Abstract
A low quadriceps slow-twitch (ST), oxidative (relative to fast-twitch) fiber proportion is prevalent in chronic diseases such Chronic Obstructive Pulmonary Disease (COPD) and is associated with exercise limitation and poor outcomes. Benefits of an increased ST fiber proportion are demonstrated in genetically modified animals. Pathway analysis of published data of differentially expressed genes in mouse ST and FT fibers, mining of our microarray data and a qPCR analysis of quadriceps specimens from COPD patients and controls were performed. ST markers were quantified in C2C12 myotubes with EGF-neutralizing antibody, EGFR inhibitor or an EGFR-silencing RNA added. A zebrafish egfra mutant was generated by genome editing and ST fibers counted. EGF signaling was (negatively) associated with the ST muscle phenotype in mice and humans, and muscle EGF transcript levels were raised in COPD. In C2C12 myotubes, EGFR inhibition/silencing increased ST, including mitochondrial, markers. In zebrafish, egfra depletion increased ST fibers and mitochondrial content. EGF is negatively associated with ST muscle phenotype in mice, healthy humans and COPD patients. EGFR blockade promotes the ST phenotype in myotubes and zebrafish embryos. EGF signaling suppresses the ST phenotype, therefore EGFR inhibitors may be potential treatments for COPD-related muscle ST fiber loss.
Subject terms: Physiology, Diseases
Introduction
Conditions such as Chronic Obstructive Pulmonary Disease (COPD), cardiac failure, type 2 diabetes, obesity and muscle wasting are major healthcare issues in ageing populations1. In these conditions, exercise capacity, whole-body metabolism and glucose homeostasis are impaired and since they are highly dependent on skeletal muscle, the proportion of slow-twitch (ST) to fast-twitch (FT) fibers in the large locomotor muscles has significant clinical impact2. This is because, in man, ST, type I, fibers are energy-efficient, mitochondria-rich, adapted for fat and glucose oxidation, and more insulin-sensitive3, resistant to fatigue4 and chronic disease-related wasting5,6 than FT oxidative/glycolytic IIa and glycolytic IIx fibers. A reduced quadriceps ST fiber proportion is prevalent in COPD (affecting more than half of patients with moderate to very severe lung disease)5,7,8, as well as in cardiac failure6, type 2 diabetes and obesity9,10. Quadriceps ST fiber proportion is an independent predictor of reduced maximal exercise capacity5 and also mortality11 in COPD. It is also an independent predictor of reduced maximal exercise capacity in patients with cardiac failure and type 2 diabetes12,13 and of insulin resistance9 and adiposity10. In COPD, the increased population of hybrid ST/IIa fibers in the quadriceps muscle indicate that ST fibers are switching to FT8. An opposite FT to ST switch can be induced in animal models with manipulation of neuromuscular stimulation patterns and forced transcription factor and co-activator expression14,15 but not with exercise in patients16. Drugs targeting relevant pathways to increase ST fibers in skeletal muscle would therefore be novel therapies for exercise limitation in COPD and metabolic disease2,17, and would be anticipated to reduce these patients’ susceptibility to muscle atrophy because of ST fibers relative resistance to atrophy.
Here we report that inhibition of an EGFR-driven pathway promotes the ST muscle phenotype in vitro and in vivo, and demonstrate that this pathway appears relevant to COPD-related ST fiber loss in the locomotor muscles.
Results
Muscle EGF transcripts are negatively associated with a ST phenotype in mice
Pathway analysis of differentially expressed genes in isolated mouse ST and FT IIb fibers data-mined from18 identified EGF signaling as the top signaling pathway negatively associated with ST phenotype, together with EGF receptor (ErbB) and mitogen-activated protein kinase/extracellular signal-related kinase (ERK) signaling, the latter a key pathway activated by EGFR in response to EGF19 and previously implicated in muscle fiber-type regulation20,21 (Fig. 1a and Supplementary Fig. E1). This led us to investigate EGF and EGFR expression in muscle from healthy humans and COPD patients.
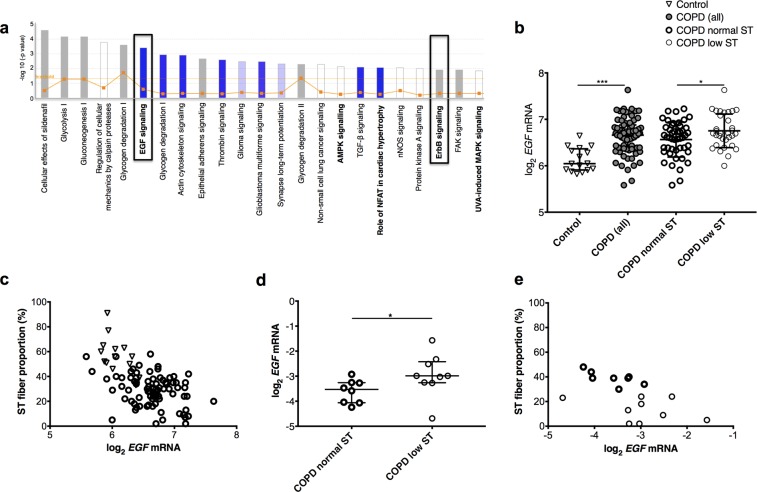
Figure 1.
Pathway analysis of mouse data and human muscle analysis. (a) Pathways overlapping with genes downregulated in mouse ST versus FT IIb fibers identified through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)48. Z-scores <0 (blue bars) and = 0 (white bars) signify more, or equal, pathway activations compared to inhibitions respectively reported in association with downregulated genes, grey indicating absence of directionality data. See methods for ratio; threshold indicates statistical significance. (b,c) Human muscle microarray data (participant characteristics in Table 1). (b) Quadriceps EGF transcripts in controls, COPD low and normal ST: patients with a ST fiber proportion <27% and ≥27% respectively. Mean (SD) and t-test results shown. (c) Quadriceps EGF transcripts versus ST fiber proportions (r = −0.69, p = 0.003 in controls, r = −0.56 in patients and −0.62 in patients with controls, both p <z0.0001). (d,e) QPCR data for small COPD cohort (Table 1). (d) Quadriceps EGF transcripts in low and normal ST groups. Median (IQR) and Mann-Whitney U-test results shown. E: Quadriceps EGF transcripts versus ST fiber proportions (r = −0.56, p = 0.019). *as for Table 1.
Muscle EGF transcript levels correlate negatively with ST fiber proportion in COPD patients and healthy controls
We have previously published a genome-wide expression analysis of quadriceps biopsies from 79 COPD patients and 16 healthy matched controls from this cohort followed by a Weighted Correlation Network Analysis (WGCNA) of the results22. This analysis revealed six modules of co-expressed transcripts, of which two (labelled ‘yellow’ and ‘turquoise’) showed the strongest correlation with type I fiber proportion and maximal aerobic exercise capacity. By conducting an exhaustive screening of all possible glm models predicting type I fiber proportion and maximal aerobic exercise capacity from the six previously defined co-expression modules, we show here that the yellow module yields the greatest overall support, and relative to the other modules, represents the most important term in predicting exercise capacity (Supplementary Fig. E2A and E2B).
Within the yellow module documented in22, EGF has the second highest absolute module membership (MM 0.81); a measure of eigengene-based connectivity widely applied for the identification of intramodular hub genes. Also showing high levels of module membership is SOX6 (MM 0.77) encoding the sex determining region y-related Box 6, a transcription factor shown elsewhere to suppress ST fiber development in mice23 and zebrafish24.
