Abstract
CaMKIIα plays an essential role in decoding Ca2+ signaling in spines by acting as a leaky Ca2+ integrator with the time constant of several seconds. However, the mechanism by which CaMKIIα integrates Ca2+ signals remains elusive. Here, we imaged CaMKIIα-CaM association in single dendritic spines using a new FRET sensor and two-photon fluorescence lifetime imaging. In response to a glutamate uncaging pulse, CaMKIIα-CaM association increases in ~0.1 s and decays over ~3 s. During repetitive glutamate uncaging, which induces spine structural plasticity, CaMKIIα-CaM association did not show further increase but sustained at a constant level. Since CaMKIIα activity integrates Ca2+ signals over ~10 s under this condition, the integration of Ca2+ signal by CaMKIIα during spine structural plasticity is largely due to Ca2+/CaM-independent, autonomous activity. Based on these results, we propose a simple kinetic model of CaMKIIα activation in dendritic spines.
Subject terms: Cellular neuroscience, Long-term potentiation
Activation of Ca2+/calmodulin-dependent kinase II (CaMKII) in dendritic spines is a key step of long-term potentiation (LTP) induction, yet the exact biochemical steps of CaMKIIα activation in dendritic spines remained elusive. In this study, the authors developed a novel imaging approach to monitor CaM interactions CaMKIIα in cultured hippocampal neurons after uncaging of glutamate. This allowed the authors to model the kinetics of CaMKIIα activation in single dendritic spines.
Introduction
Calcium (Ca2+)/calmodulin-dependent kinase II (CaMKII), a serine/threonine kinase, is critical for various forms of synaptic plasticity that underlie learning and memory. CaMKII is composed of 12 subunits, each of which is a kinase that is activated by the binding of Ca2+/calmodulin (CaM)1. The most abundant subunit in the forebrain, CaMKIIα, is required for LTP, spine structural LTP (sLTP) and spatial learning2–5. In addition to Ca2+/CaM binding, CaMKIIα activity is regulated by autophosphorylation at multiple sites. Autophosphorylation at Thr286 prolongs CaMKIIα activity6–8, permitting the integration of Ca2+ transients to facilitate the induction of spine plasticity9,10. Disruption of this phosphorylation in Camk2aT286A knock-in mice impairs LTP, sLTP, and spatial learning and memory10,11. It is known that phosphorylation at Thr286 causes an enhancement in binding affinity to Ca2+/CaM7,12 as well as induces a Ca2+/CaM-independent, autonomous kinase activity state13,14. This autonomous activity of CaMKIIα is thought to be important for the induction and the maintenance of LTP14. CaMKIIα is additionally regulated by autophosphorylation at Thr305 and Thr306, which inhibit binding of Ca2+/CaM to CaMKIIα15,16.
CaMKIIα activity in response to Ca2+ elevations in dendritic spines can be measured by a fluorescence resonance energy transfer (FRET) sensor Camuiα in combination with 2-photon fluorescence lifetime imaging (2pFLIM)6,10,17. A brief pulse of two-photon glutamate uncaging induces a transient Ca2+ elevation, lasting ~100 ms, in the stimulated spine6,10,18. This causes a rapid CaMKIIα activation, which peaks within ~0.5 s and then decays over ~10 s, in the stimulated spine10. In response to a repetitive glutamate uncaging (~0.5 Hz), which induces LTP in the stimulated spine5,6, CaMKIIα activity increases in a stepwise manner, following each uncaging pulse until plateauing within ~10 s (ref. 10). After the cessation of glutamate uncaging, CaMKIIα activity decayed with time constants of ~6 s and ~1 min (ref. 10). These experiments suggest that CaMKIIα is a leaky integrator of Ca2+ signals10.
Camuiα measures the conformation change of CaMKIIα associated with its activation by both Ca2+/CaM binding and Thr286 autophosphorylation6,17. Previous studies using this sensor suggest that the optimal integration of Ca2+ signals by CaMKIIα requires Thr286 autophosphorylation, suggesting that autonomous activity may play an important role in this process6,10. However, if an autonomous state of CaMKIIα exists in the stimulated spines, and if so, how much this state contributes to CaMKIIα activation remains elusive.
Here, we used two-photon fluorescence lifetime microscopy (2pFLIM) to probe the association between CaMKIIα and Ca2+/CaM. Our results revealed that the fraction of CaMKIIα bound to Ca2+/CaM does not continue to increase with multiple Ca2+ transients during the induction of sLTP. Taken together with our previous report showing that CaMKIIα activity integrates Ca2+ signals over ~10 s to 1 min under similar conditions10, our results suggest that the integration of Ca2+ signals depends largely on Ca2+/CaM-independent, ‘autonomous’ activity of CaMKIIα. We propose a simple kinetic scheme of CaMKIIα activation that is consistent with our experimental results both for CaMKIIα-CaM association and for CaMKIIα activation. This model highlights that autonomous activity, but not Ca2+/CaM-dependent activity, accounts for the majority of CaMKIIα activity.
Results
Sensor for association of CaMKIIα and calmodulin
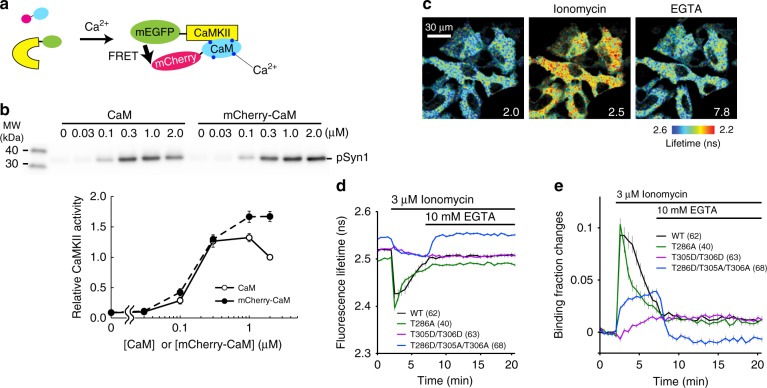
To measure the association of CaMKIIα with CaM, we developed a FRET-based biosensor made of monomeric EGFP (mEGFP)-CaMKIIα and mCherry-CaM (Fig. 1a)19. Biochemical cell-free assays showed that mCherry-CaM supports CaMKIIα activity similarly to a non-labeled CaM at a wide range of concentrations (0.03 µM < [CaM or mCherry-CaM] < 2 µM) (Fig. 1b), suggesting that mCherry fusion does not affect the affinity of CaM for CaMKIIα.
Fig. 1.
