SUMMARY
Endoplasmic reticulum (ER) stress is a pathological hallmark of numerous ischemic diseases including stroke and myocardial infarction. In these diseases, ER stress leads to activation of the unfolded protein response (UPR) and subsequent adaptation of cellular physiology in ways that dictate cellular fate following ischemia. Recent evidence highlights a protective role for the ATF6 arm of the UPR in mitigating adverse outcomes associated with ischemia/reperfusion injury in multiple disease models. This suggests that ATF6 represents a potential therapeutic target for intervening in diverse ischemia-related disorders. Here, we discuss the evidence demonstrating the importance of ATF6 signaling in protecting different tissues against ischemic damage and discuss preclinical results focused on defining the potential for pharmacologically targeting ATF6 to intervene in such diseases.
Keywords: unfolded protein response, ER stress, ATF6, stroke, myocardial infarction
Endoplasmic Reticulum (ER) Stress, the Unfolded Protein Response, and Ischemic Disease
Many diseases lead to impaired circulation, which can cause ischemic conditions in a variety of organs including the brain, heart and kidney [1]. In these settings, prolonged ischemia causes irreversible damage, which can be partially mitigated by clinical interventions that restore blood flow through reperfusion [2]. While reperfusion is necessary to mitigate the damage of continued ischemia, reperfusion itself is known to cause some additional damage, generally as a result of reactive oxygen species (ROS; see Glossary) formation [3]. The complex nature of cellular injury associated with ischemia or ischemia followed by reperfusion (I/R) has previously been shown to affect the levels and activities of numerous signaling pathways and transcription factors, including protein kinase C, protein kinase B (Akt), nitric oxide synthase, glycogen synthase kinase 3b, Nrf2, and HIF-1α [4]. One I/R-activated pathway that has been more recently studied involves the disruption of endoplasmic reticulum (ER) protein homeostasis (or proteostasis), otherwise known as ER stress [3, 5, 6]. Pathologic ER stress is associated with numerous adverse outcomes including impaired Ca2+ homeostasis, disruptions in secretory proteostasis, impaired lipid metabolism, and increased apoptotic signaling [3, 5–7]. To protect against pathological ER stress induced by I/R, tissues activate endogenous adaptive stress-responsive signaling pathways, such as the unfolded protein response (UPR).
The UPR is the primary stress-responsive signaling pathway activated in response to ER stress. In metazoans, the UPR is comprised of three stress-signaling arms activated downstream of the ER stress sensing membrane proteins PERK, IRE1, and ATF6 [8–10]. In response to ER stress, these sensors initiate well-defined signal transduction pathways that lead to translational and transcriptional remodeling of stress-responsive biologic pathways involved in diverse activities including ER proteostasis maintenance, cellular redox maintenance, and metabolite homeostasis [8–10]. The activation of these pathways is a protective mechanism to alleviate ER stress and promote cellular adaptation following an acute insult. However, severe or chronic ER stress can initiate pro-apoptotic UPR signaling, primarily as a result of activating the PERK UPR signaling arm [9, 11]. Thus, the UPR has a central role in dictating cellular physiology and fate in response to diverse types of ER insults.
All three arms of the UPR are activated in tissue-specific I/R rodent models and in post-mortem samples from humans suffering from ischemic diseases including myocardial infarction (MI) and stroke (reviewed in [4, 5]). Genetic dissection of these three UPR pathways indicates that PERK, IRE1, and ATF6 activation distinctly influence tissue physiology in response to I/R. Signaling through the PERK pathway has been proposed to promote apoptosis during I/R through mechanisms including transcriptional upregulation of pro-apoptotic factors such as CHOP [5, 6, 12]. Alternatively, signaling through the IRE1 and ATF6 pathways has been shown to protect against I/R in diverse tissues including the heart and brain. The protection afforded by IRE1 during I/R is discussed briefly in Box 1. For the remainder of this review, we specifically focus on recent genetic and pharmacologic evidence highlighting the protective benefits of ATF6 activation in ischemic disease.
Box 1. IRE1/XBP1s signaling is protective against I/R and related disorders.
When activated by ER stress, IRE1 leads to the generation of the active form of the transcription factor XBP1, referred to as spliced XBP1 or XBP1s, through a mechanism involving the IRE1-dependent regulated splicing of XBP1 mRNA [8–10, 78]. XBP1s has been shown to be protective in a number of disease models, including ischemia and I/R injury in cancer, as well as in the heart, kidney and brain [5, 11, 79–81]. While this protection is likely to be afforded through a multitude of pathways, depending on the model system, several common themes have emerged linking the XBP1s gene program to protection from ischemia and I/R, including the induction of protective chaperones, protein O-GlycNACylation, and autophagy [5, 12, 82–85]. For example, it was shown that key genes that regulate the synthesis of hexosamines, which are precursors used in the process of protein O-GlycNACylation, are direct targets of XBP1s [85]. In that study, genetic deletion of XBP1 increased I/R damage in the heart, which could be reduced by overexpression of hexosamine biosynthetic enzymes, indicating that XBP1s-dependent regulation of the hexosamine pathway is protective during I/R. XBP1s has also been shown to regulate angiogenesis in response to ischemia through a novel mechanism. In response to ischemia, XBP1s in vascular smooth muscle cells regulates the release of key microRNAs that interact with nearby endothelial cells enhancing VEGF signaling, endothelial cell migration and new blood vessel formation [86]. Thus, the IRE/XBP1s UPR signaling arm exerts broad effects on cellular processes well beyond its canonical role as a regulator of ER proteostasis that appear protective against ischemia and I/R. This suggests that these protective aspects of IRE1/XBP1s signaling could potentially be pharmacologically accessed to intervene in ischemia-related diseases using similar approaches to those described for ATF6 in this review.
The ATF6 Signaling Arm of the UPR
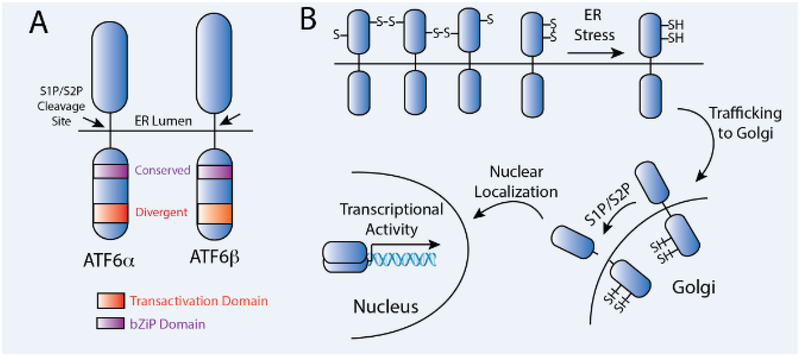
ATF6 is a type II ER transmembrane protein that consists of three functional domains: an N-terminal bZIP transcription factor domain, a transmembrane domain, and a luminal domain (Fig. 1A). Metazoans have two different isoforms of ATF6, ATF6α and ATF6β, which encode proteins with similar domain architectures [13–15](Fig. 1A). In the context of ER stress and the UPR, ATF6α is the predominant ATF6 isoform responsible for regulating the expression of ER stress responsive genes [16]. In contrast, the functional implications of ATF6β activity remain poorly defined, although it has been proposed that ATF6β can act as an endogenous repressor of ATF6α and serves to fine tune the strength and duration of ATF6α signaling during ER stress [17–20]. Genetic ablation of either Atf6α or Atf6β does not overtly impact prenatal development in mice, although genetic ablation of both ATF6 isoforms is embryonic lethal [16]. This indicates that ATF6α and ATF6β complement each other to exert functions that are required for organismal development. Since ATF6α is the predominant isoform responsible for regulating cellular physiology in response to ER stress, we primarily focus this review on the implications of signaling through the ATF6α isoform (herein referred to as ATF6 unless otherwise indicated) in ischemic diseases.