To explore these findings further we extracted EGF transcript levels from25 and assessed the relationship between its abundance, disease status and ST fiber proportion. COPD patients had higher EGF transcript levels in the quadriceps, with the highest levels in patients with a low ST fiber proportion (both p < 0.0001, Fig. 1b, normal ST proportion >27% according to reference ranges from7). Quadriceps EGF transcript abundance and ST fiber proportion were negatively correlated in patients (r = −0.56, p < 0.0001), controls (r = −0.69, p = 0.003), and in both groups when combined (r = −0.62, p < 0.0001, Fig. 1c). EGF mRNA levels were also negatively correlated with maximal aerobic exercise capacity measured as peak oxygen consumption on an incremental cycle test (Supplementary Fig. E2C). Array data was replicated by qPCR using RNA from separate residual quadriceps specimens from COPD patients with ST fiber proportions that were very low (median 13%, range 2–24%, n = 8) or within normal range (median 39%, range 30–48%, n = 9); 12 patients were in the microarray study, 5 were additional (Table 1). EGF transcript abundance was again highest in patients with a low ST fiber proportion (p = 0.036, Fig. 1d), and negatively correlated with ST fiber proportion (r = −0.56, p = 0.019, Fig. 1e) and muscle endurance (r = −0.64, p = 0.01, Supplementary Fig. E3A) in patients. Serum EGF protein levels did not differentiate patients and controls, nor patients based on ST fiber proportions (Supplementary Fig. E3D). Quadriceps EGFR mRNA levels were not different between patient groups, nor correlated with ST fiber proportion or EGF mRNA levels (Supplementary Fig. E3B,C). Together the data suggest that muscle EGF signaling as a result of EGF derived within muscle influences ST fiber proportion in humans and that increased EGF signaling in the quadriceps muscle of COPD patients may underlie ST fiber loss. We therefore investigated effects of inhibiting skeletal muscle EGF signaling in vitro.
Table 1.
COPD patient and healthy control characteristics in cohort used for microarray and PCR.
| Microarray cohort | PCR cohort | |||||
|---|---|---|---|---|---|---|
| Patients (n = 79) | Controls (n = 16) | COPD low ST (n = 32) | COPD normal ST (n = 47) | COPD low ST (n = 9) | COPD normal ST (n = 8) | |
| Age (years) | 67 (8) | 66 (7) | 66 (6) | 68 (9) | 61 (14) | 75 (11)* |
| Sex (% males) | 65 | 50 | 62 | 69 | 66 | 100 |
| FEV1 (% predicted) | 45 (18) | 109 (14)**** | 38 (14) | 50 (20) | 21 (6) | 48 (50)*** |
| TLCO (% predicted) | 43 (17) | 90 (16)**** | 35 (17) | 48 (16) | 27 (17) | 54 (18)**** |
| Quadriceps endurance (T80, s) | 80 (35) | 118 (139)* | 75 (30) | 85 (34) | 83 (64) | 90 (35) |
| Peak VO2 (ml/kg/min) | 12.3 (4.0) | 23.4 (7.6)**** | 11.6 (4.8) | 12.7 (3.4)* | 8.8 (3.0) | 10.7 (4.5) |
| % ST fibers | 30 (18) | 54 (22)*** | 18 (14) | 36 (11)**** | 13 (20) | 39 (8)*** |
| % I/IIa fibers | 3 (5) | 2 (6)* | 3 (6) | 3 (5) | 2 (9) | 2 (6) |
| % FT IIa fibers | 60 (12) | 40 (13)**** | 69 (12) | 56 (12)**** | 69 (11) | 59 (13)* |
| % FT IIx fibers | 4 (9) | 3 (6) | 8 (13) | 2 (6)*** | 8 (13) | 2 (6) |
Mean (SD) or median (IQR) shown, t-test, Mann Whitney U-test, and Fisher’s exact test as appropriate, *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001. FEV1 Forced Expiratory Volume in 1 second, TLCO carbon monoxide diffusion capacity, T80 time for force to drop to 80% of initial, VO2 oxygen consumption on maximal incremental cycle ergometry.
EGFR inhibition in differentiated muscle cells increases ST gene expression
EGF signaling blockade was investigated in C2C12 mouse myotubes using 3 approaches (i) pharmacological EGFR inhibition with AG-1478 (a selective, reversible inhibitor of the tyrosine kinase domain) (ii) EGFR knock-down using an siRNA and (iii) an EGF neutralizing antibody. Three days of AG-1478 increased transcripts of ST fiber-specific contractile components such as myosin heavy chain I (MyHC I, MYH7) and slow troponin I (TNNT1), and decreased FT fiber-specific transcripts, such as MYH2, 1 and 4 that respectively encode MyHC IIa, IIx and IIb (latter absent in man) (Fig. 2a). Increased MyHC I protein was confirmed with immunohistochemistry (Fig. 2d and Supplementary Fig. E4). AG-1478 also increased transcripts associated with mitochondrial oxidative metabolism such as citrate synthase (CS), mitochondrial uncoupling protein 3 (UCP3). Consistent with this observation, AG-1478 treatment increased MitoTracker Red staining of functional mitochondria in myotubes indicating an overall increase in mitochondrial activity (Fig. 2d and Supplementary Fig. E5). AG-1478 also increased transcripts of insulin-responsive glucose transporter 4 (GLUT4/SLC2A4), which is more highly expressed in ST muscle26 (Fig. 2a). 1 hour after AG-1478 treatment there was a very modest reduction in phosphorylated EGFR (pEGFR)/total EGFR but this was associated with reductions in downstream pERK/ERK (1 and 2) and pAkt/Akt compared to treatment with vehicle control, each time the experiment was run (n = 3, representative image in Fig. 2e and additional images and quantification in Supplementary Fig. E6A). Similar results were observed 24 hours after the final treatment (i.e. 24 hours after the third daily treatment) with AG-1478 compared to treatment with vehicle control, and also at 1 hour after treatment with AG-1478 plus 100 ng/ml EGF compared to treatment with vehicle control plus 100 ng/ml EGF (the additional EGF used to enhance the baseline signal) (Supplementary Fig. E6B,C).
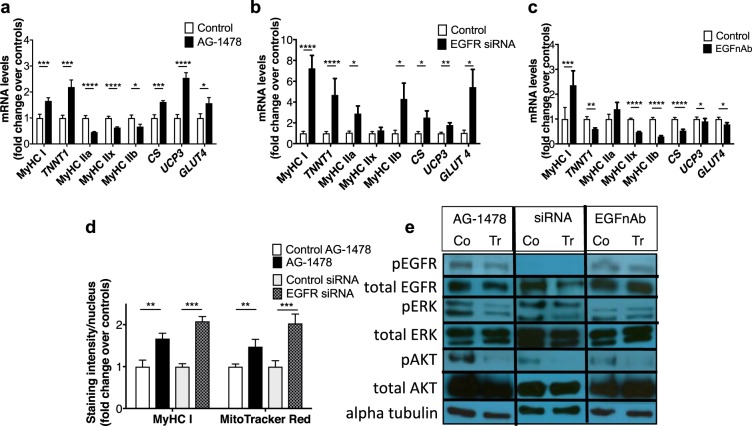
Figure 2.
EGF pathway inhibition in myotubes. (a–d) QPCR data from mouse myotubes treated with AG-1478 (a), EGFR siRNA (b), EGF neutralizing antibody (EGFnAb) (c), PD98059 (d), or respective vehicle controls. (e) MitoTracker Red and MyHC I staining intensity of myotubes treated as in (a,b). Mean(SEM) shown, n = 12/group for qPCR, n = 8/group for staining (2 experiments, 2–6 different biological replicates per experiment, 2 technical replicates per sample, t-test or Mann-Whitney U-test used depending on data distribution). * as for Table 1. Representative images of Western Blot bands from samples treated with (a) and (c) or vehicle control (Co control, Tr treated) and lysed 1-hour post-single dose, and 3 days following siRNA or control vector transfection. Phosphorylated and total protein quantifications are from the same membrane.
Knocking down EGFR with a silencing RNA increased transcripts encoding MyHC I, TNNT1, and CS, UCP3 and GLUT4, and increased MyHC I protein and staining of functional mitochondria (Figs 2b,d, Supplementary Figs E7 and E8), compared to control-transfected cells. There were additional smaller increases in transcripts encoding FT MyHC IIa and IIb (Fig. 2b) not seen with AG-1478. The siRNA knocked down EGFR protein and reduced pERK/ERK and pAkt/Akt, each time the experiment was run (n = 3, representative image in Fig. 2e and additional images and quantification in Supplementary Fig. E6A). Together with the AG-1478 data, this demonstrated that both pharmacological and genetic EGFR inhibition promoted the ST muscle phenotype in vitro.
