Abstract
Both the miR-17-92 cluster and autophagy have been suggested as critical regulators of bone development, but the potential correlation between the two factors is largely unknown. Hence, we investigated whether members of this cluster can regulate chondrogenesis through an autophagy-related signalling pathway. In this study, the expression of miR-17-92 cluster members and the level of autophagic activity were investigated during chondrogenic induction in ATDC5 cells. miR-17, miR-18a, miR-20a, and miR-92-1 showed significant changes, and the level of autophagic activity was enhanced. Among the miR-17-92 cluster members, miR-20a showed the most significant change. Histological, cellular and molecular analyses were performed after the regulation of miR-20a and autophagy. miR-20a and autophagy had the opposite effect on chondrogenic differentiation, and there was a negative correlation between them. Moreover, the expression of the autophagy regulatory gene Atg7 was inhibited by miR-20a. siRNA was then used to knock down Atg7, and the results further indicated that Atg7 might be a potential target of miR-20a in chondrogenic differentiation. In conclusion, miR-20a is a critical negative regulator of chondrogenic differentiation because it inhibits autophagy via Atg7. Other members of the miR-17-92 cluster may have a similar effect, but this hypothesis requires further investigation.
Subject terms: Macroautophagy, Bone development, Cartilage development
Introduction
Autophagy is acknowledged to play an important role in development and diseases. This process is an evolutionarily conserved intracellular catabolic mechanism that recycles nutrients and energy through the degradation of macromolecules and organelles by lysosomes1. Autophagy involves the formation of autophagosomes, phagocytosis of macromolecules and organelles, transfer of autophagosomes into lysosomes, and degradation in the autolysosomes. Generally, autophagy occurs at a low basal level, and it is rapidly upregulated when cells need to generate intracellular nutrients and energy in response to diverse stress conditions. Deregulation of autophagy has been associated with developmental disorders, bone and cartilage diseases, diseases of other organs and tissues, such as the liver, heart, muscle and nerves, and cancers2,3.
Recently, the role of autophagy in the physiology of cartilage has been intensively studied. FGF signalling, which is essential in chondrogenesis, has been reported to regulate cartilage development by targeting the conjugation of Atg5/Atg124. Lack of Atg7 results in the dysfunction of type II collagen (col2) synthesis and secretion5. Moreover, autophagy promotes the survival of hypertrophic chondrocytes in growth plates6 and prevents damage from physically or chemically induced osteoarthritis in articular cartilage7–9.
Accumulating data have correlated autophagy and the functions of microRNAs (miRNAs)5,10–12. miRNAs are small non-coding RNA molecules of approximately 22 nucleotides that function in RNA silencing and the post-transcriptional regulation of eukaryotic gene expression. By binding to the 3′-UTR of target mRNAs, miRNAs can inhibit protein translation or result in mRNA degradation. In this way, they are widely involved in physiological processes such as development, tumourigenesis, cell proliferation and apoptosis13.
The miR-17-92 cluster is expressed as a polycistronic primary transcript containing six tandem stem-loop hairpins, and it ultimately results in six mature miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a). This cluster plays pivotal roles in regulating cellular processes, including proliferation, apoptosis, inflammatory activation and autophagy14–18. miR-20a is reported to target Atg7 and Atg16L and promotes mycobacterial survival in macrophage cells19. miR-18a and miR-92a have been reported to regulate chondrogenesis, but the role of this cluster in chondrogenic differentiation requires further exploration20–23.
Considering that members of the same cluster may have synergistic effects and that signalling crosstalk may occur at autophagy-related proteins (ATGs), we speculate that the miR-17-92 cluster can regulate chondrogenic differentiation or chondrogenesis by targeting downstream ATGs such as Atg7.
Results
Expression of the miR-17-92 cluster decreases during ATDC5 chondrogenic differentiation
As the miR-17-92 cluster plays an important role in cell proliferation and differentiation in various cells, such as cardiomyocytes24, lung epithelial cells25 and palatal mesenchymal cells26, we questioned whether the miR-17-92 cluster is involved in cellular chondrogenic differentiation. After ten days of chondrogenic induction, ATDC5 cells demonstrated higher levels of proteoglycan expression (Fig. 1A), mineralization (Fig. 1B) and alkaline phosphatase activity (Fig. 1C) than control cells, as evidenced by the mRNA and protein expression of col2 (Fig. 1D–F). All six members of the miR-17-92 cluster invariably decreased during induction (Fig. 1G), suggesting a negative correlation between the miR-17-92 cluster and chondrogenesis. Specifically, the decreases in miR-18a, miR-19b, miR-20a, and miR-92-1 were statistically significant. miR-20a demonstrated the largest decrease, highlighting its potential effect on chondrogenic differentiation.
Figure 1.
