Abstract
Ligand-dependent protein degradation has emerged as a compelling strategy to pharmacologically control the protein content of cells. So far, however, only a limited number of E3 ligases have been found to support this process. Here, we use a chemical proteomic strategy that leverages broadly reactive, cysteine-directed electrophilic fragments coupled to selective ligands for intracellular proteins (e.g., SLF for FKBP12, JQ1 for BRD4) to screen for heterobifunctional degrader compounds (or PROTACs) that operate by covalent adduction of E3 ligases. This approach identified DCAF16 – a poorly characterized substrate recognition component of CUL4-DDB1 E3 ubiquitin ligases – as a target of electrophilic PROTACs that promote the nuclear-restricted degradation of proteins. We find that only a modest fraction (~10-40%) of DCAF16 needs to be modified to support protein degradation, pointing to the potential for electrophilic PROTACs to induce neo-substrate degradation without substantially perturbing the function of the participating E3 ligase.
Conventional small-molecule probes and drugs act by directly perturbing the functions of proteins (e.g., blocking enzyme catalysis or antagonizing receptor signaling). Many proteins, however, possess multiple functional domains, and therefore a compound that binds to only one of these domains may fail to fully inactivate the protein. An alternative emerging strategy uses chemical probes that direct proteins to the proteolytic degradation machinery of the cell, leading to the complete loss of protein expression1-4. This targeted protein degradation approach leverages two types of small molecules – those that form tripartite complexes with specific E3 ubiquitin ligases and neosubstrate proteins (so-called “molecular glues”5; e.g., the IMiD class of therapeutics6-10 and sulfonamides11, 12) and heterobifunctional compounds, often referred to as PROTACs (proteolysis targeting chimeras), which couple E3 ligase ligands to substrate ligands via a variably structured linker13-19.
Targeted protein degradation has the potential to act in a catalytic manner20 that may lower the drug concentrations required to produce a pharmacological effect. PROTACs also have the potential to operate with sub-stoichiometric engagement of the participating E3 ligase to avoid antagonizing its natural functions, although measuring the fractional engagement of E3 ligases by conventional (reversible) PROTACs remains technically challenging in cells. Despite these advantages, and the diverse number of proteins demonstrated to undergo ligand-induced degradation in cells21, to date, only a handful of the 600+ human E3 ligases have been found to support this process11-15, 22-24, with most studies leveraging established ligands targeting the E3 substrate recognition components cereblon (CRBN) and VHL for PROTAC development. Importantly, these E3 ligases have been found to show distinct and restricted substrate specificities25, 26, a parameter that is not yet predictable or easily controlled, underscoring the need to discover additional ligandable E3 ligases with differentiated properties to realize the full scope of targeted protein degradation as a pharmacological strategy.
Here, we present a chemical proteomics approach to discover E3 ligases that support targeted protein degradation when engaged by electrophilic PROTACs. We specifically describe a set of heterobifunctional compounds consisting of i) cysteine-directed fragment electrophiles shown in previous studies to exhibit broad proteomic reactivity27, 28 coupled through a variable length linker to ii) selective reversible ligands for protein targets of interest. We found that a subset of these compounds promote the nuclear-localized degradation of protein targets by covalently modifying DCAF16, a poorly characterized substrate recognition component of CUL4-DDB1 E3 ubiquitin ligases. We finally show using multiple complementary assays (gel-shift and mass spectrometry-based proteomics) that electrophilic PROTACs can induce protein degradation by modifying only a modest fraction (~10–40%) of DCAF16 in cells, highlighting a potential advantage of heterobifunctional compounds that operate by covalently engaging E3 ligases.
Results
An electrophilic PROTAC that degrades nuclear FKBP12.
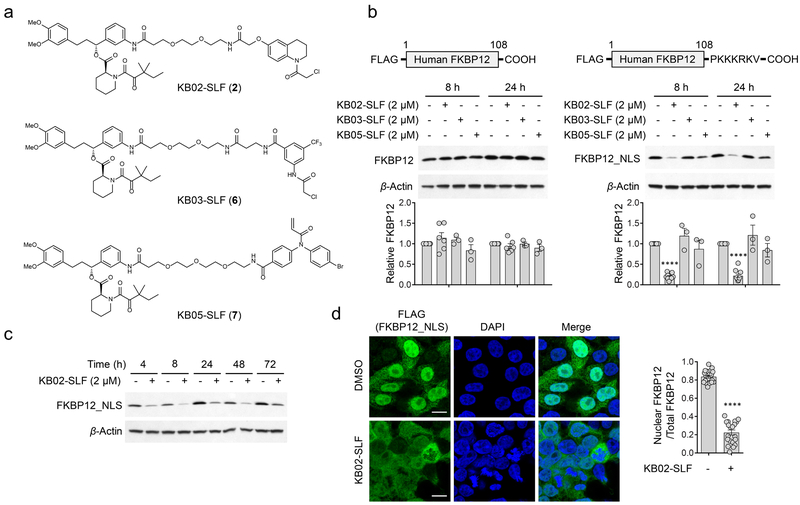
We have recently introduced chemical proteomic platforms for globally and site-specifically mapping the reactivity of electrophilic small molecules in native biological systems27-29. These experiments have identified a subset of fragment electrophiles, referred to hereafter as ‘scouts’27, 28, that engage a remarkably broad fraction of the 100s-1000s of covalent small molecule-cysteine interactions discovered so far by chemical proteomics. We surmised herein that these scout fragments, when incorporated into heterobifunctional compounds containing optimized protein ligands, may offer an expedited path to discover E3 ligases capable of supporting targeted protein degradation through covalent reactivity with cysteine residues. With this goal in mind, we fused three scout fragments – KB02, KB03, and KB05, which cover two different electrophile groups (chloroacetamide and acrylamide) and display broad cysteine reactivity in the human proteome27, 28 – to the SLF ligand that binds tightly and selectively to FKBP1230 (Fig. 1a), a cytosolic prolyl isomerase that has been frequently used to study ligand-induced protein degradation15, 31. We then stably expressed FLAG-tagged variants of FKBP12 (FLAG-FKBP12) or FKBP12 with a C-terminal nuclear localization sequence (FLAG-FKBP12_NLS) in HEK293T cells to provide cell models for evaluating cytosolic- and nuclear-localized E3-mediated degradation pathways, respectively (Fig. 1b and Supplementary Fig. 1a).
Figure 1. Discovery of an electrophilic PROTAC that promotes the loss of nuclear FKBP12.
a, Structures of electrophilic bifunctional compounds, or PROTACs, containing scout fragments (KB02, KB03, and KB05) coupled to the FKBP12 ligand SLF. b, Western blot using anti-FLAG antibody of cytosolic (FLAG-FKBP12) and nuclear (FLAG-FKBP12_NLS) FKBP12 proteins expressed by stable transduction in HEK293T cells following 8 or 24 h of treatment with electrophilic PROTACs (2 µM). Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments for KB03-SLF and KB05-SLF-treated cells, n = 6 biologically independent experiments for DMSO and KB02-SLF-treated cells expressing FLAG-FKBP12, n = 10 biologically independent experiments for DMSO and KB02-SLF-treated cells expressing FLAG-FKBP12_NLS). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. ****P < 0.0001. P values were 6.5×10−18 and 4.0×10−13. Full images of blots are shown in Supplementary Fig. 14. c, Time-dependent degradation of FLAG-FKBP12_NLS in HEK293T cells treated with KB02-SLF (2 µM). The result is a representative of two experiments (n = 2 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. d, Immunofluorescence using anti-FLAG antibody of FLAG-FKBP12_NLS in HEK293T cells following treatment with DMSO or KB02-SLF (2 µM, 8 h). Bar graph (right) represents quantification of the relative nuclear to whole cell immunostaining for DMSO- and KB02-SLF-treated samples. Data represent mean values ± SEM (n = 20 from two biologically independent experiments. 10 images were collected from each biological replicate). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. ****P < 0.0001. P value was 1.3×10−21. Scale bar, 10 µm.
We first confirmed by anti-FLAG western blotting that both cytosolic and nuclear FKBP12 are degraded by lenalidomide-SLF (1) (Supplementary Fig. 1b, c), a bifunctional molecule comprised of SLF coupled to lenalidomide, a ligand for the E3 ligase cereblon (CRBN)6, 8, 9, 15, 31. We next evaluated the electrophilic scout fragment-SLF bifunctional compounds for effects on FKBP12 degradation in HEK293T cells. Under the treatment conditions (2 µM, 8 or 24 h), none of the compounds altered cytosolic FKBP12 (Fig. 1b, left blots). On the other hand, the KB02-SLF (2) compound promoted a substantial reduction in nuclear FKBP12 (Fig. 1b, right blots) that was sustained across a 4–72 h time frame (Fig. 1c). Cell imaging studies confirmed the selective loss of nuclear-localized FKBP12 in KB02-SLF-treated cells (Fig. 1d). These imaging studies, along with western blotting experiments (Supplementary Fig. 1a), pointed to a fraction of FKBP12 that remained cytosolically localized in FLAG-FKBP12_NLS-transfected cells and was consequently unaffected by KB02-SLF treatment (Fig. 1d).