Next, we reduced the concentrations of EGF in the cell culture medium (present as a result of the addition of horse serum27 and any secretion from cells). Addition of EGF neutralizing antibody (EGFnAb) to the medium increased transcripts encoding MyHC I but decreased all other mRNAs measured (Fig. 2c). Addition of the antibody did not reduce pERK1/ERK or pERK2/ERK2 but did cause a modest reduction in pEGFR/EGFR and pAkt/Akt 1 hour after treatment, each time the experiment was run (n = 3, representative image in Fig. 2e and additional images and quantification in Supplementary Fig. E6A). 24 hours after the final treatment (i.e. 24 hours after the third daily treatment) 24 hours after the final treatment (the third day of treatment) there was a modest reduction in pEGFR/EGFR but no reduction in markers of downstream signaling (Supplementary Fig. E6B and E6C). Therefore EGF depletion from medium decreased EGFR phosphorylation status but did not suppress downstream ERK signaling as EGFR blockade had, nor did it increase ST gene expression. This may relate to the presence of alternative EGFR ligands, which are known to have different downstream signaling patterns28, in the medium that maintain suppression of the ST muscle phenotype despite reduction in EGF concentrations.
egfra depletion increases ST fiber development and mitochondrial content in zebrafish
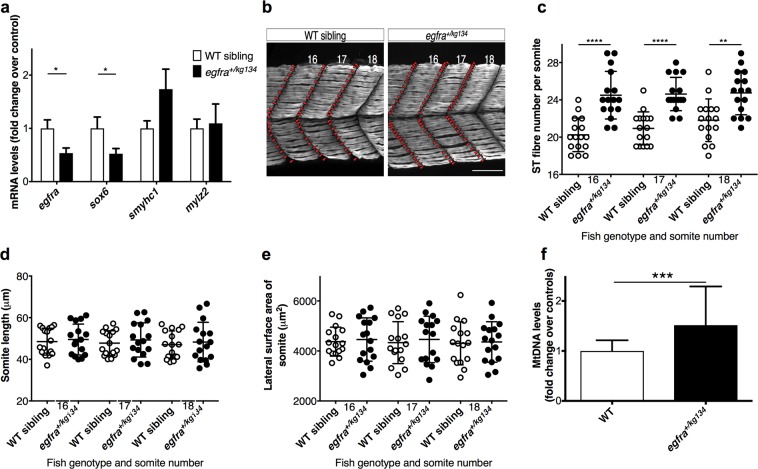
After demonstrating that EGFR inhibition promotes the ST phenotype in vitro, we investigated EGFR signaling in vivo. We confirmed that egfra is widely expressed in larvae 2 dpf29 using mRNA in-situ hybridization (Supplementary Fig. E9), then generated an early nonsense mutation in egfra (egfrakg134) in zebrafish embryos with CRISPR/Cas9 (see Supplementary Figs E10 and E11 and Supplementary Information for details). Homozygous egfrakg134 zebrafish larvae died early, consistent with Egfr-knockout lethality in mice30. At 2 dpf, heterozygous egfra+/kg134 larvae expressed approximately 50% less egfra transcript (Fig. 3a), and had a significantly greater (16 ± 2%) number of ST fibers per somite, without somite size differing, compared to wild-type (WT) siblings (Fig. 3b–e, Supplementary Video E12). Consistent with this, transcripts of sox6, a suppressor of ST fiber development in zebrafish24 were (50%) lower (Fig. 3a), while mitochondrial DNA content was 50% greater, in heterozygotes than WT siblings (Fig. 3f). egfra depletion therefore promotes the ST muscle phenotype in this in vivo model.
Figure 3.
egfra depletion on ST fiber development and mitochondrial content. Analysis of 2 dpf egfr+/kg134 larvae and WT siblings. (a) QPCR data (n = 4 (pool of 5 larvae)/group, from 2 outcrosses). Smyhc1 and mylz2 encode slow MyHC and fast myosin light chain respectively. (b) Maximum intensity projections of immunostained ST fibers (scale bar = 50 µm). (c–e) ST fiber number (C), length (D) and lateral surface area (E) of somites 16–18. n = 16/group (5–6 larvae, 3 crosses, Mann-Whitney U-test). (f) Mitochondrial DNA content n = 12/group, from 2 outcrosses. Mean (SEM) and t-test results shown. *as for Table 1.
Discussion
Our results, from human muscle and varying in vitro and in vivo experiments using pharmacological and genetic approaches, suggest that increased EGF-EGFR signaling contributes to ST fiber loss in COPD patients and demonstrate that EGFR signaling blockade promotes the development and maintenance of the ST muscle phenotype. EGF, and ErbB receptor, signaling was first identified from pathway analysis of mouse data before a consistent negative association between quadriceps EGF transcripts and ST fiber proportions was demonstrated in healthy elderly humans and in COPD patients, and EGF expression was found to be higher in patients than controls, and higher in patients with an abnormally low ST fiber proportion compared to patients with a ST fiber proportion within normal range. Consistent with EGF signaling being a regulator of fiber type, EGFR blockade in differentiated mouse muscle cells in culture increased ST (including mitochondrial) gene and protein expression, effects not recapitulated by reducing EGF levels in the medium. Furthermore, in an in vivo model using another vertebrate species, the zebrafish, and in the context of development, we found a similar role of EGFR signaling blockade again: egfra depletion resulted in an increased number of ST fibers. The data suggest that the EGF-EGFR signaling mechanism is an important pathway in regulating skeletal muscle fiber type which has been conserved across species. Moreover, it raises the possibility that EGFR blockade will have some therapeutic value in COPD patients with a low quadriceps ST fiber proportion, where this is associated with exercise limitation and mortality and may also predispose patients to muscle wasting as FT fibers are more prone to COPD-related atrophy than ST fibers5,31. EGFR inhibitors, such as gefitinib and erlotinib, are in the clinic as cancer treatments. Since our data suggests that modest EGFR blockade modifies muscle phenotype, lower, and hence more tolerable, doses than those used in cancer treatment regimens (which aim for total downstream signaling blockade32,33) may be efficacious in COPD patients. Further data supporting this hypothesis could be gained from investigating muscle ST fiber proportion in patients receiving EGFR inhibitor treatment for cancer.
The data is novel because no previous data implicates EGFR activation in skeletal muscle fiber-type specification nor identifies a signaling mechanism related to ST fiber loss in COPD that is targetable by clinically available pharmacotherapies. Our results are, however, in keeping with the finding that Scube2 proteins, which contain EGF-like repeats, are pivotal for ST fiber development in zebrafish34, and previous literature implicating ERK1/2 signaling (which is activated by EGFR signaling19) in fiber-type specification. Consistent with our results, Shi et al. have reported greater activity of ERK in mouse FT, compared to ST, muscles35, and that ERK inhibition increases slow myosin expression in vitro and in non-regenerating mouse and rat FT muscles20. However, ERK1/2 activation (rather than inhibition) has been implicated in re-establishing the slow muscle programming in rat soleus muscle (but not fast muscle) following denervation-induced ST to FT switch21. This suggests the context and presence of additional regulators dictates fiber-type effects in response to ERK signaling, and is consistent with the fact that ERK signaling is activated by a number of upstream pathways, not only EGFR signaling. For example, insulin growth factor 1 signaling activates ERK in skeletal muscle36 but has not previously been described to induce fiber-type specific gene expression37. Our data suggest a reduction in pAkt in response to EGFR inhibition, raising the possibility that treatment with an EGFR inhibitor would reduce muscle protein synthesis and thereby muscle mass. How likely such an outcome would be, in a clinical context, is open to conjecture, as is any impact on physiology. Multiple regulators signal via Akt in vivo each with their own subset of outcomes. Inhibiting one of these regulatory pathways (in this case the EGFR pathway) may have minimal effect on overall Akt activity but a specific impact on the outcomes related to that pathway. As IGF-1 appears to be the major regulator of Akt in muscle influencing protein synthesis, EGFR may not have a significant impact on muscle mass. Furthermore, the positive effect on oxidative phenotype may outweigh any reductions in muscle mass on clinically relevant endpoints like exercise performance on submaximal exercise tests remains to be seen.