Design and characterization of CaMKIIα-CaM association sensor. a Design of a FRET sensor for CaMKIIα-CaM association. Monomeric EGFP (mEGFP) and monomeric Cherry (mCherry) fluorescent protein are fused to the N-terminus of CaMKIIα and the N-terminus of CaM, respectively. b mCherry-CaM activates CaMKIIα to the degree similar to non-labeled CaM at different concentrations of CaM in a cell-free system. Upper panel: western blot of phosphorylated Synapsin1 peptide (pSyn1) fused to mCherry. Lower panel: quantification of pSyn1 signal from 4 experiments, normalized with the pSyn1 signal at 2 µM non-labeled CaM. c Fluorescence lifetime images of CaMKIIα-CaM association sensor expressed in HeLa cells. d Time courses of fluorescence lifetime of CaMKIIα-CaM association sensor and its mutants (T286A, T305D/T306D and T286D/T305A/T306A) in response to bath application of ionomycin (3 µM) and EGTA (10 mM). e Time courses of changes in CaMKIIα-CaM association calculated from d. All data are shown in mean ± sem
We further characterized the CaMKIIα-CaM association sensor in HeLa cells (Fig. 1c–e). To do so, we bath-applied an ionophore (3 µM ionomycin) to elevate intracellular [Ca2+], and then subsequently added EGTA to reverse the reaction. In response to the ionophore application, the CaMKIIα-CaM association sensor first showed a rapid increase in FRET signal, which decayed over a few minutes. This signal further decayed in response to extracellular EGTA application, which chelates extracellular [Ca2+] (and thus decreasing intracellular [Ca2+]) (Fig. 1c–e). However, we observed a residual CaMKIIα-CaM association, which persisted more than 20 min.
Next, we characterized the association of CaM with various CaMKIIα phosphorylation mutants. We first introduced phospho-mimic mutations of inhibitory autophosphorylation sites (T305D/T306D) to inhibit the interaction of CaM to the regulatory domain of CaMKIIα2,15. We found that the mutation largely inhibited the rapid CaM binding, consistent with the previous reports15. However, there was a small and persistent increase in FRET signal, whose amplitude and time scale are similar to that of the persistent component of the FRET signal of wildtype (WT) CaMKIIαWT-CaM association. Since the regulatory domain of T305D/T306D mutant does not have the capability to bind Ca2+/CaM2,15, the observed persistent CaMKIIαWT-CaM association is unlikely due to the association of CaM to the regulatory domain of CaMKIIα. Phospho-dead mutation at Thr286 (T286A), the site important for autonomous CaMKIIα activation8, accelerated the decay of FRET signal, consistent with a role phosphorylation at this site to prolong CaMKIIα activation10. A small population of CaMKIIαT286A mutant also exhibited persistent CaM binding, suggesting that this component is related to neither CaM binding to the regulatory domain nor T286 autophosphorylation, and thus perhaps not related to the regulation of CaMKIIα activation. Finally, we measured the binding of CaM with a phospho-mimic mutation at Thr286 (T286D). Since this mutation is known to cause inhibitory autophosphorylation at T305/T306, which inhibits Ca2+/CaM binding15,16, we introduced T305A/T306A mutation in addition to T286D (CaMKIIαT286D/T305A/T306A)20. In response to ionophore application, CaMKIIαT286D/T305A/T306A-CaM association displayed a persistent increase, which was reversed by EGTA application.
Association of CaMKIIα-CaM in dendritic spines
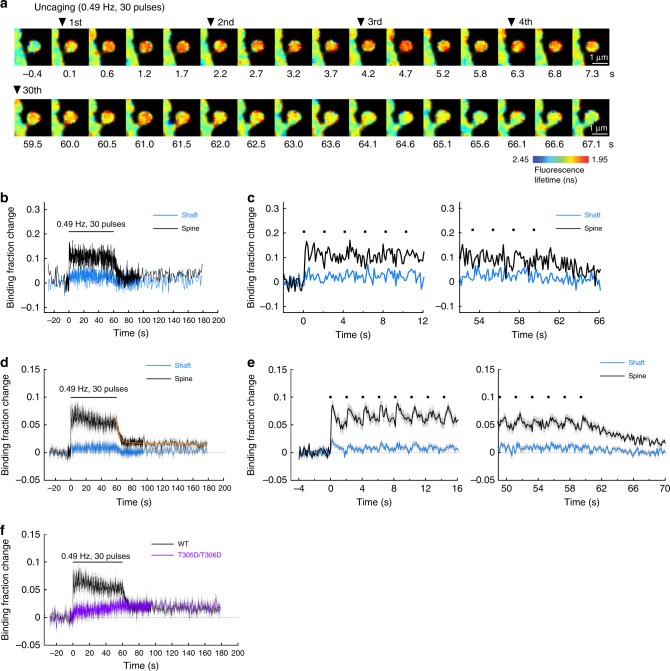
To measure the association of CaMKIIα-CaM during the induction of spine plasticity, we biolistically transfected organotypic hippocampal slice cultures of mice with the CaMKIIα-CaM association sensor and imaged CA1 pyramidal neurons with 2pFLIM. Structural LTP (sLTP) was induced in a single spine by applying repetitive pulses (0.49 Hz, 30 pulses) of two-photon glutamate uncaging to the spine in the absence of extracellular Mg2+ (refs. 5,6).
We first measured CaMKIIα-CaM association during sLTP induction with a temporal resolution of 128 ms/frame (Fig. 2a). Binding of CaM to CaMKIIα occurs rapidly within 1 frame (128 ms) in the stimulated spine. The binding plateaued with the first glutamate uncaging pulse, and subsequent uncaging pulses did not result in a higher level of CaMKIIα-CaM association (Fig. 2b–e). The fractional change in binding of CaMKIIα to Ca2+/CaM during sLTP induction was independent of the overexpression level (Supplementary Fig. 1). After cessation of glutamate uncaging, CaM dissociated from CaMKIIα with the time constant of 3.2 ± 0.7 s. In addition to the fast decay, we observed a persistent component after cessation of uncaging (Fig. 2d, e). This component appeared to be not related to the binding of Ca2+/CaM to the CaM-binding domain of CaMKIIα, since a CaMKIIα mutant without binding capability (CaMKIIαT305D/T306D) also showed this persistent component (Fig. 2f), similarly to the results in HeLa cells (Fig. 1e).
Fig. 2.
