Summary
Invasion of intestinal epithelial cells by Campylobacter jejuni is a critical step during infection of the intestine by this important human pathogen. In this study we investigated the role played by DNA supercoiling in the regulation of invasion of epithelial cells and the mechanism by which this could be mediated. A significant correlation between more relaxed DNA supercoiling and an increased ability of C. jejuni strains to penetrate human epithelial cells was demonstrated. Directly inducing relaxation of DNA supercoiling in C. jejuni was shown to significantly increase invasion of epithelial cells. Mutants in the fibronectin binding proteins CadF and FlpA still displayed an increased invasion after treatment with novobiocin suggesting these proteins were not essential for the observed phenotype. However, a large increase in protein secretion from multiple C. jejuni strains upon relaxation of DNA supercoiling was demonstrated. This increase in protein secretion was not mediated by outer membrane vesicles and appeared to be dependent on an intact flagellar structure. This study identifies relaxation of DNA supercoiling as playing a key role in enhancing C. jejuni pathogenesis during infection of the human intestine and identifies proteins present in a specific invasion associated secretome induced by relaxation of DNA supercoiling.
Introduction
Campylobacter jejuni infection is the leading cause of bacterial gastroenteritis. Although many putative virulence factors have been identified, relatively little is known about the molecular mechanisms responsible for the pathogenesis of infection. An integral step during C. jejuni human disease is invasion of intestinal epithelial cells, which does not occur during the commensal infection of chickens. A greater understanding of the mechanisms utilized by C. jejuni to invade human epithelial cells and the genetic regulation of this process is thus essential in order to further our understanding of how this organism causes disease and reduce the burden of C. jejuni gastroenteritis on society (Young et al., 2007; Ó Cróinín and Backert, 2012).
A number of C. jejuni virulence factors have been shown to contribute to C. jejuni adhesion and invasion in-vitro (Ó Cróinín and Backert, 2012). It is clear that production of Peb1 and Peb4 are important for the ability of C. jejuni to adhere to the epithelial cell boundary but their influence on C. jejuni adhesion/invasion appears to be indirect (Pei et al., 1991; Kervella et al., 1993; Burucoa et al., 1995; Pei et al., 1998; Kale et al., 2011). JlpA is a 42 kDa C. jejuni specific adhesin, which is surface exposed. Using strain TGH9011 (ATCC 43431) it was shown that a jlpA mutant associates with HEp-2 cells at a lower rate than that of the parent strain (Jin et al., 2001). However, two subsequent studies failed to find significant reductions in adhesion or invasion using jlpA mutants in C. jejuni strains 81-176 or F38011 (Flanagan et al., 2009; Novik and Galán, 2010), indicating that the importance of jlpA may be isolate-specific. CadF ‘Campylobacter adhesion to fibronectin’ and FlpA ‘fibronectin like protein A’ are among the best characterised C. jejuni virulence factors and have consistently been shown to contribute to pathogenicity in a variety of studies (Konkel et al., 1997; Moser and Salnikow, 1997; Ziprin et al., 1999). CadF is a 37 kDa outer-membrane protein that can bind to purified fibronectin (Konkel et al., 1997). FlpA was initially identified as a potential virulence factor when 2D proteomic analysis of a fresh human isolate strain JHH1 revealed a two fold up-regulation in a protein encoded by cj1279c compared to a reference strain ATCC 700297 (Cordwell et al., 2008). FlpA also plays a role during colonization of chickens. An F38011 flpA- mutant strain associates with chicken epithelial cells at roughly two thirds that of the wild type (Flanagan et al., 2009). Reduced adhesion to INT-407 cells also was shown using an flpA- strain and the attenuated interaction was suggested to be due to a reduced ability to bind extracellular fibronectin (Konkel et al., 2010).
Multiple studies have shown that the C. jejuni flagellum functions as a secretory organelle. A number of non-flagellar proteins appear to be dependent upon correct flagellar formation for their delivery to the extracellular environment or for translocation to the intracellular environment of cell lines in-vitro. These include the Cia (Campylobacer invason antigen) proteins CiaB, CiaC, CiaD, CiaI, FlaC and FspA (Konkel et al., 1999; Konkel et al., 2004; Song et al., 2004; Poly et al., 2007; Christensen et al., 2009; Buelow et al., 2011; Samuelson et al., 2013). CiaC and CiaI have also been shown to alter host cell signalling once translocated across the epithelial cell boundary (Buelow et al., 2011; Neal-McKinney and Konkel, 2012). In the absence of a classical T3SS injectosome, the secretion of non-flagellar proteins via the C. jejuni flagellum has been implicated as a possible mechanism used by the bacteria to inject effector proteins to mediate bacterial internalization of in-vitro cell lines.
Mechanisms of C. jejuni gene regulation are poorly defined when compared to other enteric pathogens, although recent strides have been made in the genetic regulation of C. jejuni flagellar formation and function (Lertsethtakarn et al., 2011). The regulation of Campylobacteriales DNA supercoiling is also poorly defined compared to other pathogens. Campylobacteriales do not contain a number of well characterized nucleoid-associated proteins such as FIS, IHF and H-NS that are found in Escherischia coli and Salmonella enterica (Kelly et al., 2004; Mangan et al., 2006; Mangan et al., 2011). In Helicobacter pylori it has been shown that bacterial growth phase strongly influences DNA supercoiling profiles in a manner similar to that seen in E. coli and S. enterica. Relaxation of negative DNA supercoiling in H. pylori results in a large decrease in expression of important flagellar genes flaA and flgR as well as the DNA gyrase genes, gyrA and gyrB (Ye et al., 2007). In a recent study we revealed that relaxation of DNA supercoiling has a similar effect on motility and flagellar gene expression in C. jejuni and that supercoiling levels were affected by growth in the presence of mucus (Shortt et al., 2016). This finding implicated a possible role for DNA supercoiling in the regulation of virulence genes in C. jejuni during infection of the host.
DNA supercoiling has previously been shown to have a significant influence on the ability of pathogens to invade epithelial cells and survive within macrophages. Salmonella and Shigella species in particular utilise the regulatory influence of DNA supercoiling for the secretion of proteins to increase their pathogenicity and survival (Dorman et al., 1990; Ó Cróinín et al., 2006). For example in S. enterica increased expression of the SPI-I gene, invA, occurs during growth of cultures containing high salt, a condition known to cause increased levels of negative DNA supercoiling (Galán and Curtiss, 1990). Intracellular growth of S. enterica within the J774A.1 macrophage cell line has also been shown to result in the opposite effect, a reduction of negative DNA supercoiling. This observation was shown to correlate with increased expression of the ssrA and ssaG promoters, indicative of expression of the SPI-II T3SS (Ó Cróinín et al., 2006).
The aim of this study was to determine whether changes in rates of C. jejuni DNA supercoiling results in an altered ability for the bacterium to invade in-vitro cell lines as has been previously described for Salmonella and Shigella species. Furthermore, the effect of DNA supercoiling on known adhesins and secretion systems in C. jejuni was studied to identify the mechanism by which any change in the ability of the bacteria to invade epithelial cells could be mediated.