The cell culture differences resulting from varied disruption of EGF signaling warrant discussion. AG-1478 treatment suppressed expression of FT IIb myosin transcripts unlike the EGFR siRNA, suggesting that this may have resulted from effects of AG-1478 on ErbB2-4 receptors or off-target effects. The relevance to humans, given that they do not express MyHC IIb is debatable. EGFnAb in the medium decreased expression of ST and mitochondrial markers, apart from ST myosin, in contrast to EGFR blockade. An inadequate decrease in EGF concentrations from the medium is one possible explanation of the findings. If the main EGF source for the muscle was not accessible to the EGFnAb, which could occur if cell-derived EGF was retained in the matrix, is one possible explanation. This suggestion is consistent with sequestration of muscle-derived EGF within the extracellular matrix during regeneration without circulating EGF rising38 and with our human data correlating muscle, but not circulating, EGF with ST fiber proportions. Alternatively, or additionally, the EGFnAb experiment results could reflect non-EGF mediated EGFR (and other ErbB receptor) activation, released by the removal of competition of these ligands with EGF. Since signaling downstream of EGFR is specific to the particular ligand28,39 it would not be surprising that the phenotype produced differs to that expected with reduced EGF-induced EGFR activation. In any case, this experiment suggests that the effects of EGFR blockade and systemically administered EGF neutralizing antibody on skeletal muscle phenotype differ.
Limitations
The study focused on the role of EGF signaling in fiber-type specification, and the therapeutic potential of inhibiting it, hence the focus on loss-of-function experiments. The focus was on describing the clinical phenotype of increased ST gene expression/fibers with EGFR inhibition, rather than describing mechanistic aspects such as which of the known signaling mechanisms downstream of EGFR responsible for the phenotype change, as this is not relevant to translation to clinical studies of EGFR inhibitors. However, we appreciate that these mechanistic studies and also gain-of-function studies would be interesting and produce more indepth insights. Cell culture has limitations in modelling what happens in vivo, regardless of cell type. We used C2C12 myotubes rather than primary cells from humans because this was simpler, associations between EGF and fiber type were the same in mouse and human, and C2C12 myotubes beyond 6 days of differentiation express more FT than ST myosin40 resembling the pattern in patients. We used AG-1478 rather than an EGFR inhibitor in the clinic, because the likely required dose ranges were well-outlined and the mechanism of action of AG-1478 and the commercial inhibitors are identical41. We appreciate that there were differences in the experimental design of the EGFR inhibitor and siRNA cell culture experiments, necessitated by the difficulty of introducing genetic material into fully differentiated myotubes, but were reassured by the similarities between findings despite this. The zebrafish was a practical model to address our question because of ease of breeding and tracking ST fibers which lateralize within the somite early in development42. Had we performed the experiment in mice, this would have necessitated generating a muscle-specific inducible knockout of EGFR. This is because of the pre- and post-natal lethality of early EGFR knockout and also the ubiquitous role of EGFR in many tissues, such that that knockout in non-muscle tissues may have significant secondary effects on muscle. The use of a zebrafish rather than a mammalian model, and a developmental/myogenesis model rather than an adult in vivo model, could be viewed as limitations. However, the observation that inhibition of EGF signaling promotes the ST phenotype in this different context reassured us of the consistent role of EGF in ST fiber specification. It could also be argued that there is a more direct relevance of our developmental/myogenesis data. Fast-to-slow fiber type transitions in adult animals, at least when induced by changes in electrical stimulation, have been shown to involve recruitment of satellite cells, such that ablation of satellite cells by irradiation attenuates the fiber type transitions43,44. In quadriceps of COPD patients there is evidence of increased satellite cell recruitment to regenerating muscle fibers than in controls, in the form of increased numbers of fibers with centralised nuclei45–47. Therefore, if EGFR inhibition promotes commitment of satellite cells to a ST muscle phenotype this would promote FT to ST fiber-type switching in adult muscle. We didn’t replicate the work in a mammalian model as the data already gathered appears to be sufficient to proceed to the next step -a prospective observational study of muscle fiber type changes in cancer patients treated with EGFR inhibitors.
In conclusion, we describe that EGFR inhibition promotes the ST muscle phenotype, a mechanism that appears conserved across species and scenarios of both myogenesis and fiber maintenance, and that increased EGF signaling is associated with a loss of the ST phenotype in COPD patients as well as being negatively associated with the ST muscle phenotype in mice. These findings suggest that studies of muscle fiber type in patients receiving EGFR inhibitor treatment for cancer would be valuable to determine whether EGFR inhibitors have this effect in humans in vivo, and therefore whether EGFR inhibitors may have therapeutic potential for patients, for example with COPD, with a locomotor muscle ST to FT fiber-type.
Methods
Pathway analysis
Data relating to differentially expressed genes in isolated mouse ST and FT IIb fibers taken from18 were analyzed through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis)48 to identify ST phenotype-associated pathways (see Supplementary Information).
COPD patient and healthy control study
Vastus lateralis samples were collected for the study in5 which details clinical phenotyping, biopsy technique, and fiber typing. Studies were approved by the Royal Brompton and Harefield NHS Foundation Trust and Ealing and West London Mental Health Trust Research Ethics Committees (Studies 06/Q0404/35 and 06/Q0410/54) and all participants gave informed consent. Samples were stored in accordance with the Human Tissue Authority guidelines. The microarray analysis is described in25 and RNA extraction, cDNA synthesis and real-time quantitative PCR and quantification of serum EGF concentrations by ELISA are detailed in the Supplementary Information.
Cell culture experiments
C2C12 mouse myoblasts49 were differentiated for 7 days into multinucleate myotubes, which co-express ST and FT myosins within single cells. From day 8 myotubes were treated daily for 3 days with 100 ng/ml EGF neutralizing antibody (EGFnAb) or 200 nM tyrphostin/AG-1478 or the respective vehicle control. 200 nM AG-1478 reportedly elicits preferential EGFR inhibition (IC50 of 3 nM compared to >100 uM for ErbB2 and PDGF receptors50 in vitro) and downstream MAPK/extracellular related kinase (ERK) signaling inhibition in different cells51,52. Myoblasts were also transfected with 1.5 μM siRNA to EGFR or scrambled negative control, differentiated for 3 days then lysed for RNA or stained with anti-MyHC I antibody (DSHB Cat# BA-F8, RRID: AB_10572253) or MitoTracker Red (ThermoFisher Scientific). qPCR was performed with HPRT as the housekeeping gene. (This approach was used because transfection efficiency of genetic material into fully differentiated myotubes is poor without electroporation53). Protein was extracted from myotubes treated as above and from myotubes treated with same AG-1478 and EGFnAb concentrations and lysed 1-hour post dose. Extracts were immunoblotted for phosphorylated EGFR (phosphorylated at tyrosine 1068, 175 kDa), and for markers of downstream EGFR signaling, pERK [pERK1 phosphorylated at Thr202 and/or Tyr204 (44 kDa), pERK2 phosphorylated at Thr185 and/or Tyr187 (42 kDa)] and pAkt (Akt 1, 2 and 3 phosphorylated at serine 473, 60 kDA), then stripped and re-probed for total proteins (for the 1-hour post dose samples). Details are in the Supplementary Information.