CaMKIIα-CaM association during sLTP induction. a Representative fluorescence lifetime images of CaMKIIα-CaM association sensor during glutamate uncaging at 0.49 Hz. Warmer colors indicate lower fluorescence lifetime, corresponding to a higher binding fraction of mCherry-CaM to mEGFP-CaMKIIα. Scale bar, 1 µm. b Time course of CaMKIIα-CaM association in a stimulated spine (black) and nearby dendrite (blue). Analyzed from images in a. Black dots represent uncaging pulses. c Expanded view of the rising phase (left) and the decay phase (right) of b. d Averaged change in CaMKIIα-CaM association in stimulated spines (black) and nearby dendrite (blue) (n = 27 spines/9 neurons). The orange curve indicates the decay of binding fraction change obtained by curve fitting of a double-exponential function: B(t) = B0 [Pfast·exp(–t/τfast) + Pslow·exp(–t/τslow)], where B0 is the initial binding fraction change, τfast and τslow are the fast and slow decay time constants and Pfast and Pslow are the respective populations. The time constants are obtained as τfast = 3.2 ± 0.6 s (Pfast = 71%) and τslow = 572 ± 843 s (Pslow = 29%). e Expanded view of the rising phase (left) and the decay phase (right) of d. f Average time course of CaMKIIα-CaM association for a mutant mEGFP-CaMKIIαT305D/T306D in which the Thr305 and Thr306 are mutated to aspartate. The mutation precludes Ca2+/CaM binding in the stimulated spine during glutamate uncaging at 0.49 Hz (purple; n = 34 spines/5 neurons). The data for CaMKIIαWT (black) are from c for the comparison. All data are shown in mean ± sem, and sem of time constants is obtained by bootstrapping
The above experiments were performed at room temperature (25–27 °C). At a near physiological temperature (34–35 °C), Ca2+/CaM dissociated faster (τ = 0.4 ± 0.5 s; Supplementary Fig. 2). The temperature coefficient of the dissociation kinetics was determined to be Q10 = 10.3.
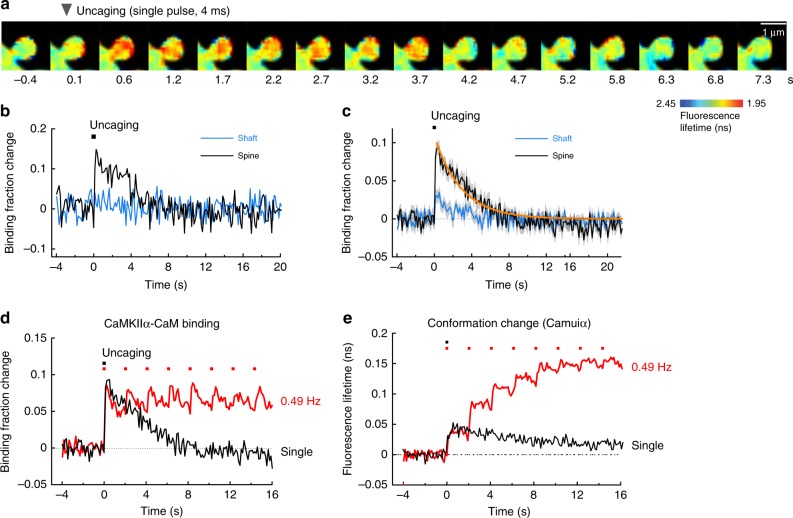
To determine whether CaMKIIα-CaM association showed the integration of multiple uncaging pulses, we compared the binding induced during sLTP (trains of pulses) with the CaMKIIα-CaM association in response to a single glutamate uncaging pulse (Fig. 3). Binding of Ca2+/CaM to CaMKIIα increased rapidly in response to a single uncaging pulse, to a magnitude similar to sLTP-inducing stimulations and then decayed (Fig. 3a–c). The dissociation time constant was obtained as τ = 2.9 ± 0.3 s (Fig. 3c), a value similar to that obtained after the cessation of sLTP induction (Fig. 2e). The fraction of CaMKIIα binding to CaM was similar during trains of glutamate uncaging and in response to a single glutamate uncaging pulse (Fig. 3d). This is a sharp contrast to measurements of the active conformation CaMKIIα in spines (Fig. 3e), which shows a slower decay in response to a single uncaging pulse (6.4 ± 0.7 s (74%) and 92.6 ± 50.7 s (26%)), and accumulates to higher levels during trains of uncaging pulses10.
Fig. 3.
CaMKIIα-CaM association in response to a single glutamate uncaging pulse. a Representative fluorescence lifetime images of CaMKIIα-CaM association in response to a single glutamate uncaging pulse. Warmer colors indicate lower fluorescence lifetime, corresponding to a higher binding fraction of mCherry-CaM to mEGFP-CaMKIIα. Scale bar, 1 µm. b Time course of CaMKIIα-CaM association in a stimulated spine (black) and nearby dendritic (blue). Inset is an expanded view of the rising phase. Black squares denote uncaging pulses. Analyzed from images in a. c Averaged changes in CaMKIIα-CaM association in spines and nearby dendrite (n = 28 spines/4 neurons). The orange curve indicates the decay of binding fraction change obtained by curve fitting of an exponential function: B(t) = B0 exp(–t/τ), where B0 is the initial binding fraction change, τ is the dissociation time constant. The time constant is obtained as τ = 2.9 ± 0.3 s. d Comparison of CaMKIIα-CaM association in response to a single pulse (c) and to a train of glutamate uncaging (Fig. 2d, e). e CaMKIIα conformation change measured with Green-Camuiα in response to a single pulse and a train of glutamate uncaging. Data from our previous publication10. All data are shown in mean ± sem, and sem of time constants is obtained by bootstrapping
Role of Thr286 phosphorylation in CaMKIIα-CaM association
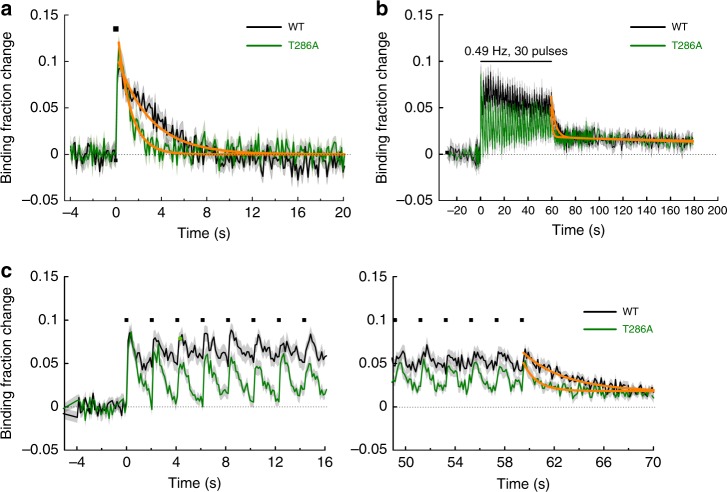
It has been reported that the binding affinity of CaMKIIα for Ca2+/CaM increases by Thr286 phosphorylation7. To examine to what degree Thr286 phosphorylation affects the decay kinetics of CaMKIIα-CaM interaction, we used a CaMKIIαT286A mutant sensor (Fig. 4). To minimize the effects of inter-subunit FRET between mEGFP-CaMKIIαT286A and mCherry-CaM bound to the adjacent endogenous wildtype CaMKII, we used hippocampal slices from Camk2aT286A knock-in mice. Thus, in this scheme, all the Thr286 residues in CaMKIIα subunits in a holoenzyme are mutated to Ala. We first compared the activation of the T286A mutant to that of mEGFP-CaMKIIαWT in response to a single glutamate uncaging pulse (Fig. 4a). We observed that the binding fraction increased to a level similar to that of wildtype, but the dissociation was faster by ~3 fold (τ = 1.2 ± 0.1 s).