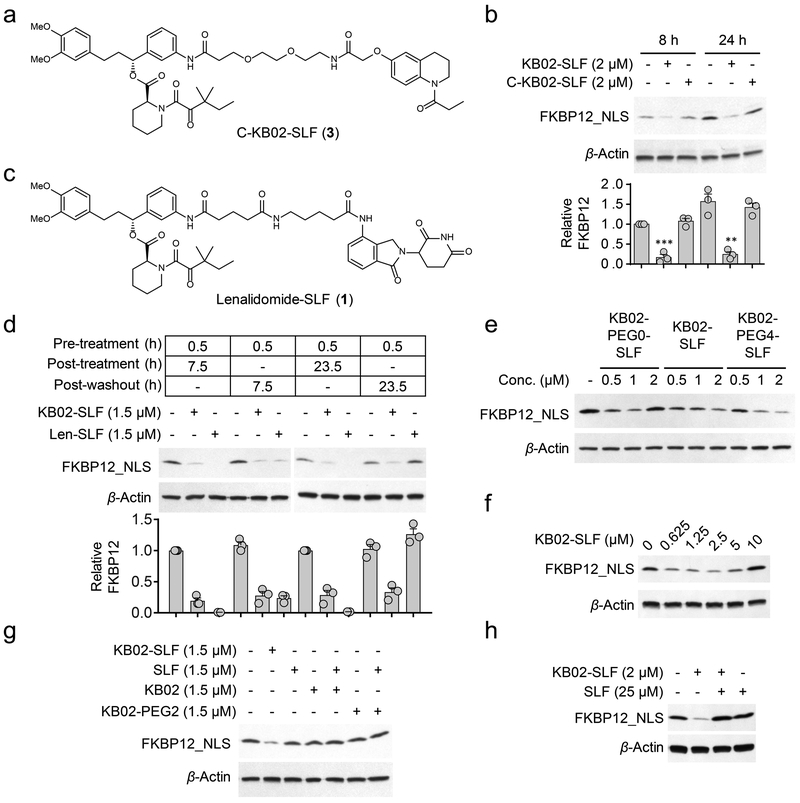
Reductions in FKBP12_NLS were not observed with an analogue of KB02-SLF where the electrophilic alpha-chloroacetamide group was replaced with an unreactive acetamide (C-KB02-SLF (3); Fig. 2a, b), indicating that the mechanism of action of KB02-SLF involved covalent modification of one or more proteins. Washing of cells to remove free heterobifunctional compound did not affect the durability of KB02-SLF-mediated degradation of FKBP12_NLS evaluated ~24 h post-washout, but led to full recovery of FKBP12_NLS in cells treated with the non-covalent PROTAC lenalidomide-SLF (Fig. 2c, d). KB02-SLF, as well as analogues of this compound with different linker lengths (Supplementary Fig. 2), promoted the loss of FKBP12_NLS across a concentration of ~0.5–5 µM (Fig. 2e, f), but showed varied reductions in activity at higher concentrations (Fig. 2f). Such parabolic concentration-dependence is a feature of PROTACs, where binary complexes gain prevalence over ternary complexes at higher cellular concentrations of compound16, 20. However, drawing this firm conclusion in our studies was more difficult because KB02-SLF showed evidence of cytotoxicity at higher test concentrations (> 10 µM, see below), which could also impair protein degradation pathways. Nonetheless, we did not observed reductions in FKBP12_NLS in cells treated with the two separate components (KB02 (4) and SLF (5)) of the KB02-SLF bifunctional compound (Fig. 2g and Supplementary Fig. 2), which is consistent with a ternary complex mode of action. Pre-treatment with SLF did, on the other hand, block the KB02-SLF-induced loss of FKBP12_NLS (Fig. 2h).
Figure 2. KB02-SLF promotes the loss of nuclear FKBP12.
a, Structure of non-electrophilic control compound (C-KB02-SLF). b, Western blot of FLAG-FKBP12_NLS in HEK293T cells treated with KB02-SLF or C-KB02-SLF (2 µM, 8 or 24 h). Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. **P < 0.01; ***P < 0.001. P values were 0.00036 and 0.0027. Full images of blots are shown in Supplementary Fig. 14. c, Structure of lenalidomide-SLF. d, Western blot of FLAG-FKBP12_NLS in HEK293T cells pre-treated with 1.5 µM of KB02-SLF or lenalidomide (Len)-SLF for 0.5 h, washed with PBS, and resuspended in fresh media for additional 7.5 or 23.5 h. Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. e, Western blot of FLAG-FKBP12_NLS following treatment of HEK293T cells with KB02-PEG0-SLF, KB02-SLF, or KB02-PEG4-SLF (10) (0.5-2 µM, 8 h). The result is a representative of three experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. f, Concentration-dependent degradation of FLAG-FKBP12_NLS by KB02-SLF in HEK293T cells (24 h treatment with indicated concentrations of KB02-SLF). The result is a representative of four experiments (n = 4 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. g, Western blot of FLAG-FKBP12_NLS in HEK293T cells following treatment of HEK293T cells with KB02, KB02-PEG2 (11), SLF, or the combination of SLF and KB02 or KB02-PEG2 (1.5 µM, 8 h). The result is a representative of two experiments (n = 2 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. h, Degradation of FLAG-FKBP12_NLS by KB02-SLF (2 µM, 8 h) is blocked by excess SLF (25 µM). The result is a representative of three independent experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14.
DCAF16 mediates nuclear FKBP12 degradation by KB02-SLF.
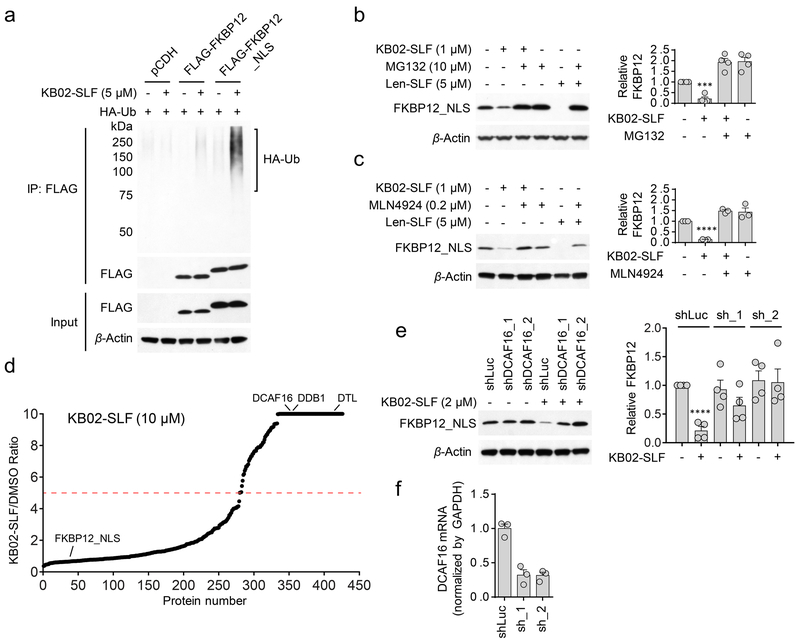
We found that KB02-SLF induced polyubiquitination of nuclear FKBP12_NLS (Fig. 3a and Supplementary Fig. 3), but not cytosolic FKBP12 (Fig. 3a) and KB02-SLF-mediated loss of FKBP12_NLS was blocked by the proteasome inhibitor MG132 (Fig. 3b) and neddylation inhibitor MLN492432 (Fig. 3c). Similar effects of KB02-SLF on FKBP12_NLS were observed in a second human cell line (MDA-MB-231) (Supplementary Fig. 4a-c). Since MLN4924 selectively inhibits E3 ligases of the Cullin-RING ubiquitin ligase (CRL) class, the activity of which depends on neddylation of Cullin proteins33-35, these data supported that KB02-SLF promoted the proteasomal degradation of nuclear-localized FKBP12 via the action of a CRL.
Figure 3. KB02-SLF promotes proteasomal degradation of FKBP12 via the action of Cullin-RING ubiquitin ligases.
a, KB02-SLF mediates polyubiquitination of nuclear (FLAG-FKBP12_NLS), but not cytosolic (FLAG-FKBP12) FKBP12 in HEK293T cells. HEK293T cells stably expressing FLAG-FKBP12 or FLAG-FKBP12_NLS were transiently transfected with HA-Ubiquitin (HA-Ub) for 24 h and then treated with DMSO or KB02-SLF (5 µM) in the presence of the proteasome inhibitor MG132 (10 µM) for 2 h. The result is a representative of three experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. b, c, KB02-SLF-mediated FLAG-FKBP12_NLS degradation is blocked by MG132 (b) and the neddylation inhibitor MLN4924 (c). HEK293T cells stably expressing FLAG-FKBP12_NLS were co-treated with KB02-SLF (1 µM) and MG132 (10 µM) or MLN4924 (0.2 µM) for 8 h. Lenalidomide (Len)-SLF was used as a positive control. Bar graph (right) represents quantification of the relative FKBP12 protein, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 4 biologically independent experiments for (b), n = 3 biologically independent experiments for (c)). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples without MG132 or MLN4924. ***P < 0.001; ****P < 0.0001. P values were 0.00016 (b) and 1.8×10−7 (c). Full images of blots are shown in Supplementary Fig. 14. d, SILAC heavy/light (KB02-SLF/DMSO) ratio values of proteins identified in anti-FLAG affinity enrichment experiments (outlined in Supplementary Fig. 5a), where a high ratio indicates proteins selectively enriched from cells treated with KB02-SLF (10 µM). The red dashed line marks a 5-fold ratio value, which was used as a threshold for designating proteins that were substantially enriched by KB02-SLF. The results shown are average ratios from three experiments (n = 3 biologically independent experiments). See method for detailed criteria of data filtering. e, Western blot of stably expressed FLAG-FKBP12_NLS in HEK293T cells transiently transduced with shRNAs targeting DCAF16 (sh_1 and sh_2) or a control shRNA (shLuc) followed by treatment with KB02-SLF (2 µM, 8 h). Right, quantification of the relative FKBP12 protein content, with DMSO-treated cells expressing shLuc set to a value of 1. Data represent mean values ± SEM (n = 4 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. ****P < 0.0001. P value was 0.000036. Full images of blots are shown in Supplementary Fig. 14. f, DCAF16 mRNA was measured by qPCR. Data represent mean values ± SEM (n = 3 biologically independent experiments).