Results
C. jejuni DNA supercoiling is correlated with invasion of human cell lines
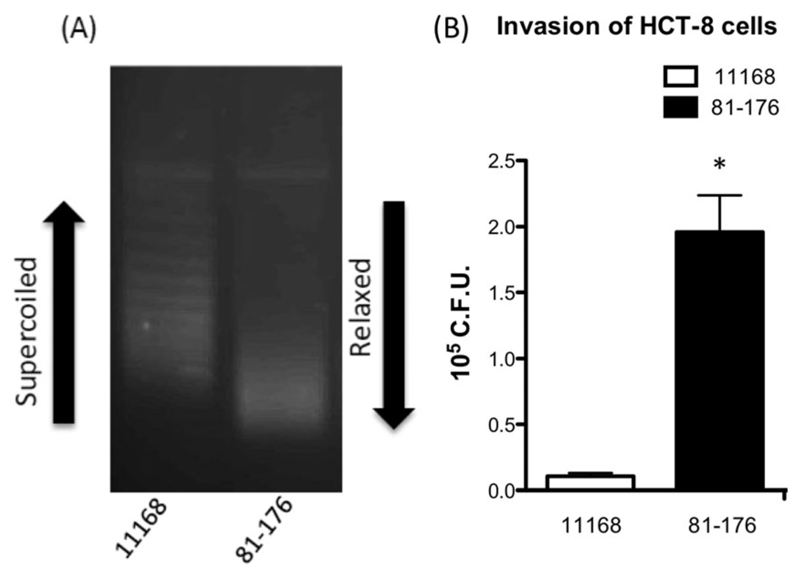
To measure relative rates of DNA supercoiling, pRY107 was extracted from NCTC11168 and 81-176 grown to mid log phase followed by analysis using chloroquine gel electrophoresis. At 10 μg/ml chloroquine, relaxed topoisomers of pRY107 migrate further than those more negatively supercoiled. Figure 1A demonstrates that strain NCTC11168 maintained pRY107 in a more negatively supercoiled state than plasmid extracted from strain 81-176. This increased rate of negative DNA supercoiling by NCTC11168 correlated with lower numbers of HCT-8 cell internalized bacteria when compared to the more relaxed, and significantly more invasive strain 81-176 after 5 h as observed in Fig. 1B.
Fig. 1.
Correlation exists between DNA supercoiling profiles and invasion of epithelial cells.
A. Chloroquine gel analysis revealing resting supercoiling profiles of reporter plasmid pRY107 in strains NCTC11168 and 81-176 after overnight growth.
B. The topologically more relaxed strain 81-176 shows significantly greater numbers of invasion of HCT-8 cells after 5 h than the more negatively supercoiled strain NCTC11168. * denotes a P value < 0.05.
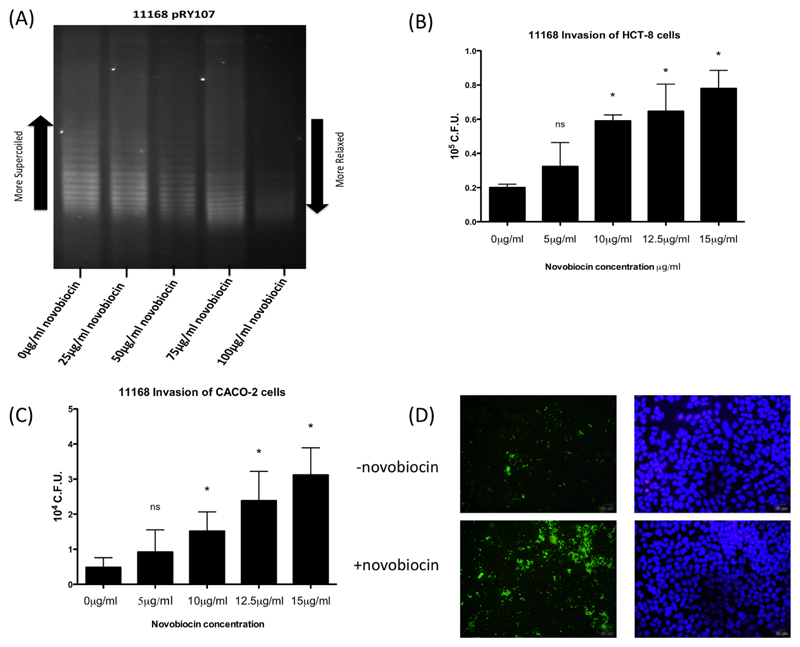
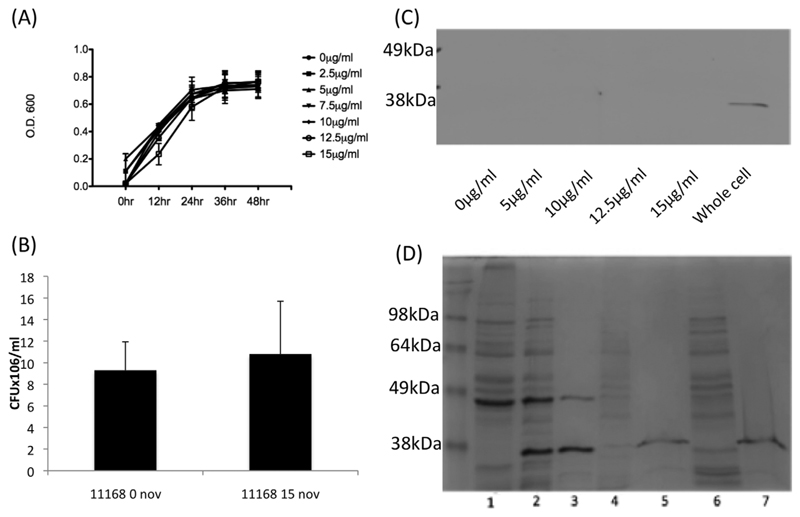
To investigate if reduced negative C. jejuni supercoiling altered rates of epithelial cell invasion, the poorly invasive, highly negatively supercoiled NCTC11168 was cultured using sub-inhibitory concentrations of the antibiotic novobiocin. Incubation of NCTC11168 with novobiocin to artificially manipulate DNA topology confirmed a dose dependant relaxation of pRY107 supercoiling (Fig. 2A) without affecting viability as previously described (Shortt et al., 2016). Addition of novobiocin to NCTC11168 cultures also resulted in a large and dose dependent increase in the ability of NCTC11168 to penetrate the HCT-8 (Fig. 2B) and CACO-2 cell lines (Fig. 2C) after 5 h. When invading bacteria were calculated as a percentage of total associated bacteria a similar trend was observed suggesting that greater numbers of associated bacteria were invading as DNA relaxation increased (Supporting Information Fig. S1A and B). This increased invasion of novobiocin treated bacteria with HCT-8 cells was confirmed and visualized using immunofluorescent microscopy (Fig. 2D).
Fig. 2.
A. Treatment of NCTC11168 with the DNA gyrase inhibitor novobiocin results in dose dependent DNA relaxation as observed in mid log bacteria. Treatment of NCTC11168 overnight cultures with sub-inhibitory concentrations of novobiocin results in a dose dependent increase in bacterial invasion of the HCT-8 (B) and Caco2 (C) cell lines after 5 h. * denotes a P value <0.05 compared to the untreated strain. (D) Immunofluorescence microscopy confirms a higher rate of HCT-8 cell interaction following culture of NCTC11168 in the presence of novobiocin.