Fig. 4.
CaMKIIαT286A-CaM association during sLTP induction. a Averaged change in CaMKIIαT286A-CaM association in a stimulated spine (green; n = 18 spines/4 neurons) in response to a single glutamate uncaging pulse (black square). The orange curve on CaMKIIαT286A is obtained by curve fitting of an exponential function: B(t) = B0·e−t/τ. The dissociation time constant is obtained as τ = 1.2 ± 0.1 s. The data and fitted curve for CaMKIIαWT are from Fig. 3c for the comparison. b Averaged change in CaMKIIαT286A-CaM association in stimulated spines (green; n = 24 spines/7 neurons) during glutamate uncaging at 0.49 Hz. The orange curve indicates the decay of binding fraction change obtained by curve fitting of a double-exponential function: B(t) = B0 [Pfast·exp(–t/τfast) + Pslow·exp(–t/τslow)]. The time constants for CaMKIIαT286A are obtained as τfast = 1.0 ± 0.2 s (Pfast = 63%) and τslow = 356 ± 221 s (Pslow = 37%). The data and fitted curve for CaMKIIαWT (black) are from Fig. 2d for the comparison. c Expanded view of the initial phase (left) and the late (right) phase of plot in b. All data are shown in mean ± sem, and sem of time constants is obtained by bootstrapping
Next, we measured CaMKIIαT286A-CaM association during sLTP induction (glutamate uncaging at 0.49 Hz) (Fig. 4b, c). Unlike the association of CaMKIIαWT with CaM, which plateaued after the first uncaging pulse, the association of CaMKIIαT286A with CaM decayed after each uncaging pulse, showing a sawtooth-shaped pattern. However, the peak level of the binding fraction change of CaMKIIαT286A-CaM was similar to that of CaMKIIαWT-CaM. The dissociation time constant of the CaM-CaMKIIαT286A interaction was obtained as τ = 1.0 ± 0.2 s (0.3 ± 0.1 at 34–35 °C, Supplementary Fig. 2b, Q10 = 4.1).
We again observed a persistent component in the decay of CaMKIIαT286A-CaM association after the train (Fig. 4b). Overall, this component requires neither T286 phosphorylation nor CaM binding to the regulatory domain of CaMKIIα.
Inhibitory phosphorylations accelerate CaMKIIα inactivation
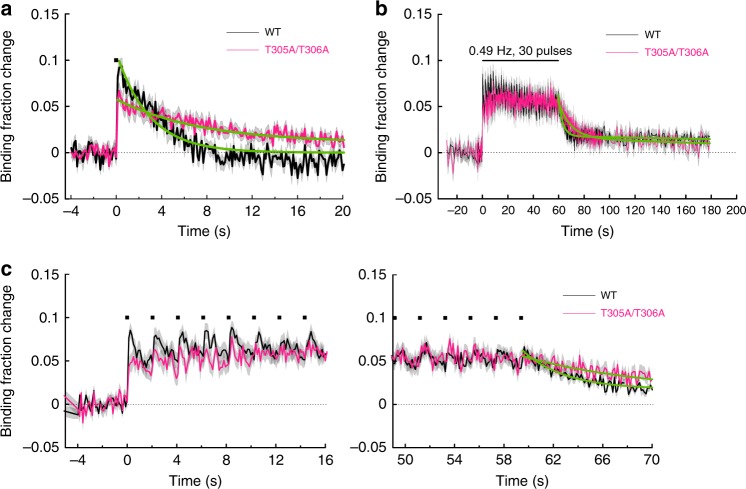
Next, we asked how the inhibitory phosphorylation at Thr305 and Thr306 may influence the Ca2+/CaM association during spine plasticity induction16,21. To do so, we mutated these phosphorylation sites from Threonine to Alanine and measured CaMKIIαT305A/T306A association with Ca2+/CaM in response to glutamate uncaging. Following a single uncaging pulse, the binding fraction change of CaMKIIαT305A/T306A with Ca2+/CaM increased to a level similar to that of CaMKIIαWT but with a slightly slower decay (τ = 7.5 ± 1.1 s; Fig. 5a). During repetitive glutamate uncaging at 0.49 Hz (sLTP protocol), Ca2+/CaM binding to CaMKIIαT305A/T306A increased to the level similar to that of CaMKIIαWT (Fig. 4b) and decayed with the time constant of τ = 9.3 ± 1.8 s (Fig. 5b, c), which was, again, slower than that of CaMKIIαWT (τ ~ 3 s). These results suggested that inhibitory phosphorylation at Thr305/Thr306 dynamically occurs during CaMKIIα activation, which inhibits the rebinding of Ca2+/CaM on CaMKIIα. However, preventing this regulation during the induction of sLTP (enhancing binding affinity to Ca2+/CaM) did not result in a higher level of Ca2+/CaM binding.
Fig. 5.
Association of CaMKIIαT305A/T306A-CaM during sLTP induction. a Averaged change in CaMKIIαT305A/T306A-CaM association in a stimulated spine (magenta; n = 34 spines/6 neurons) in response to a single glutamate uncaging pulse (black square). The green curve on CaMKIIαT305A/T306A is obtained by curve fitting of an exponential function: B(t) = B0 exp(–t/τ). The dissociation time constant is obtained as τ = 7.5 ± 1.1 s. Inset is a expanded view. The data and fitted curve for CaMKIIαWT are from (Fig. 3c) for the comparison. b Averaged change in CaMKIIαT305A/T306A-CaM association (n = 27 spines/8 neurons) during glutamate uncaging at 0.49 Hz. The green curve indicates the decay of binding fraction change obtained by curve fitting of a double-exponential function: B(t) = B0 [Pfast·exp(–t/τfast) + Pslow·exp(–t/τslow)]. The time constants are obtained as τfast = 9.3 ± 1.8 s (Pfast = 71%) and τslow = 249 ± 229 s (Pslow = 29%). The data and fitted curve for CaMKIIαWT are from (Fig. 2d) for the comparison. c Expanded view of the initial phase (left) and the late phase (right) in b
A kinetic model of CaMKIIα activation
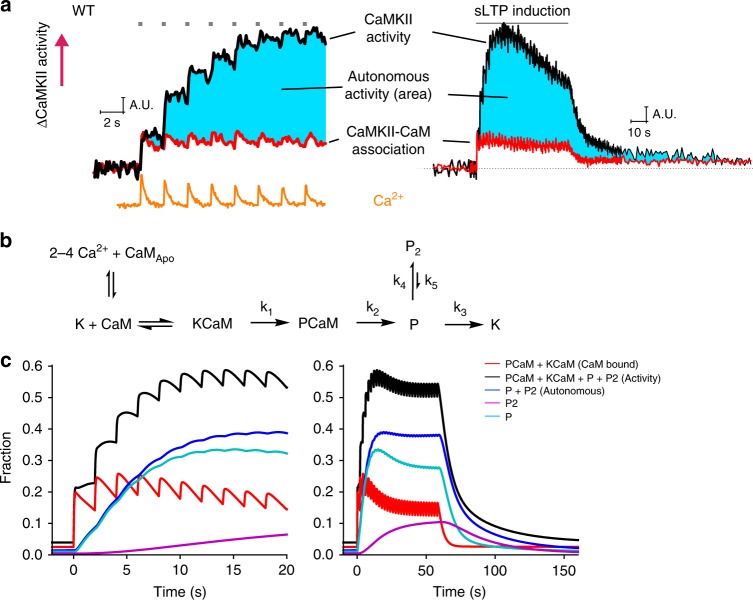
Our results indicated that CaMKII-CaM association was rapidly activated by a single glutamate uncaging pulse, but did not show any integration over repetitive glutamate uncaging (Fig. 3d). In contrast, CaMKIIα activity measured with Camuiα under similar conditions showed a high degree of integration10 (Figs. 3e, 6a). The plot assumes that the active population of CaMKIIα is equal to the fraction of CaMKIIα bound to Ca2+/CaM after the first pulse of glutamate. Since CaMKIIα autonomous activation is defined by active CaMKIIα without CaM binding, this population should be obtained by subtracting the fraction of CaMKIIα bound to CaM from CaMKIIα activity (cyan area, Fig. 6a). This suggests that CaMKIIα activity during sLTP is almost entirely supported by autonomous CaMKIIα activation.
