoff values may not be generalizable to more homogenous populations. We used MRI determined volumes to calculate density normalized PSA derivatives rather than TRUS volumes at the time of the biopsy; however, there is an excellent concordance between these methods [29, 30]. Finally, the method of biopsy selection was by urologist preference and we were unable to determined differences in selection preference among urologists; although, the difference in positive biopsy samples using NFUS and FUS methods in this study was similar to that observed in a randomized controlled trial comparing biopsy methods [27].
Conclusion
In conclusion, our findings demonstrate that the choice of serum PSA and PSA derivatives for predicting positive cancer cores is a function of the biopsy method. Also, PSAD and PHID appear to have greater value when software-guided fusion targeted biopsy methods are used. Further studies with larger cohorts are warranted to validate these findings and to establish standardized cutoff values for these serum tests.
Acknowledgments
Disclosures: No funding or grant support was used in the preparing of this manuscript.
Footnotes
CONFLICT OF INTEREST
Manuscript #:URL-D-18–02466
Title: “The Utility of Prostate Specific Antigen Density, Prostate Health Index and Prostate Health Index Density in Predicting Positive Prostate Biopsy Outcome is Dependent on the Prostate Biopsy Methods”
Corresponding Author: Dr. Frank H Miller
Remaining Authors: Camila Lopes Vendrami, MD; Robert J McCarthy, Pharm D; Argha Chatterjee, MD; David Casalino, MD; Edward M Schaeffer, MD; William J Catalona, MD;
Submitted to: UROLOGY
Please copy this Conflict of Interest form and paste into your word processing software, type in each author and either indicate “no conflict” or specify any conflicts; it will be required that you submit the completed form (with all authors indicated) with the revised manuscript. Please list only conflicts of interest specific to this manuscript.
Examples of Conflict of Interest:
(a) Source of Funding
(b) Paid consultant to Sponsor
(c) Study Investigator Funded by Sponsor
(d) Employee of Sponsor
(e) Board Membership with Sponsor
(f) Stock Holder for Mentioned Product/Company
(g) Patent Inventor for Mentioned Product
(h) Any Financial Relationship to Competitors of Mentioned Product
(i) Other (please specify)
This information will be kept confidential. The Editor will discuss the method of disclosure of any potential conflict of interest with the corresponding author on an individual basis.
AuthorSpecify “No Conflict” or indicate specific conflict
e.g. John Doeno conflict
e.g. Jane Doepaid consultant to company “x”
Camila Lopes Vendrami- No Conflict
Robert J McCarthy- No Conflict
Argha Chatterjee- No Conflict
David Casalino- No Conflict
Edward M Schaeffer- No Conflict
William J Catalona- No Conflict
Frank H. Miller- No Conflict
I accept the responsibility for the completion of this document and attest to its validity on behalf of the co-authors.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Watson MJ, George AK, Maruf M, et al. Risk stratification of prostate cancer: integrating multiparametric MRI, nomograms and biomarkers. Future Oncol 2016; 12:2417–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberlin DT, Casalino DD, Miller FH, Meeks JJ. Dramatic increase in the utilization of multiparametric magnetic resonance imaging for detection and management of prostate cancer. Abdom Radiol (NY) 2017; 42:1255–1258 [DOI] [PubMed] [Google Scholar]
- 4.Al-Khalil S, Ibilibor C, Cammack JT, de Riese W. Association of prostate volume with incidence and aggressiveness of prostate cancer. Res Rep Urol 2016; 8:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aminsharifi A, Howard L, Wu Y, et al. Prostate Specific Antigen Density as a Predictor of Clinically Significant Prostate Cancer When the Prostate Specific Antigen is in the Diagnostic Gray Zone: Defining the Optimum Cutoff Point Stratified by Race and Body Mass Index. J Urol 2018; 200:758–766 [DOI] [PubMed] [Google Scholar]
- 6.Brassetti A, Lombardo R, Emiliozzi P, et al. Prostate-specific antigen density is a good predictor of upstaging and upgrading, according to the new grading system: the keys we are seeking may be already in our pocket. Urology 2018; 111:129–135 [DOI] [PubMed] [Google Scholar]
- 7.Tosoian JJ, Druskin SC, Andreas D, et al. Prostate health index density improves detection of clinically significant prostate cancer. BJU Int 2017; 120:793–798 [DOI] [PubMed] [Google Scholar]
- 8.Magheli A, Hinz S, Hege C, et al. Prostate specific antigen density to predict prostate cancer upgrading in a contemporary radical prostatectomy series: a single center experience. J Urol 2010; 183:126–131 [DOI] [PubMed] [Google Scholar]
- 9.Catalona WJ, Partin AW, Sanda MG, et al. A multicenter study of [−2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ml prostate specific antigen range. J Urol 2011; 185:1650–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filella X, Gimenez N. Evaluation of [−2] proPSA and prostate health index (PHI) for the detection of prostate cancer: a systematic review and meta-analysis. Clin Chem Lab Med 2013; 51:729–739 [DOI] [PubMed] [Google Scholar]