CadF and FlpA proteins are not necessary for increased NCTC11168 invasion following DNA relaxation
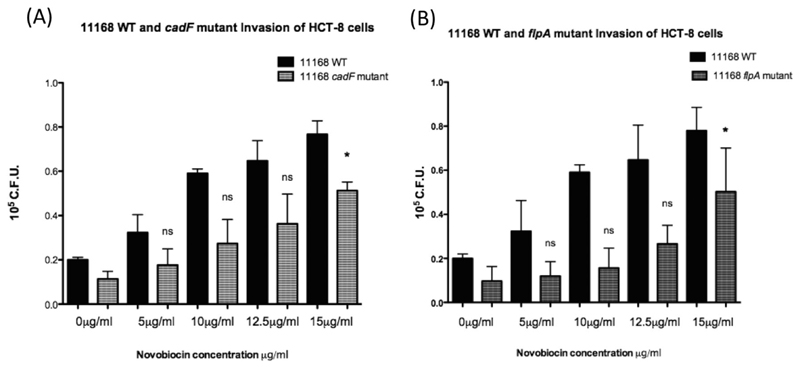
To investigate if the observed increase in NCTC11168 invasion following DNA relaxation was mediated by CadF or FlpA, insertional cadF and flpA mutants were transformed into strain NCTC11168. These strains were then cultured in a range of novobiocin concentrations and used as individual inocula for infection of HCT-8 cells (Fig. 3). As shown in Fig. 3, disruption of either cadF or flpA resulted in lower levels of invasion than NCTC11168 WT, as has been previously reported (Moser and Salnikow, 1997; Konkel et al., 2010). Figure 3 also shows that neither the CadF nor FlpA proteins are required to mediate an increase in NCTC11168 invasion following DNA relaxation. This trend was also observed when results were expressed as invading bacteria as a percentage of total associated bacteria (Supporting Information Fig. S1C).
Fig. 3.
Disruption of either cadF or flpA genes does not prevent increased invasion of NCTC11168 following growth in the presence of novobiocin. Invasion of the HCT-8 cell line using NCTC11168 with a disrupted (A) cadF or (B) flpA gene resulted in increased invasion following DNA relaxation but to a lesser extent observed than the wild type for both mutants. * denotes a P value <0.05 compared to the untreated strain.
DNA relaxation is correlated with increased protein within bacterial supernatants
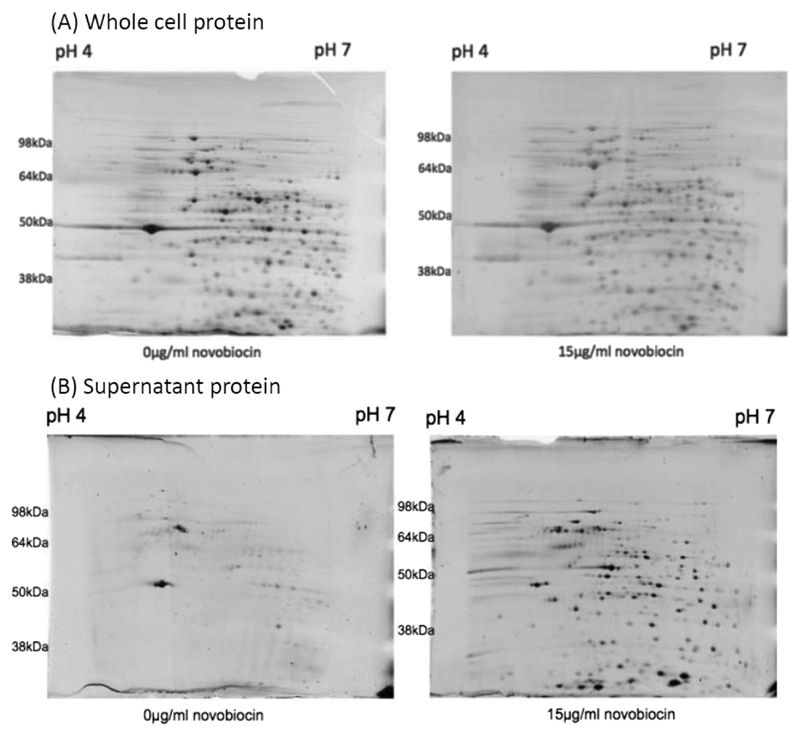
Protein secretion to both the extracellular environment and translocation across epithelial cell membranes has been proposed as a mechanism by which C. jejuni can enhance its ability to cause enteric disease (Ó Cróinín and Backert, 2012). Figure 4A shows that growth of NCTC11168 overnight in the presence and absence of 15 μg/ml novobiocin, resulted in a minimal effect on total cell protein production. In contrast, the addition of 15 μg/ml novobiocin resulted in a large increase in the amount of protein detected within concentrated bacterial supernatants (Fig. 4B). Strikingly, this induction in protein secretion begins at a concentration of 10 μg/ml novobiocin (Supporting Information Fig. S1D), the concentration at which invasion of HCT-8 and CACO-2 cells became statistically significant (Fig. 2).
Fig. 4.
Reduced negative DNA supercoiling induced by 15 ug/ml novobiocin does not lead to large changes in (A) whole cell protein production but leads to a large increase in (B) protein found within concentrated bacterial supernatants.
Increased extracellular protein following DNA relaxation is not mediated by cell lysis or through outer membrane vesicles (OMV’s)
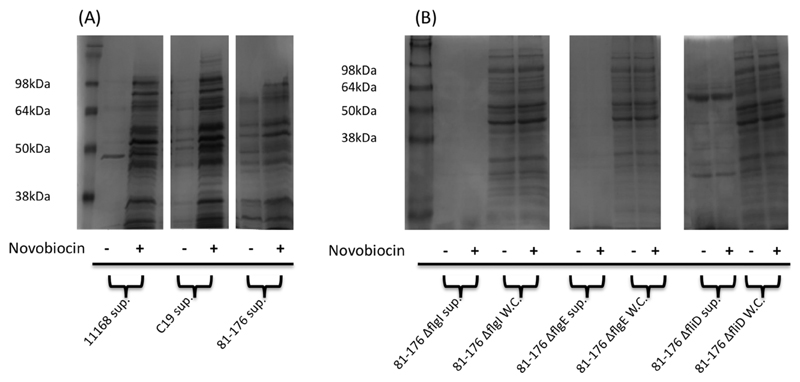
In a previous study we have shown that sub inhibitory concentrations of novobiocin had no significant effect on cell growth or viability (Shortt et al., 2016). Growth curve analysis and viability counts under the conditions used in this study again revealed that no evidence of cell lysis could be observed (Fig. 5A and B). Furthermore, supernatant fractions were probed with antibodies raised against the outer membrane protein CadF (Fig. 5C) and the protein could not be detected in the supernatant from novobiocin treated cultures suggesting that increasing quantities of protein within the bacterial supernatant was not due to a general decreased membrane integrity following novobiocin treatment. One potential mechanism for protein export to supernatants involves the production of outer membrane vesicles (OMV’s) (Elmi et al., 2012; Jang et al., 2014). Due to our method of concentrating supernatants by centrifugal filtration it was unlikely that we would capture OMVs but to confirm this a number of tests were carried out. The absence of CadF in supernatants diminished the possibility that OMV’s mediated the observed increase in protein secretion based on data from previous studies (Elmi et al., 2012; Jang et al. 2014). In addition, supernatant proteins were sensitive to proteinase K treatment (Fig. 5D), further indicating that OMV export was unlikely to mediate the observed novobiocin stimulated protein secretion. Increased protein secretion following DNA relaxation was also observed in the C. jejuni chicken isolate C19 and strain 81-176, suggesting this phenotype was not strain specific (Fig. 6A). Furthermore, the more invasive 81-176 which has a more relaxed DNA supercoiling resting level (Fig. 1B) was shown to have a greater number of proteins secreted into the supernatant even in the absence of novobiocin although a further increase was observed after greater relaxation of DNA supercoiling.
Fig. 5.
The observed increased secretion is not mediated by cell lysis or outer membrane vesicles.