Working under a model where KB02-SLF induced the degradation of FKBP12_NLS by covalently reacting with one or more CRL(s), we first attempted to identify the recruited CRL(s) using a chemical proteomic method termed isoTOP-ABPP (isotopic tandem orthogonal proteolysis-activity-based protein profiling), where ligand-engaged cysteines in cells are mapped by competitive blockade of reactivity with an iodoacetamide (IA)-alkyne probe36. However, we did not observe any cysteines on CRL(s) that were strongly engaged (> 75%) by KB02-SLF (10 µM, 2 h) in HEK293T cells by isoTOP-ABPP (Supplementary Dataset 1). We alternatively considered that KB02-SLF may show low stoichiometry engagement of a CRL, which would then function catalytically to drive FKBP12_NLS degradation. Such low fractional occupancy ligand-protein interactions are not easily mapped by competitive profiling methods like isoTOP-ABPP, as they would minimally perturb IA-alkyne-cysteine reactions. We therefore employed an alternative proteomic approach, in which FLAG-mediated affinity enrichment was used to identify proteins that associated with FKBP12_NLS in a KB02-SLF-dependent manner (Supplementary Fig. 5a). This experimental set up identified two substrate receptor components of CRLs – DCAF16 and DTL – that were substantially enriched (> 5-fold) in HEK293T cells treated with two different concentrations of KB02-SLF (2 and 10 µM) compared to DMSO-treated control cells (Fig. 3d, Supplementary Fig. 5b, c, and Supplementary Dataset 2). Neither DCAF16 nor DTL were enriched by KB02-SLF in an additional control experiment performed with mock-transduced cells lacking FKBP12_NLS (Supplementary Fig. 5b, d and Supplementary Dataset 2). We also observed KB02-SLF-dependent enrichment of DCAF16 in MDA-MB-231 cells (Supplementary Fig. 4d, e and Supplementary Dataset 2).
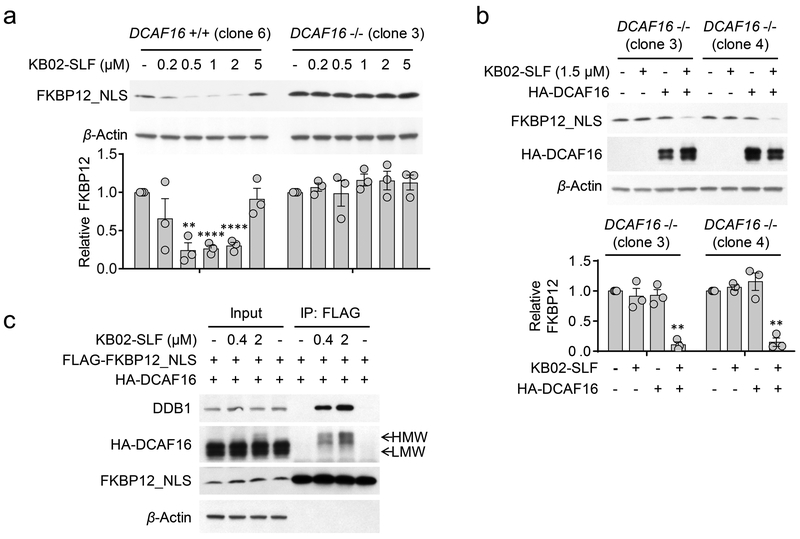
Notably, both DCAF16 and DTL are predicted to be nuclear proteins (https://psort.hgc.jp/). DTL (also known as CDT2 and DCAF2) plays a key role in cell cycle control and DNA damage response37-39. DCAF16, on the other hand, remains poorly characterized and shares negligible homology with other DDB1- and CUL4-associated factor (DCAF) proteins that participate in CRLs40. Consistent with KB02-SLF engaging functional CRL complexes potentially containing DCAF16 and/or DTL, we also observed enrichment of additional CRL components – DDB1 and CUL4B – both of which are found in the nuclear and cytoplasmic compartments of cells41 – in KB02-SLF-treated cells (Fig. 3d, Supplementary Fig. 4e and 5b, c). We next used small hairpin RNA (shRNA)-mediated knockdown to find that reductions in DCAF16, but not DTL, substantially prevented KB02-SLF-mediated degradation of FKBP12_NLS (Fig. 3e, f and Supplementary Fig. 5e). shRNA-knockdown of DCAF16 also abolished KB02-SLF-induced polyubiquitination of FKBP12_NLS (Supplementary Fig. 5f). We further confirmed the nuclear localization of an HA-tagged DCAF16 in transfected HEK293T cells (Supplementary Fig. 5g, h). Next, we verified the involvement of DCAF16 in KB02-SLF-mediated degradation of FKBP12_NLS by CRISPR/Cas9 genetic knockout42, 43. We used genomic sequencing and mass spectrometry-based proteomics to confirm the genetic disruption of DCAF16 in three independent clones (DCAF16−/−) compared to three wild type DCAF16 clones (DCAF16+/+) (Supplementary Fig. 6a-c and Supplementary Dataset 3). KB02-SLF supported the degradation of FKBP12_NLS in all three DCAF16+/+ clones, but not in any of the three DCAF16−/− clones (Fig. 4a and Supplementary Fig. 6d). Moreover, recombinant expression of HA-tagged DCAF16 restored KB02-SLF-induced FKBP12_NLS degradation in DCAF16−/− cells (Fig. 4b) and further enhanced the extent of KB02-SLF-induced FKBP12_NLS degradation in DCAF16+/+ cells (Supplementary Fig. 6e). We did not observe the degradation of HA-DCAF16 itself following treatment of KB02-SLF (Fig. 4b and Supplementary Fig. 6f). These data, taken together, demonstrate that the degradation of FKBP12_NLS induced by KB02-SLF is mediated by DCAF16.
Figure 4. DCAF16 mediates KB02-SLF-induced degradation of FKBP12.
a, Concentration-dependent degradation of stably expressed FLAG-FKBP12_NLS in DCAF16+/+ (clone 6) and DCAF16−/− (clone 3) HEK293 cells following treatment with KB02-SLF (0.2–5 µM) for 8 h. Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. **P < 0.01; ****P < 0.0001. P values were 0.0015, 0.000059 and 0.000075. Full images of blots are shown in Supplementary Fig. 14. b, Expression of HA-DCAF16 in DCAF16−/− cells restored KB02-SLF-mediated degradation of FLAG-FKBP12_NLS. DCAF16−/− cells (clone 3 and 4) were transiently transfected with FLAG-FKBP12_NLS and either HA-DCAF16 or empty pRK5 vector as a control for 24 h and then treated with KB02-SLF (1.5 µM, 8 h). Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. **P < 0.01. P values were 0.0016 and 0.0033. Full images of blots are shown in Supplementary Fig. 14. c, A higher molecular weight (HMW) form of HA-DCAF16 is observed in HEK293T cells treated with KB02-SLF (0.4 or 2 µM, 2 h in the presence of 10 µM MG132), and this HMW form, but not the lower molecular weight (LMW) form of HA-DCAF16 co-immunoprecipitated with FLAG-FKBP12_NLS. The result is a representative of three experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14.
In further support of ternary complex formation involving a DCAF16-CRL, we found that HA-DCAF16 and DDB1 co-immunoprecipitated with FLAG-FKBP12_NLS in the presence of KB02-SLF (Fig. 4c) or its linker analogues (Supplementary Fig. 7a). In the course of performing these co-immunoprecipitation experiments, we noticed that KB02-SLF-treated cells showed a higher molecular weight (HMW) form of HA-DCAF16, consistent with covalent modification of this protein by KB02-SLF (Fig. 4c and Supplementary Fig. 7a-f). Only a modest fraction of HA-DCAF16 was converted to this HMW across a concentration range of KB02-SLF that supports FKBP12_NLS degradation (0.2–5 µM, Fig. 4c and Supplementary Fig. 7b), while treating cells with higher concentrations of KB02-SLF (50–100 µM) led to substantial formation (~80%) of the HMW of HA-DCAF16. (Supplementary Fig. 7g). Notably, co-immunoprecipitations with FKBP12_NLS exclusively pulled down the HMW, but not LMW form of DCAF16 (Fig. 4c and Supplementary Fig. 7f), supporting a ternary complex model where KB02-SLF is covalently and non-covalently bound to DCAF16 and FKBP12_NLS, respectively. Also consistent with this model, DCAF16 was not co-immunoprecipitated from cells treated with the non-electrophilic control compound C-KB02-SLF (Supplementary Fig. 7c) or with the other two electrophilic bifunctional compounds (KB03-SLF (6) and KB05-SLF (7)) that did not support FKBP12_NLS degradation (Supplementary Fig. 7d, e). Finally, to provide further evidence that the HMW form of DCAF16 represented a covalent adduct with KB02-SLF, we performed a far-western blot experiment44 using recombinant His-tagged FKBP12 protein and an anti-His antibody, which revealed that FKBP12 bound to the KB02-SLF-induced HMW form of DCAF16, but not the LMW forms of this protein (Supplementary Fig. 8).
Evaluation of cysteine residues in DCAF16.