Expression of miR-17-92 cluster members decreases during ATDC5 chondrogenic differentiation. (A–C) Alcian blue, alizarin red and ALP staining of ATDC5 cells. Scale bar = 100 μm. (D) mRNA expression of col2 analysed by PCR. (E,F) Western blotting results of col2 after culture. Signal intensities were quantified using ImageJ software (version 1.80) (G) Expression of miR-17-92 cluster members during ATDC5 chondrogenic differentiation. ALP = alkaline phosphatase, col2 = collagen type-II; the error bars indicate the standard deviation, *p < 0.05, **p < 0.01, ns: not significant.
miR-20a suppresses ATDC5 chondrogenic differentiation
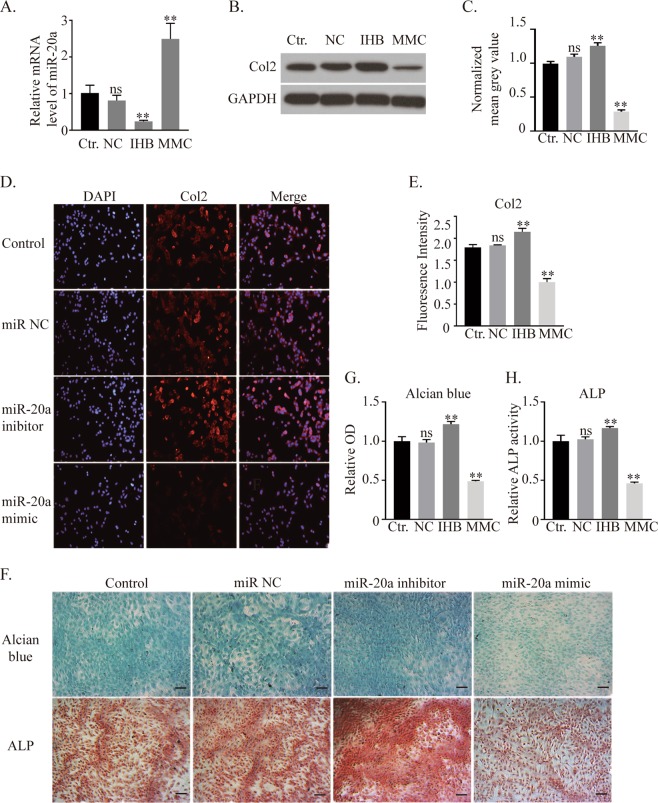
To test whether miR-20a inhibits chondrogenic differentiation, a miR-20a mimic, miR-20a inhibitor and miRNA NC (negative control) were transfected into ATDC5 cells. qRT-PCR analyses demonstrated miR-20a suppression in the miR-20a inhibitor group and miR-20a overexpression in the miR-20a mimic group, indicating successful transfection (Fig. 2A). According to the western blot results, the expression of col2 was not significantly different between the miRNA NC group and the control group, but it was significantly decreased by 72% in the miR-20a mimic group and significantly increased by 28% in the miR-20a inhibitor group (Fig. 2B,C). A col2 immunofluorescent assay showed similar results (Fig. 2D,E). Alcian blue and ALP histochemical staining (Fig. 2F–H) also demonstrated higher proteoglycan levels and alkaline phosphatase activity in the groups with less miR-20a. These data suggested that miR-20a inhibited chondrogenic differentiation in ATDC5 cells.
Figure 2.
Regulation of miR-20a affects the chondrogenic differentiation of ATDC5 cells. (A) Expression of miR-20a after miRNA negative control, miR-20a inhibitor and miR-20a mimic transfection. (B,C) Western blot results of col2 and its quantification. (D,E) Immunohistochemistry analysis of col2 after regulating the expression of miR-20a. (F–H) Alcian blue and ALP staining and quantitative analysis. Scale bar = 100 μm. Ctr. = control, NC = miRNA negative control, IHB = miR-20a inhibitor, MMC = miR-20a mimic. *p < 0.05, **p < 0.01, ns: not significant.
Autophagy is enhanced during ATDC5 chondrogenic differentiation
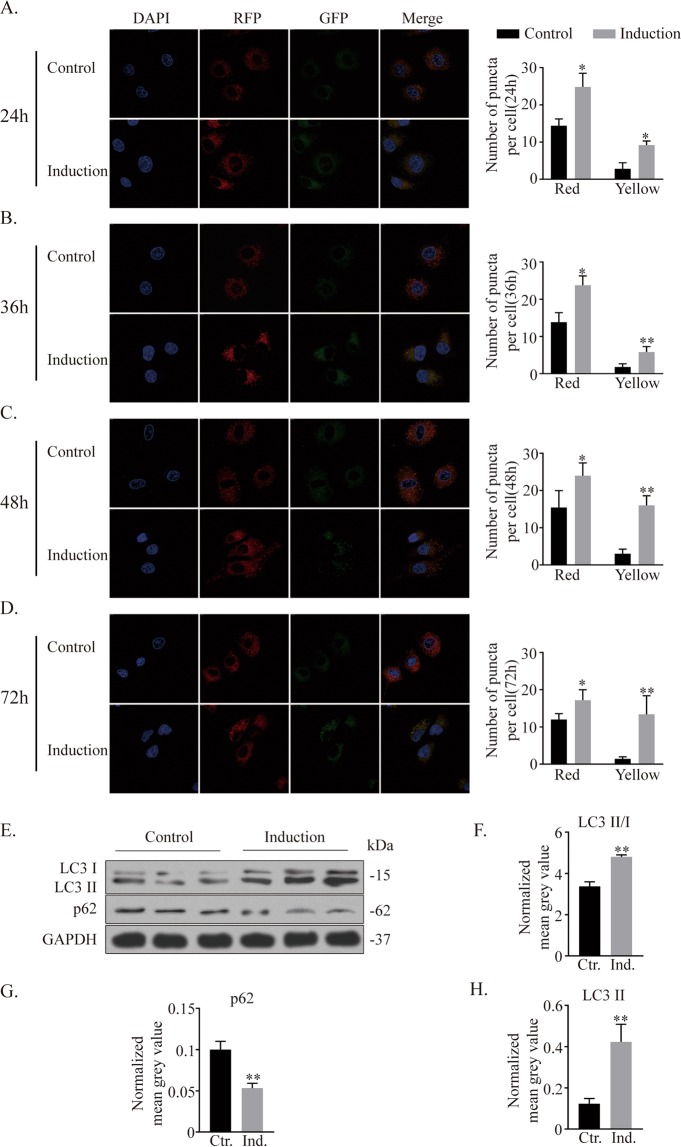
To investigate the level of autophagy during chondrogenic differentiation, ATDC5 cells were transfected with an mRFP-GFP-LC3 adenoviral vector, and autophagic flux was measured at multiple time points because autophagy is a dynamic process (Fig. 3A–D). At all the time points, the numbers of both red and yellow puncta significantly increased in the induction group, indicating that autophagic activity was higher in the chondrogenic induction group27.