Fig. 6.
Simulated CaMKIIα activation during spine plasticity induction. a The comparison of CaMKIIα activity measured with Green-Camuiα (data from ref. 10), Ca2+ measured with Fluo-4FF (data from ref. 10), and CaMKIIα-CaM association measured in this study. Autonomous activity is the subtraction of CaMKIIα-CaM association from CaMKIIα activation (cyan). The first time point right after uncaging is matched for CaMKIIα-CaM and CaMKIIα activity, assuming that there is no autonomous CaMKIIα at the time point. b Reaction scheme of CaMKIIα activation. K is the inactive state of CaMKIIα (closed form), CaMApo is the inactive form of calmodulin without bound Ca2+, CaM is the active form of calmodulin with 2–4 bound Ca2+ ions, P and P2 are the two different states of Thr286-phosphorylated CaMKIIα. c Simulated CaMKIIα activation based on the proposed reaction scheme. Black: concentration of total active CaMKIIα (KCaM + PCaM + P + P2). Red: concentration of KCaM and PCaM. Green: concentration of Thr286-phosphorylated CaMKIIα (P + P2). Light blue: concentration of P state of CaMKIIα. Navy: concentration of P2 state of CaMKIIα
To further clarify this point, we created a simple kinetic model of CaMKIIα (Fig. 6b, c). We constructed a set of rate equations to describe CaMKIIα biochemical reactions based on the proposed model (Table 1), and simulated the reaction in response to repetitive glutamate uncaging induced Ca2+ transients at 0.49 Hz, our standard sLTP protocol (Fig. 6b, c). For Ca2+-CaM-CaMKIIα interaction, we used a model previously established based on biochemical experiments22 (Table 1). When two adjacent subunits are activated, CaMKIIα subunit (K) undergoes phosphorylation (P). We assume that the rate of phosphorylation (k1) is proportional to the chance that the adjacent subunit is active: the fraction of CaM-bound, unphosphorylated form (KCaM), plus CaM-bound, phosphorylated form (PCaM), plus phosphorylated subunit (P and P2; see below for the explanation of the P2 state). The maximum rate has been reported to be 6.3 s−1 (ref. 23), but we found that a two-fold higher value (12.6 s−1) fits our data better. Following previous kinetic models22,23, we assume that CaM rebinding to the P state (P → PCaM) and dephosphorylation while the subunit bound to CaM (PCaM → KCaM) do not occur. The rate of CaM dissociation from PCaM (k2) was measured to be 1/3 s−1 (or time constant of 3 s) in this study (Fig. 1), and the rate of dephosphorylation of CaMKIIα (k3) has been previously measured to be 1/6 s−1 (or time constant of 6 s; Chang et al.). In addition, we assume that the persistent component of the FRET signal of CaMKIIα-CaM association is not related to the activation of CaMKIIα, as it is sensitive neither to T286A mutation nor to T305D/T306D mutations. However, the slow component of CaMKIIα activation measured with Camuiα (time constant ~1 min) depends on T286 phosphorylation, and thus likely represents the autophosphorylation state of CaMKIIα10. To explain this component of decay in CaMKIIα activation, we included a slow phosphorylation state (P2) with the time constant of 1 min (k5 = 1/60 s−1). The fraction of slow component of CaMKIIα activation (~25%; Chang et al.) can be approximated by the ratio of k3 and k4, we set k3 to be 0.25 k3 (k4 = 0.25/6 s−1). Overall, we obtained most of the kinetic parameters necessary for simulating the reaction (parameters for CaM association and k1–k5 in Fig. 6b) from this and previous studies10,22 (Table 1).
Table 1.