Zebrafish experiments
Zebrafish on the AB background were housed and manipulated under UK Home Office Licence and standard procedures54. Whole-mount in situ hybridization for egfra was performed to confirm egfra expression as previously55. The CRISPR/Cas9 system was used to generate exon 1 egfra gene mutations and F3 generation allele number kg134 heterozygotes and their WT siblings were analysed (see Supplementary Information for details). RNA and DNA were extracted from pools of 5 larvae at 2 days post-fertilisation (dpf) for qPCR (normalization to geomean of beta-2 microglobulin and TATA-binding protein transcripts) and from single larva for genotyping. F2 egfra+/kg134 males were crossed to tagged ß-actin: GFP Tg(Ola.Actb:Hsa.HRAS-EGFP)vu119 line56 females. Resulting GFP-positive egfra+/kg134 larvae and WT siblings were immunostained with anti-myosin antibody (DSHB Cat# A4.1025, RRID: AB_528356) at 2 dpf and ST fibers which exist at the outer somite border were counted in a standard somite sample (16–18) by an investigator blind to genotype.
Statistical analysis
Exhaustive glm model comparison was carried out using the model selection and multimodel inference package glmulti, applying the Akaike information criterion. Continuous data was compared with a two-tailed t-test or Mann Whitney U-test depending on normality. Categorical data and correlations were analysed using Fisher’s exact test and calculation of Pearson’s coefficient respectively. A p-value < 0.05 was deemed statistically significant.
Supplementary information
Supplementary information including figures
Supplementary Information - Full Blots for All Figures
Supplementary Video E12 of wild-type zebrafish sibling
Supplementary Video E12 of egfra +kg134 zebrafish sibling
Supplementary information excluding figures
Acknowledgements
S.M.H. is a Medical Research Council Scientist with Programme Grant G1001029 and MR/N021231/1 support. M. Ciano was funded by a by a Biomedicine and Bioengineering in Osteoarthritis (BBOA) MRes/PhD scholarship at Imperial College London, M. Connolly was supported by a BHF studentship. This work was also supported by funding from the Wellcome Trust (clinical study and specimens), Medical Research Council (MRC) COPDMAP, the British Heart Foundation (BHF), and also the National Institute for Health Research (NIHR) Respiratory Disease Biomedical Research Unit at the Royal Brompton and Harefield NHS Foundation Trust and Imperial College London who part fund MIP’s salary. The views expressed in this publication are those of the authors and not necessarily those of the NHS, NIHR or the Department of Health. We would like to thank G. Bergamin, other members of the Hughes lab, King’s College London zebrafish facility staff and Imperial College London FILM facility staff for their help.
Author Contributions
The majority of the experiments were performed by M.C., G.M. and S.A.N. Clinical data and human biopsies were collected by S.A.N., supervised by M.I.P., microarray data were acquired and analysed by S.W.-O., M.M. and W.C. Data analysis was performed by S.A.N., P.R.K. and M.C. M.P.-C. conducted the IPA analysis. The study was designed by S.A.N., P.R.K. and S.M.H. S.A.N. designed the study concept, wrote the manuscript with the help of all authors, particularly J.A.M., S.M.H. and P.R.K., and had final approval of the manuscript.
Competing Interests
Dr S.A. Natanek and Dr P. Kemp have acted in an advisory capacity for Edgewise Therapeutics Inc. but have not received financial compensation for this, and the company does not have interest in this manuscript nor had any input into this publication. Materials, data and associated protocols from this manuscript are freely available by contacting the corresponding author.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-45567-4.
References
- 1.Kassebaum Nicholas J, Arora Megha, Barber Ryan M, Bhutta Zulfiqar A, Brown Jonathan, Carter Austin, Casey Daniel C, Charlson Fiona J, Coates Matthew M, Coggeshall Megan, Cornaby Leslie, Dandona Lalit, Dicker Daniel J, Erskine Holly E, Ferrari Alize J, Fitzmaurice Christina, Foreman Kyle, Forouzanfar Mohammad H, Fullman Nancy, Gething Peter W, Goldberg Ellen M, Graetz Nicholas, Haagsma Juanita A, Hay Simon I, Johnson Catherine O, Kemmer Laura, Khalil Ibrahim A, Kinfu Yohannes, Kutz Michael J, Kyu Hmwe H, Leung Janni, Liang Xiaofeng, Lim Stephen S, Lozano Rafael, Mensah George A, Mikesell Joe, Mokdad Ali H, Mooney Meghan D, Naghavi Mohsen, Nguyen Grant, Nsoesie Elaine, Pigott David M, Pinho Christine, Rankin Zane, Reinig Nikolas, Salomon Joshua A, Sandar Logan, Smith Alison, Sorensen Reed J D, Stanaway Jeffrey, Steiner Caitlyn, Teeple Stephanie, Troeger Chris, Truelsen Thomas, VanderZanden Amelia, Wagner Joseph A, Wanga Valentine, Whiteford Harvey A, Zhou Maigeng, Zoeckler Leo, Abajobir Amanuel Alemu, Abate Kalkidan Hassen, Abbafati Cristiana, Abbas Kaja M, Abd-Allah Foad, Abraham Biju, Abubakar Ibrahim, Abu-Raddad Laith J, Abu-Rmeileh Niveen M E, Achoki Tom, Ackerman Ilana N, Adebiyi Akindele Olupelumi, Adedeji Isaac Akinkunmi, Adsuar José C, Afanvi Kossivi Agbelenko, Afshin Ashkan, Agardh Emilie Elisabet, Agarwal Arnav, Agarwal Sanjay Kumar, Ahmed Muktar Beshir, Kiadaliri Aliasghar Ahmad, Ahmadieh Hamid, Akseer Nadia, Al-Aly Ziyad, Alam Khurshid, Alam Noore K M, Aldhahri Saleh Fahed, Alegretti Miguel Angel, Aleman Alicia V, Alemu Zewdie Aderaw, Alexander Lily T, Ali Raghib, Alkerwi Ala'a, Alla François, Allebeck Peter, Allen Christine, Alsharif Ubai, Altirkawi Khalid A, Martin Elena Alvarez, Alvis-Guzman Nelson, Amare Azmeraw T, Amberbir Alemayehu, Amegah Adeladza Kofi, Amini Heresh, Ammar Walid, Amrock Stephen Marc, Anderson Gregory M, Anderson Benjamin O, Antonio Carl Abelardo T, Anwari Palwasha, Ärnlöv Johan, Arsenijevic Valentina S Arsic, Artaman Al, Asayesh Hamid, Asghar Rana Jawad, Avokpaho Euripide Frinel G Arthur, Awasthi Ashish, Quintanilla Beatriz Paulina Ayala, Azzopardi Peter, Bacha Umar, Badawi Alaa, Balakrishnan Kalpana, Banerjee Amitava, Barac Aleksandra, Barker-Collo Suzanne L, Bärnighausen Till, Barregard Lars, Barrero Lope H, Basu Sanjay, Bayou Tigist Assefa, Beardsley Justin, Bedi Neeraj, Beghi Ettore, Bell Brent, Bell Michelle L, Benjet Corina, Bennett Derrick A, Bensenor Isabela M, Berhane Adugnaw, Bernabé Eduardo, Betsu Balem Demtsu, Beyene Addisu Shunu, Bhala Neeraj, Bhansali Anil, Bhatt Samir, Biadgilign Sibhatu, Bienhoff Kelly, Bikbov Boris, Abdulhak Aref A Bin, Biryukov Stan, Bisanzio Donal, Bjertness Espen, Blore Jed D, Borschmann Rohan, Boufous Soufiane, Bourne Rupert R A, Brainin Michael, Brazinova Alexandra, Breitborde Nicholas J K, Brugha Traolach S, Buchbinder Rachelle, Buckle Geoffrey Colin, Butt Zahid A, Calabria Bianca, Campos-Nonato Ismael Ricardo, Campuzano Julio Cesar, Carabin Hélène, Carapetis Jonathan R, Cárdenas Rosario, Carrero Juan Jesus, Castañeda-Orjuela Carlos A, Rivas Jacqueline Castillo, Catalá-López Ferrán, Cavalleri Fiorella, Chang Jung-Chen, Chiang Peggy Pei-Chia, Chibalabala Mirriam, Chibueze Chioma Ezinne, Chisumpa Vesper