Figure 1. ATF6 is activated in response to ER stress through a process involving regulated proteolysis.
A. Illustration showing the domain organization of ATF6α and ATF6β highlighting the conserved and divergent features of these two ATF6 genes.
B. Illustration showing the mechanism of ATF6 activation during ER stress. In the absence of ER stress, ATF6 is retained within the ER in disulfide bond-linked oligomers. In response to ER stress, these disulfides are reduced to yield reduced ATF6 monomers that traffic to the Golgi where they are proteolyzed by the site 1 (S1P) and site 2 (S2P) proteases. This proteolysis releases the cytosolic, N-terminal ATF6 transcription factor domain that localizes to the nucleus, homo- or hetero-dimerizes, and promotes ATF6-regulated transcriptional activity.
Of the three arms of the UPR, the mechanism for ER stress-dependent ATF6 activation remains the least well understood. In response to ER stress, ATF6 is trafficked from the ER to the Golgi where it is cleaved by site 1 and site 2 proteases at specific sites near or within the transmembrane domain (Fig. 1B) [21, 22]. This proteolysis liberates the cytosolic N-terminal domain of ATF6, allowing it to localize to the nucleus where it acts as a transcription factor to regulate expression of stress-responsive genes (e.g., BiP) that constitute the ATF6 transcriptional program [21, 22]. The details of the key step in this activation mechanism, the regulated trafficking from the ER to the Golgi, are currently poorly defined, but appear to involve regulation of both ATF6 redox status and oligomerization. In the absence of ER stress, ATF6 is retained within the ER as disulfide-linked oligomers bound to the ER chaperone BiP, a configuration that favors ER localization and prevents trafficking to the Golgi [23, 24]. In response to ER stress, these disulfide bonds are reduced, facilitating the dissociation of ATF6 oligomers to reduced monomers that can traffic to the Golgi and undergo proteolytic processing by Site 1 and Site 2 proteases (S1P and S2P, respectively) [23, 24]. Although the molecular factors responsible for regulating ATF6 redox status in the absence or presence of ER stress are not well defined, protein disulfide isomerases (PDIs), such as PDIA5, have been implicated in the ER stress-dependent reduction of ATF6 disulfides [25, 26], suggesting a critical role for PDIs for regulating ATF6 trafficking. While the reduction and subsequent dissociation of ATF6 disulfide bonds is required for both trafficking to the Golgi and proteolytic activation, genetic disruption of ATF6 disulfide bonds alone was not sufficient to promote complete ATF6 activation [23], indicating that other mechanisms are also involved in dictating activation of this UPR signaling pathway. For example, other molecular events including dissociation of the ER chaperone BiP from the ATF6 luminal domain [27, 28] or alterations in ATF6 N-linked glycosylation [29] have been implicated in ER stress-dependent ATF6 activation and likely contribute to the regulation of ATF6 activation during ER stress. However, how, and if, these other events integrate with the regulation of ATF6 disulfides remains an open question.
Upon proteolytic activation, the cleaved version of ATF6 homodimerizes and binds to promoter regions of target genes containing canonical ATF6 binding motifs such as the ER stress element (ERSE) [30]. The ATF6 transcriptional program has been elucidated using multiple genetic and chemical genetic approaches, all of which demonstrate that this pathway has a central role in regulating the composition of biological pathways involved in diverse functions including ER proteostasis maintenance, protein degradation, and cellular redox regulation [30–33]. Unlike the other arms of the UPR regulated by PERK and IRE1, the ATF6 arm has not been substantially linked to pro-apoptotic signaling [9, 11]. Instead, ATF6 primarily functions in the so-called adaptive UPR designed to promote protective, adaptive remodeling of cellular physiology and recovery following acute physiologic and pathologic insults. As part of the adaptive UPR, ATF6 integrates with multiple other stress-responsive signaling pathways to sensitively adapt cellular physiology to diverse types of ER insults. In the context of the UPR, this integration can be achieved through heterodimerization of ATF6 with other UPR-regulated bZIP transcription factors such as XBP1s (a bZIP transcription factor activated downstream of IRE1) or ATF6β. ATF6/XBP1s heterodimers promote expression of select subsets of genes involved in biologic functions such as ER-associated degradation (ERAD) [16, 31]. Alternatively, as mentioned above, heterodimerization of ATF6α and ATF6β bZIP transcription factors provides a mechanism to ‘fine tune’ the extent and timing of the ATF6 stress response [17–20]. ATF6 also has the potential to heterodimerize with other bZIP transcription factors similarly regulated through a mechanism involving S1P/S2P-dependent proteolysis such as CREB-H[34, 35], providing additional mechanisms to sensitively adapt ATF6 signaling during ER stress. Apart from heterodimerization, ATF6 signaling also integrates with other stress-responsive signaling pathways such as PGC1α-dependent mitochondrial biogenesis and mTOR signaling (see Box 2). The capacity for ATF6 to integrate with other signaling pathways through multiple mechanisms reflects a unique potential for this UPR signaling arm to coordinate protective cellular responses, in addition to ER proteostasis remodeling, to a wide range of pathologic insults that induce ER stress, including I/R.
Box 2. ATF6 activity integrates with diverse signaling pathways to coordinate cellular physiology during stress.
Recent studies have revealed that in addition to its roles as a regulator of genes that fortify ER proteostasis, ATF6 influences the expression of genes that affect cellular physiology more broadly. This comes in part through integration with other stress-responsive signaling pathways. For example, ATF6 has been found to induce genes related to growth of skeletal [87] and cardiac muscle [88], the latter of which has been attributed to ATF6-specific induction of mTOR-activating proteins, which coordinates upregulation of ER protein folding resources with increased protein folding demand during growth. The ATF6 gene program in a given cell or tissue type differs depending upon the stimulus. For example, oxidative stress leads to ATF6-dependent induction of antioxidant genes, but not growth promoting genes, while growth stimuli lead to induction of growth promoting genes but not antioxidant genes [88]. Additionally, lipotoxic stress activates lipid metabolism genes but not ER proteostasis genes, while proteotoxic stress induces proteostasis genes, but not lipid metabolism genes [89]. Such differential ATF6 target gene induction by treatments that all activate ATF6 suggests that there are yet-to-be-described regulatory layers that fine-tune the ATF6 gene program to best adapt to specific conditions. Some possible mechanisms that could contribute to this differential expression are beginning to emerge, as it has been shown that ATF6 can functionally interact with other transcription factors, such as NRF1, PGC1α, PPARα and ERRγ [66, 87, 90, 91], which changes the transcriptional programming in ways that sensitively adapt ATF6-dependent cellular responses to diverse types of pathologic insults.