Figure 3.
Autophagy is activated during the chondrogenic differentiation of ATDC5 cells. (A–D) Images taken at 24 h, 36 h, 48 h, and 72 h to monitor autophagic flux. Autophagic activity is reflected by the quantification of autophagic puncta. (E–H) Western blotting results of autophagy-related proteins after chondrogenic induction and quantitative analysis using ImageJ software. *p < 0.05, **p < 0.01.
Western blot results showed evident increases in both LC3-I and LC3-II (with increased LC3-I/LC3-II conversion) and decreases in the autophagosome cargo protein p62 after ten days of induction culture compared to control conditions (Fig. 3E–H). These data suggested that autophagy was enhanced during ATDC5 cell chondrogenic differentiation.
miR-20a suppresses autophagy during ATDC5 cell chondrogenic differentiation
The potential correlation between miR-20a and autophagy was further tested by exogenous delivery of miR-20a inhibitor/mimic and an autophagy inhibitor (3-MA). PCR analyses verified the effectiveness of both the miR-20a inhibitor and mimic in manipulating miR-20a expression (Fig. 4A). No significant difference in the expression of miR-20a was detected among the control, 3-MA, miRNA NC and miRNA NC + 3-MA groups. Likewise, there was no significant difference in miR-20a expression between the miR-20a inhibitor group and the miR-20a inhibitor + 3-MA group and between the miR-20a mimic group and the miR-20a mimic + 3-MA group, indicating that autophagy inhibition by 3-MA did not influence the expression of miR-20a.
Figure 4.
miR-20a inhibits autophagy activity during chondrogenic differentiation. (A) mRNA expression of miR-20a in eight groups after transfection of miRNA reagents, addition of an autophagy inhibitor or their combination under inducing culture. (B,C) Western blotting results of autophagy-related proteins, including LC3-II/I, p-Bcl-2, and p62, and the chondrogenic marker protein col2. Comparisons between each of the two groups were analysed using Dunnett’s test. (D,E) Immunofluorescent assay of col2 and quantification by ImageJ. *p < 0.05, **p < 0.01. *Without a connecting line is compared to the control group.
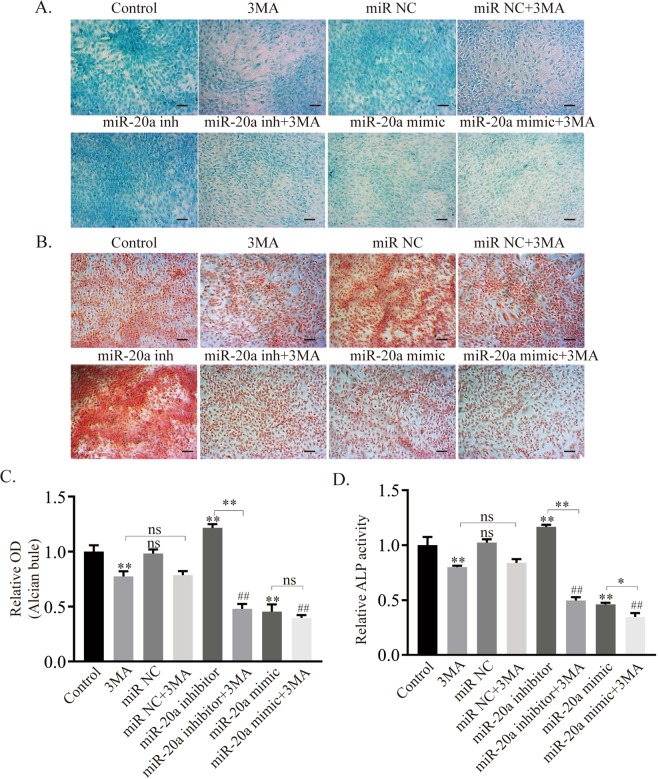
At the protein level (Fig. 4B,C), the expression of LC3-II/I, p-Bcl-2 and col2 decreased, while that of p62 increased in the groups treated with 3-MA or miR-20a mimic. The addition of 3-MA and the miR-20a inhibitor induced a dramatic shift from autophagy activation to autophagy suppression. When 3-MA was added to the miR-20a mimic group, the suppression of autophagy was aggravated. The immunofluorescent results for col2 showed that its expression was positively correlated with the autophagy level and negatively correlated with miR-20a expression (Fig. 4D,E). At the same time, alcian blue and ALP (Fig. 5A–D) staining showed that the synthesis of proteoglycans and the activity of alkaline phosphatase were inhibited under the conditions of miR-20a overexpression and autophagy suppression.