A. NCTC11168 grown in the presence of subinhibitory concentrations of novobiocin shows no significant effect on growth.
B. NCTC11168 grown overnight in the presence of novobiocin shows no significant difference in numbers of colony forming units.
C. NCTC11168 supernatant from bacteria grown in the presence of novobiocin does not react with antibodies raised against the outer membrane protein CadF.
D. NCTC11168 protein secreted when grown in the presence of novobiocin is not resistant to proteinase K treatment. 1-Whole cell lysate, 2-Whole cell lysate, Prot. K treated, 3-Whole cell lysate boiled, Prot. K treated, 4-Uninduced secreted fraction, 5-Uninduced secreted fraction, Prot. K treated, 6-Induced fraction, 7-Induced fraction, Prot. K treated.
Fig. 6.
DNA relaxation leads to increased protein found within the supernatants of multiple C. jejuni strains and novobiocin induced secretion is inhibited by mutations in the flagellar genes flgI, flgE and fliD.
A. Coomassie staining of concentrated bacterial supernatants from C. jejuni strains 11168, C19 and 81-176 grown in the absence and presence of novobiocin.
B. No increase in protein within supernatants or whole cell lysates (WC) of strain 81-176 containing mutations in genes flgI, flgE or fliD is observed following DNA relaxation.
Increased extracellular protein following DNA relaxation is dependent on an intact flagellar structure
To investigate if the increase in extracellular protein following novobiocin treatment was dependent upon a functional flagellar export apparatus, isogenic mutations in the flagellar genes flgI, flgE and fliD were tested. Based on previous studies, disruption of flgI or flgE are predicted to inhibit flagellar protein secretion (Poly et al., 2007; Neal-McKinney and Konkel, 2012; Barrero-Tobon and Hendrixson, 2014). Since strain 81-176 has a more relaxed resting DNA topology it is naturally more sensitive to novobiocin and so a lower concentration of 5 μg/ml was used for induction. This more relaxed strain showed much greater secretion even in the absence of novobiocin strongly supporting the relationship between increased secretion and the more relaxed DNA topology. Treatment with novobiocin again led to an increase in secreted proteins. In contrast, supernatants from 81-176 flgI-, 81-176 flgE- showed very low amounts of protein and treatment with novobiocin resulted in no detectable increase in protein secretion, Fig. 6B. Cultures of 81-176 fliD-showed a higher level of protein in the supernatant, consistent with previous reports (Barrero-Tobon and Hendrixson, 2014), but again no detectable increase upon treatment with novobiocin was observed. This result strengthens the evidence that this is a specific secretome and implicates the flagellar export apparatus as a mechanism utilized to mediate this increased protein secretion following relaxation of C. jejuni DNA supercoiling.
LC-MS of bacterial supernatants reveals scale of protein secretion following DNA relaxation
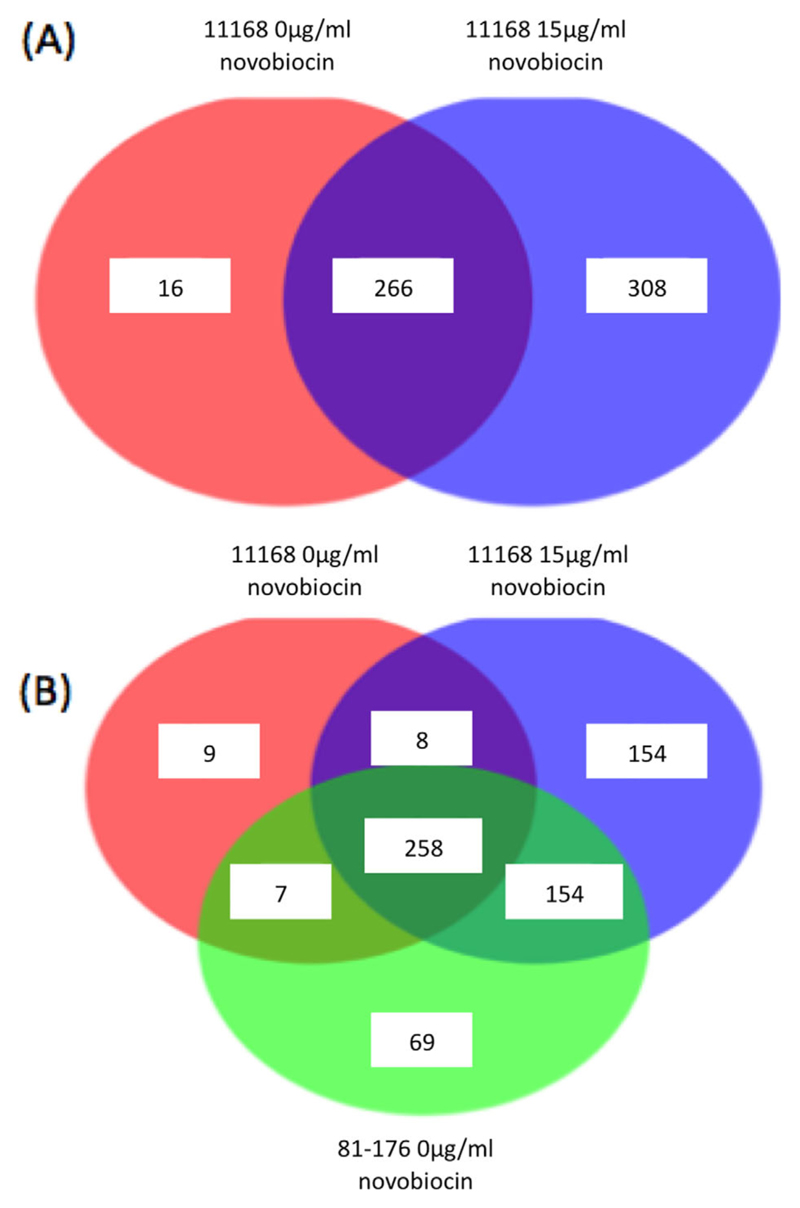
To investigate the composition of protein within NCTC11168 bacterial supernatant both before and after novobiocin treatment, concentrated supernatants were analyzed by LC-MS. In total, 16 proteins were found only in the supernatant of NCTC11168 exclusively prior to DNA relaxation while 308 proteins were found present only in the novobiocin treated supernatant, Fig. 7A. The identities of the top 10 proteins found within the novobiocin treated supernatants are shown in Table 1. Subtraction of this dataset with proteins detected in the supernatant of NCTC11168 prior to novobiocin is also shown in Table 2. These datasets contribute evidence that Peb4, DnaK, a putative periplasmic protease Cj0511, SodB and KatA are all found within NCTC11168 supernatants at a higher rate following DNA relaxation. The identities of proteins found in the supernatant of the naturally more relaxed and invasive strain 81-176 were also compared with NCTC11168 samples. As observed in Fig. 6A, LC-MS results revealed that a higher number of proteins were present within 81-176 supernatant than that of NCTC11168, of which a large number were also found exclusively within NCTC11168 supernatants following DNA relaxation, 154 in total. Full protein lists have been supplied in Supplementary Data.
Fig. 7.
LC-MS of bacterial supernatants grown in the absence and presence of novobiocin.
A. Numbers of proteins found within the supernatant of strain NCTC11168 grown in the absence and presence of novobiocin reveals the extent of increased protein secretion following DNA relaxation.
B. Many of the proteins found within the supernatant of NCTC11168 following DNA relaxation are also secreted by the less negatively supercoiled strain 81-176 in the absence of novobiovin.