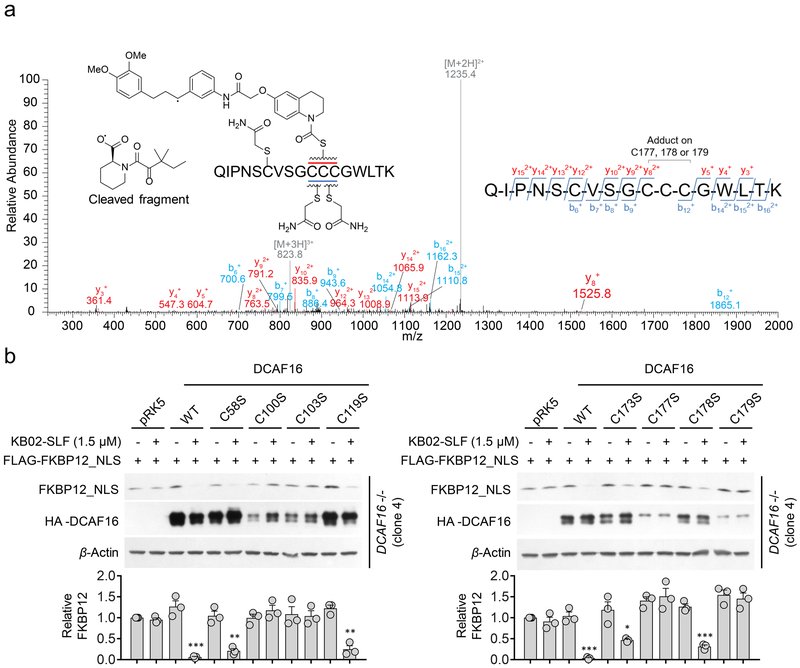
DCAF16 is a 216 aa protein that is highly conserved across mammals (e.g., human and rabbit DCAF16 share 97% identity), but absent from rodents. The DCAF16 protein has eight cysteine residues, including a cluster of four cysteines between amino acids (aa) 173–179 (Supplementary Fig. 9a). We next set out to map KB02-PEG0-SLF (8)-reactive cysteine(s) in DCAF16 by treating HA-DCAF16-transfected HEK293T cells with DMSO or KB02-PEG0-SLF (10 µM, 2 h), affinity purifying HA-DCAF16, and subjecting the protein to trypsin digestion and LC-MS/MS analysis (Supplementary Fig. 9a). We searched the MS1 (parent ion) profiles for the m/z values of unmodified and KB02-PEG0-SLF-modified DCAF16 tryptic peptides and identified a KB02-PEG0-SLF-modified form of the tryptic peptide (aa 168–184) containing C173 and C177–179 (Supplementary Fig. 9b). In contrast, we did not identify KB02-PEG0-SLF-modified forms for tryptic peptides containing C100/C103 (aa 97–106) or C119 (aa 107–133) (Supplementary Fig. 9c, d). We did not detect the tryptic peptide containing C58, likely because it is a very small in length (four aa). Tandem (MS/MS) analysis of the KB02-PEG0-SLF-modified DCAF16 peptide indicated that the most likely modified residue(s) was C177, C178, and/or C179 (Fig. 5a). Our attempts to evaluate the contribution of C177–179 and other cysteine residues to KB02-SLF-induced degradation of FKBP12_NLS by site-directed mutagenesis were hindered, in part, by the dramatic reduction in expression of cysteine-to-serine mutants for multiple cysteine residues (e.g., C100, C103, C177, C179) (Fig. 5b and Supplementary Fig. 10a). These mutagenesis studies did reveal that the C58S-, C119S-, C173S-, and C178S-DCAF16 mutants were expressed at near-wild type levels and supported KB02-SLF-induced degradation of FKBP12_NLS (Fig. 5b), indicating that C58, C119, C173, and C178 are unlikely to constitute primary sites of engagement for KB02-SLF that support target degradation. We additionally analyzed whether cysteine-to-serine mutants of DCAF16 could form a ternary complex with KB02-SLF and FKBP12_NLS in cells pre-treated with the proteasome inhibitor MG132, which we found to normalize the expression of certain DCAF16 mutant proteins (Supplementary Fig. 10a). Among the eight mutants tested, C58S-, C119S-, C173S- and C178S-DCAF16, but not C100S-, C103S-, C177S- and C179S-DCAF16, co-immunoprecipitated with FKBP12_NLS in KB02-SLF-treated cells (Supplementary Fig. 10b, c). While our data, taken in total, support C177 and/or C179 in DCAF16 as possible sites of modification responsible for KB02-SLF-induced degradation of FKBP12_NLS, we should qualify this interpretation by noting the potential for improper folding of the C177S- and C179S-DCAF16 mutants, which may prevent productive reactivity with KB02-SLF and/or interactions with FKBP12_NLS.
Figure 5. Evaluation of DCAF16 cysteines involved in KB02-SLF-induced degradation of FKBP12.
a, MS/MS spectrum of KB02-PEG0-SLF-modified, triply charged DCAF16 peptide (amino acids 168–184; see Supplementary Fig. 9b). The m/z 1235.4 ion represents a doubly charged DCAF16 peptide (168–184) with a cleaved probe adduct that occurs in MS/MS. The b- and y-ions are shown along with the peptide sequence. The experiment is repeated twice independently with similar results. b, Western blot of FLAG-FKBP12_NLS in DCAF16−/− cells (clone 4) expressing WT or C58S, C100S, C103S, C119S, C173S, C177S, C178S, or C179S mutants of HA-DCAF16 (or empty pRK5 vector control) following treatment with KB02-SLF (1.5 µM, 8 h). Bar graph (bottom) represents quantification of the relative FKBP12 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n =3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-SLF-treated samples. *P < 0.05; **P < 0.01; ***P < 0.001. P values were 0.00098, 0.0024, 0.0011, 0.00057, 0.019 and 0.00028. Full images of blots are shown in Supplementary Fig. 14.
Protein degradation by low fractional DCAF16 engagement.
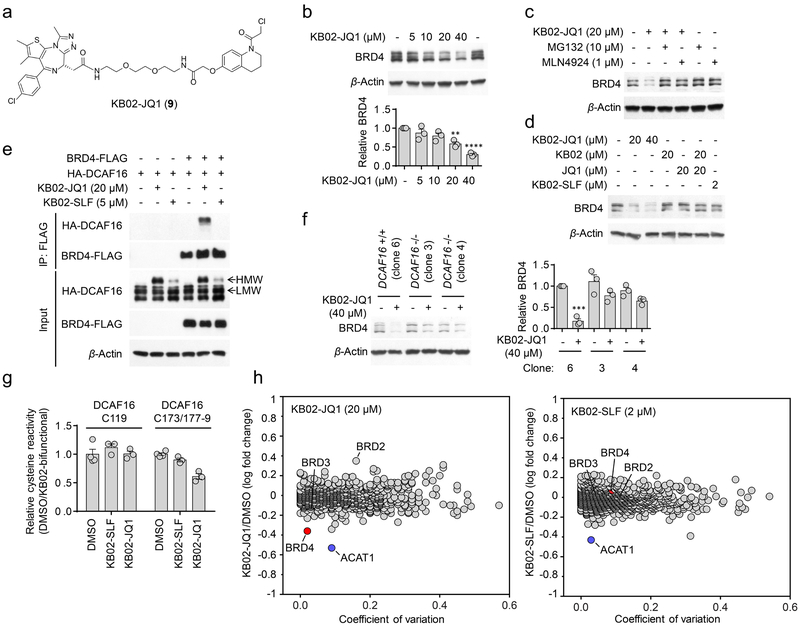
We next investigated whether covalent modification of DCAF16 could support the degradation of another nuclear protein. For these studies, we selected BRD4 as the nuclear protein, as it has a potent and selective ligand JQ1 that has been successfully coupled to other E3 ligands to promote degradation13, 15, 45-47. We found that a KB02-JQ1 (9) bifunctional compound (Fig. 6a) promoted, in a concentration-dependent manner, the degradation of BRD4 in HEK293T cells (Fig. 6b), and this effect was blocked by MG132 or MLN4924 (Fig. 6c). BRD4 was not degraded in cells treated with either the two separate components (KB02 and JQ1) of KB02-JQ1 or the FKBP12-directed bifunctional compound KB02-SLF (Fig. 6d). Consistent with DCAF16 mediating the degradation of BRD4 by KB02-JQ1, we found that the KB02-JQ1-induced, but not the KB02-SLF-induced HMW form of DCAF16 co-immunoprecipitated with BRD4 (Fig. 6e). KB02-JQ1-induced degradation of BRD4 was also substantially blocked in DCAF16−/− cells (Fig. 6f).