Hichilombwe, Choi Jee-Young Jasmine, Choudhury Lincoln, Christensen Hanne, Ciobanu Liliana G, Colistro Valentina, Colomar Mercedes, Colquhoun Samantha M, Cortinovis Monica, Crump John A, Damasceno Albertino, Dandona Rakhi, Dargan Paul I, das Neves José, Davey Gail, Davis Adrian C, Leo Diego De, Degenhardt Louisa, Gobbo Liana C Del, Derrett Sarah, Jarlais Don C Des, deVeber Gabrielle A, Dharmaratne Samath D, Dhillon Preet K, Ding Eric L, Doyle Kerrie E, Driscoll Tim R, Duan Leilei, Dubey Manisha, Duncan Bruce Bartholow, Ebrahimi Hedyeh, Ellenbogen Richard G, Elyazar Iqbal, Endries Aman Yesuf, Ermakov Sergey Petrovich, Eshrati Babak, Esteghamati Alireza, Estep Kara, Fahimi Saman, Farid Talha A, Farinha Carla Sofia e Sa, Faro André, Farvid Maryam S, Farzadfar Farshad, Feigin Valery L, Fereshtehnejad Seyed-Mohammad, Fernandes Jefferson G, Fernandes Joao C, Fischer Florian, Fitchett Joseph R A, Foigt Nataliya, Fowkes F Gerry R, Franklin Richard C, Friedman Joseph, Frostad Joseph, Fürst Thomas, Futran Neal D, Gabbe Belinda, Gankpé Fortuné Gbètoho, Garcia-Basteiro Alberto L, Gebrehiwot Tsegaye Tewelde, Gebremedhin Amanuel Tesfay, Geleijnse Johanna M, Gibney Katherine B, Gillum Richard F, Ginawi Ibrahim Abdelmageem Mohamed, Giref Ababi Zergaw, Giroud Maurice, Gishu Melkamu Dedefo, Giussani Giorgia, Godwin William W, Gomez-Dantes Hector, Gona Philimon, Goodridge Amador, Gopalani Sameer Vali, Gotay Carolyn C, Goto Atsushi, Gouda Hebe N, Gugnani Harish, Guo Yuming, Gupta Rahul, Gupta Rajeev, Gupta Vipin, Gutiérrez Reyna A, Hafezi-Nejad Nima, Haile Demewoz, Hailu Alemayehu Desalegne, Hailu Gessessew Bugssa, Halasa Yara A, Hamadeh Randah Ribhi, Hamidi Samer, Hammami Mouhanad, Handal Alexis J, Hankey Graeme J, Harb Hilda L, Harikrishnan Sivadasanpillai, Haro Josep Maria, Hassanvand Mohammad Sadegh, Hassen Tahir Ahmed, Havmoeller Rasmus, Hay Roderick J, Hedayati Mohammad T, Heredia-Pi Ileana Beatriz, Heydarpour Pouria, Hoek Hans W, Hoffman Daniel J, Horino Masako, Horita Nobuyuki, Hosgood H Dean, Hoy Damian G, Hsairi Mohamed, Huang Hsiang, Huang John J, Iburg Kim Moesgaard, Idrisov Bulat T, Innos Kaire, Inoue Manami, Jacobsen Kathryn H, Jauregui Alejandra, Jayatilleke Achala Upendra, Jeemon Panniyammakal, Jha Vivekanand, Jiang Guohong, Jiang Ying, Jibat Tariku, Jimenez-Corona Aida, Jin Ye, Jonas Jost B, Kabir Zubair, Kajungu Dan K, Kalkonde Yogeshwar, Kamal Ritul, Kan Haidong, Kandel Amit, Karch André, Karema Corine Kakizi, Karimkhani Chante, Kasaeian Amir, Katibeh Marzieh, Kaul Anil, Kawakami Norito, Kazi Dhruv S, Keiyoro Peter Njenga, Kemp Andrew Haddon, Kengne Andre Pascal, Keren Andre, Kesavachandran Chandrasekharan Nair, Khader Yousef Saleh, Khan Abdur Rahman, Khan Ejaz Ahmad, Khang Young-Ho, Khoja Tawfik Ahmed Muthafer, Khubchandani Jagdish, Kieling Christian, Kim Cho-il, Kim Daniel, Kim Yun Jin, Kissoon Niranjan, Kivipelto Miia, Knibbs Luke D, Knudsen Ann Kristin, Kokubo Yoshihiro, Kolte Dhaval, Kopec Jacek A, Koul Parvaiz A, Koyanagi Ai, Defo Barthelemy Kuate, Kuchenbecker Ricardo S, Bicer Burcu Kucuk, Kuipers Ernst J, Kumar G Anil, Kwan Gene F, Lalloo Ratilal, Lallukka Tea, Larsson Anders, Latif Asma Abdul, Lavados Pablo M, Lawrynowicz Alicia Elena Beatriz, Leasher Janet L, Leigh James, Leung Ricky, Li Yichong, Li Yongmei, Lipshultz Steven E, Liu Patrick Y, Liu Yang, Lloyd Belinda K, Logroscino Giancarlo, Looker Katharine J, Lotufo Paulo A, Lucas Robyn M, Lunevicius Raimundas, Lyons Ronan A, Razek Hassan Magdy Abd El, Mahdavi Mahdi, Majdan Marek, Majeed Azeem, Malekzadeh Reza, Malta Deborah Carvalho, Marcenes Wagner, Martinez-Raga Jose, Masiye Felix, Mason-Jones Amanda J, Matzopoulos Richard, Mayosi Bongani M, McGrath John J, McKee Martin, Meaney Peter A, Mehari Alem, Melaku Yohannes Adama, Memiah Peter, Memish Ziad A, Mendoza Walter, Meretoja Atte, Meretoja Tuomo J, Mesfin Yonatan Moges, Mhimbira Francis Apolinary, Millear Anoushka, Miller Ted R, Mills Edward J, Mirarefin Mojde, Mirrakhimov Erkin M, Mitchell Philip B, Mock Charles N, Mohammad Karzan Abdulmuhsin, Mohammadi Alireza, Mohammed Shafiu, Monasta Lorenzo, Hernandez Julio Cesar Montañez, Montico Marcella, Moradi-Lakeh Maziar, Mori Rintaro, Mueller Ulrich O, Mumford John Everett, Murdoch Michele E, Murthy Gudlavalleti Venkata Satyanarayana, Nachega Jean B, Naheed Aliya, Naldi Luigi, Nangia Vinay, Newton John N, Ng Marie, Ngalesoni Frida Namnyak, Nguyen Quyen Le, Nisar Muhammad Imran, Pete Patrick Martial Nkamedjie, Nolla Joan M, Norheim Ole F, Norman Rosana E, Norrving Bo, Obermeyer Carla Makhlouf, Ogbo Felix Akpojene, Oh In-Hwan, Oladimeji Olanrewaju, Olivares Pedro R, Olusanya Bolajoko Olubukunola, Olusanya Jacob Olusegun, Oren Eyal, Ortiz Alberto, Ota Erika, Oyekale Abayomi Samuel, PA Mahesh, Park Eun-Kee, Parsaeian Mahboubeh, Patten Scott B, Patton George C, Pedro João Mário, Pereira David M, Perico Norberto, Pesudovs Konrad, Petzold Max, Phillips Michael Robert, Piel Frédéric B, Pillay Julian David, Pishgar Farhad, Plass Dietrich, Polinder Suzanne, Popova Svetlana, Poulton Richie G, Pourmalek Farshad, Prasad Noela M, Qorbani Mostafa, Rabiee Rynaz H S, Radfar Amir, Rafay Anwar, Rahimi Kazem, Rahimi-Movaghar Vafa, Rahman Mahfuzar, Rahman Mohammad Hifz Ur, Rahman Sajjad Ur, Rai Dheeraj, Rai Rajesh Kumar, Rajsic Sasa, Raju Murugesan, Ram Usha, Ranganathan Kavitha, Refaat Amany H, Reitsma Marissa B, Remuzzi Giuseppe, Resnikoff Serge, Reynolds Alex, Ribeiro Antonio L, Ricci Stefano, Roba Hirbo Shore, Rojas-Rueda David, Ronfani Luca, Roshandel Gholamreza, Roth Gregory A, Roy Ambuj, Sackey Ben Benasco, Sagar Rajesh, Sanabria Juan R, Sanchez-Niño Maria Dolores, Santos Itamar S, Santos João Vasco, Sarmiento-Suarez Rodrigo, Sartorius Benn, Satpathy Maheswar, Savic Miloje, Sawhney Monika, Schmidt Maria Inês, Schneider Ione J C, Schutte Aletta E, Schwebel David C, Seedat Soraya, Sepanlou Sadaf G, Servan-Mori Edson E, Shahraz Saeid, Shaikh Masood Ali, Sharma Rajesh, She Jun, Sheikhbahaei Sara, Shen Jiabin, Sheth Kevin N, Shibuya Kenji, Shigematsu Mika, Shin Min-Jeong, Shiri Rahman, Sigfusdottir Inga Dora, Silva Diego Augusto Santos, Silverberg Jonathan I, Simard Edgar P, Singh Abhishek, Singh Jasvinder A, Singh Prashant Kumar, Skirbekk Vegard, Skogen Jens Christoffer, Soljak Michael, Søreide Kjetil, Sorensen Reed J D, Sreeramareddy Chandrashekhar T, Stathopoulou Vasiliki, Steel Nicholas, Stein Dan J, Stein Murray B, Steiner Timothy J, Stovner Lars Jacob, Stranges Saverio, Stroumpoulis Konstantinos, Sunguya Bruno F, Sur Patrick J, Swaminathan Soumya, Sykes Bryan L, Szoeke Cassandra E I, Tabarés-Seisdedos Rafael, Tandon Nikhil, Tanne David, Tavakkoli Mohammad, Taye Bineyam, Taylor Hugh R, Ao Braden J Te, Tegegne Teketo Kassaw, Tekle Dejen Yemane, Terkawi Abdullah Sulieman, Tessema Gizachew Assefa, Thakur J S, Thomson Alan J, Thorne-Lyman Andrew L, Thrift Amanda G, Thurston George D, Tobe-Gai Ruoyan, Tonelli Marcello, Topor-Madry Roman, Topouzis Fotis, Tran Bach Xuan, Truelsen Thomas, Dimbuene Zacharie Tsala, Tsilimbaris Miltiadis, Tura Abera Kenay, Tuzcu Emin Murat, Tyrovolas Stefanos, Ukwaja Kingsley N, Undurraga Eduardo A, Uneke Chigozie Jesse, Uthman Olalekan A, van Gool Coen H, van Os Jim, Vasankari Tommi, Vasconcelos Ana Maria Nogales, Venketasubramanian Narayanaswamy, Violante Francesco S, Vlassov Vasiliy Victorovich, Vollset Stein Emil, Wagner Gregory R, Wallin Mitchell T, Wang Linhong, Weichenthal Scott, Weiderpass Elisabete, Weintraub Robert G, Werdecker Andrea, Westerman Ronny, Wijeratne Tissa, Wilkinson James D, Williams Hywel C, Wiysonge Charles Shey, Woldeyohannes Solomon Meseret, Wolfe Charles D A, Won Sungho, Xu Gelin, Yadav Ajit Kumar, Yakob Bereket, Yan Lijing L, Yano Yuichiro, Yaseri Mehdi, Ye Pengpeng, Yip Paul, Yonemoto Naohiro, Yoon Seok-Jun, Younis Mustafa Z, Yu Chuanhua, Zaidi Zoubida, Zaki Maysaa El Sayed, Zeeb Hajo, Zodpey Sanjay, Zonies David, Zuhlke Liesl Joanna, Vos Theo, Lopez Alan D, Murray Christopher J L. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet. 2016;388(10053):1603–1658. doi: 10.1016/S0140-6736(16)31460-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiological reviews. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 3.Daugaard JR, et al. Fiber type-specific expression of GLUT4 in human skeletal muscle: influence of exercise training. Diabetes. 2000;49:1092–1095. doi: 10.2337/diabetes.49.7.1092. [DOI] [PubMed] [Google Scholar]
- 4.Kugelberg E, Edstrom L. Differential histochemical effects of muscle contractions on phosphorylase and glycogen in various types of fibres: relation to fatigue. Journal of neurology, neurosurgery, and psychiatry. 1968;31:415–423. doi: 10.1136/jnnp.31.5.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natanek SA, et al. Heterogeneity of quadriceps muscle phenotype in chronic obstructive pulmonary disease (Copd); implications for stratified medicine? Muscle & nerve. 2013;48:488–497. doi: 10.1002/mus.23784. [DOI] [PubMed] [Google Scholar]
- 6.Drexler H, et al. Alterations of skeletal muscle in chronic heart failure. Circulation. 1992;85:1751–1759. doi: 10.1161/01.CIR.85.5.1751. [DOI] [PubMed] [Google Scholar]
- 7.Gosker HR, Zeegers MP, Wouters EF, Schols AM. Muscle fibre type shifting in the vastus lateralis of patients with COPD is associated with disease severity: a systematic review and meta-analysis. Thorax. 2007;62:944–949. doi: 10.1136/thx.2007.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosker HR, et al. Skeletal muscle fibre-type shifting and metabolic profile in patients with chronic obstructive pulmonary disease. The European respiratory journal. 2002;19:617–625. doi: 10.1183/09031936.02.00762001. [DOI] [PubMed] [Google Scholar]
- 9.Oberbach A, et al. Altered fiber distribution and fiber-specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes care. 2006;29:895–900. doi: 10.2337/diacare.29.04.06.dc05-1854. [DOI] [PubMed] [Google Scholar]
- 10.Helge JW, et al. Interrelationships between muscle fibre type, substrate oxidation and body fat. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity. 1999;23:986–991. doi: 10.1038/sj.ijo.0801030. [DOI] [PubMed] [Google Scholar]
- 11.Patel MS, et al. Vastus lateralis fiber shift is an independent predictor of mortality in COPD. American journal of respiratory and critical care medicine. 2014;190:350–352. doi: 10.1164/rccm.201404-0713LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaufelberger M, Eriksson BO, Grimby G, Held P, Swedberg K. Skeletal muscle fiber composition and capillarization in patients with chronic heart failure: relation to exercise capacity and central hemodynamics. Journal of cardiac failure. 1995;1:267–272. doi: 10.1016/1071-9164(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 13.Segerstrom AB, et al. Exercise capacity in relation to body fat distribution and muscle fibre distribution in elderly male subjects with impaired glucose tolerance, type 2 diabetes and matched controls. Diabetes research and clinical practice. 2011;94:57–63. doi: 10.1016/j.diabres.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Windisch A, Gundersen K, Szabolcs MJ, Gruber H, Lomo T. Fast to slow transformation of denervated and electrically stimulated rat muscle. The Journal of physiology. 1998;510(Pt 2):623–632. doi: 10.1111/j.1469-7793.1998.623bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YX, et al. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS biology. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maltais F, et al. Skeletal muscle adaptation to endurance training in patients with chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 1996;154:442–447. doi: 10.1164/ajrccm.154.2.8756820. [DOI] [PubMed] [Google Scholar]
- 17.Zierath JR, Hawley JA. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS biology. 2004;2:e348. doi: 10.1371/journal.pbio.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chemello F, et al. Microgenomic analysis in skeletal muscle: expression signatures of individual fast and slow myofibers. PloS one. 2011;6:e16807. doi: 10.1371/journal.pone.0016807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, et al. Modulation of skeletal muscle fiber type by mitogen-activated protein kinase signaling. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2008;22:2990–3000. doi: 10.1096/fj.07-097600. [DOI] [PubMed] [Google Scholar]
- 21.Murgia M, et al. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nature cell biology. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 22.Willis-Owen SAG, et al. COPD is accompanied by co-ordinated transcriptional perturbation in the quadriceps affecting the mitochondria and extracellular matrix. Scientific reports. 2018;8:12165. doi: 10.1038/s41598-018-29789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quiat D., Voelker K. A., Pei J., Grishin N. V., Grange R. W., Bassel-Duby R., Olson E. N. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor Sox6. Proceedings of the National Academy of Sciences. 2011;108(25):10196–10201. doi: 10.1073/pnas.1107413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100 H. Developmental dynamics: an official publication of the American Association of Anatomists. 2005;234:301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- 25.Saffron, A. G. et al. COPD is accompanied by co-ordinated transcriptional perturbation in the quadriceps affecting the mitochondria and extracellular matrix. Scientific reports, in press (2018). [DOI] [PMC free article] [PubMed]
- 26.Daugaard JR, Richter EA. Relationship between muscle fibre composition, glucose transporter protein 4 and exercise training: possible consequences in non-insulin-dependent diabetes mellitus. Acta physiologica Scandinavica. 2001;171:267–276. doi: 10.1046/j.1365-201x.2001.00829.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang, Z. & Xiong, H.-R. Culture Conditions and Types of Growth Media for Mammalian Cells, B. T. C., Dr. Ceccherini-Nelli, Luca (Ed.), InTech, 10.5772/52301. Available from, https://mts.intechopen.com/books/biomedical-tissue-culture/culture-conditions-and-types-of-growth-media-for-mammalian-cellsCulture Conditions and Types of Growth Media for Mammalian Cells, Biomedical Tissue Culture (2012).