Table 1. The identities of the top 10 proteins found within the concentrated supernatant of NCTC11168 treated with Novobiocin.
| Accession | Gene | Name | Product | % Coverage | #Peptides | #Unique | Mass (Da) |
|---|---|---|---|---|---|---|---|
| Q9PPD9 | Cj0780 | napA | nitrate reductase catalytic subunit | 79 | 94 | 94 | 104,944 |
| QOPAS1 | Cj0596 | peb4cbf2 | major antigenic peptide PEB-cell binding factor | 89 | 76 | 75 | 30,518 |
| 046108 | Cj0193c | tig | trigger factor | 87 | 75 | 74 | 50,968 |
| QOPAYS | Cj0531 | icd | isocitrate dehydrogenase | 75 | 69 | 68 | 82,331 |
| Q9PI16 | Cj0493 | fusA | elongation factor G | 86 | 67 | 67 | 76,719 |
| QOPA55 | Cj0835c | acnB | bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | 62 | 61 | 60 | 92,793 |
| 069298 | Cj0759 | dnaK | molecular chaperone DnaK | 79 | 58 | 58 | 67,419 |
| 069303 | Cj0470 | tuf | elongation factor Tu | 99 | 50 | 50 | 43,594 |
| QOPBP1 | Cj0264c | - | molybdopterin containing oxidoreductase | 62 | 47 | 47 | 93,278 |
| QOP8A3 | Cj1516 | - | oxidoreductase | 67 | 46 | 46 | 59,077 |
Table 2. The identities of the top 10 proteins found within the concentrated supernatant of NCTC11168 treated with Novobiocin but absent in the concentrated supernatants of untreated NCTC11168.
| Accession | Gene | Name | Product | %10 Coverage | #Peptides | #Unique | Mass (Da) |
|---|---|---|---|---|---|---|---|
| QOPCA1 | Cj0024 | nrdA | ribonucleotide-diphosphate reductase subunit alpha | 35 | 35 | 35 | 90,052 |
| QOPC5O | Cj0087 | aspA | aspartate ammonia-lyase | 54 | 32 | 32 | 51,766 |
| QOPC85 | Cj0041 | fliK | flagellar hook-length control protein | 39 | 24 | 23 | 67,841 |
| QOPBR4 | Cj0240c | iscS | cysteine desulfurase | 60 | 22 | 22 | 43,053 |
| Q0PB04 | Cj0511 | - | protease | 49 | 21 | 20 | 48,876 |
| Q59296 | Cj1385 | katA | catalase | 47 | 22 | 22 | 58,416 |
| Q9PMKS5 | Cj1455 | prf13 | peptide chain release factor 2 | 54 | 21 | 21 | 41,068 |
| QOPBW9 | Cj0169 | sodB | superoxide dismutase | 70 | 20 | 20 | 24,813 |
| QOPBZ1 | Cj0146c | trxB | thioredoxin reductase | 61 | 20 | 20 | 33,118 |
| Q9PNCO | Cj1175c | argS | arginyl-tRNA synthetase | 36 | 20 | 19 | 60,190 |
Discussion
DNA supercoiling has been shown to impact the regulation of process’ involved with pathogenesis of multiple bacterial species. Here we have shown that levels of DNA supercoiling also have a profound effect on the ability of C. jejuni to penetrate human cell lines and secrete protein to the extracellular environment.
Observing rates of DNA supercoiling and invasion of HCT-8 cells using strains NCTC11168 and 81-176 implied a potential correlation between reduced rates of negative supercoiling and an increased ability to invade HCT-8 cells. This correlation was further demonstrated by the growth of strain NCTC11168 in the presence of increasing novobiocin concentrations. The ability of NCTC11168 to invade HCT-8 and CACO-2 cell lines was significantly increased following dose dependent DNA relaxation. The relationship between decreased negative supercoiling and increased invasion of epithelial cells is an inverse one to that previously described for S. enterica, in which increased rates of negative supercoiling result in an increased ability to invade epithelial cells (Galan et al., 1990) while more relaxed DNA supercoiling is associated with survival in the more stressful environment of the macrophage (Ó Cróinín et al., 2006). In the case of C. jejuni we have previously shown that increased negative supercoiling is associated with increased motility (Shortt et al., 2016) while this study suggests that more relaxed DNA supercoiling is associated with the invasive phenotype for this pathogen. This is an intriguing observation as both of these pathogens cause disease in similar niches. One possible explanation would be that the human intestine is a more stressful environment for C. jejuni than the chicken intestine leading to a more relaxed DNA supercoiling profile and subsequent increased invasion. Further work would be required to support this hypothesis and to identify the environmental stimuli that lead to relaxation of DNA supercoiling in vivo.
Following the observations that reduced negative supercoiling correlates with increased invasion, any potential role the fibronectin binding proteins CadF and FlpA may play to mediate this increase in invasion was investigated. Increased invasion following novobiocin treatment of NCTC11168 cadF- or NCTC11168 flpA- strains revealed that neither of these proteins are necessary to observe an increase in invasion following novobiocin treatment. Although this was the case, increased invasion only became statistically significant at the highest concentration of novobiocin tested, 15 μg/ml, with disruption of the flpA gene resulting in a greater defect in invasion following novobiocin treatment. This may point to a co-operative role for these fibronectin binding proteins with other processes such as flagellar secretion to enhance rates of invasion during conditions of DNA relaxation as has been described by another research group (Eucker and Konkel, 2012). Future research is required to establish the transcriptional influence of reduced negative supercoiling on global C. jejuni gene transcription and investigation into its specific influence on genes involved in pathogenesis such as cadF and flpA.
Strikingly the large increase in protein secretion observed following DNA relaxation occurred at the same concentration in which rates of increased cell line invasion became statistically significant, 10 μg/ml. This increase in protein secretion was also shown to occur when using strains C. jejuni C19 and 81-176. Interestingly both baseline levels of protein secretion and the scale by which protein secretion may increase following novobiocin treatment was strain specific. It was also noted that the highly invasive strain 81-176 secreted at a much higher baseline rate than that of strains NCTC11168 and the chicken isolate C19. The specific increase in proteins found within the concentrated supernatant of strains grown in the presence of novobiocin was deemed not to be due to increased rates of OMV release. This was confirmed by probing supernatant samples with antibodies raised against the outer-membrane bound CadF protein. Evidence for this was further strengthened as protein released following novobiocin treatment was not resistant to proteinase K, revealing it appears not to be contained within a lipid bilayer. Multiple studies have now established the C. jejuni flagellum as a mechanism for protein secretion (Poly et al., 2007; Neal McKinney and Konkel, 2012; Barrero-Tobon and Hendrixson, 2014). Due to this, strains carrying mutations in flagellar genes which have previously been documented to inhibit the flagellar secretion were analyzed. Novobiocin treatment of strain 81-176 with directed mutations in the flagellar genes flgI, flgE or fliD did not increase protein secretion in the manner observed using the wild type counterpart. This provides strong evidence that the observed increase in protein secretion following reduced negative supercoiling is dependent on a wild type flagellum structure. When taken together with our recent observation that more relaxed DNA supercoiling leads to a reduction in motility without a visible effect on the structure of the flagellum (Shortt et al., 2016) these observations raise the possibility that DNA relaxation provides a key signal to change the function of the flagellar structure from motility to secretion. Previous studies have required induction of secretion from the flagellar structure using environmental stimuli such as bile salts (Rivera-Amill et al., 2001). The secretome identified in this study was induced by novobiocin and thus it would be interesting to test a variety of environmental stimuli to see if these could affect DNA supercoiling and induce secretion.