ATF6 Protects against I/R Damage in Multiple Tissues
Consistent with a protective role for ATF6 in regulating cellular physiology during pathologic insults, ATF6 protects against I/R-associated damage in numerous tissues. One of the first examples of this protection was demonstrated using a chemical genetic strategy to regulate the activity of ATF6 in the heart. This strategy employed a transgenic mouse line that expresses a tamoxifen-activated form of ATF6; administration of tamoxifen to these transgenic mice induced robust activation of ATF6 in the heart, evidenced by increased expression of multiple ATF6 target genes [32]. Tamoxifen-dependent ATF6 activation in vivo significantly improved the recovery of cardiac contractility following ex vivo I/R and reduced markers of damage, such as infarct size and apoptotic signaling in response to in vivo I/R [32]. Consistent with these findings, inhibiting ATF6 activation using S1P inhibitors or a dominant negative ATF6 increased cardiac damage in a mouse model of myocardium infarction (MI), reduced cardiac function, and reduced 14 day survival following MI [36]. The importance of ATF6 in protecting the heart during I/R was further confirmed using whole body and cardiac myocyte-specific Atf6−/− mice, both of which showed increased sensitivity to cardiac I/R damage both ex vivo and in vivo [37, 38]. In both cases, exogenous overexpression of ATF6 ameliorated this cardiac sensitivity to I/R damage and improved cardiac function, both in wild-type and Atf6−/− mice [36–38]. Apart from cardiac ischemia, Atf6-deletion also sensitized the brain to middle cerebral artery occlusion (MCAO) models of ischemic stroke [37–39], and ATF6 overexpression in the brain ameliorated neurologic defects associated with this model [40]. These results indicate that ATF6 can protect multiple tissues against ischemic insults.
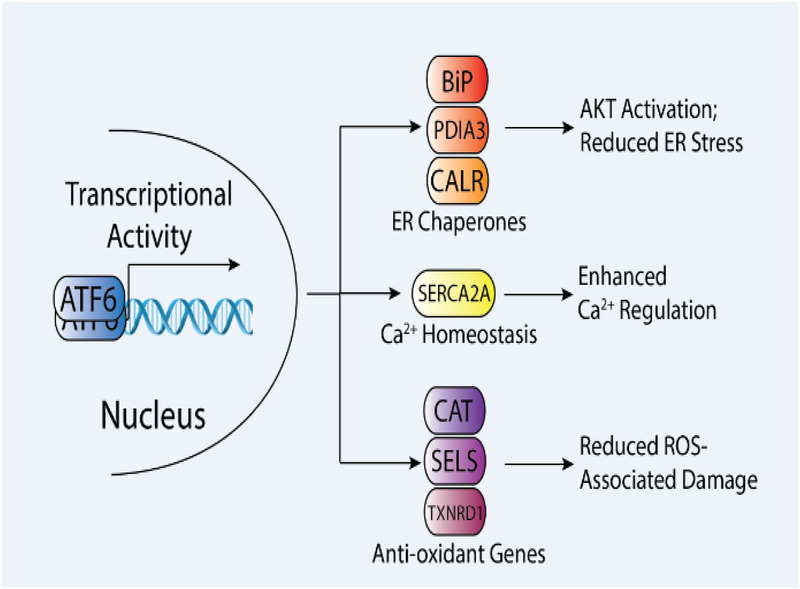
The molecular mechanism by which ATF6 protects against I/R damage in different tissues appears to be tightly linked to ATF6-reguated stress-responsive genes involved in diverse biologic functions (Fig. 2). ATF6-dependent increases in ER chaperones, such as the ATP-dependent chaperone BiP are likely implicated in ATF6 protection against I/R in tissues such as the heart. I/R-dependent increases in BiP are blocked in Atf6-deficient mice, confirming the specific regulation of this chaperone by ATF6 [37, 38]. Mimicking ATF6-dependent increases in BiP by overexpression of this chaperone reduced ROS-associated damage and improved heart function in mice subjected to cardiac I/R [41]. Similarly, BiP has been shown to be protective in both in vitro and in vivo mouse models of ischemic stroke [42–44]. The specific mechanism by which BiP protects against I/R-associated damage remains to be fully defined. However, cellular models of simulated I/R using neonatal rat ventricular myocytes (NRVM) indicate that the protection afforded by BiP overexpression results from activation of AKT – a central kinase involved in regulating cell survival during I/R [45–48] – potentially through a mechanism involving increased localization of BiP to the cell surface [41]. Alternatively, BiP overexpression in astrocytes improves Ca2+ regulation and mitochondrial function in cell culture models of ischemia, suggesting that BiP could protect cells against I/R through mitochondrial regulation [44]. Increases in BiP also likely protect by improving ER proteostasis and attenuating ER stress-associated proapoptotic signaling through the UPR. Consistent with this, overexpression of other ATF6-regulated ER chaperones and folding enzymes including GRP94, PDIA3, and PDIA6, also have been shown to protect cells and tissues against I/R associated injuries [49–51]. Despite the evidence indicating that these chaperones are protective against I/R, the specific dependence of ATF6-mediated protection on the increased expression of these different chaperones and folding enzymes remains to be determined.
Figure 2.
Molecular mechanisms by which ATF6 activation protect tissues against ischemia and ischemia/reperfusion (I/R). Illustration showing three types of protective, stress-responsive genes induced by ATF6 including endoplasmic reticulum (ER) chaperones (e.g., BiP, PDIA3, CALR) and Ca2+ pumps (SERCA2A), which are involved in canonical ER proteostasis pathways, and anti-oxidant genes (e.g., Cat, SELS, TXNRD1), which are examples of non-canonical targets of ATF6. The specific impact of these different genes on cellular physiology during I/R is highlighted.
ATF6 also transcriptionally induces the expression of the Ca2+ pump SERCA2a, which is involved in regulating intracellular cytosolic contractile Ca2+ in the heart (Fig. 2)[52]. Increased expression of SERCA2a afforded by ATF6 could mitigate dysregulation of Ca2+ levels in the cytosol, which are associated with adverse outcomes during cardiac I/R [53]. Consistent with this, SERCA2a overexpression improved myocardial function, decreased apoptotic signaling, and reduced damage in numerous models of cardiac I/R [54, 55]. However, like the ER chaperones, the specific contributions of ATF6-dependent increases in SERCA2a on the protection afforded by ATF6 have not been defined.
Interestingly, recent evidence suggested that ATF6 could attenuate I/R injury by directly decreasing ROS. Transcriptional profiling of ventricles from mice overexpressing the active ATF6 transcription factor in the heart showed that ATF6 induced expression of multiple anti-oxidant genes that encode proteins residing outside of the ER, such as catalase (Cat), SelS, and thioredoxin reductase 1 (Txnrd1) (Fig. 2)[37]. The increased expression of these anti-oxidant genes could explain the reduction in ROS and decreased ROS-associated I/R damage afforded by ATF6 activation [37]. Consistent with this, overexpression of one of the anti-oxidant genes, catalase, rescued the impaired cardiac function observed in Atf6-deficient hearts subjected to I/R [37], indicating that ATF6 activation protects against cardiac I/R, at least partly through increased anti-oxidant gene expression.
The finding that ATF6 regulates the expression of multiple genes whose overexpression protects against I/R damage in preclinical models indicates that ATF6 likely confers protection through multiple complementary mechanisms. This suggests that pharmacologically activating ATF6 could provide multiple avenues for improving tissue outcomes in diverse models of I/R damage. However, the development of this approach hinges on both the safety of targeting ATF6 for disease intervention and the potential for developing compounds that selectively activate ATF6 signaling in affected tissues with limited off-target effects. We address these two specific points below in the following sections.