List of parameters used for simulation
| Name | Meaning | Value (Rate constant or concentration) | Note |
|---|---|---|---|
| CaM | Calmodulin | ||
| CaMApo | Calmodulin without bound Ca2+ | ||
| Ca(n)CaM-C | Calmodulin binding n Ca2+ on its C-lobe | ||
| Ca(n)CaM-N | Calmodulin binding n Ca2+ on its N-lobe | ||
| Ca4CaM | Calmodulin binding 4 Ca2+ | ||
| KCaM | CaMKIIα without T286 phosphorylation bound to CaM | ||
| K | CaMKIIα without T286 phosphorylation | ||
| P | CaMKIIα with Thr286 phosphorylation | ||
| P2 | A different form of CaMKIIα with Thr286 phosphorylation | ||
| KCaM | K associated with CaM | ||
| PCaM | P associated with CaM | ||
| F | Fraction of active CaMKII subunits, KCaM + PCaM + P + P2 | ||
| CaM1Con | Ca2+ + CaMApo → CaCaM-C | 5 × 106 M−1s−1 | Values from Pepke et al.22 |
| CaM1Coff | CaCaM-C → Ca2+ + CaMApo | 50 s−1 | |
| CaM2Con | Ca2+ + CaCaM-C → Ca2CaM-C | 10 × 106 M−1s−1 | |
| CaM2Coff | Ca2CaM-C → Ca2+ + CaCaM-C | 10 s−1 | |
| CaM1Non | Ca2+ + CaMApo → CaCaM-N | 100 × 106 M−1s−1 | |
| CaM1Noff | CaCaM-N → Ca2+ + CaM-N | 2 × 103 s | |
| CaM2Non | Ca2+ + CaCaM-N → Ca2CaM-N | 200 × 106 M−1s−1 | |
| CaM2Noff | Ca2CaM-N → Ca2+ + Ca2CaM | 500 s−1 | |
| KCaM1Con | Ca2+ + KCaMApo → KCaCaM-C | 44 × 106 M−1s−1 | |
| KCaM1Coff | KCaCaM-C → Ca2+ + KCaMApo | 33 s−1 | |
| KCaM2Con | Ca2+ + KCaCaM-C → KCa2CaM-C | 44 × 106 M−1s−1 | |
| KCaM2Coff | KCa2CaM-C → Ca2+ + KCaCaM-C | 0.8 s−1 | |
| KCaM1Non | Ca2+ + KCaMApo → KCaCaM-N | 76 × 106 M−1s−1 | |
| KCaM1Noff | KCaCaM-N → Ca2+ + KCaMApo | 300 s−1 | |
| KCaM2Non | Ca2+ + KCaCaM-N → KCa2CaM-N | 76 × 106 M−1s−1 | |
| KCaM2Noff | KCa2CaM-N → Ca2+ + KCaCaM-N | 20 s−1 | |
| R1 | 2 Ca2+ + CaMApo → Ca2CaM-C | Coarse grained model by Pepke et al.22 for R1–R24 | |
| R2 | Ca2CaM-C → 2 Ca2+ + CaMApo | ||
| R3 | 2 Ca2+ + CaMApo → Ca2CaM-N | ||
| R4 | Ca2CaM-N → 2 Ca2+ + CaMApo | ||
| R5 | 2 Ca2+ + Ca2CaM-C → Ca4CaM | Same as R3 | |
| R6 | Ca4CaM → 2 Ca2+ + Ca2CaM-C | Same as R4 | |
| R7 | 2 Ca2+ + Ca2CaM-N → Ca4CaM | Same as R1 | |
| R8 | Ca4CaM → 2 Ca2+ + Ca2CaM-N | Same as R2 | |
| R9 | 2 Ca2+ + KCaMApo → KCa2CaM-C | ||
| R10 | KCa2CaM-C → 2 Ca2+ + KCaMApo | ||
| R11 | 2 Ca2+ + KCaMApo → KCa2CaM-N | ||
| R12 | KCa2CaM-N → 2 Ca2+ + KCaMApo | ||
| R13 | 2 Ca2+ + KCa2CaM-C → KCa4CaM | Same as R11 | |
| R14 | KCa4CaM → 2 Ca2+ + KCa2CaM-C | Same as R12 | |
| R15 | 2 Ca2+ + KCa2CaM-N → KCa4CaM | Same as R9 | |
| R16 | KCa4CaM → 2 Ca2+ + KCa2CaM-N | Same as R10 | |
| R17 | K + CaMApo → KCaMApo | 3.8 × 103 M−1s−1 | |
| R18 | KCaMApo → K + CaMApo | 5.5 s−1 | |
| R19 | K + Ca2CaM-C → KCa2CaM-C | 0.92 × 103 M−1s−1 | |
| R20 | KCa2CaM-C → K + Ca2CaM-C | 6.8 s−1 | |
| R21 | K + Ca2CaM-N → KCa2CaM-N | 0.12 × 103 M−1s−1 | |
| R22 | KCa2CaM-N → K + Ca2CaM-N | 1.7 s−1 | |
| R23 | K + Ca4CaM → KCa4CaM | 30 × 103 M−1s−1 | |
| R24 | KCa4CaM → K + Ca4CaM | 1.5 s−1 | |
| R25 | KCaM → PCaM | k1: F × 12.6 s−1 | 6.3 according to Lucic et al.;23 F is the fraction of active CaMKII subunits |
| R26 | PCaM → P + CaM | k2: 0.33 s−1 | Decay of Ca2+-CaMKII association, 3 s. |
| R27 | P → P2 | k4: 0.041 s−1 | k3/k4 = 1/4: the fraction of slow component |
| R28 | P2 → P | k5: 0.017 s−1 | Slow decay of CaMKII: 60 s Chang et al.10 |
| R29 | P → K | k3: 0.17 s−1 | Fast decay of CaMKII activity: 6 s (ref. 10) |
| R30 – R33 | Same as R17, R19, R21, R23, with K replaced by P | 0.1 × R17, R19, R21, R23 for the model in Fig. S3a and 0 for the model in Fig. 6a | Ca2+/CaM binding to phosphorylated CaMKII (P) |
| R34 – R41 | Same as R9 – R16, with K replaced by P | Same as R9 – R16. | Ca2+ binding to CaM on P |
| [Ca2+]peak | Peak [Ca2+] | 4 µM for uncaging, 0.8 µM for back-propagating action potential (bAP), 2.4 µM for bAP paired with synaptic stimulation. | Evans et al.;39 Chang et al.;10 Sabatini et al.27 |
| τ Ca | Decay of Ca2+ | 100 ms for uncaging, 20 ms for bAP and bAP paired with synaptic stimulation. | Evans et al.;39 Chang et al.;10 Sabatini et al.27 |
| [Ca2+]0 | Resting [Ca2+] | 50 nM | Evans et al.;39 Chang et al.;10 Sabatini et al.27 |
| CaMT | Total calmodulin concentration | 30 µM | Pepke et al.;22 Kakiuchi et al.41 |
| CaMKIIT | Total CaMKII subunit concentration | 70 µM | Pepke et al.;22 Lee et al.6 |
This reaction scheme recapitulates several key features of CaMKIIα activation and CaM-CaMKIIα binding in single dendritic spines: (1) decay kinetics of CaMKIIα activation with two time constants10, (2) integration of CaMKIIα activation in response to each pulse10, (3) no accumulation of CaMKIIα-CaM interaction during repetitive Ca2+ pulses, (4) decay of CaMKIIα activity in response to a single pulse (~10 s), which is longer than that following a train of pulses (~6 s)10, (5) time course of CaM binding to T286A mutant, simulated by removing the effects of phosphorylation (setting k1 to 0). The model shows that autonomous CaMKIIα activation (P + P2) increases over time, and becomes the dominant population after ~3–4 uncaging pulses.