LC-MS of supernatant fractions from NCTC11168 indicated the extent of increased protein secretion following reduced negative supercoiling. It also revealed that protein secretion induced from NCTC11168 following novobiocin treatment contains many proteins secreted from the more relaxed strain 81-176 in the absence of novobiocin. Intriguingly, peptides found within the supernatant of NCTC11168 treated with 15 μg/ml novobiocin contained multiple chaperone proteins. SodB, KatA and cj0511 were also among the proteins secreted from NCTC11168 only after DNA relaxation. The serine peptidase encoded by cj0511 has been identified as being essential for colonization of chickens and has been implicated in the degradation of Occludin and E-cadherin leading to enhanced invasion (Karlyshev et al., 2014; Elmi et al., 2016). The presence of SodB and KatA is also interesting given that reactive oxygen species have recently been implicated as important mucosal defences against pathogens including C. jejuni suggesting that secretion of these proteins could be a mechanism of protecting C. jejuni from these reactive species (Corcionivoschi et al., 2012; Alvarez et al., 2016). It is also of note that the induction of proteins involved in protection from reactive oxygen species would also be observable during survival of S. enterica in the macrophage, an environment which has also been shown to induce more relaxed DNA supercoiling.
This work reveals a significant influence of DNA supercoiling on the ability of C. jejuni to invade human cell lines. It has also begun to define the mechanisms by which DNA relaxation increases rates of cell line invasion. We have established that increased invasion of the HCT-8 and CACO-2 cell lines occurs using NCTC11168 following DNA relaxation and this can occur independently of the CadF or FlpA proteins. We have also described a large increase in protein secretion from multiple C. jejuni strains following DNA relaxation, which correlates strongly with the observed increased rates of invasion and appears to be dependent on wild type flagellum formation. In doing this we have identified a novel mechanism by which C. jejuni regulates the invasion of epithelial cells and a possible global regulator of virulence in this important human pathogen.
Experimental procedures
Bacterial strains and growth conditions
C. jejuni 81-176 is a well characterized sequenced strain used widely in infection studies, C. jejuni NCTC11168 is the type strain of C. jejuni. Strain C19 was isolated from a chicken and was acquired from the laboratory of Professor Seamus Fanning. NCTC11168 cadF- and flpA- were acquired from Professor Steffen Backert and Dr Nick Dorrell respectively. 81-176 flgI-, 81-176 flgE- and 81-176 fliD- as well as the pRY107 plasmid were acquired from Professor Patricia Guerry. All C. jejuni strains were cultured on Mueller Hinton (MH) agar (Oxoid) at 37°C under microaerophilic conditions created using Campygen gas packs (Oxoid). C. jejuni stock cultures were maintained using MH broth (Oxoid) supplemented with 20% glycerol and stored at −80°C. To revive stocks of C. jejuni strains, bacteria were streaked with a single use sterile inoculation loop (Sarstedt) on MH agar and incubated under microaerophilic conditions for 48 h at 37°C. For liquid cultures, C. jejuni strains were equalized to specific optical densities in MH broth and incubated under microaerophilic conditions at 37°C, shaking at 200 rpm. NCTC11168 cadF-, NCTC11168 flpA- or strains transformed with plasmid pRY107 were cultured with the addition of 50 μg/ml kanamycin. 81-176 flgI-, 81-176 flgE- and 81-176 fliD- were cultured with the addition of 10 μg/ml chloramphenicol.
Chloroquine gel electrophoresis
To visualize the supercoiling profile of plasmid DNA, chloroquine gel electrophoresis was carried out on plasmids isolated from strains grown to mid log phase in the presence and absence of novobiocin. All chloroquine gels comprised of 0.8% agarose made with 2X Tris borate EDTA (TBE) buffer. Addition of 10 μg/ml chloroquine salt solution to both the gel and running buffer allows relaxed circular DNA to migrate further while supercoiled circular DNA is retarded in the gel remaining higher. All chloroquine gels were run for 24 h at 100 V. At this point the gel was washed approximately 10 times with water for 30-minute periods to remove the chloroquine salt. Ethidium bromide solution at a concentration of 10 μg/ml was then added for two hours to visualize plasmid DNA
Treatment of C. jejuni strains with novobiocin to relax DNA
To achieve relaxation of DNA, the aminocoumarin antibiotic novobiocin was used at sub-inhibitory concentrations to C. jejuni growth. For overnight cultures of C. jejuni containing novobiocin, strains were equalized at an O.D.600 of 0.02 and incubated under microaerophilic conditions in MH broth containing indicated concentrations of novobiocin for 18 h at 37°C, shaking at 200 rpm. To demonstrate the effect of novobiocin on DNA supercoiling in C. jejuni, chloroquine gel analysis was carried out on mid log phase cultures. For this, C. jejuni strains were equalized at an O.D.600 of 0.2 and incubated for 4 h under microaerophilic conditions at 37°C, shaking at 200 rpm. At this point novobiocin was added to the culture and incubated for a further 3 h.
Culture of in-vitro cell lines
The human ileocecal adenocarcinoma HCT-8 and CACO-2 cell lines were purchased from the ATCC (CC-L244, HTB-37). HCT-8 cells were grown using RPMI 1640 (Gibco) medium supplemented with 10% FBS, 1% sodium pyruvate (Sigma) and 1% Glutamax (Gibco). CACO-2 cells were grown using DMEM (life technologies) supplemented with 10% FBS and 1% non-essential amino acids. Cells were grown in 75 cm2 tissue culture flasks and incubated at 37°C with 5% CO2 in a humidified atmosphere. Cells were grown until roughly 80% confluence was observed after being seeded with 2 × 105 cells.
Total association and gentamycin protection assays
HCT-8 cells were seeded at 2 × 105 cells on 12 well plates (Corning) for 48 h until 80% confluence was observed. The multiplicity of infection for all total association or invasion assays was bacteria:cells, 100:1. C. jejuni strains to be used for total association and gentamycin protection assays were prepared as follows. Strains were equalized at an O.D.600 of 0.02 in MH broth with or without relevant concentration of novobiocin for 18 h at 37°C, shaking at 200 rpm under microaerophilic conditions. For total association assays infected cells were incubated at 37°C in a microaerophilic conditions for 5 h in the presence or absence of novobiocin. Wells were then washed three times with PBS and cells were lysed with 0.1% Triton-X-100 in PBS for 15 min. After this, serial dilutions of the cell lysate were carried out using MH broth and plated on MH agar. To determine the number of internalized bacteria in gentamycin protection assays, infected HCT-8 or CACO-2 cells were incubated at 37°C under microaerophilic conditions for 3 h in the presence or absence of novobiocin. After 3 h, the media overlying the infected HCT-8 or CACO-2 cells was changed to complete RPMI/DMEM media containing 400 μg/ml gentamycin sulphate (Lonza) and infected HCT-8 cells were incubated at 37°C under microaerophilic conditions for a further 2 h. After this, cells were washed 3 times with PBS and lysed with 0.1% Triton X-100 in PBS for 15 min. Serial dilutions of the cell lysates were carried out using MH broth and plated on MH agar. Dilutions of mutant C. jejuni strains were carried out on MH agar plates containing 50 μg/ml kanamycin. All MH plates were incubated for 72 h under microaerophilic conditions using Campygen gas packs (Oxoid) at 37°C before colony counting took place.