ATF6 as a Therapeutic Target for Human Disease
Recent human genetic data highlights the therapeutic potential for targeting ATF6 to intervene in diseases such as ischemic disorders. Polymorphisms within ATF6α have been implicated in altered glucose homeostasis and plasma cholesterol levels in certain human populations, although the specific dependence of these altered phenotypes on ATF6 mutations remain to be established [56–59]. More recently, three independent groups identified mutations in ATF6α that are clinically associated with the congenital retinal disease achromatopsia [60–62]. Achromatopsia is an autosomal recessive blinding disease caused by the selective dysfunction of cone photoreceptors. Disease-relevant ATF6α mutations were identified by next generation whole-exome sequencing and shown to cause missense, non-sense, splice site, and single nucleotide perturbations throughout the entire ATF6 coding region [60–62]. These mutations can be clustered into three distinct classes based on their functional impact on ATF6 activity [63]. Class 1 and 3 ATF6α mutations localize to either the ATF6 luminal domain or the DNA binding domain. In both cases, these mutations attenuate ATF6 activation in response to ER stress by either disrupting ER-to-Golgi trafficking or directly impairing transcriptional activity of the active ATF6 transcription factor, respectively [63]. However, class 2 ATF6α mutations introduce a premature stop codon at the end of the ATF6 transcription factor domain. This leads to the production of the active ATF6 transcription factor independent of the transmembrane and luminal regulatory domains. Patients expressing these class 2 mutations have moderate levels of constitutive ATF6 activity [63]. This lower level of ATF6 activation likely reflects nonsense mediated decay of the mutant ATF6α mRNA, although this remains to be formally tested [63]. Despite presenting with achromatopsia – a developmental disorder specific to the retina – patients expressing class 2 ATF6α mutations have not been reported to present with systemic or neurologic defects, even though ATF6 is constitutively active in every tissue. Similarly, patients expressing class 1 and 3 ATF6α mutations do not present with any severe systemic or neurologic defect. Although the number of achromatopsia patients expressing constitutively-active ATF6α mutants are low, these results suggest that alterations in ATF6 signaling could be well-tolerated in adult humans.
These human genetic results are consistent with significant evidence from animal models showing that alterations in ATF6 activity do not induce global organismal defects. In mice, deletion of ATF6α does not impair organismal development [16], although ATF6α-deficient mice do show progressive rod and cone dysfunction in the eye as they age [60]. Apart from the eyes, ATF6α-deficient mice do not show defects in aging, although they are more sensitive to ER stress-associated insults [16, 64]. There are also numerous reports that overexpressing constitutively-active or ligand-regulated ATF6 in diverse tissues of model organisms increases signaling through the ATF6 arm of the UPR to physiologic levels, but does not induce global organismal toxicity. For example, while high levels of ATF6 activity in zebrafish can induce adverse outcomes such as hepatosteatosis, lower, physiologically-relevant levels of ATF6 activity do not [65]. Furthermore, increasing ATF6 activity is not generally associated with adverse outcomes in mouse tissues such as the liver, heart, brain, or kidney [32, 37, 38, 40, 66, 67]. Instead, increasing the activity of ATF6 in these tissues is often associated with protection (see above). These results, in combination with the genetic evidence highlighting the potential for ATF6 to ameliorate I/R related disorders in preclinical models likely through multiple mechanisms (see previous section), suggests ATF6 as a potential target to intervene in ischemic diseases.
Pharmacologic Targeting of ATF6
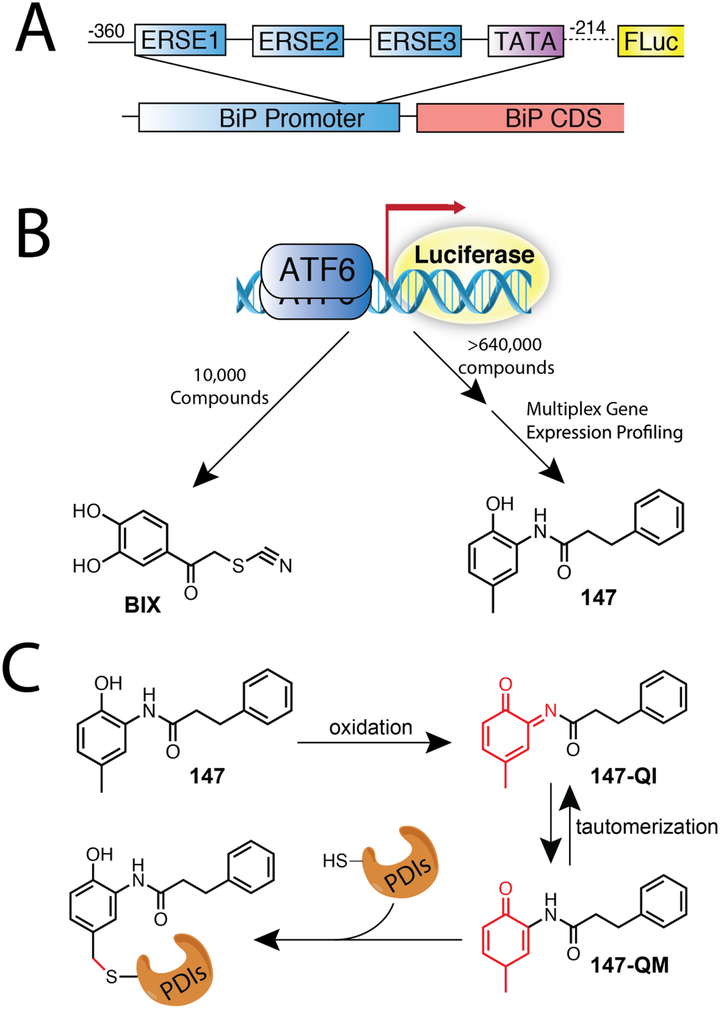
ATF6 is a very challenging target protein due to a lack of knowledge concerning its activation mechanism and the absence of known small molecule binding sites on ATF6 that can be pharmacologically targeted. As such, strategies to identify pharmacologic activators of ATF6 have largely relied on phenotypic assays using cell-based reporters. One widely-used reporter of ATF6 activity monitors expression of firefly luciferase expressed downstream of a promoter region isolated from the well-studied ATF6 target gene BiP (Fig. 3A). The first effort to identify an ATF6 activating compound used this reporter in a high throughput screen (HTS) of a ~10,000 compound library to identify those that increased luciferase expression [68] (Fig. 3B, left). This HTS identified a small molecule called BiP inducer X (BiX), which was shown to increase expression of the ATF6 target gene BiP in multiple cell models and in vivo in tissues, such as the brain and kidney without increasing the expression of genes regulated by the IRE1 or PERK arms of the UPR [68–71]. BiX-dependent induction of BiP was ablated in cells deficient in both ATF6α and ATF6β [68]. This demonstrated that BiX induces BiP through an ATF6-dependent mechanism, although the mechanistic significance of BiX-dependent ATF6 activation on these two ATF6 isoforms remains to be defined. Despite the dependence on ATF6, administration of BiX did not significantly induce expression of other well studied ATF6 target genes, such as GRP94, indicating that this compound is selective for ATF6-dependent induction of BiP and does not robustly activate the entire ATF6 gene program [68]. While the mechanism by which BiX preferentially induces ATF6-dependent expression of BiP remains to be delineated, the identification of this compound provided the first indication that ATF6 could be pharmacologically targeted, motivating future efforts to identify compounds with improved potential for global activation of the ATF6 transcriptional program.
Fig. 3. Screening strategies to identify pharmacologic ATF6 activators.