Figure 5.
Increased expression of miR-20a and inhibition of autophagy impair the chondrogenic process of ATDC5 cells. (A,C) Alcian blue staining and quantification. (B,D) ALP staining and quantification. Scale bar = 100 μm, *p < 0.05, **p < 0.01, *Without a connecting line is compared to the control group, ## is compared to the 3-MA group.
Collectively, miR-20a suppressed the level of autophagy during the chondrogenic differentiation of ATDC5 cells.
miR-20a suppresses autophagy by inhibiting Atg7 during chondrogenic differentiation
Since miR-20a was reported to target Atg719,28, which has been associated with col2 synthesis and secretion and cartilage development4,5, we questioned whether miR-20a inhibited autophagy via Atg7 during chondrogenic differentiation. Hence, Atg7 siRNA was introduced into the in vitro culture system to explore this mechanism.
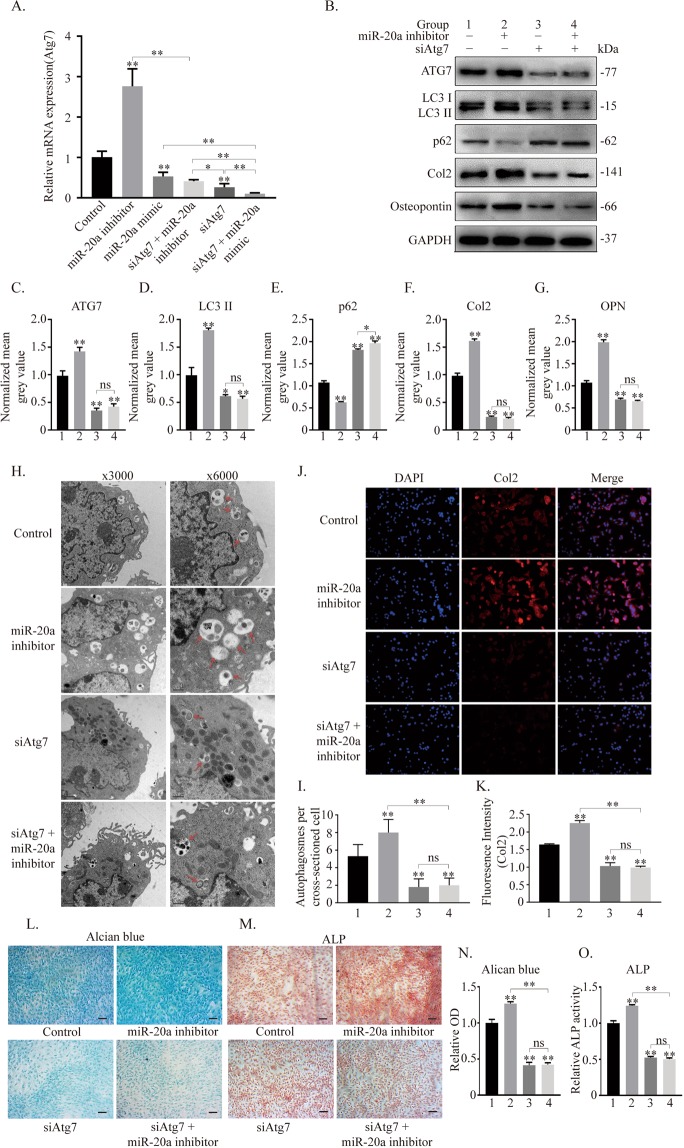
According to PCR analyses, the expression of Atg7 was found to be significantly upregulated when miR-20a was inhibited and significantly downregulated when miR-20a was overexpressed. Transfection of siAtg7 effectively inhibited Atg7 expression in ATDC5 cells (Fig. 6A). The expression level of ATGs, including Atg7, p62 and LC3 conversion, significantly increased in the miR-20a inhibitor group but decreased in the groups treated with siAtg7. The protein expression levels of col2 and osteopontin were basically in accordance with the level of autophagy (Fig. 6B–G). The number of autophagic vacuoles was detected by transmission electron microscopy (TEM). This number was increased in the miR-20a inhibitor group but decreased in the siAtg7 groups when compared to the that in the control group. Moreover, there was no significant difference between the other two groups (Fig. 6H,I). The immunofluorescent assay showed that the expression of col2 increased in the miR-20a inhibitor group, which supports the suppression of miR-20a. The expression of col2 decreased in the siAtg7 groups, even with miR-20a inhibitor treatment, but there was still no difference between them (Fig. 6J,K). Chondrogenic differentiation was enhanced in the miR-20a inhibitor group and was decreased in the groups treated with siAtg7 as demonstrated by alcian blue and ALP staining (Fig. 6L–O). Taken together, these data suggest that Atg7 is a potential target through which miR-20a suppresses autophagy during chondrogenic differentiation.