Statistical analysis
Tests for statistical analysis in total association and invasion assays were carried out using the Student’s t-test (two-tailed distribution). Statistically different values between samples possessed P-values <0.05. All total association and invasion experiments were performed in duplicate, with at least three biological replicates. The average number of bacteria counted post total association or invasion assay was counted and error bars represent the standard deviation for each sample condition.
Immunofluorescent microscopy
All immunofluorescent microscopy was done using HCT-8 cells grown on microscope coverslips within 6 well tissue culture plates (Corning) seeded with HCT-8 cells at 2 × 105 and grown for 2 days at 37°C in a humidified atmosphere. Infection of the cells was carried out as explained for infection assays. Once bacteria had been incubated with the cells, the wells were washed in triplicate with PBS and cells were fixed with 2% formaldehyde for 10 min. Cells were again washed in triplicate and permeabilized with 0.025% Triton-X-100 for 10 min. Once washed for a third time, a blocking solution of PBS containing 10% goat serum and 1% BSA was applied and left overnight. The following day cells were incubated with a primary antibody (Anti-C. jejuni 1:400) for 30 min and again washed in triplicate with PBS. A secondary antibody was then added for a further 30 min (AlexaFluor 488 conjugate Anti-Rabbit, Thermo Scientific 1:500). DAPI stain also was added to visualize nuclei of HCT-8 cells (1:5,000). Finally the coverslips were washed once more in triplicate with PBS and mounted on microscope slides using fluorescent mounting medium (Dako) prior to being examined using a fluorescent microscope.
Immunoblotting
Ten percent SDS polyacrylamide gels were transferred to nitrocellulose membranes at 100 V for one hour. All blocking steps were done overnight at 4°C shaking at 30 rpm using 3% fat free skimmed milk (Marvel). Anti-CadF primary antibodies for whole cell and supernatant samples were used at 1:10,000. This antibody was provided by Prof. Steffan Backert. Secondary antibody anti-rabbit IgG conjugated to HRP was used at 1:10,000.
Isolation of secreted protein from C. jejuni liquid cultures
C. jejuni cultures for the isolation of secreted proteins were inoculated at 0.02 O.D. in 10 ml in duplicate under microaerophilic conditions, shaking at 200 rpm. Duplicates for each sample were combined to compose 20 ml cultures equalized to an O.D.600 of 0.6. Each 20 ml culture was centrifuged at 4000 × g for 20 min, the supernatant was transferred to a fresh tube and the centrifuge step was repeated. Supernatants were then passed through a syringe with a 0.22 μm filter to remove any remaining whole bacteria. Once this was carried out supernatants were transferred to Amicon Ultra centrifugal filter units with a nominal molecular weight limit of 30 K (Millipore) and centrifuged at 4000 × g for 20 min. Resulting concentrated supernatant was aliquoted and stored at −20°C for future use.
Two-dimensional polyacrylamide gel electrophoresis
Total cell protein was collected from C. jejuni overnight liquid cultures in the presence/absence of novobiocin at concentrations indicated. Bacteria were pelleted, re-suspended in PBS and protein concentration was calculated. Total proteins of concentrated supernatants were precipitated using methanol/chloroform (Wessel and Flugge, 1984), dried and stored for future use at −20°C. Precipitated protein samples were re-suspended for 16 h in rehydration buffer (7M urea, 2M thiourea and 1% ABS-15). DTTand pH 4-7 IPG buffer (GE) were then added to a final concentration of 30 mM and 0.5% respectively. 1 μl of 1% bromophenol blue was also added and sample was made up to 126 μl for 7 cm immobiline drystrip (GE) rehydration. Once strips were rehydrated, isoelectric focusing was carried out under the following conditions: 300 V for 30 min, a gradient to 1,000 V for 30 min, a gradient to 5,000 V for 90 min and 5,000 V for a further 25 min. Following isoelectric focusing, strips were washed for 10 min in equilibration buffer (6 M Urea, 75 mM Tris-HCL, pH 8.8, 29.3% Glycerol, 2% (w/v) SDS, 0.002% Bromophenol blue) containing 1% DTT shaking at 60 rpm. A second wash of each strip using equilibration buffer containing 2.5% iodoacetamide was then carried out. Strips were then placed on top of 10% polyacrylamide gels and sealed into place using an agarose seal solution (0.5% agarose, 0.002% bromophenol blue, made to 100 ml with 1xSDS electrophoresis buffer). Gels were run at 160 V for approximately 90 min and stained using Sypro ruby protein stain (Life Technologies) as per the manufacturer’s instructions.
Treatment of secreted protein samples with proteinase K
Proteinase K was added to concentrated supernatant samples to a final concentration of 100 μg/ml in PBS. Samples were then incubated in a water bath at 37°C for 1 h. PMSF was then added to each sample at a final concentration of 5 mM and left for 10 min at room temperature. Reducing SDS-PAGE sample buffer was then added to each sample and boiled for 5 min prior to loading on a 12.5% polyacrylamide SDS-PAGE gel.
Liquid chromatography-mass spectrometry
Bacterial supernatant samples analysed by LC-MS were concentrated and precipitated as described above. The samples were denatured and reduced using 8 M urea, 50 mM ammonia bicarbonate and 10 mM dithiothreitol, alkylated using 15 mM iodoacetamide and digested with 1 ug of trypsin (sequencing grade Trypsin, Promega) at 37°C for 18 h on the thermomixer at 350 rpm. Prior to LC-MS/MS analysis possible contaminants were removed using C18 Ziptips (Millipore). The samples were run on a Thermo Scientific Q Exactive mass spectrometer connected to a Dionex Ultimate 3000 (RSLCnano) chromatography system. Tryptic peptides were resuspended in 0.1% formic acid. Each sample was loaded onto a fused silica emitter (75 μm ID, pulled using a laser puller (Sutter Instruments P2000), packed with Reprocil Pur C18 (1.9 μm) reverse phase media and was separated by an increasing acetonitrile gradient over 40 or 60 min at a flow rate of 250 nL/min. The mass spectrometer was operated in positive ion mode with a capillary temperature of 320°C, and with a potential of 2,300 V applied to the frit. All data was acquired with the mass spectrometer operating in automatic data dependent switching mode. A high resolution (70,000) MS scan (300–1600 m/z) was performed using the Q Exactive to select the 12 most intense ions prior to MS/MS analysis using HCD. Proteins identified were present within two biological replicates of novobiocin treatment. Using the peaks 7 software, data filtration was set to ≥2 unique peptides per protein and a false discovery rate of 1%. The parent mass tolerance was 10 ppm and the fragment mass error tolerance was 0.03 Da. The raw data was de novo sequenced and searched against the C. jejuni NCTC11168 Uniprot unreviewed database (http://www.uniprot.org/taxonomy/1385724) using the search engine PEAKS Studio 7 (Bioinformatics Solutions), for peptides cleaved with trypsin. Each peptide used for protein identification met specific Peaks parameters, i.e. only peptide scores that corresponded to a false discovery rate (FDR) of ≤1% were accepted from the Peaks PTM database search.
Supplementary Material
Additional Supporting Information may be found in the online version of this article at the publisher’s website.