Figure 6. KB02-JQ1 degrades BRD4 by sub-stoichiometric modification of DCAF16.
a, Structure of KB02-JQ1. b, Concentration-dependent degradation of endogenous BRD4 in HEK293T cells following treatment with KB02-JQ1 for 24 h. Bar graph (bottom) is quantification of the relative BRD4 level. The BRD4 level from DMSO-treated cells is set to 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-JQ1-treated samples. **P < 0.01; ****P < 0.0001. P values were 0.0014 and 0.000012. Full images of blots are shown in Supplementary Fig. 14. c, KB02-JQ1-mediated BRD4 degradation is blocked by proteasome inhibitor MG132 and neddylation inhibitor MLN4924. HEK293T cells were preincubated with 10 µM MG132 or 1 µM MLN4924 for 4 h, followed by 20 h of treatment with 20 µM KB02-JQ1 and 10 µM MG132 or 1 µM MLN4924. The result is a representative of three experiments (n = 3 biologically independent experiments). Full images of blots are shown in Supplementary Fig. 14. d, Western blot of BRD4 in HEK293T cells following treatment with the indicated concentrations of KB02-JQ1, KB02, JQ1, KB02-SLF, or the combination of KB02 and JQ1 (24 h). The result is a representative of two experiments (n = 2 biologically independent experiments). The experiment is repeated twice independently with similar results. Full images of blots are shown in Supplementary Fig. 14. e, FLAG-tagged BRD4 co-immunoprecipitated with HA-DCAF16 in the presence of KB02-JQ1, but not KB02-SLF. HEK293T cells were co-transfected with BRD4-FLAG and HA-DCAF16 or pRK5 vector for 24 h and treated with KB02-JQ1 (20 µM), KB02-SLF (5 µM), or DMSO in the presence of MG132 (10 µM) for 2 h. The experiment is repeated four times independently with similar results. Full images of blots are shown in Supplementary Fig. 14. f, Degradation of BRD4 in HEK293 DCAF16+/+ (clone 6) and −/− (clone 3 and 4) cells following 24 h of treatment with 40 µM KB02-JQ1. Bar graph (right) represents quantification of the relative BRD4 protein content, with DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent experiments). Statistical significance was calculated with unpaired two-tailed Student’s t-tests comparing DMSO- to KB02-JQ1-treated samples. ***P < 0.001. P value was 0.00014. Full images of blots are shown in Supplementary Fig. 14. g, Competitive ABPP results measuring relative reactivity (DMSO/KB02-bifunctional) of C119- and C173/C177–179-containing DCAF16 tryptic peptides from DMSO-, 2 µM KB02-SLF-, or 20 µM KB02-JQ1-treated HEK293T cells, with the peptide signals from DMSO-treated cells set to a value of 1. Data represent mean values ± SEM (n = 3 biologically independent samples). See Supplementary Fig. 12 for more details on the experimental protocol and plots of global cysteine reactivity. h, Log10 fold-change in protein abundance between heavy- and light-isotopically labeled HEK293T cells treated with KB02-JQ1 (20 µM, heavy), KB02-SLF (2 µM, heavy) or DMSO (light) for 24 h. The y-axis and x-axis correspond to the average relative log10 abundance (KB02-JQ1/DMSO or KB02-SLF/DMSO) and coefficient of variation, respectively, from two experiments (n = 2 biologically independent experiments). The average relative log10 abundance of each protein is normalized to the average log10 abundance of proteins in control DMSO/DMSO samples (to account for slight deviations in heavy isotope incorporation; Supplementary Dataset 5).
We noted that much higher concentrations of KB02-JQ1 (20–40 µM; Fig. 6b) were required to degrade BRD4 compared to the degradation of FKBP12_NLS by KB02-SLF (0.5–2 µM; Fig. 2e, f). This reduction in potency may reflect a combination of factors, including differential cellular uptake of the two KB02 bifunctional compounds (KB02-JQ1 showed a rightward shift in cytotoxicity (IC50 > 50 µM) compared to KB02-SLF (IC50 = 14 ± 1.1 µM) (Supplementary Fig. 11)) and less efficient DCAF16-mediated degradation of BRD4 compared to FKBP12. To explore the latter possibility, we measured the relative cellular engagement of DCAF16 by KB02-JQ1 and KB02-SLF. Specifically, we used competitive ABPP to quantitatively map the fractional blockade of IA-alkyne-modified cysteines on endogenously expressed DCAF16 in HEK293T cells treated with concentrations of KB02-SLF (2 µM) or KB02-JQ1 (20 µM) that support FKBP12 and BRD4 degradation, respectively. These chemical proteomic data revealed that KB02-SLF and KB02-JQ1 produced ~10% and 40% engagement, respectively, of the DCAF16 peptide (aa 168–184) containing cysteines 173 and 177–179 (Fig. 6g, Supplementary Fig. 12, and Supplementary Dataset 4). Another IA-alkyne-reactive cysteine, C119, showed no evidence of engagement in these chemical proteomic experiments (Fig. 6g, Supplementary Fig. 12, and Supplementary Dataset 4). We measured a similar pattern of engagement by comparing the HMW and LMW forms of recombinantly expressed DCAF16 in HEK293T cells treated with KB02-SLF (5 µM, 13% engagement) and KB02-JQ1 (20 µM, 35% engagement) (Supplementary Fig. 13). Taken together, these data indicate that differential amounts of DCAF16 engagement are required to support FKBP12 and BRD4 degradation by their respective electrophilic PROTACs, but, in neither case is a substantial fraction (> 50%) of DCAF16 diverted to a PROTAC-modified state.
Finally, MS-based proteomic analysis of KB02-JQ1-treated HEK293T cells revealed selective degradation of BRD4, but not BRD2 or BRD3 (Fig. 6h and Supplementary Dataset 5). BRD2 appeared to be stabilized by KB02-JQ1, as has been found for JQ1 itself48. One additional protein across the proteome – ACAT1 – displayed substantially decreased abundance in KB02-JQ1-treated cells (Fig. 6h and Supplementary Dataset 5). Control cells treated with KB02-SLF also showed reductions in ACAT1, but not BRD4 (Fig. 6h and Supplementary Dataset 5). Interestingly, ACAT1 harbors a highly reactive cysteine (C126)27, 28, 36 that was fully engaged by KB02-JQ1 or KB02-SLF in HEK293T cells (Supplementary Fig. 12b and Supplementary Dataset 4). These data suggest that covalent modification of C126 by KB02-containing compounds could lead to the degradation of ACAT1, possibly by disrupting homo-oligomeric forms of the enzyme49. While BRD2 was the only protein that showed a > two-fold increase in abundance in KB02-JQ1-treated cells, a handful of additional proteins (nine total) showed more modest elevations (~1.5–1.8-fold) (Supplementary Dataset 5). None of these proteins were altered in KB02-SLF-treated cells (Supplementary Dataset 5). Whether these proteins represent potential endogenous substrates of DCAF16 that are stabilized by the higher cellular engagement of this E3 ligase by KB02-JQ1 versus KB02-SLF in cells, or, alternatively, are stabilized by direct modification of KB02-JQ1 represents an important topic for future investigation.
Discussion
In this study, we have used a chemical proteomic strategy to discover that the E3 ligase subunit DCAF16 supports targeted protein degradation when engaged by electrophilic PROTACs. DCAF16 may offer advantages as a new addition to the emerging subset of E3 ligases that promote ligand-induced protein degradation. First, our data suggest that DCAF16 exclusively promotes the degradation of nuclear proteins, which may provide a way to improve the specificity of ligand-induced protein degradation by, for instance, avoiding cytosolic proteins that engage a bifunctional compound. Second, DCAF16 appears capable of supporting ligand-induced protein degradation at low fractional engagement (~10% for KB02-SLF and ~40% for KB02-JQ1), which we attribute, at least in part, to covalent interaction with electrophilic PROTACs, which enables ligand-induced protein degradation to proceed via a pseudo-binary complex between the substrate and the PROTAC-E3 ligase covalent adduct (in contrast to the mechanism of action of conventional PROTACs, which form ternary complexes that must accommodate two reversibly bound interfaces16, 25, 26 to support protein degradation). Also, by converting a sub-stoichiometric portion of DCAF16 to a ‘neo-E3 ligase’, electrophilic PROTACs may support the degradation of target proteins while minimally perturbing endogenous substrates of DCAF16, which could presumably be degraded by the still predominantly unmodified fraction of this E3 ligase in cells. Finally, the covalent adduct formed between electrophilic PROTACs and DCAF16 may improve the durability of protein degradation in biological systems, as loss of degradation activity should correlate with turnover of the DCAF16 protein versus clearance of the PROTAC compound. Our washout experiments provide support for this hypothesis, as KB02-SLF, but not lenalidomide-SLF, maintained degradation activity ~24 h-post washing of cells to remove free PROTAC compounds (Fig. 2d).
Our findings also underscore the value of broadly reactive scout fragments as tools to discover ligandable and functional sites on poorly characterized proteins like DCAF16. We should note, however, that scout fragments, even when active as electrophilic PROTACs at sub-µM concentrations, still modify several other proteins in cells, which limits the types of biological studies that can be performed with these early stage compounds. The optimization of ligands for DCAF16-dependent protein degradation therefore represents an important future objective. Here, structural information on DCAF16, in particular, adducted to covalent PROTACs, would be valuable, as our chemical proteomic data point to a site(s) of modification that contains several clustered cysteines. We also wonder whether this feature indicates the potential for endogenous electrophiles and/or oxidative stress to modify DCAF16 and shape its substrate scope in vivo. Finally, from a broader perspective, we believe that our chemical proteomic studies provide an attractive framework for the pursuit of additional E3 ligases capable of supporting protein degradation mediated by electrophilic PROTACs. In this regard, we note that more than 562 cysteines on 211 E3 ligases distributed across all major subclasses (CRL, RING, UBR, HECT, UBOX) have been quantified in aggregate isoTOP-ABPP experiments performed to date (Supplementary Dataset 1)27, 28, 50. The extent to which E3 ligase cysteines can be engaged by heterobifunctional, or even molecular glue, compounds to promote ligand-induced protein degradation should represent a fertile area of future investigation.