Figure 6.
miR-20a suppresses autophagy by downregulating Atg7 during the chondrogenic process of ATDC5 cells. (A) mRNA expression of Atg7. (B–G) Expression of ATG7, LC3-I, LC3-II, p62, col2 and osteopontin evaluated by western blotting and analysed according to the mean grey value. (H,I) TEM analysis of autophagic vesicles within ATDC5 cells. The red arrow indicates autophagic vesicles, including autophagosomes and autolysosomes. Magnification: 3000×, 1 μm; and 6000×, 0.5 μm for close-up images. The numbers of autophagic puncta were counted per cross-sectioned cell and compared using Student’s t-test. (J,K) Immunofluorescent assay of col2 among groups and quantification of fluorescence intensity. (L–O) Histochemical alcian blue and ALP staining and corresponding quantitative analysis. Scale bar = 100 μm. *p < 0.05, **p < 0.01.
Discussion
Due to their extensive and rapid proliferation, homogeneity and chondrogenic capacity, ATDC5 cells have been recognized as a good model to study chondrogenesis in vitro. Evidence has shown that ATDC5 cells have properties that replicate the multiple steps of chondrocyte differentiation. ATDC5 cells undergo cellular condensation and sequential chondrogenic differentiation with characteristic proteoglycan synthesis and col2 expression when treated with insulin28–30.
In the miR-17-92 cluster, miR-18a and miR-92a have been correlated with chondrogenesis in previous studies20–23. miR-18a suppresses the expression of CCN2/connective tissue growth factor (CCN2/CTGF), causing the suppression of chondrogenic differentiation20. miR-92a is highly enriched in chondrogenic progenitors, and its inactivation results in the loss of pharyngeal cartilage elements. miR-92a sustains chondrogenesis through BMP signalling by suppressing noggin321. Moreover, miR-92a directly targets histone deacetylase 2 (HDAC2) to mediate the suppression of cartilage-specific gene expression in human chondrocytes22. miR-92a also regulates the expression of aggrecanase-1 and aggrecanase-2 in human chondrocytes and may be involved in the development of osteoarthritis23. All of these studies demonstrate the correlation between the miR-17-92 cluster and chondrogenesis. For the first time, our data suggested that miR-20a is another member of the cluster that acts as an important regulator of chondrogenic differentiation. During the chondrogenic differentiation of ATDC5 cells, most members of the miR-17-92 cluster, including miR-18a, miR-19b, miR-20a and miR-92-1, showed a significant decrease, indicating that these four members may all be involved in the regulation of chondrogenic differentiation. This study focused on miR-20a because it had the biggest change. Inhibition and overexpression experiments clarified the potential role of miR-20a in regulating chondrogenic differentiation. The synthesis of col2 and proteoglycans, the activity of alkaline phosphatase, and the extent of mineralization were all enhanced when miR-20a was downregulated. These changes were suppressed when miR-20a was upregulated. However, the extent of increase in chondrogenesis in the miR-20a inhibitor group was not the same as the extent of reduction in the miR-20a mimic group, which indicated a potential synergistic effect from other members of the cluster.
Previous studies have shown differences in the expression of ATGs in chondrogenic differentiation. The expression of beclin1 and LC3 increases during chondrogenic differentiation in adipose stem cells31 and decreases during differentiation in mesenchymal stem cells32. The difference between these studies may be due to the different cell lineages and observations at different differentiation stages, but the results indicate the role of autophagy in drastic morphological and structural cellular changes. Moreover, multiple studies report that chondrogenic differentiation can be downregulated by decreasing the expression of ATGs31,33. Our study confirmed this regulatory role of autophagy in chondrogenic differentiation. In the present study, LC3-II and p-Bcl-2 increased, p62 decreased, and the LC3-II/I ratio increased, indicating enhanced autophagy activity during chondrogenic differentiation in ATDC5 cells. Considering that autophagy is a dynamic process, we employed a tandem monomeric RFP-GFP-tagged LC3 system (mRFP-GFP-LC3) to detect the autophagy flux. The increases in the numbers of yellow puncta and red puncta indicated enhanced autophagy compared to the control group, which supported the western blot results.
The members of the miR-17-92 cluster have been reported to potentially regulate autophagy34. The miR-17-92 cluster and its paralogs, including miR-106a-92 and miR-106b-25, share the same seed sequence (AAAGUGC), which is highly expressed in myeloid progenitors, and SQSTM1/p62 has been identified as a target for these miRNAs35. miR-20a negatively regulates autophagy by targeting ATG16L1 in hypoxia-induced osteoclast differentiation36 and ULK1 in C2C12 myoblasts37. Moreover, miR-20a can target Atg7 to suppress tumour proliferation in neuroblastoma38 and to promote mycobacterial survival in macrophage cells19. These studies confirm that the miR-17-92 cluster can regulate autophagy by targeting various ATGs. In our study, miR-20a upregulation reduced the autophagy level, while miR-20a downregulation increased the autophagy level, indicating that miR-20a had an inhibitory effect on autophagy. The overexpression and inhibition of miR-20a affected the expression of ATGs, while the use of an autophagy inhibitor did not significantly change the expression of miR-20a, implying that the autophagy pathway was downstream of miR-20a and that there was no obvious feedback regulation. The expression of Atg7 significantly increased in the miR-20a inhibitor group and decreased in the miR-20a mimic or siAtg7 groups. After siAtg7 was added, the expression of ATGs and chondrogenic marker proteins in the miR-20a inhibitor group was reversed. Accordingly, the quantification of autophagosomes per cellular cross-section by TEM, immunofluorescent and histochemical analyses supported these results. However, there was no significant difference between the siAtg7 group and the siAtg7 + miR-20a inhibitor group, indicating that the miR-20a inhibitor did not ameliorate the autophagy inhibition caused by siAtg7 and chondrogenic differentiation through other potential targets or pathways. That is, Atg7 is likely to be the target through which miR-20a regulates chondrogenic differentiation in ATDC5 cells.