A. Illustration showing the ATF6-selective luciferase reporter used to identify compounds that preferentially activate the ATF6 UPR signaling pathways. This reporter was prepared using a promoter fragment from the ATF6-regulated gene BiP including 3 ER stress element (ERSE) binding sites.
B. Illustration showing the strategies employed to identify the ATF6-activating compounds BiX (left; [68]) and 147 (right; [26]).
C. Illustration showing the mechanism by which compound 147-selectively activates ATF6. Compound 147 is metabolically activated by oxidases to yield a reactive quinone-imine (QI) and/or quinone-methide (QM). These electrophilic compounds selectively label free cysteines within ER PDIs involved in regulating ATF6 redox status. This modification increases ER populations of reduced, monomeric ATF6 that can traffic to the Golgi for proteolytic activation (see Fig. 1B). This panel is adapted from [73].
A more recent HTS of >640,000 compounds that used the same luciferase reporter employed to identify BiX identified numerous new compounds found to selectively, and more globally, activate the ATF6 arm of the UPR (Fig. 3B, right) [26]. An important addition employed in this screening strategy was the incorporation of multiplexed transcriptional profiling as part of the screening pipeline to increase the potential for identifying compounds that globally activate the ATF6 transcriptional program. Prioritized hits from this screen included compound 147, which was further shown using both RNAseq transcriptional profiling and whole cell quantitative proteomics to preferentially activate the ATF6 transcriptional program independent of other arms of the UPR or other stress-responsive signaling pathways [26]. Compound 147 selectively activates the ATF6 arm of the UPR in multiple cell culture models and embryonic stem cells [26, 38, 72]. Furthermore, intravenous (IV) injection of compound 147 in mice selectively induced ATF6 target genes, including BiP and CAT in multiple tissues such as the heart, liver, kidney, and brain [38] – the latter indicating that this compound can cross the blood brain barrier. Importantly, genes that are downstream of other UPR signaling arms were not activated in mice injected with compound 147 [38]. The maximal level of ATF6 activation that can be achieved by compound 147 is ~40% that observed for the chemical ER stressor thapsigargin (Tg) [26, 72]. This 40% level of activation is similar to the level of ATF6 activation observed in achromatopsia patients expressing constitutively-active class 2 ATF6 mutants [63], suggesting that 147-dependent ATF6 activation to these levels will not induce systemic or neurologic defects in adults. Consistent with this, administration of compound 147 to mice for two weeks did not show any adverse outcomes in the heart, brain, kidney, or liver [38], reflecting the potential safety of this compound in mammals.
Genetic disruption of ATF6, or pharmacologic inhibition of site 1 protease, blocked 147-dependent activation of ATF6 target genes [26], indicating that this compound activates ATF6 through a mechanism involving the canonical ATF6 activation mechanism. Using a combination of medicinal chemistry and genetics, it was shown that compound 147 promoted ATF6 activation through a mechanism involving both metabolic activation and selective modification of a subset of ER protein disulfide isomerases (PDIs) [73] (Fig. 3C). This selective modification of PDIs appeared to disrupt PDI-dependent maintenance of ATF6 disulfide bonds within the ER luminal domain, which increases the levels of reduced ATF6 monomers that can traffic to the Golgi (Fig. 1B). Increasing populations of reduced ATF6 monomers, on its own, are not predicted to fully activate signaling through this pathway [23]. However, increased populations of reduced ATF6 monomers could sensitize this pathway to homeostatic changes in the ER to allow low levels of activation, providing a potential mechanism to explain the moderate ATF6 activation observed upon treatment with compound 147. While the complete mechanism by which compound 147 activates ATF6 remains to be fully established, these data indicate that it involves the disruption of PDI-dependent maintenance of ATF6 disulfide bonds. Importantly, despite the fact that compound 147 appears to influence ATF6 activation by targeting PDI activity, administration of this compound did not impact the secretion of wild-type proteins or the endogenous secreted proteome [26], indicating that 147-dependent alterations in PDI activity do not cause a global impairment of secretory proteostasis.
Apart from ATF6 activating compounds, highly-selective ATF6 inhibitors have also recently been reported. These compounds, referred to as ceapins, were identified through a HTS screen focused on identifying compounds that prevented the ER stress-dependent activation of an ATF6-selective luciferase reporter similar to that shown in Fig. 3A [74]. These compounds inhibited ATF6 activation by stabilizing ATF6 oligomers in the ER, preventing the oligomer dissociation required for ATF6 trafficking to the Golgi [74, 75]. Importantly, these compounds did not influence the regulated trafficking and proteolysis of other proteins regulated through a mechanism similar to ATF6 such as ATF6β or SREBP [74], highlighting the selectivity of ceapins for ATF6α. While the specific underlying molecular mechanisms by which ceapins stabilize ATF6 oligomers within the ER remains to be elucidated, it is clear that these compounds block ATF6 activation at a very early step in its activation mechanism. Consistent with this, administration of an active ceapin compound blocked 147-dependent ATF6 activation [73]. While genetic evidence indicates that inhibiting ATF6 signaling will be detrimental for ischemic disease, these compounds provide a powerful, highly-selective tool to pharmacologically inhibit ATF6 signaling that can be used to further define the specific involvement of ATF6 signaling at various times after I/R – a clinically relevant issue that had only been previously accessible using inhibitors of S1P that disrupt signaling through multiple other stress-responsive transcription factors that employ a similar regulated proteolysis activation mechanism such as SREBP [21].
Ameliorating I/R Injury in vivo Through Pharmacologic ATF6 Activation
The establishment of compounds that selectively activate ATF6 (e.g., BiX and 147) provides a unique opportunity to both further define the importance of ATF6 activation in protecting tissues against I/R and to define the potential for pharmacologic ATF6 activation to ameliorate pathologic phenotypes in preclinical models of ischemic disease. BiX protected neuronal cells in culture against ER stress, suggesting that this compound could protect neurons during ER stress induced by ischemic stroke [68]. Consistent with this, intracerebroventricular (ICV) injection of BiX protected hippocampal neurons against degeneration in a middle cerebral artery occlusion (MCAO) surgical model of stroke in both mice and gerbils [68, 69]. BiX was protective in this model even when administrated 3h post ischemia, resulting in both reduced infarct size and improved neurological function [71]. This indicates that protection against stroke using pharmacologic compounds such as BiX does not require pretreatment or immediate administration after the ischemic event. Apart from animal models of stroke, intraperitoneal or sub-renal capsule injection of BiX also reduced renal cell death and improved kidney function in mouse models of kidney I/R, demonstrating that BiX protects multiple tissues from I/R-associated damage [70]. While the mechanistic basis and dependence on ATF6 activation for BiX-dependent protection in these models remains undefined, this compound likely functions through ATF6-dependent regulation of the ER chaperone BiP, mirroring the benefits for increased BiP activity in protecting tissues against I/R injury [41–44].
Like BiX, compound 147 has similarly been shown to protect against I/R-associated insults. Treatment with compound 147 improved survival of primary cardiomyocytes treated with either ER stress or in vitro simulated I/R [38]. RNAi-depletion of ATF6 blocked this protection, confirming its dependence on 147-dependent ATF6 activation. Interestingly, genetic depletion of ATF6-regulated genes such as BiP or Cat differentially influenced 147-dependent protection against ER stress or simulated I/R [38]. This demonstrates that pharmacologic ATF6 activation can protect against different types of stress through mechanisms dependent on distinct subsets of protective ATF6-regulated genes. (see Fig. 2).