References
- Alvarez LA, Kovacic L, Rodriguez J, Gosemann JH, Kubica M, Pircalabioru GG, et al. NADPH oxidase-derived H2O2 subverts pathogen signaling by oxidative phosphotyrosine conversion to PB-DOPA. Proc Natl Acad Sci USA. 2016;113:10406–10411. doi: 10.1073/pnas.1605443113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Tobon AM, Hendrixson DR. Flagellar biosynthesis exerts temporal regulation of secretion of specific Campylobacter jejuni colonization and virulence determinants. Mol Microbiol. 2014;93:957–974. doi: 10.1111/mmi.12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow DR, Christensen JE, Neal-McKinney JM, Konkel ME. Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol Microbiol. 2011;80:1296–1312. doi: 10.1111/j.1365-2958.2011.07645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burucoa C, Frémaux C, Pei Z, Tummuru M, Blaser MJ, Cenatiempo Y, et al. Nucleotide sequence and characterization of peb4A encoding an antigenic protein in Campylobacter jejuni. Res Microbiol. 1995;146:467–476. doi: 10.1016/0923-2508(96)80292-0. [DOI] [PubMed] [Google Scholar]
- Christensen JE, Pacheco SA, Konkel ME. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol. 2009;73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcionivoschi N, Alvarez LA, Sharp TH, Strengert M, Alemka A, Mantell J, et al. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe. 2012;12:47–59. doi: 10.1016/j.chom.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordwell SJ, Len ACL, Touma RG, Scott NE, Falconer L, Jones D, et al. Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics. 2008;8:122–139. doi: 10.1002/pmic.200700561. [DOI] [PubMed] [Google Scholar]
- Dorman CJ, Bhriain NN, Higgins CF. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature. 1990;344:789–792. doi: 10.1038/344789a0. [DOI] [PubMed] [Google Scholar]
- Elmi A, Watson E, Sandu P, Gundogdu O, Mills DC, Inglis NF, et al. Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect Immun. 2012;80:4089–4098. doi: 10.1128/IAI.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi A, Nasher F, Jagatia H, Gundogdu O, Bajaj-Elliott M, Wren B, Dorrell N. Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occluding. Cell Microbiol. 2016;18:561–572. doi: 10.1111/cmi.12534. [DOI] [PubMed] [Google Scholar]
- Eucker TP, Konkel ME. The cooperative action of bacterial fibronectin-binding proteins and secreted proteins promote maximal Campylobacter jejuni invasion of host cells by stimulating membrane ruffling. Cell Microbiol. 2012;14:226–238. doi: 10.1111/j.1462-5822.2011.01714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, Curtiss R., 3rd Expression of Salmonella Typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990 Jun;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun. 2009;77:2399–2407. doi: 10.1128/IAI.01266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K-S, Sweredoski MJ, Graham RLJ, Hess S, Clemons WM., Jr Comprehensive proteomic profiling of outer membrane vesicles from Campylobacter jejuni. J Proteomics. 2014;98:90–98. doi: 10.1016/j.jprot.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Joe A, Lynett J, Hani EK, Sherman P, Chan VL. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol Microbiol. 2001;39:1225–1236. doi: 10.1111/j.1365-2958.2001.02294.x. [DOI] [PubMed] [Google Scholar]
- Kale A, Phansopa C, Suwannachart C, Craven CJ, Rafferty JB, Kelly DJ. The virulence factor PEB4 (Cj0596) and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni. J Biol Chem. 2011;286:21254–21265. doi: 10.1074/jbc.M111.220442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlyshev AV, Thacker G, Jones MA, Clements MO, Wren BW. Campylobacter jejuni gene cj0511 encodes a serine peptidase essential for colonization. FEBS Open Bio. 2014;9:468–472. doi: 10.1016/j.fob.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A, Goldberg MD, Carroll Ronan K, Danino V, Hinton JCD, Dorman CJ. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology. 2004;150:2037–2053. doi: 10.1099/mic.0.27209-0. [DOI] [PubMed] [Google Scholar]
- Kervella M, Pagès JM, Pei Z, Grollier G, Blaser MJ, Fauchère JL. Isolation and characterization of two Campylobacter glycine-extracted proteins that bind to HeLa cell membranes. Infect Immun. 1993;61:3440–3448. doi: 10.1128/iai.61.8.3440-3448.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Garvis SG, Tipton SL, Anderson JDE, Cieplak JW. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol Microbiol. 1997;24:953–963. doi: 10.1046/j.1365-2958.1997.4031771.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mam-malian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, et al. Secretion of Virulence Proteins from Campylobacter jejuni Is Dependent on a Functional Flagellar Export Apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Larson CL, Flanagan RC. Campylobacter jejuni FlpA Binds Fibronectin and Is Required for Maximal Host Cell Adherence. J Bacteriol. 2010;192:68–76. doi: 10.1128/JB.00969-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan M, Lucchini S, Danino VÓ, Cróinín T, Hinton JCD, Dorman CJ. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:1831–1847. doi: 10.1111/j.1365-2958.2006.05062.x. [DOI] [PubMed] [Google Scholar]
- Mangan M, Lucchini SÓ, Cróinín T, Fitzgerald S, Hinton JCD, Dorman CJ. Nucleoid-associated protein HU controls three regulons that coordinate virulence, response to stress and general physiology in Salmonella enterica serovar Typhimurium. Microbiology. 2011;157:1075–1087. doi: 10.1099/mic.0.046359-0. [DOI] [PubMed] [Google Scholar]
- Moser ISM, Salnikow J. Campylobacter jejuni major outer membrane protein and a 59-kDa protein are involved in binding to fibronectin and INT 407 cell membranes. FEMS Microbiol Lett. 1997;157:233–238. doi: 10.1111/j.1574-6968.1997.tb12778.x. [DOI] [PubMed] [Google Scholar]
- Neal-McKinney JM, Konkel M. The Campylo-bacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front Cell Infect Microbiol. 2012;202:31. doi: 10.3389/fcimb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik VHD, Galán JE. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect Immun. 2010;78:3540–3553. doi: 10.1128/IAI.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Cróinín T, Carroll RK, Kelly A, Dorman CJ. Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;62:869–882. doi: 10.1111/j.1365-2958.2006.05416.x. [DOI] [PubMed] [Google Scholar]
- Ó Cróinín T, Backert S. Host epithelial cell invasion by Campylobacter jejuni: trigger or zipper mechanism? Front Cell Infect Microbiol. 2012;2:25. doi: 10.3389/fcimb.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZH, Ellison RT, Blaser MJ. Identification, purification, and characterization of major antigenic proteins of Campylobacter jejuni. J Biol Chem. 1991;266:16363–16369. [PubMed] [Google Scholar]
- Pei Z, Burucoa C, Grignon B, Baqar S, Huang X-Z, Kopecko DJ, et al. Mutation in the peb1A Locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect Immun. 1998;66:938–943. doi: 10.1128/iai.66.3.938-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, et al. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence–associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- Samuelson DR, Eucker TP, Bell JA, Dybas L, Mansfield LS, Konkel ME. The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease. Cell Commun Signal. 2013;11:79. doi: 10.1186/1478-811X-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortt C, Scanlan E, Hilliard A, Cotroneo CE, Bourke B, Croinin TO. DNA supercoiling regulates the motility of Campylobacter jejuni and is altered by growth in the presence of chicken mucus. mBio. 2016 Sep;7:13. doi: 10.1128/mBio.01227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YC, Jin S, Louie H, Ng D, Lau R, Zhang Y, et al. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol Microbiol. 2004;53:541–553. doi: 10.1111/j.1365-2958.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochemis. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Ye FBT, Niehus E, Drlica K, Josenhans C, Suerbaum S. Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. Int J Med Microbiol. 2007;297:65–81. doi: 10.1016/j.ijmm.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. doi: 10.1038/nrmicro1718. [DOI] [PubMed] [Google Scholar]
- Ziprin RL, Young CRL, Stanker H, Hume ME, Konkel ME. The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 1999;43:586–589. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.