Because the ATDC5 cell line derives from mouse teratocarcinoma fibroblastic cells, it can provide fundamental biological insights, but it cannot replace normal cell lineages such as MSCs, so it would be better to verify these results in MSCs and ECSs. Due to the incomplete effectiveness of siAtg7, Atg7 expression was knocked down by only 60~70% instead of being completely knocked out, and the regulatory effect of miR-20a on autophagy during chondrogenesis also needs to be verified in knockout mice. 3-MA effectively blocks autophagy at the early stage by inhibiting class III PI3K, but its specificity is questionable. It is suggested that 3-MA also inhibits class I PI3K and can thus promote autophagy at suboptimal concentrations. However, 3-MA does not block beclin1-independent autophagy.
In conclusion, our study suggests that miR-20a is a negative regulator of chondrogenesis by regulating Atg7, which is a possible intervention target for chondrogenic differentiation.
Materials and Methods
Cell culture
The murine ATDC5 cell line was purchased from Bang-yi Biotechnology (Shanghai, China) and cultured at 37 °C in a humidified atmosphere of 5% CO2 in Dulbecco’s modified Eagle’s medium: Nutrient Mixture F-12 (F-12/DMEM (1:1), Gibco, USA) supplemented with 5% foetal bovine serum (FBS, Gibco, USA). For chondrogenesis induction, the medium was supplemented with 5% FBS, 1% insulin-transferrin-selenium (ITS), vitamin C (50 µg/ml) and ascorbate 2-phosphate (37.5 µg/ml).
Cell transfection
Murine ATDC5 cells were allowed to reach 50–70% confluence at the time of transfection, and they were transfected with miRNA negative control, miR-20a mimic, miR-20a inhibitor (GenePharma, China) and ATG7 siRNA using Lipofectamine 2000 (Invitrogen, USA) for 24 h before experimental culture in accordance with the manufacturer’s protocol. The dose of each miR-20a-related reagent was 50 nM, and that of ATG7 siRNA (sense: GGAGUCACAGCUCUUCCUUTT, antisense: AAGGAAGAGCUGUGACUCCTT) was 50 pmol. The mRFP-GFP-LC3 adenovirus was purchased from HanBio Technology (Shanghai, China) and was used to transfect ATDC5 cells 48 h before induction culture at an MOI (multiplicity of infection) of 100 according to the manufacturer’s instructions to monitor autophagic flux.
Quantitative real-time PCR
Total RNA from transfected cells was collected, placed in Trizol reagent (Invitrogen), homogenized for 20 seconds and incubated on ice for 5 min. Then, chloroform, isopropanol and absolute ethanol were sequentially added and centrifuged at 12000 r.p.m. after each addition for ten minutes at 4 °C. The precipitate was obtained and dried at room temperature for 10 min and then added to the reverse transcription reaction system. Quantitative real-time PCR (qRT-PCR) was performed to detect relative mRNAs using a PrimeScript™ RT Reagent Kit (Takara, Japan). For the miRNAs, cDNA was synthesized using a MiR-X™ miRNA First-Strand Synthesis Kit (Takara), and qRT-PCR was performed using a TB Green™ Kit (Takara). qRT-PCR was performed in triplicate using a LightCycler 480 II quantitative PCR system (Roche, Switzerland). A comparative Ct (2−ΔΔCT) method was used for analysis, and U6 and GAPDH served as internal reference genes. The primers for qRT-PCR are shown in Supplemental Table S1.
Western blot analysis
For western blot analysis, cells were washed three times with PBS and then added to lysis buffer. Cell lysates were incubated on ice for 30 min; then, the soluble fraction was isolated by centrifugation at 12,000 r.p.m. for 10 min at 4 °C. Total protein concentrations of the cellular extracts were measured using a colourimetric BCA protein assay kit (Beyotime). Protein extracts were then loaded onto 10% SDS-PAGE gels and transferred onto a polyvinylidene difluoride membrane (PVDF, Millipore, USA). After blocking with 5% nonfat milk, the membranes were probed with primary antibodies at 4 °C with gentle shaking overnight. Next, the membranes were shaken at room temperature for 30 min and washed with TBST three times, followed by incubation with secondary antibodies at 37 °C for 1 h. Finally, after washing three times, the chemiluminescent signals on the membranes were detected using an ECL reagent (Thermo, USA). Densitometry values were normalized to the intensity of the corresponding bands for GAPDH. Quantitative analysis of western blotting was performed using Adobe Photoshop CS6 and ImageJ. The primary antibodies used in this study are listed in Supplemental Table S2.