Intravenous (IV) administration of compound 147 to mice increased expression of multiple protective, ATF6-regulated genes in the heart including BiP, Serca2a, and Cat, highlighting the potential for this compound to protect the heart against I/R through multiple mechanisms (see Fig. 2). Consistent with this, IV injection of compound 147 improved cardiac recovery in an ex vivo model of I/R [38]. Furthermore, IV administration of compound 147, either 24 h prior to ischemic insult or following ischemic insult during reperfusion, reduced cardiac infarct size, decreased cardiac damage, reduced heart hypertrophy, and improved cardiac function for at least 1 week [38]. Importantly, IV administration of 147 did not show cardioprotection from I/R in cardiac-specific Atf6–knockout mice [38], indicating that the protection afforded by this treatment required ATF6. Apart from the heart, IV-administration of compound 147 to mice also protected the kidney and brain against I/R damage [38]. While the specific dependence of this protection in other tissues on ATF6 and specific ATF6-regulated genes remains to be defined, these results, combined with those discussed above using BiX, clearly demonstrate that pharmacologic ATF6 activation is broadly protective against I/R damage in multiple preclinical models of tissue-specific ischemic disease.
Concluding Remarks
The central importance of ATF6 in protecting diverse tissues against I/R damage in the context of ischemic diseases, such as myocardial infarction and stroke, has just begun to be defined and many new questions remain (see Outstanding Questions). However, it is now clear that ATF6 activation integrates the expression of numerous protective genes, beyond those involved in the canonical ER stress response, to mitigate damage associated with I/R in multiple tissues. The establishment of new pharmacologic approaches to both activate and inhibit ATF6 signaling provides new opportunities to carefully dissect the timing and extent of ATF6 signaling involved in protecting different tissues against I/R. As these and new experimental approaches are applied, the central importance of ATF6 signaling in tissue protection during ischemia and I/R will continue to be defined, revealing new mechanisms by which this pathway functions and integrates with other stress-responsive signaling pathways to influence tissue damage and survival in response to ischemia-associated disorders (see Box 2). In addition, the establishment of highly specific ATF6 activating compounds will facilitate further preclinical studies aimed at determining the potential for pharmacologic ATF6 activation to improve tissue outcomes in rodent and larger animal models of ischemic disease. Although many previous pharmacological strategies aimed at ameliorating I/R have failed [76, 77], the apparent safety of constitutive ATF6 activity in mice and humans, combined with the ability of ATF6 to provide protection through multiple mechanisms in various cellular locations (Fig. 2) suggest that compounds that activate ATF6 should be tested in further pre-clinical models of I/R damage. However, a significant amount of work remains to define the translational potential for this approach to mitigate I/R associated pathology in humans. As we and others continue to develop new ATF6 activating compounds with increased potency and in vivo activity, new opportunities will emerge to continue testing this potential in larger animal models (e.g. pigs) that better reflect disease pathology in patients and further define the therapeutic potential for targeting ATF6 in ischemia-related and other diseases.
Clinician’s Corner
The unfolded protein response (UPR) is an endogenous stress-responsive signaling pathway activated during ischemia. The UPR is comprised of three integrated signaling pathways activated downstream of the stress-sensing proteins PERK, IRE1, and ATF6. These pathways are activated in response to ER stress and function to both adapt cellular physiology and dictate cell survival in response to diverse physiologic and pathologic insults. The ATF6 arm of the UPR has been shown to contribute to tissue survival in response to ischemia/reperfusion (I/R) in preclinical models. Genetic depletion of Atf6 in the hearts or brains of mice increases I/R-associated damage in these tissues, while increasing ATF6 activity protects these tissues against I/R.
In response to ER stress, ATF6 activation leads to increased expression of multiple, protective stress-responsive genes involved in diverse biological functions. The induction of ATF6 during I/R leads to increases in proteins known to reduce I/R damage. These include ER chaperones that can alleviate I/R-associated ER stress, Ca2+ pumps that can normalize imbalances in cellular Ca2+ homeostasis associated with I/R, and antioxidants that can attenuate oxidative-stress associated damage that accumulates during I/R.
Clinical and pre-clinical evidence suggests that ATF6 is an attractive therapeutic target that could correct pathologic defects associated with diverse diseases including ischemic diseases. Genetic activation of ATF6 in different mouse tissues does not induce any overt phenotype, but instead protects against numerous types of tissue damage and cellular stress. Furthermore, mutations in ATF6 that render this pathway constitutively active have been identified in patients suffering from the congenital disorder achromatopsia, caused by defects in retinal development. However, no systemic or neurologic defects have been reported in these patients to date. Although the number of achromatopsia patients is low, the lack of systemic or neurologic phenotypes does suggest that ATF6 activation could be well tolerated in adult humans.
Small molecules that selectively activate ATF6 alleviate I/R-associated damage in preclinical mouse models of ischemic diseases such as stroke and myocardial infarction. Administration of such pharmacologic ATF6 activators either prior to or following ischemia reduces tissue damage and improves tissue function in these models. This suggests that pharmacologic ATF6 activation is a potential strategy that could be used to mitigate I/R damage in diverse tissues. However, significant work in disease models that more closely recapitulate ischemic disease in humans is required to determine whether pharmacologic ATF6 activation has translational potential to intervene in ischemic disease.
Highlights.
ATF6 activation is protective against ischemia and ischemia/reperfusion in cellular and animal models of diverse ischemic diseases including myocardial infarction and stroke.
ATF6 activation ameliorates ischemia-associated damage through the transcriptional upregulation of genes involved in mulltiple, protective biologic activities including ER proteostasis maintenance, Ca2+ regulation, and anti-oxidant response.
Human genetic and animal studies indicate that moderate levels of ATF6 activation is well tolerated in adults and does not induce systemic or neurological defects.
Recently established pharmacologic ATF6 activating compounds are protective against ischemia-associated damage in multiple tissues both in vitro and in vivo.
Outstanding Questions.
Does ATF6 activation integrate with other stress-responsive signaling pathways to ameliorate I/R damage in models of ischemic disease?
Does ATF6 activation induce tissue-specific remodeling to selectively protect different tissues against I/R damage?
Can ATF6 activating compounds be used in combination with compounds that influence other aspects of UPR signaling (e.g., IRE1 activating compounds; see Box 1) to synergistically promote recovery following ischemic or I/R?
What is the translational potential for pharmacologic ATF6 activating compounds such as BiX and Compound 147 for ischemic disease?
Can ATF6 activation be used to mitigate ER stress and/or oxidative damage associated with the pathogenesis of other diseases, such as Alzheimer’s and ALS?
Acknowledgements.
We apologize to all of our colleagues whose work we were unable to cite due to space limitations. We thank Justine Lebeau for critical reading of this manuscript and Julia Grandjean for help with figure preparation. This work was supported by the NIH (NS092829 and DK107604 to RLW and HL141463, HL135983 and HL127439 to CCG).