Online Methods
Common reagents and antibodies
The anti-HA (3724 and 2367), FLAG (14793 and 8146), His-HRP (9991), DDB1 (6998), BRD4 (13440S), Lamin A/C (2032), K48-linked polyubiquitin (4289), HRP-linked rabbit IgG (7074) and HRP-linked mouse IgG (7076) antibodies were purchased from Cell Signaling Technology. The anti-GAPDH (G8795), anti-FLAG HRP antibody (A8592), anti-FLAG affinity gel (A2220) and anti-HA agarose (A2095) were purchased from Sigma-Aldrich. The anti-β-Actin antibody (sc-47778), polybrene (sc-134220) and desthiobiotin azide (sc-496162) were purchased from Santa Cruz Biotechnology. Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) secondary antibody (A-11001) and Alexa Fluor 568-conjugated goat anti-rabbit IgG (H+L) secondary antibody (A-11011) were purchased from Invitrogen. Recombinant human FKBP12 protein (NBC1–21651) was purchased from Novus Biologicals. FuGene 6 (E2692) transfection reagent and sequencing grade modified trypsin (V5111) were purchased from Promega. Enzyme-linked chemiluminescence (ECL) (32106), ECL plus (32132) western blotting detection reagents, Streptavidin agarose (20349), Pierce high pH reversed-phase peptide fractionation kit (84868) and TMT10plex isobaric label reagent (90110) were purchased from Thermo Scientific. MG132 (S2619) was purchased from Selleck Chemicals. MLN4924 (15217), SLF (10007974) and JQ1 (11187) were purchased from Cayman Chemical. Polyethylenimine (PEI, MW 40,000, 24765–1) was purchased from Polysciences, Inc. Iodoacetamide-alkyne and isotopically-labeled TEV-tags were synthesized as previously described28.
Cell lines
Human Embryonic Kidney (HEK) 239T and MDA-MB-231 cells were obtained from ATCC and cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning) with 10% (v/v) fetal bovine serum (FBS, Omega Scientific) and L-glutamine (2mM). For SILAC experiments, heavy and light labeled cell lines were cultured in DMEM medium with [13C6, 15N2]-L-lysine and [13C6, 15N4]-L-arginine (heavy labeled cells, 100 mg/mL each), or L-lysine and L-arginine (light labeled cells, 100 mg/mL each) for five generations. Both heavy and light SILAC medium were also supplemented with 10% (v/v) dialyzed FBS (Gemini), penicillin, streptomycin, and L-glutamine (2 mM). Human Embryonic Kidney (HEK) 239 cells with DCAF16 gRNA CRISPR editing were purchased from Synthego and cultured in DMEM with 10% FBS and L-glutamine (2 mM). All the cell lines were tested negative for mycoplasma contamination.
Cloning and mutagenesis
Human FKBP12 cDNA with N-terminal FLAG tag or N-terminal FLAG tag and C-terminal nuclear localization sequence (NLS, PKKKRKV) was obtained by reverse transcription polymerase chain reaction (RT-PCR) amplification of a cDNA pool extracted from HEK293T cells and subcloned via EcoRI and BamHI sites into pCDH-CMV-MCS-EF1-Puro vector. Human DCAF16 cDNA with N-terminal HA tag was obtained by RT-PCR amplification of a cDNA pool extracted from HEK293T cells and subcloned via SalI and NotI sites into pRK5 vector. Human BRD4 with C-terminal FLAG tag was inserted into pRK5 vector by Gateway cloning technology. The expression vectors for DCAF16 mutants were generated by QuikChange site-directed mutagenesis.
Generation of FLAG-FKBP12 and FLAG-FKBP12_NLS stably expressed HEK293T cell lines by lentivirus transduction
FLAG-FKBP12 or FLAG-FKBP12_NLS lentivirus was generated by co-transfection of FLAG-FKBP12 or FLAG-FKBP12_NLS, pCMV-dR8.2 and pMD2.G into HEK 293T cells using FuGene 6 transfection reagent. Virus-containing medium were collected 48 h after transfection, filtered with 0.45 µM filter, and used to transduce HEK293T and MDA-MB-231 cells in the presence of 10 µg/mL polybrene. 72 h after transduction, puromycin (2 µg/mL) was added to cells. HEK293T cells stably expressing FLAG-FKBP12 or FLAG-FKBP12_NLS were obtained after puromycin selection for 7 days. HEK293T cells stably expressing pCDH empty vector were generated in parallel as control.
Generation of DTL and DCAF16 knockdown in HEK293T cell lines
shRNA lentivirus was generated by co-transfection of shRNA-containing vector, pCMV-dR8.2 and pMD2.G into HEK293T cells using FuGene 6 transfection reagent. Virus-containing medium were collected 48 h after transfection and used to transiently transduce HEK293T cells stably expressing FLAG-FKBP12_NLS for 48 h. Lentiviral shRNAs targeting human DCAF16 or DTL were in pLKO.1 vector and purchased from Sigma. The sequences of shRNAs are described below.
shLuc (SHC007, Sigma): CCGGCGCTGAGTACTTCGAAATGTCCTCGAGGACATTTCGAAGTACTCAGCGTTTTT
shDTL_1 (TRCN0000118815, Sigma): CCGGCTGGTGAACTTAAACTTGTTACTCGAGTAACAAGTTTAAGTTCACCAGTTTTTG
shDTL_2 (TRCN0000118813, Sigma): CCGGGCCTAGTAACAGTAACGAGTACTCGAGTACTCGTTACTGTTACTAGGCTTTTTG
shDCAF16_1 (TRCN0000122576, Sigma): CCGGCAAACAGCTAAGCCGAACATTCTCGAGAATGTTCGGCTTAGCTGTTTGTTTTTTG
shDCAF16_2 (TRCN0000369937, Sigma): CCGGTCCTGGTTGTATCATGCTAAACTCGAGTTTAGCATGATACAACCAGGATTTTTG
Generation of CRISPR-mediated knockout in HEK293 cell lines
DCAF16 CRISPR knockout HEK293 cell pools were generated by Synthego using nucleofection of Cas9-gRNA ribonucleoprotein (RNP) complex. Editing efficiency 48 h post nucleofection of cell pool was 88%. HEK293 editing cell pools were subjected to single cell sorting in The Flow Cytometry Core Facility at Scripps Research. Individual cell clone was grown in 96-well plate until the cells were confluent. To confirm the editing in DCAF16 gene, genomic DNAs from each clone were extracted using PureLink genomic DNA mini kit (K182001, Invitrogen). DCAF16 gene was amplified by PCR and confirmed by DNA sequencing. Indel analysis was performed using ICE analysis from Synthego (https://ice.synthego.com/#/). The sequences of DCAF16 gRNA and sequencing primers is described blow.
DCAF16 gRNA: TCTGACAAGTGGTCAGGAGA
DCAF16 sequencing primer (forward): GCCAGGATTTGAAGGAGATACTCT
DCAF16 sequencing primer (reverse): TATTCAGGTATGGGAGTGGCTCTA
To confirm DCAF16 knockout at the protein level, lentivirus containing FLAG-FKBP12_NLS was used to infect three wild type DCAF16 clones (clone 6, 17, 18) and three DCAF16 knockout clones (clone 3, 4, 22). Clone 6 stably expressing FLAG-FKBP12_NLS was grown in heavy SILAC DMEM medium for 5 generations. The other clones stably expressing FLAG-FKBP12_NLS were grown in light SILAC DMEM medium for 5 generations. All 6 clones were treated with 5 µM KB02-SLF and 10 µM MG132 for 2 h. Cells were collected and analyzed for FLAG-FKBP12_NLS associating proteins using the same method described below. Proteins from clone 6 (heavy labeled) were combined with the other clones (light labeled) separately and identified by LC-MS/MS.
Cell lysis and Western blot
Cells were collected and lysed in 1% NP-40 lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 10% glycerol, 1% Nonidet P-40) with cOmplete protease inhibitor cocktail (Roche). Cells were vortexed and sonicated for 5 pulses (40%, 4). The supernatant was collected after centrifugation at 16,000g for 10 min at 4 °C. Protein concentration was determined by DC assay (5000112, Bio-Rad). Protein lysate was heated at 95 °C for 5 min in 1X Laemmli sample buffer. Proteins were resolved by 12% or 14% Novex Tris-Glycine mini gels (Invitrogen) and transferred to polyvinylidene fluoride (PVDF) membrane (0.2 µM, 1620177, Bio-Rad). The membrane was blocked with 5% BSA in TBST buffer (0.1% Tween 20, 20 mM Tris-HCl 7.6, 150 mM NaCl) at room temperature for 1 h. The antibody was diluted with fresh 5% BSA in TBST buffer (1:10000 dilution for FLAG, HA and β-Actin, 1:1000 dilution for others) and incubated with membrane (1 h room temperature for FLAG, HA and β-Actin, overnight (14 h) 4 °C for others). Membrane was washed three times with TBST buffer and incubated with secondary antibody (1:5000 dilution in 5% BSA in TBST) at room temperature for 1 h. Membrane was washed three times with TBST buffer. The chemiluminescence signal in membrane was recorded after developing in ECL or ECL plus western blotting detection reagent using CL-XPosure film (Thermo Scientific). Relative band intensities were quantified using ImageJ.
Subcellular fractionation
HEK293T cells stably expressing FLAG-FKBP12 or FLAG-FKBP12_NLS were harvested. Cell pellets were re-suspended in subcellular fraction buffer (250 mM Sucrose, 20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, protease inhibitor cocktail) and homogenized on ice by 20 passes through a 25-gauge syringe needle. The pellets after centrifugation (720 g, 5 min) were homogenized on ice by 10 passes through a 25-gauge syringe needle. Nuclear fraction was collected as the pellets after centrifuging at 720 g for 5 min. The supernatant was ultracentrifuge at 100,000 g for 1 h. The resulting supernatant was the cytosol fraction. Equivalent portions of the cytosol and nuclear fractions were then subjected to Western blot analyses.
Immunoprecipitations
Cells were collected and lysed in 1% NP-40 lysis buffer with cOmplete protease inhibitor cocktail on ice for 10 min, followed by 5 pulses of sonication (40%, 4). After centrifugation at 16,000 g for 10 min at 4 °C, the supernatant was collected. FLAG affinity gel (20 µL slurry per sample) was incubated with protein lysates at 4 °C for 1 h and washed four times with IP washing buffer (0.2% NP-40, 25 mM Tris-HCl pH 7.4, 150 mM NaCl). The affinity gel was heated at 95 °C for 10 min in 2X Laemmli sample buffer, followed by western blot analysis.