Fluorescence microscopy
ATDC5 cells were seeded onto slides and cultured for 12 h. Then, 6-Bio was used to treat the cells for 24 h, and an equal amount of DMSO was added to the control group. The cells were washed quickly with PBS 2 times, fixed with 4% paraformaldehyde in PBS for 20 min on ice and permeabilized with 0.5% Triton. The slides were then washed again with PBS, incubated with a blocking solution of 5% FBS for 1 h and exposed to an antibody against col2 at 4 °C overnight. After being washed three times with blocking solution, the slides were incubated with secondary antibody. Finally, the cells counterstained with DAPI were observed, and images were collected using a fluorescence microscope (Olympus IX73, Japan).
Analysis of autophagic flux
After transfection with mRFP-GFP-LC3 adenoviral vectors for 48 h, ATDC5 cells underwent chondrogenic induction culture and were then collected and fixed with 4% paraformaldehyde in PBS for 30 min on ice. After washing with PBS three times and staining with DAPI, the cells were observed and recorded at four time points (24 h, 36 h, 48 h, 72 h) using confocal microscopy (Leica TCS SP8, Germany) to monitor autophagic flux. This assay is based on the different pH stabilities of green and red fluorescent proteins. When autophagosomes are transferred into the acidic conditions (pH < 5) of lysosomes, the fluorescent EGFP signal will be quenched, while the mRFP fluorescent signal will be sustained. Therefore, colocalization of both GFP and mRFP fluorescence indicates a compartment that has not fused with a lysosome, such as phagophore or autophagosome; a mRFP signal without GFP corresponds to an amphisome or autolysosome. In green and red images, autophagy flux is increased when the numbers of both yellow and red puncta are increased, while it is blocked when there are more yellow puncta with no change in the number of red puncta or when the numbers of both yellow and red puncta are decreased in cells39,40. Quantification of the numbers of red and yellow puncta per cell determines autophagic flux.
Histochemistry assay
After 10 days of culture, cells were collected and stained. For alizarin red staining and quantitative analysis, the cells were rinsed with PBS, fixed in 4% paraformaldehyde for 15 min, washed with ddH2O three times, and then stained with Alizarin Red S Kit (Solarbio, China) for mineralized nodules. The cells were washed again before taking photos. After that, 10% cetylpyridinium chloride was used to dissolve the nodules, and the OD (optical density) was measured at 562 nm. For ALP staining and activity analysis, cells were rinsed with PBS and treated with 1% Triton X-100. The ALP activity of the cell lysates was tested with an ALP Activity Assay Kit (Beyotime, China). The protein concentrations were examined using an Enhanced BCA Protein Assay Kit (Beyotime) following the manufacturer’s instructions. For alcian blue staining and analysis, cells were also fixed in 4% paraformaldehyde for 10 min and then washed and stained with an Alcian Blue Staining Kit (ScienceCell, USA). After staining, the cells were washed with diH2O 3 times, and 6 mol/L guanidine hydrochloride was added and incubated for 2 h at room temperature. The OD was measured at 630 nm, and the cells were observed and captured under an inverted microscope (Olympus IX73, Japan).
TEM
TEM was used to detect autophagosomes. ATDC5 cells were transfected with a miR-20a inhibitor, Atg7 siRNA or both and then cultured for eight days. The cells were then collected and fixed with 2.5% glutaraldehyde in 0.05 M sodium phosphate buffer. After washing at least 5 times, the samples were post-fixed in 1% w/v oso4 in 0.1 M sodium cacodylate buffer for 2 h, followed by dehydration with an increasing concentration gradient of ethanol and epon; then, the cells were embedded within pure Epon812. Next, the samples were cut into 50-nm sections and stained with 3% uranyl acetate and lead citrate. Images were acquired using a JEM-1230 electron microscope (JEOL, JAPAN) at 3000× and 6000× magnification. For each experimental group, at least 10 cellular cross-sections were scanned for the quantification of autophagosomes.
Statistical analysis
All experiments were repeated three times. Student’s t-test was used to evaluate the statistical significance of the results (GraphPad Prism, La Jolla, CA, USA). All P-values represented 2-sided tests of statistical significance (*P < 0.05, **P < 0.01, ns = not significant).
Supplementary information
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China to J.L. (No. 81500829) and X.Y. (No. 81801019).
Author Contributions
B.S. and J.L. participated in the design and coordination of this study. R.X. and Y.W. performed the experiments, acquired the data and performed the statistical analysis. R.X. prepared all the figures and drafted the paper. J.L. and X.Y. critically revised the manuscript. All the authors reviewed the manuscript.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/21/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45502-7.