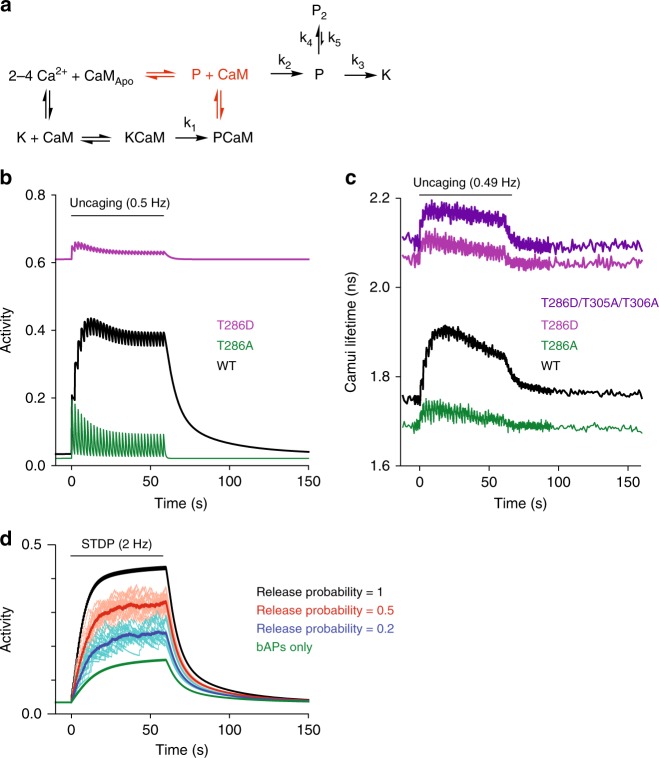
Previously our and other groups examined the effects of Thr286 dephosphorylation on CaMKIIα activity using Camuiα sensor with T286D mutation or wildtype Camuiα sensor in the presence of phosphatase inhibitor10,24. These studies showed a high basal level of CaMKIIα activity before glutamate uncaging, consistent with this study that T286 phosphorylation accounts for most of CaMKIIα activation. Interestingly, both studies showed that there exists a smaller, rapid increase of active CaMKIIα which decays rapidly after cessation of glutamate uncaging. Since the phosphorylation state of T286 is constantly in “on” state under this condition, this rapid activation must be due to the association/dissociation of Ca2+/CaM from CaMKIIαT286D.
To simulate the activation of CaMKIIαT286D, we slightly modified the above model. First, we allowed phosphorylated CaMKIIα binds to CaM (P → PCaM) with 10% of the association rate of non-phosphorylated K-CaM association (K → KCaM). Second, we assumed that CaMKIIα activity in the autonomous state (P or P2) is 60% as high as that when binding with CaM (KCaM or PCaM), as measured previously with FRET sensors6,17 and substrate phosphorylation25 (Fig. 7a). This model produced a time course of CaMKIIαWT activation similar to that produced by the original model (Fig. 7b). Importantly, when we set dephosphorylation rate to zero to simulate T286D mutation, we recapitulated all above features of CaMKIIαT286D activity10,24 (Fig. 7b, c), including high basal activity and a rapid activation and inactivation due to CaM binding and unbinding, respectively. The same model also reproduced the activity profile of T286A mutation (set the rate of phosphorylation to 0), showing smaller basal activity, smaller activation, and faster decay10 (Fig. 7b, c).
Fig. 7.
Modified model of CaMKIIα activation during spine plasticity induction. a Reaction scheme of CaMKIIα activation that includes binding of CaM to phosphorylated CaMKIIα (P state). The difference from Fig. 6b is highlighted in red. b Simulated activation of CaMKIIα with mutations at T286 based on the proposed reaction scheme (a). Black: wildtype, green: T286A mutant, purple: T286D mutant, dark blue: T286D/T305A/T306A mutant. T305A/T306A mutations are to prevent inhibition of CaM binding to T286D mutant by inhibitory T305/T306 phosphorylations24. c Activation of CaMKIIα and its mutants in dendritic spines measured with Green-Camuiα (data from ref. 10). d Simulated CaMKIIα during a protocol to induce spike-timing-dependent plasticity (2 Hz pairing of synaptic stimulation and back-propagating action potentials)
Finally, we examined how this model predicts CaMKIIα activation during spike-timing-dependent plasticity (STDP), in which LTP can be induced by pairing synaptic stimulation with back-propagating action potentials (bAP) with slight delay26. We assumed that bAPs produce Ca2+ transient with the peak concentration of 0.8 µM and the decay time constant of 20 ms27,28. When paired with synaptic release at the synapse, ~3 times more Ca2+ is produced29. In this model, bAPs alone produced little CaMKIIα activation (Fig. 7d). However, when paired with synaptic activity, the stimulation activated CaMKIIα to a higher level, reaching the level similar to that produced by glutamate uncaging, particularly at high presynaptic release probability (Fig. 7d).
Discussion
The fraction of CaMKIIα bound to Ca2+/CaM remains constant during repetitive uncaging pulses, and does not increase with each additional Ca2+ transient. This temporal pattern is sharply contrasted by the stepwise activation of CaMKIIα observed with the conformational sensor, Camuiα10. This suggests that CaM-independent CaMKIIα activation, i.e., autonomous activation, is the dominant mechanism that causes the accumulation of CaMKIIα activity during the induction of sLTP. These results highlight the important role of autonomous activation by the phosphorylation of Thr286 plays in the induction of synaptic plasticity10.
In the absence of Thr286 phosphorylation (T286A), the association of CaMKIIαT286A-CaM showed a transient binding during sLTP induction, which was similar to CamuiαT286A activation10. Thus, the activation of CaMKIIαT286A is mediated by the transient binding of Ca2+/CaM (τ ~ 1 s). In addition, from the decay rate, we found that the decay time constant between T286A is ~3 times faster than wildtype, suggesting that Thr286 phosphorylation slows down the dissociation rate. It has been reported that the binding affinity of CaMKIIα for Ca2+/CaM is enhanced by orders of magnitude upon Thr286 phosphorylation in cuvette7,12. However, the obtained decay rates suggest that the enhancement is only a few folds in the spine.
In addition to Thr286, CaMKIIα undergoes autophosphorylation at Thr305 and Thr306 upon its activation. Phosphorylation of these sites is known to inhibit CaM binding to CaMKII16,21. Consistent with these previous studies, our imaging results indicate that the dissociation of CaM from CaMKIIα is slower when this phosphorylation is prevented by mutations of Thr305 and Thr306 to Ala. Transgenic CaMKIIαT305V/T306A mice have been shown to have a lower threshold for hippocampal LTP2. The longer activity of CaMKIIαT305A/T306A suggests that there might be a less stringent window in LTP stimulation frequency required for LTP induction in transgenic CaMKIIαT305V/T306A mice.
Taken together with our previous studies of CaMKIIα activation during repetitive Ca2+ pulses in the spine6,10, CaMKIIα activation, but not CaMKIIα-CaM binding, integrates Ca2+ pulses. This suggests that most of the active CaMKIIα population is in a CaM-independent, autonomous activation state. Our kinetic model also predicts that the CaMKIIα bound to CaM accounts for only a small fraction of CaMKIIα activity (~1/4), and most of the activity is from autonomous activation.
We propose a slow state in Thr286-phosphorylated CaMKIIα (P2) to explain the minor population (~25%) with a long decay time of CaMKIIα activity (~60 s)10. However, there would be different ways to explain this fraction. For example, it could also possibly originate from two different types of phosphatases which target different populations of CaMKIIα30,31. Further experiments are required to disentangle these two different states.