Far-western blot
HEK293T cells were transfected with HA-DCAF16 plasmid by PEI transfection reagent for 24 h and treated with DMSO or KB02-SLF (5 µM) for 2 h. Cells were collected and lysed in 1% NP-40 lysis buffer with complete protease inhibitor cocktail. HA immunoprecipitation (20 µL slurry anti-HA agarose, 4 °C, 2 h) was performed with 2 mg of total protein lysates to purify HA-DCAF16. After washing the HA resin four times with IP washing buffer, HA affinity gel was heated at 95 °C for 10 min in 2X Laemmli sample buffer to elute HA-DCAF16 protein. Proteins were resolved by 14% Novex Tris-Glycine mini gels and transferred to PVDF membrane. The membrane was blocked with 5% BSA in TBST buffer at room temperature for 1 h. The recombinant His-tagged FKBP12 protein was diluted with fresh 5% BSA in TBST buffer (final concentration: 2 µg/mL) and incubated with membrane at 4 °C overnight (14 h). Membrane was washed three times with TBST buffer and incubated with anti-His-HRP antibody (1:1000 dilution in 5% BSA in TBST) at room temperature for 4 h. Membrane was washed three times with TBST buffer. The chemiluminescence signal in membrane was recorded after developing in ECL plus western blotting detection reagent using CL-XPosure film. The same protein samples were resolved by another 14% Novex Tris-Glycine mini gel and analyzed by anti-HA Western blot in parallel.
Immunofluorescence
Cells were seeded and grown in 35 mm glass bottom dish (MatTek). After compound treatment, cells were rinsed twice with PBS and fixed in 1 mL of 4% paraformaldehyde (v/v in PBS) for 15 min at room temperature. The fixed cells were rinsed twice with PBS, permeabilized and blocked with 0.1% Triton X-100 (v/v in 5% BSA in PBS) for 30 min at room temperature. The cells were incubated overnight (14 h) at 4°C with FLAG or HA antibody at 1/100 dilution (in 0.1% Triton X-100/5% BSA in PBS). Cells were washed with 0.1% Triton X (in PBS) three times and incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) secondary antibody (for FLAG tag) or Alexa Fluor 568-conjugated goat anti-rabbit IgG (H+L) secondary antibody (for HA tag) at 1/1000 dilution (in 0.1% Triton X-100/5% BSA in PBS) at room temperature in dark for 1 h. Cells were washed with 0.1% Triton X (in PBS) three times and mounted with ProLong Gold Antifade Mountant with DAPI (P36931, Invitrogen). Cells were imaged with Zeiss LSM780 in The Core Microscopy Facility at Scripps Research. Images were processed in ImageJ software. To quantify the degree of nucleus-localized fluorescence signal, background was subtracted. The nuclear and whole cell area were selected and quantified for each cell examined. Relative nucleus with respect to whole cell fluorescence intensity was presented.
Identification of FLAG-FKBP12_NLS interacting proteins
HEK293T light and heavy SILAC cells stably expressing FLAG-FKBP12_NLS were treated with DMSO and KB02-SLF (2 or 10 µM) for 2 h, respectively, in the presence of 10 µM MG132. Heavy and light cells were collected and lysed in 1% NP-40 lysis buffer with complete protease inhibitor cocktail. FLAG immunoprecipitation (20 µL slurry per sample) was performed with 2 mg of total protein lysates to enrich FLAG-FKBP12_NLS from light and heavy cell lysates. After washing the FLAG resin four times with IP washing buffer, FLAG resin from light and heavy samples were combined and washed once with PBS. FLAG-FKBP12_NLS and its associating proteins were eluted by heating at 65 ºC for 10 min with 8 M urea in PBS, then reduced with 12.5 mM DTT at 65 ºC for 15 min and alkylated with 25 mM iodoacetamide at 37 ºC for 30 min. The protein solution was diluted with PBS to 2 M urea and digested with 2 µg trypsin at 37 ºC for 6 h. Tryptic peptides were acidified with 5% formic acid and loaded onto a silica capillary column (250 µm) packed with 3 cm of C18 resin (Aqua 5 µm, Phenomenex). Peptides were analyzed on LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific) coupled with Thermo UltiMate 3000 UHPLC system. Peptides were separated on a capillary column packed with 3 cm of strong cation exchange (SCX) resin (Luna 5 µm, Phenomenex), 10 cm of C18 resin (Aqua 5 µm, Phenomenex) and a 5 µm tip. A five-step MudPIT method and CIMAGE software were used to analyze the peptides as previously described51. To generate the plot in Fig. 3d and Supplementary Fig. 5c, the following quality filters were applied: (1) proteins must have at least two quantified peptides; (2) proteins must be absence in control SILAC (light and heavy amino acid-labeled HEK293T cells stably expressing pCDH empty vector were treated for 2 h with DMSO or 10 µM KB02-SLF, respectively in the presence of 10 µM MG132). (3) proteins must be quantified in at least two replicates; (4) the coefficient of variation of heavy/light ratio from three replicates is < 0.5.
isoTOP-ABPP
HEK293T cells were treated with DMSO or 10 µM KB02-SLF for 2 h. Cells were collected and subjected to isoTOP-ABPP sample preparation using the same protocol as previously described28.
Quantitative ABPP
HEK293T cells were treated with DMSO (four biological replicates), 2 µM KB02-SLF (three biological replicates) or 20 µM KB02-JQ1 (three biological replicates) for 1.5 h (ten samples in total). Cells were collected, washed once with PBS and lysed in PBS by sonication (40%, 4, 20 pulses). Samples were clarified by centrifugation for 10 min at 8,000 g. Lysate was adjusted to 1.0 mg/mL in 500 µL. Samples were labeled with 100 µM iodoacetamide alkyne (5 µL of 10 mM stock in DMSO) at room temperature for 1 h, conjugated by CuACC to desthiobiotin azide (10 µL of 5 mM stock in DMSO, final concentration = 100 µM), tris(2-carboxyethyl)phosphine hydrochloride (TCEP, 10 µL of 50 mM stock in H2O, final concentration = 1 mM), TBTA (30 µL of 1.7 mM stock in 4:1 tBuOH:DMSO, final concentration = 100 µM) and CuSO4 (10 µL of 50 mM stock in H2O, final concentration = 1 mM). The samples were allowed to react for 1 h at room temperature. Proteins were precipitated by adding 3.5 mL of H2O, 4mL of methanol and 1.5 mL of chloroform, followed by centrifugation at 5,000 g for 15 min (4 ºC). After aspirating the top layer, the protein pellets with bottom layer were washed with 8 mL cold methanol by centrifugation at 5,000 g for 15 min (4 ºC). Protein pellets were resuspended in 90 µL of buffer containing 9 M urea, 10 mM DTT and 50 mM tetramethylammonium bicarbonate (pH 8.5), sonicated (40%, 4, 5 pulses) and heated at 65 ºC for 20 min. Samples were cooled to room temperature and alkylated with 50 mM iodoacetamide (10 µL of 500 mM stock in H2O) at 37 ºC for 30 min. Samples were diluted with 350 µL of 50 mM tetramethylammonium bicarbonate (pH 8.5) and digested with 2 µg trypsin at 37 ºC for 4 h. 40 µL of streptavidin-agarose beads slurry was washed in IP washing buffer and added to each sample. Beads with peptide samples were rotated for 1 h at room temperature, filtered off by micro Bio-Spin chromatography column (7326204, Bio-Rad) and washed with 1 mL of IP washing buffer (three times), 1 mL of PBS (three times) and 1 mL of H2O. Peptides were eluted by adding 300 µL of 50% acetonitrile with 0.1% of formic acid and dried in SpeedVac vacuum concentrator.
Peptides were resuspended in 100 µL of 30% acetonitrile in 200 mM EPPS (pH 8) and labeled with TMT10plex isobaric reagent following the manufacturer’s instructions. 0.15% of hydroxylamine (3 µL of 5% stock in H2O) was added to each sample to quench the reaction (incubate for 15 min). After adding 5 µL of formic acid to each sample, 10 peptide samples were combined and dried in SpeedVac vacuum concentrator. Peptides were further fractionated into 21 fractions using Pierce high pH reversed-phase peptide fractionation kit following the manufacturer’s instructions. The 21 resulting fractions were pooled into 7 fractions by combing every 7th fraction (eg. fraction 1, 8 and 15, fraction 2, 9 and 16, etc.) for subsequent mass spectrometry analysis.
The samples were analyzed in Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) coupled to UltiMate 3000 HPLC system (Thermo Scientific). Peptides were separated on a capillary column packed with 30 cm of C18 resin (ACQUITY UPLC BEH, 1.7 µm, Waters) and a 5 µm tip. The MS1 scan parameters were set up as described below: 1) Orbitrap resolution: 120,000, 2) Scan range: m/z 400–1700, 3) RF lens (%): 60, 4) AGC target: 2×105, 5) Maximum injection time: 50 ms. The MS2 data were acquired in the ion trap with the parameter settings as described below: 1) CID collision energy (%): 35, 2) AGC target: 1.8×104, 3) Maximum injection time: 120 ms, 4) Activation Q: 0.25. The MS3 data were acquired in the Orbitrap with the parameter settings as described below: 1) Orbitrap resolution: 50,000, 2) Number of SPS precursors: 10, 3) First mass: m/z 100, 4) HCD collision energy (%): 55, 5) AGC target: 1.5×105, 6) maximum injection time: 120 ms. The raw data was acquired in Xcalibur operation software.