GLOSSARY
- Activating Transcription Factor 6 (ATF6)
An unfolded protein response (UPR)-associated stress-sensing protein that is activated in response to ER stress. ATF6 activation involves a regulated proteolysis event that releases the ATF6 N-terminal domain containing an active transcription factor. This active transcription factor induces expression of multiple stress-responsive genes shown to be protective against I/R. Endoplasmic Reticulum (ER) Stress: A cellular condition induced by physiologic and pathologic insults that disrupt ER proteostasis and function
- Ischemia/Reperfusion (I/R)
A condition defined by the initial restriction of blood flow to a specific tissue (ischemia) followed by the restoration of blood flow to that tissue (reperfusion)
- Myocardial Infarction (MI)
A condition involving restriction of blood flow to the heart (cardiac ischemia) and the resulting damage associated with the heart
- O-GlycNACylation
The process of adding a single O-linked N-acetylglucosamine to proteins at serine or threonine residues of proteins
- Proteolysis
A biologic process involving the breakdown of proteins into smaller fragments or amino acids
- Proteostasis
The condition of an organism, tissue, or cell having sufficiently properly folded proteins to maintain essential physiologic functions while minimizing the accumulation of toxic misfolded protein species
- Reactive Oxygen Species (ROS)
Reactive chemical species including oxygen atoms. These include peroxide and superoxide
- Unfolded Protein Response (UPR)
A cellular pathway that functions to regulate cellular adaptation and survival in response to physiologic and pathologic insults that induce ER stress
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Benjamin EJ et al. (2018) Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation 137 (12), e67–e492. [DOI] [PubMed] [Google Scholar]
- 2.Hausenloy DJ and Yellon DM (2016) Ischaemic conditioning and reperfusion injury. Nat Rev Cardiol 13 (4), 193–209. [DOI] [PubMed] [Google Scholar]
- 3.Murphy E and Steenbergen C (2008) Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev 88 (2), 581–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heusch G et al. (2015) Remote ischemic conditioning. J Am Coll Cardiol 65 (2), 177–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X et al. (2018) The unfolded protein response in ischemic heart disease. J Mol Cell Cardiol 117, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W and Paschen W (2016) Unfolded protein response in brain ischemia: A timely update. J Cereb Blood Flow Metab 36 (12), 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yellon DM and Hausenloy DJ (2007) Myocardial reperfusion injury. N Engl J Med 357 (11), 1121–35. [DOI] [PubMed] [Google Scholar]
- 8.Han J and Kaufman RJ (2017) Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes Dev 31 (14), 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hetz C and Papa FR (2018) The Unfolded Protein Response and Cell Fate Control. Mol Cell 69 (2), 169–181. [DOI] [PubMed] [Google Scholar]
- 10.Walter P and Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 (6059), 1081–6. [DOI] [PubMed] [Google Scholar]
- 11.Sano R and Reed JC (2013) ER stress-induced cell death mechanisms. Biochim Biophys Acta 1833 (12), 3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C et al. (2017) Unfolded protein response plays a critical role in heart damage after myocardial ischemia/reperfusion in rats. PLoS One 12 (6), e0179042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshida H et al. (2000) ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20 (18), 6755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haze K et al. (2001) Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J 355 (Pt 1), 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida H et al. (2001) Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol 21 (4), 1239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto K et al. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 13 (3), 365–76. [DOI] [PubMed] [Google Scholar]
- 17.Thuerauf DJ et al. (2007) Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J Biol Chem 282 (31), 22865–78. [DOI] [PubMed] [Google Scholar]
- 18.Thuerauf DJ et al. (2004) Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem 279 (20), 21078–84. [DOI] [PubMed] [Google Scholar]
- 19.Forouhan M et al. (2018) Paradoxical roles of ATF6alpha and ATF6beta in modulating disease severity caused by mutations in collagen X. Matrix Biol 70, 50–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieper LA et al. (2017) ATF6beta-based fine-tuning of the unfolded protein response enhances therapeutic antibody productivity of Chinese hamster ovary cells. Biotechnol Bioeng 114 (6), 1310–1318. [DOI] [PubMed] [Google Scholar]
- 21.Ye J et al. (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6 (6), 1355–64. [DOI] [PubMed] [Google Scholar]
- 22.Haze K et al. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell 10 (11), 3787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadanaka S et al. (2007) Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol 27 (3), 1027–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadanaka S et al. (2006) Reduction of disulfide bridges in the lumenal domain of ATF6 in response to glucose starvation. Cell Struct Funct 31 (2), 127–34. [DOI] [PubMed] [Google Scholar]
- 25.Higa A et al. (2014) Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol 34 (10), 1839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plate L et al. (2016) Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen J et al. (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3 (1), 99–111. [DOI] [PubMed] [Google Scholar]
- 28.Shen J et al. (2005) Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol 25 (3), 921–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong M et al. (2004) Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem 279 (12), 11354–63. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K et al. (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J Biochem 136 (3), 343–50. [DOI] [PubMed] [Google Scholar]
- 31.Shoulders MD et al. (2013) Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep 3 (4), 1279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martindale JJ et al. (2006) Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res 98 (9), 1186–93. [DOI] [PubMed] [Google Scholar]
- 33.Adachi Y et al. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33 (1), 75–89. [DOI] [PubMed] [Google Scholar]
- 34.Asada R et al. (2011) The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem 149 (5), 507–18. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K et al. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124 (3), 587–99. [DOI] [PubMed] [Google Scholar]
- 36.Toko H et al. (2010) ATF6 is important under both pathological and physiological states in the heart. J Mol Cell Cardiol 49 (1), 113–20. [DOI] [PubMed] [Google Scholar]
- 37.Jin JK et al. (2017) ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ Res 120 (5), 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackwood EA et al. (2019) Pharmacologic ATF6 Activation Confers Global Protection in Widespread Disease Models by Reprogramming Cellular Proteostasis. Nature Communications 10 (1), 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa A et al. (2015) Deletion of Atf6alpha impairs astroglial activation and enhances neuronal death following brain ischemia in mice. J Neurochem 132 (3), 342–53. [DOI] [PubMed] [Google Scholar]
- 40.Yu Z et al. (2017) Activation of the ATF6 branch of the unfolded protein response in neurons improves stroke outcome. J Cereb Blood Flow Metab 37 (3), 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bi X et al. (2018) Endoplasmic Reticulum Chaperone GRP78 Protects Heart From Ischemia/Reperfusion Injury Through Akt Activation. Circ Res 122 (11), 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouyang YB et al. (2012) miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol Dis 45 (1), 555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P et al. (2015) Micro-RNA-30a regulates ischemia-induced cell death by targeting heat shock protein HSPA5 in primary cultured cortical neurons and mouse brain after stroke. J Neurosci Res 93 (11), 1756–68. [DOI] [PubMed] [Google Scholar]
- 44.Ouyang YB et al. (2011) Overexpressing GRP78 influences Ca2+ handling and function of mitochondria in astrocytes after ischemia-like stress. Mitochondrion 11 (2), 279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita K et al. (2001) Reperfusion-activated Akt kinase prevents apoptosis in transgenic mouse hearts overexpressing insulin-like growth factor-1. Circ Res 88 (6), 609–14. [DOI] [PubMed] [Google Scholar]