- 28.Knudsen SL, Mac AS, Henriksen L, van Deurs B, Grovdal LM. EGFR signaling patterns are regulated by its different ligands. Growth factors (Chur, Switzerland) 2014;32:155–163. doi: 10.3109/08977194.2014.952410. [DOI] [PubMed] [Google Scholar]
- 29.Goishi K, et al. Inhibition of zebrafish epidermal growth factor receptor activity results in cardiovascular defects. Mechanisms of development. 2003;120:811–822. doi: 10.1016/S0925-4773(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 30.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science (New York, N.Y.) 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 31.Gosker HR, et al. Muscle fiber type IIX atrophy is involved in the loss of fat-free mass in chronic obstructive pulmonary disease. The American journal of clinical nutrition. 2002;76:113–119. doi: 10.1093/ajcn/76.1.113. [DOI] [PubMed] [Google Scholar]
- 32.Efferth T. Signal transduction pathways of the epidermal growth factor receptor in colorectal cancer and their inhibition by small molecules. Current medicinal chemistry. 2012;19:5735–5744. doi: 10.2174/092986712803988884. [DOI] [PubMed] [Google Scholar]
- 33.Gao Q, Lei T, Ye F. Therapeutic targeting of EGFR-activated metabolic pathways in glioblastoma. Expert opinion on investigational drugs. 2013;22:1023–1040. doi: 10.1517/13543784.2013.806484. [DOI] [PubMed] [Google Scholar]
- 34.Hollway GE, et al. Scube2 mediates Hedgehog signalling in the zebrafish embryo. Developmental biology. 2006;294:104–118. doi: 10.1016/j.ydbio.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 35.Shi H, et al. Extracellular signal-regulated kinase pathway is differentially involved in beta-agonist-induced hypertrophy in slow and fast muscles. American journal of physiology. Cell physiology. 2007;292:C1681–1689. doi: 10.1152/ajpcell.00466.2006. [DOI] [PubMed] [Google Scholar]
- 36.Fuentes EN, et al. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. American journal of physiology. Regulatory, integrative and comparative physiology. 2011;300:R1532–1542. doi: 10.1152/ajpregu.00535.2010. [DOI] [PubMed] [Google Scholar]
- 37.Philippou A, Maridaki M, Halapas A, Koutsilieris M. The role of the insulin-like growth factor 1 (IGF-1) in skeletal muscle physiology. In vivo (Athens, Greece) 2007;21:45–54. [PubMed] [Google Scholar]
- 38.Ceafalan LC, et al. Interstitial Outburst of Angiogenic Factors During Skeletal Muscle Regeneration After Acute Mechanical Trauma. Anatomical record (Hoboken, N.J.: 2007) 2015;298:1864–1879. doi: 10.1002/ar.23254. [DOI] [PubMed] [Google Scholar]
- 39.Krall JA, Beyer EM, MacBeath G. High- and low-affinity epidermal growth factor receptor-ligand interactions activate distinct signaling pathways. PloS one. 2011;6:e15945. doi: 10.1371/journal.pone.0015945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown DM, Parr T, Brameld JM. Myosin heavy chain mRNA isoforms are expressed in two distinct cohorts during C2C12 myogenesis. Journal of muscle research and cell motility. 2012;32:383–390. doi: 10.1007/s10974-011-9267-4. [DOI] [PubMed] [Google Scholar]
- 41.Jimeno A, Hidalgo M. Pharmacogenomics of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors. Biochimica et biophysica acta. 2006;1766:217–229. doi: 10.1016/j.bbcan.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Pownall ME, Gustafsson MK, Emerson CP., Jr. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annual review of cell and developmental biology. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 43.Martins KJ, et al. Effect of satellite cell ablation on low-frequency-stimulated fast-to-slow fibre-type transitions in rat skeletal muscle. The Journal of physiology. 2006;572:281–294. doi: 10.1113/jphysiol.2005.103366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martins KJ, et al. Satellite cell ablation attenuates short-term fast-to-slow fibre type transformations in rat fast-twitch skeletal muscle. Pflugers Archiv: European journal of physiology. 2009;458:325–335. doi: 10.1007/s00424-008-0625-z. [DOI] [PubMed] [Google Scholar]
- 45.Theriault ME, Pare ME, Lemire BB, Maltais F, Debigare R. Regenerative defect in vastus lateralis muscle of patients with chronic obstructive pulmonary disease. Respiratory research. 2014;15:35. doi: 10.1186/1465-9921-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis A, et al. Increased expression of H19/miR-675 is associated with a low fat-free mass index in patients with COPD. Journal of cachexia, sarcopenia and muscle. 2016;7:330–344. doi: 10.1002/jcsm.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theriault ME, Pare ME, Maltais F, Debigare R. Satellite cells senescence in limb muscle of severe patients with COPD. PloS one. 2012;7:e39124. doi: 10.1371/journal.pone.0039124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (Oxford, England) 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 50.Osherov N, Levitzki A. Epidermal-growth-factor-dependent activation of the src-family kinases. European journal of biochemistry. 1994;225:1047–1053. doi: 10.1111/j.1432-1033.1994.1047b.x. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, et al. Stimulation of the mitogen-activated protein kinase cascade and tyrosine phosphorylation of the epidermal growth factor receptor by hepatopoietin. The Journal of biological chemistry. 2000;275:37443–37447. doi: 10.1074/jbc.M004373200. [DOI] [PubMed] [Google Scholar]
- 52.Levitzki A, Gazit A. Tyrosine kinase inhibition: an approach to drug development. Science (New York, N.Y.) 1995;267:1782–1788. doi: 10.1126/science.7892601. [DOI] [PubMed] [Google Scholar]
- 53.Sandri M, Bortoloso E, Nori A, Volpe P. Electrotransfer in differentiated myotubes: a novel, efficient procedure for functional gene transfer. Experimental cell research. 2003;286:87–95. doi: 10.1016/S0014-4827(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 54.Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th edition edn, (University of Oregon Press, Eugene, 2000).
- 55.Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development (Cambridge, England) 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cooper MS, et al. Visualizing morphogenesis in transgenic zebrafish embryos using BODIPY TR methyl ester dye as a vital counterstain for GFP. Developmental dynamics: an official publication of the American Association of Anatomists. 2005;232:359–368. doi: 10.1002/dvdy.20252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information including figures
Supplementary Information - Full Blots for All Figures
Supplementary Video E12 of wild-type zebrafish sibling
Supplementary Video E12 of egfra +kg134 zebrafish sibling
Supplementary information excluding figures