The MS2 and MS3 spectra data were extracted and analyzed in IP2 Integrated Proteomics Pipeline (http://goldfish.scripps.edu/). MS2 spectra were searched using the ProLuCID algorithm (http://fields.scripps.edu/downloads.php) and a reverse concatenated, nonredundant variant of the Human UniProt database (release-2012_11). Cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146) and up to four differential modification for desthiobiotin-IA (+494.3207). Lysine residues and peptide N-termini were searched with a static modification for TMT-tag labeling (+229.162932). Methionine residues were searched up to four differential modification for oxidation (+15.9949). MS3 quantification was performed using 10plex TMT analysis parameters (m/z 126.127726, 127.124761, 127.131081, 128.128116, 128.134436, 129.131471, 129.13779, 130.134825, 130.141145 and 131.13818) with the mass tolerance of 30 ppm.
Proteome-wide identification of KB02-JQ1- or KB02-SLF-induced protein degradation
HEK293T light and heavy SILAC cells were treated with DMSO and KB02-JQ1 (20 µM) or KB02-SLF (2 µM) for 24 h, respectively. Light and heavy cells were collected and lysed in 1% NP-40 lysis buffer with cOmplete protease inhibitor cocktail. Cells were vortexed and sonicated for 5 pulses (40%, 4). The supernatant was collected after centrifugation at 16,000 g for 10 min at 4 °C. Protein concentration was determined by DC assay. 50 µg proteome from light and heavy samples were mixed, followed by methanol/chloroform precipitation. Protein pellets were heated at 65 ºC for 10 min with 8 M urea in PBS, then reduced with 12.5 mM DTT at 65 ºC for 15 min and alkylated with 25 mM iodoacetamide at 37 ºC for 30 min. The protein solution was diluted with PBS to 2 M urea and digested with 2 µg trypsin at 37 ºC for 6 h. Tryptic peptides were acidified with 5% formic acid. 5 µg peptides were loaded onto a silica capillary column (250 µm) packed with 3 cm of C18 resin. The same MudPIT method and CIMAGE software as described above were used to analyze the peptides. The median SILAC ratios from quantified peptides was used as measures of protein abundance. The following quality filters were further applied to generate the plot in Fig. 6h: (1) proteins must have at least two quantified peptides; (2) proteins must be present in both replicates; (3) for each protein, the standard deviation of measured ratios of peptides must be < 1.0.
Cell viability assay
HEK293T cells were seeded in 96-well clear bottom white plate (Corning) at 3×104 cells per well in 100 µL of DMEM medium and grown for 24 h. The cells were treated with 0.625, 1.25, 2.5, 5, 10, 20 and 40 µM of KB02-SLF or KB02-JQ1 (DMSO stock, final DMSO concentration is 0.1% (v/v)) in 100 µL of DMEM medium for 24 h. 50 µL of Cell Titer Glo reagent (Promega) was added to each well and incubate for 10 min at room temperature. The luminescence was read on CLARIOstar (BMG LABTECH).
qPCR analysis
Total RNA was extracted from the cells using RNeasy mini kit (74104, Qiagen). cDNA was synthesized using iScript Reverse Transcription Supermix (1708840, Bio-Rad). Amplification of DCAF16, DTL and GAPDH genes was performed using SYBR Select Master Mix (4472908, Applied Biosystems) on an ABI Real Time PCR System (Applied Biosystems). The sequences of qPCR primers are described below. Relative DCAF16 and DTL gene expression was normalized to the GAPDH gene.
GAPDH primer (forward): CTGGGCTACACTGAGCACC
GAPDH primer (reverse): AAGTGGTCGTTGAGGGCAATG
DCAF16 primer (forward): AGTCTTGCCTGGCAGGTTAAG
DCAF16 primer (reverse): GGGACTTGTAAGAGGCTTTTGAA
DTL primer (forward): TCACTGGAATGCCGTCTTTGA
DTL primer (reverse): CTCACCAGCTTTTACGTCCC
LC-MS/MS detection of KB02-PEG0-SLF-modified tryptic peptides on DCAF16
HEK293T cells were transfected with HA-DCAF16 plasmid by PEI transfection reagent for 24 h and treated with DMSO or 10 µM KB02-PEG0-SLF for 2 h. Cells were collected and lysed in 1% NP-40 lysis buffer with complete protease inhibitor cocktail. HA immunoprecipitation (40 µL slurry anti-HA agarose, 4 °C, 2 h) was performed with 10 mg of total protein lysates to purify HA-DCAF16. After washing the HA resin three times with IP washing buffer and once with PBS, HA-DCAF16 protein was eluted by heating at 65 ºC for 10 min with 8 M urea in PBS, then reduced with 12.5 mM DTT at 65 ºC for 15 min and alkylated with 25 mM iodoacetamide at 37 ºC for 30 min. The protein solution was diluted with PBS to 2 M urea and digested with 2 µg trypsin at 37 ºC for 6 h. Tryptic peptides were acidified with 5% formic acid and loaded onto a silica capillary column (250 µm) packed with 3 cm of C18 resin (Aqua 5 µm, Phenomenex). Peptides were analyzed on LTQ-Orbitrap Elite mass spectrometer (Thermo Scientific) coupled with a Thermo UltiMate 3000 UHPLC system. Peptides were separated on a capillary column packed with 10 cm of C18 resin (Aqua 5 µm, Phenomenex) and a 5 µm tip. MS parameters were set as previously described51. The raw data was acquired in Xcalibur operation software.
Statistical analysis
Quantitative data were expressed in scatter plots with mean ± SEM (standard error of the mean, shown as error bar) shown. Differences between two groups were examined using unpaired two-tailed Student’s t test. The P values were indicated (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001). P values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgements.
This work was supported by the NIH (CA087660 (B.F.C.), CA231991 (B.F.C.), CA211526 (M.M.D.), CA212467 (V.M.C.)) and the Damon-Runyon Cancer Research Foundation (X.Z. DRG-2341–18).
Footnotes
Competing Financial Interests. Dr. Cravatt is a founder and scientific advisor to Vividion Therapeutics, a biotechnology company interested in developing small-molecule therapeutics.
Data availability. All data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
References
- 1.Cromm PM & Crews CM Targeted Protein Degradation: from Chemical Biology to Drug Discovery. Cell chemical biology 24, 1181–1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lebraud H & Heightman TD Protein degradation: a validated therapeutic strategy with exciting prospects. Essays Biochem 61, 517–527 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Bondeson DP & Crews CM Targeted Protein Degradation by Small Molecules. Annu Rev Pharmacol Toxicol 57, 107–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins I, Wang H, Caldwell JJ & Chopra R Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway. Biochem J 474, 1127–1147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes SJ & Ciulli A Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem 61, 505–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu G et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito T & Handa H Cereblon and its downstream substrates as molecular targets of immunomodulatory drugs. Int J Hematol 104, 293–299 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Girona A et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia 26, 2326–2335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito T et al. Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Matyskiela ME et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Uehara T et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat Chem Biol 13, 675–680 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Han T et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Lu J et al. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem Biol 22, 755–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto KM et al. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A 98, 8554–8559 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13, 514–521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin C et al. Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression. J Med Chem 61, 6685–6704 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B et al. Discovery of a Small-Molecule Degrader of Bromodomain and Extra-Terminal (BET) Proteins with Picomolar Cellular Potencies and Capable of Achieving Tumor Regression. J Med Chem 61, 462–481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zorba A et al. Delineating the role of cooperativity in the design of potent PROTACs for BTK. Proc Natl Acad Sci U S A 115, E7285–E7292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bondeson DP et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 11, 611–617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai AC & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okuhira K et al. Specific degradation of CRABP-II via cIAP1-mediated ubiquitylation induced by hybrid molecules that crosslink cIAP1 and the target protein. FEBS Lett 585, 1147–1152 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Schneekloth JS Jr. et al. Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc 126, 3748–3754 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Schneekloth AR, Pucheault M, Tae HS & Crews CM Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg Med Chem Lett 18, 5904–5908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bondeson DP et al. Lessons in PROTAC Design from Selective Degradation with a Promiscuous Warhead. Cell Chem Biol 25, 78–87 e75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang HT et al. A Chemoproteomic Approach to Query the Degradable Kinome Using a Multi-kinase Degrader. Cell Chem Biol 25, 88–99 e86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Peled L et al. Chemical Proteomics Identifies Druggable Vulnerabilities in a Genetically Defined Cancer. Cell 171, 696–709 e623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backus KM et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Weerapana E, Blewett MM & Cravatt BF A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods 11, 79–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amara JF et al. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci U S A 94, 10618–10623 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nabet B et al. The dTAG system for immediate and target-specific protein degradation. Nat Chem Biol 14, 431–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soucy TA et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Kawakami T et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J 20, 4003–4012 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakata E et al. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol 14, 167–168 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Deshaies RJ, Emberley ED & Saha A Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem 54, 41–56 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Weerapana E et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin J, Arias EE, Chen J, Harper JW & Walter JC A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23, 709–721 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Sansam CL et al. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev 20, 3117–3129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higa LA et al. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Lee J & Zhou P DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 26, 775–780 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Guerrero-Santoro J et al. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res 68, 5014–5022 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Kim S, Kim D, Cho SW, Kim J & Kim JS Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y, Li Q & Chen XZ Detecting protein-protein interactions by Far western blotting. Nat Protoc 2, 3278–3284 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Filippakopoulos P et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raina K et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 113, 7124–7129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka M et al. Design and characterization of bivalent BET inhibitors. Nat Chem Biol 12, 1089–1096 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan J et al. Tetrameric Acetyl-CoA Acetyltransferase 1 Is Important for Tumor Growth. Mol Cell 64, 859–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blewett MM et al. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci Signal 9, rs10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 51.Weerapana E, Speers AE & Cravatt BF Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc 2, 1414–1425 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.