- 46.Matsui T et al. (2001) Akt activation preserves cardiac function and prevents injury after transient cardiac ischemia in vivo. Circulation 104 (3), 330–5. [DOI] [PubMed] [Google Scholar]
- 47.Fujio Y et al. (2000) Akt promotes survival of cardiomyocytes in vitro and protects against ischemia-reperfusion injury in mouse heart. Circulation 101 (6), 660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fayard E et al. (2005) Protein kinase B/Akt at a glance. J Cell Sci 118 (Pt 24), 5675–8. [DOI] [PubMed] [Google Scholar]
- 49.Vekich JA et al. (2012) Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J Mol Cell Cardiol 53 (2), 259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo DY et al. (2019) Protein disulfide-isomerase A3 significantly reduces ischemia-induced damage by reducing oxidative and endoplasmic reticulum stress. Neurochem Int 122, 19–30. [DOI] [PubMed] [Google Scholar]
- 51.Vitadello M et al. (2003) Overexpression of the stress protein Grp94 reduces cardiomyocyte necrosis due to calcium overload and simulated ischemia. FASEB J 17 (8), 923–5. [DOI] [PubMed] [Google Scholar]
- 52.Thuerauf DJ et al. (2001) Sarco/endoplasmic reticulum calcium ATPase-2 expression is regulated by ATF6 during the endoplasmic reticulum stress response: intracellular signaling of calcium stress in a cardiac myocyte model system. J Biol Chem 276 (51), 48309–17. [DOI] [PubMed] [Google Scholar]
- 53.Kranias EG and Hajjar RJ (2012) Modulation of cardiac contractility by the phospholamban/SERCA2a regulatome. Circ Res 110 (12), 1646–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xin W et al. (2011) Improved cardiac function after sarcoplasmic reticulum Ca(2+)-ATPase gene transfer in a heart failure model induced by chronic myocardial ischaemia. Acta Cardiol 66 (1), 57–64. [DOI] [PubMed] [Google Scholar]
- 55.Xin W et al. (2011) Attenuation of endoplasmic reticulum stress-related myocardial apoptosis by SERCA2a gene delivery in ischemic heart disease. Mol Med 17 (3–4), 201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meex SJ et al. (2007) Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch Caucasians. J Clin Endocrinol Metab 92 (7), 2720–5. [DOI] [PubMed] [Google Scholar]
- 57.Meex SJ et al. (2009) The ATF6-Met[67]Val substitution is associated with increased plasma cholesterol levels. Arterioscler Thromb Vasc Biol 29 (9), 1322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu WS et al. (2007) Activating transcription factor 6 (ATF6) sequence polymorphisms in type 2 diabetes and pre-diabetic traits. Diabetes 56 (3), 856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thameem F et al. (2006) Association of amino acid variants in the activating transcription factor 6 gene (ATF6) on 1q21-q23 with type 2 diabetes in Pima Indians. Diabetes 55 (3), 839–42. [DOI] [PubMed] [Google Scholar]
- 60.Kohl S et al. (2015) Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet 47 (7), 757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ansar M et al. (2015) Mutation of ATF6 causes autosomal recessive achromatopsia. Hum Genet 134 (9), 941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skorczyk-Werner A et al. (2017) Autosomal recessive cone-rod dystrophy can be caused by mutations in the ATF6 gene. Eur J Hum Genet 25 (11), 1210–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiang WC et al. (2017) Achromatopsia mutations target sequential steps of ATF6 activation. Proc Natl Acad Sci U S A 114 (2), 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu J et al. (2007) ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell 13 (3), 351–64. [DOI] [PubMed] [Google Scholar]
- 65.Howarth DL et al. (2014) Activating transcription factor 6 is necessary and sufficient for alcoholic fatty liver disease in zebrafish. PLoS Genet 10 (5), e1004335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X et al. (2016) Hepatic ATF6 Increases Fatty Acid Oxidation to Attenuate Hepatic Steatosis in Mice Through Peroxisome Proliferator-Activated Receptor alpha. Diabetes 65 (7), 1904–15. [DOI] [PubMed] [Google Scholar]
- 67.Ozcan L et al. (2016) Hepatocyte DACH1 Is Increased in Obesity via Nuclear Exclusion of HDAC4 and Promotes Hepatic Insulin Resistance. Cell Rep 15 (10), 2214–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kudo T et al. (2008) A molecular chaperone inducer protects neurons from ER stress. Cell Death Differ 15 (2), 364–75. [DOI] [PubMed] [Google Scholar]
- 69.Oida Y et al. (2008) Induction of BiP, an ER-resident protein, prevents the neuronal death induced by transient forebrain ischemia in gerbil. Brain Res 1208, 217–24. [DOI] [PubMed] [Google Scholar]
- 70.Prachasilchai W et al. (2009) The protective effect of a newly developed molecular chaperone-inducer against mouse ischemic acute kidney injury. J Pharmacol Sci 109 (2), 311–4. [DOI] [PubMed] [Google Scholar]
- 71.Oida Y et al. (2010) Post-treatment of a BiP inducer prevents cell death after middle cerebral artery occlusion in mice. Neurosci Lett 484 (1), 43–6. [DOI] [PubMed] [Google Scholar]
- 72.Kroeger H et al. (2018) The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci Signal 11 (517). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paxman R et al. (2018) Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gallagher CM et al. (2016) Ceapins are a new class of unfolded protein response inhibitors, selectively targeting the ATF6alpha branch. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gallagher CM and Walter P (2016) Ceapins inhibit ATF6alpha signaling by selectively preventing transport of ATF6alpha to the Golgi apparatus during ER stress. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bulluck H et al. (2016) Reducing myocardial infarct size: challenges and future opportunities. Heart 102 (5), 341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossello X and Yellon DM (2016) A critical review on the translational journey of cardioprotective therapies! Int J Cardiol 220, 176–84. [DOI] [PubMed] [Google Scholar]
- 78.Glimcher LH (2010) XBP1: the last two decades. Ann Rheum Dis 69 Suppl 1, i67–71. [DOI] [PubMed] [Google Scholar]
- 79.Koumenis C (2006) ER stress, hypoxia tolerance and tumor progression. Curr Mol Med 6 (1), 55–69. [DOI] [PubMed] [Google Scholar]
- 80.Jiang D et al. (2015) Targeting the IRE1alpha-XBP1 branch of the unfolded protein response in human diseases. Semin Cancer Biol 33, 48–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmitz ML et al. (2018) The Crosstalk of Endoplasmic Reticulum (ER) Stress Pathways with NF-kappaB: Complex Mechanisms Relevant for Cancer, Inflammation and Infection. Biomedicines 6 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prachasilchai W et al. (2008) A protective role of unfolded protein response in mouse ischemic acute kidney injury. Eur J Pharmacol 592 (1–3), 138–45. [DOI] [PubMed] [Google Scholar]
- 83.Liu Y et al. (2018) O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. Cell Death Dis 9 (5), 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang M et al. (2017) XBP1 (X-Box-Binding Protein-1)-Dependent OGlcNAcylation Is Neuroprotective in Ischemic Stroke in Young Mice and Its Impairment in Aged Mice Is Rescued by Thiamet-G. Stroke 48 (6), 1646–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang ZV et al. (2014) Spliced X-box binding protein 1 couples the unfolded protein response to hexosamine biosynthetic pathway. Cell 156 (6), 1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y et al. (2016) XBP1 splicing triggers miR-150 transfer from smooth muscle cells to endothelial cells via extracellular vesicles. Sci Rep 6, 28627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu J et al. (2011) The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab 13 (2), 160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blackwood EA et al. (2018) ATF6 Regulates Cardiac Hypertrophy by Transcriptional Induction of the mTORC1 Activator, Rheb. Circ Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tam AB et al. (2018) The UPR Activator ATF6 Responds to Proteotoxic and Lipotoxic Stress by Distinct Mechanisms. Dev Cell 46 (3), 327–343 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Misra J et al. (2013) Transcriptional cross talk between orphan nuclear receptor ERRgamma and transmembrane transcription factor ATF6alpha coordinates endoplasmic reticulum stress response. Nucleic Acids Res 41 (14), 6960–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baird L et al. (2017) A Homeostatic Shift Facilitates Endoplasmic Reticulum Proteostasis through Transcriptional Integration of Proteostatic Stress Response Pathways. Mol Cell Biol 37 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]