- 11.Loeb S, Sanda MG, Broyles DL, et al. The prostate health index selectively identifies clinically significant prostate cancer. J Urol 2015; 193:1163–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Druskin SC, Tosoian JJ, Young A, et al. Combining prostate health index density, magnetic resonance imaging and prior negative biopsy status to improve the detection of clinically significant prostate cancer. BJU Int 2018; 121:619–626 [DOI] [PubMed] [Google Scholar]
- 13.Sarkar S, Verma S. MR imaging-targeted prostate biopsies. Radiol Clin North Am 2018; 56:289–300 [DOI] [PubMed] [Google Scholar]
- 14.Friedl A, Stangl K, Bauer W, et al. Prostate-specific antigen parameters and prostate health index enhance prostate cancer prediction with the in-bore 3-T magnetic resonance imaging-guided transrectal targeted prostate biopsy after negative 12-core biopsy. Urology 2017; 110:148–153 [DOI] [PubMed] [Google Scholar]
- 15.Porpiglia F, Cantiello F, De Luca S, et al. In-parallel comparative evaluation between multiparametric magnetic resonance imaging, prostate cancer antigen 3 and the prostate health index in predicting pathologically confirmed significant prostate cancer in men eligible for active surveillance. BJU Int 2016; 118:527–534 [DOI] [PubMed] [Google Scholar]
- 16.Porpiglia F, Russo F, Manfredi M, et al. The roles of multiparametric magnetic resonance imaging, PCA3 and prostate health index-which is the best predictor of prostate cancer after a negative biopsy? J Urol 2014; 192:60–66 [DOI] [PubMed] [Google Scholar]
- 17.Tan TW, Png KS, Lee CH, et al. MRI fusion-targeted transrectal prostate biopsy and the role of prostate-specific antigen density and prostate health index for the detection of clinically significant prostate cancer in southeast asian men. J Endourol 2017; 31:1111–1116 [DOI] [PubMed] [Google Scholar]
- 18.Tosoian JJ, Druskin SC, Andreas D, et al. Use of the prostate health index for detection of prostate cancer: results from a large academic practice. Prostate Cancer Prostatic Dis 2017; 20:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuya K, Kawahara T, Narahara M, et al. Measurement of serum isoform [−2]proPSA derivatives shows superior accuracy to magnetic resonance imaging in the diagnosis of prostate cancer in patients with a total prostate-specific antigen level of 2–10 ng/ml. Scand J Urol 2017; 51:251–257 [DOI] [PubMed] [Google Scholar]
- 20.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate imaging - reporting and data system: 2015, Version 2. Eur Urol 2016; 69:16–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol 2016; 40:244–252 [DOI] [PubMed] [Google Scholar]
- 22.Oberlin DT, Casalino DD, Miller FH, et al. Diagnostic value of guided biopsies: fusion and cognitive-registration magnetic resonance imaging versus conventional ultrasound biopsy of the prostate. Urology 2016; 92:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wysock JS, Rosenkrantz AB, Huang WC, et al. A prospective, blinded comparison of magnetic resonance (MR) imaging-ultrasound fusion and visual estimation in the performance of MR-targeted prostate biopsy: the PROFUS trial. Eur Urol 2014; 66:343–351 [DOI] [PubMed] [Google Scholar]
- 24.Delongchamps NB, Escourou C, Cornud F. Integrated US-MR fusion images and MR targeted biopsies. What are their role and value in the detection and follow-up of prostate cancer. Arch Esp Urol 2015; 68:349–353 [PubMed] [Google Scholar]
- 25.Wegelin O, van Melick HHE, Hooft L, et al. Comparing three different techniques for magnetic resonance imaging-targeted prostate biopsies: a systematic review of in-bore versus magnetic resonance imaging-transrectal ultrasound fusion versus cognitive registration. is there a preferred technique? Eur Urol 2017; 71:517–531 [DOI] [PubMed] [Google Scholar]
- 26.Verma S, Choyke PL, Eberhardt SC, et al. The current state of MR imaging-targeted biopsy techniques for detection of prostate cancer. Radiology 2017; 285:343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasivisvanathan V, Rannikko AS, Borghi M, et al. MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 2018; 378:1767–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohler JL, Armstrong AJ, Bahnson RR, et al. Prostate cancer, version 1.2016. J Natl Compr Canc Netw 2016; 14:19–30 [DOI] [PubMed] [Google Scholar]
- 29.Weiss BE, Wein AJ, Malkowicz SB, Guzzo TJ. Comparison of prostate volume measured by transrectal ultrasound and magnetic resonance imaging: is transrectal ultrasound suitable to determine which patients should undergo active surveillance? Urol Oncol 2013; 31:1436–1440 [DOI] [PubMed] [Google Scholar]
- 30.Tewari A, Indudhara R, Shinohara K, et al. Comparison of transrectal ultrasound prostatic volume estimation with magnetic resonance imaging volume estimation and surgical specimen weight in patients with benign prostatic hyperplasia. J Clin Ultrasound 1996; 24:169–174 [DOI] [PubMed] [Google Scholar]