References
- 1.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–30. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro IM, Layfield R, Lotz M, Settembre C, Whitehouse C. Boning up on autophagy: the role of autophagy in skeletal biology. Autophagy. 2014;10:7–19. doi: 10.4161/auto.26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, et al. FGFR3/fibroblast growth factor receptor 3 inhibits autophagy through decreasing the ATG12-ATG5 conjugate, leading to the delay of cartilage development in achondroplasia. Autophagy. 2015;11:1998–2013. doi: 10.1080/15548627.2015.1091551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cinque L, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272–5. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 6.Bohensky J, et al. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–14. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki H, et al. Autophagy modulates osteoarthritis-related gene expression in human chondrocytes. Arthritis Rheum. 2012;64:1920–8. doi: 10.1002/art.34323. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, et al. Enhancement of chondrocyte autophagy is an early response in the degenerative cartilage of the temporomandibular joint to biomechanical dental stimulation. Apoptosis. 2013;18:423–34. doi: 10.1007/s10495-013-0811-0. [DOI] [PubMed] [Google Scholar]
- 9.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy Is a Protective Mechanism in Normal Cartilage, and Its Aging-Related Loss Is Linked With Cell Death and Osteoarthritis. Arthritis And Rheumatism. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao C, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nature Cell Biology. 2010;12:781–U38. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 11.Frankel LB, et al. microRNA-101 is a potent inhibitor of autophagy. Embo Journal. 2011;30:4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33:2018–25. doi: 10.1093/carcin/bgs266. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang IK, et al. MiR-20a-5p mediates hypoxia-induced autophagy by targeting ATG16L1 in ischemic kidney injury. Life Sci. 2015;136:133–41. doi: 10.1016/j.lfs.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Tazi MF, et al. Elevated Mirc1/Mir17-92 cluster expression negatively regulates autophagy and CFTR (cystic fibrosis transmembrane conductance regulator) function in CF macrophages. Autophagy. 2016;12:2026–2037. doi: 10.1080/15548627.2016.1217370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L, et al. microRNA-20a Inhibits Autophagic Process by Targeting ATG7 and ATG16L1 and Favors Mycobacterial Survival in Macrophage Cells. Front Cell Infect Microbiol. 2016;6:134. doi: 10.3389/fcimb.2016.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohgawara T, et al. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009;583:1006–10. doi: 10.1016/j.febslet.2009.02.025. [DOI] [PubMed] [Google Scholar]
- 21.Ning GZ, Liu XL, Dai MM, Meng AM, Wang Q. MicroRNA-92a Upholds Bmp Signaling by Targeting noggin3 during Pharyngeal Cartilage Formation. Developmental Cell. 2013;24:283–295. doi: 10.1016/j.devcel.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Mao G, et al. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthritis Cartilage. 2017;25:521–532. doi: 10.1016/j.joca.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Mao G, et al. MicroRNA-92a-3p Regulates Aggrecanase-1 and Aggrecanase-2 Expression in Chondrogenesis and IL-1beta-Induced Catabolism in Human Articular Chondrocytes. Cell Physiol Biochem. 2017;44:38–52. doi: 10.1159/000484579. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Shi JY, Zhu GQ, Shi B. MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem. 2012;113:1235–44. doi: 10.1002/jcb.23457. [DOI] [PubMed] [Google Scholar]
- 27.Yu T, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548–563 e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atsumi T, Miwa Y, Kimata K, Ikawa Y. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ Dev. 1990;30:109–16. doi: 10.1016/0922-3371(90)90079-C. [DOI] [PubMed] [Google Scholar]
- 29.Shukunami C, et al. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J Bone Miner Res. 1997;12:1174–88. doi: 10.1359/jbmr.1997.12.8.1174. [DOI] [PubMed] [Google Scholar]
- 30.Yao Y, Wang Y. ATDC5: an excellent in vitro model cell line for skeletal development. J Cell Biochem. 2013;114:1223–9. doi: 10.1002/jcb.24467. [DOI] [PubMed] [Google Scholar]
- 31.Marycz K, Kornicka K, Grzesiak J, Smieszek A, Szlapka J. Macroautophagy and Selective Mitophagy Ameliorate Chondrogenic Differentiation Potential in Adipose Stem Cells of Equine Metabolic Syndrome: New Findings in the Field of Progenitor Cells Differentiation. Oxid Med Cell Longev. 2016;2016:3718468. doi: 10.1155/2016/3718468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver L, Hue E, Priault M, Vallette FM. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev. 2012;21:2779–88. doi: 10.1089/scd.2012.0124. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Fang X, Zhao W, Liang Q. The transcriptional coactivator YAP1 is overexpressed in osteoarthritis and promotes its progression by interacting with Beclin-1. Gene. 2019;689:210–219. doi: 10.1016/j.gene.2018.11.068. [DOI] [PubMed] [Google Scholar]
- 34.Comincini S, et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–86. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu JZ, Wang YF, Tan XR, Jing HJ. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy. 2012;8:873–882. doi: 10.4161/auto.19629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun KT, et al. MicroRNA-20a regulates autophagy related protein-ATG16L1 in hypoxia-induced osteoclast differentiation. Bone. 2015;73:145–53. doi: 10.1016/j.bone.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179–86. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Yu Y, et al. MiR-20a-5p suppresses tumor proliferation by targeting autophagy-related gene 7 in neuroblastoma. Cancer Cell Int. 2018;18:5. doi: 10.1186/s12935-017-0499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou CH, et al. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy. 2012;8:1215–1226. doi: 10.4161/auto.20284. [DOI] [PubMed] [Google Scholar]
- 40.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.