For the simulation of T286D mutant (or phosphor-mimic) form of CaMKIIα, we needed to modify the model so that it incorporates the binding of CaM to the phosphorylated form of CaMKIIα. While some of the previously developed models ignore this reaction32, it would be valid since our experiments in HeLa cells clearly shows CaM binding to T286D mutant. In addition, we incorporated the previous measurements suggesting that the CaM-bound form is higher than autonomous activity6,17,25. This modified model recapitulated the reported time course of CaMKIIαT286D in single spines: high basal binding, rapid activation, and rapid inactivation10,24. Importantly, the rapid inactivation of CaMKIIαT286D has been used to challenge the idea that the decay of CaMKIIα is due to dephosphorylation of CaMKIIα24. However, our simulation indicates that, while the decay of CaMKIIαT286D is due to unbinding of CaM, that of wildtype CaMKIIα is limited mostly by dephosphorylation of the autonomous form.
Finally, the model does not explicitly incorporate several factors including caching of CaM by neurogranin, cooperativity between subunits and inhibitory autophosphorylation at Thr305/Thr30623,32–34. Perhaps the more detailed model based on CaMKIIα structure and biochemical data together with our imaging results in dendritic spines would improve our understanding of CaMKIIα activation in dendritic spines in response to Ca2+ elevation33,35.
Methods
Experimental animals
Mice from BL6/C57 strain (purchased from Charles River Laboratories) were used for CaMKIIαWT-CaM association measurements in 2pFLIM imaging. Camk2aT286A knock-in mice (gift from Dr. Giese) were used for CaMKIIαT286A-CaM association measurements. All experimental animals were bred in-house under the guidelines of Institutional Animal Care and Use Committee (IACUC) of Duke University Medical Center and Max Planck Florida Institute for Neuroscience.
Organotypic slices
Organotypic cultured hippocampal slices were prepared from postnatal 4–7 day mice36. The isolated hippocampus was sliced with a tissue chopper (McIlwain Tissue Chopper, Ted Pella Inc). The slices were plated on cell culture inserts (hydrophilic PTFE, 0.4 µm, Millipore) and maintained in tissue medium (minimum essential medium Eagle (MEM) 8.4 mg/ml, horse serum 20%, L-glutamine 1 mM, CaCl2 1 mM, MgSO4 2 mM, D-glucose 12.9 mM, NaHCO3 5.2 mM, HEPES 30 mM, insulin 1 µg/ml, ascorbic acid 0.075%) at 37 °C supplemented with 5% CO2 until experiments (DIV 12–19). Hippocampal slices were biolistically transfected with plasmids at DIV 5–10 (12 mg gold particle, size: 1 µm, 45–112 µg plasmid). Preparation of slice cultures was in accordance with the guidelines of the Institutional Animal Care and Use Committee of Duke University Medical Center and Max Planck Florida Institute for Neuroscience.
Cell lines
HeLa cells (ATCC, Cat#CCL-2) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. Cells were transfected with mCherry-CaM and mEGFP-CaMKIIα (or its mutant) using Lipofectamine-2000 for 24–48 h, and subjected to fluorescence lifetime imaging in a solution containing (in mM) 130 NaCl, 20 HEPES, 2 NaHCO3, 25 D-glucose, 2.5 KCl, 1.25 NaH2PO4, 0.8 MgCl2 and 1.8 mM CaCl2 (pH 7.3). Cells were treated with 3 µM ionomycin (Tocris) and then 5 min later 10 mM EGTA (Sigma).
Protein purification
His-tagged mCherry-synapsin 1 peptide (a gift from Dr. Murakoshi)37, His-tagged mCherry-CaM and His-tagged calmodulin were cloned into pRSET bacterial expression vector (Thermo Fisher Scientific) and expressed in T7 Express lysY Competent Escherichia Coli (New England BioLabs Inc.), purified with a Ni-NTA column (HisTrap™ HP; GE Healthcare) and desalted with PD-10 column (GE Healthcare). The purified protein concentrations were measured by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). The purity of each fraction was confirmed by SDS-PAGE and Coomassie staining.
Kinase assay
Standard kinase assays were performed for the indicated time at room temperature with 20 nM purified full-length recombinant human CaMKIIα (#PV3142; Thermo Fisher Scientific), 2 µM mCherry-Syn1, 0.03–2 µM calmodulin or mCherry-CaM, 200 µM CaCl2 and 500 µM ATP in a reaction buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 2 mM DTT). The reactions were stopped at 10 min by adding SDS sample buffer and then analyzed by Western blotting. The following antibodies were used: Phospho-(Ser/Thr) PKA Substrate Antibody (#9621; Cell Signaling Technology) for phosphorylated mCherry-Syn1 detection; Goat Anti-Rabbit IgG (H + L)-HRP Conjugate (#1706515; Bio-Rad). We repeated the experiment four times from one preparation of the samples. Original images of the blots are in Source Data.
Plasmid construction for CaMKIIα-CaM association imaging
We inserted cDNA sequence of calcium/calmodulin-dependent protein kinase II alpha (Camk2a) from Rattus norvegicus into the C-terminus of mEGFP containing pCAG plasmid, and calmodulin 1 (calm1) from Mus musculus into the C-terminus of mCherry containing pCAG plasmid. Molecular cloning and mutations were carried out using QuikChange site-directed mutagenesis kit (Agilent Technologies) and InFusion cloning kit (Clontech) for mEGFP-CaMKIIαT286A, mEGFP-CaMKIIαT305A/T306A, mEGFP-CaMKIIαT305D/T306D, mEGFP-CaMKIIαT286A/305D/T306D. The amount of transfected plasmids in the specified experiments are as follows: 1) mEGFP-CaMKIIαWT/or mEGFP-CaMKIIαT286A (20 µg), and mCherry-CaM (40 µg); 2) mEGFP-CaMKIIαT305D/T306D or mEGFP-CaMKIIαT305A/T306A (20 µg), mCherry-CaM (40 µg), and pCAG-Cre recombinase (12 µg).
Microscope
The fluorescent lifetime of mEGFP-CaMKIIα was measured by a home-built two-photon fluorescence lifetime imaging microscopy (2pFLIM). mEGFP-CaMKIIα was excited with a Ti:Sapphire laser tuned at 920 nm (Coherent, Chameleon) with laser power measured under the water immersion objective (Olympus, NA = 1.0, ×60) in the range of 1–1.5 mW19,38. A second Ti:Sapphire laser at 720 nm (laser power measured under the objective: 2.5–3 mW), pulse duration of 4–6 ms was used to photolysis MNI-caged L-glutamate5.
CaMKIIα-CaM association imaging
Hippocampal slices were bathed in artificial cerebrospinal fluid (ACSF) bubbled with carbogen (95% O2/ 5% CO2) during the image recordings. Final ion concentrations (in mM) in imaging solution: NaCl 127, NaHCO3 25, d-glucose 25, KCl 2.5, NaH2PO4 1.25, supplemented with CaCl2 4, MNI-caged L-glutamate (Tocris) 4, TTX 0.001, Trolox (Sigma) 1. Between DIV 12–19, we imaged individual transfected CA1 pyramidal neurons. Dendritic spines on the secondary and tertiary apical dendrites were used for imaging. Images were acquired by a home-built 2pFLIM microscope controlled by custom software (MatLab or C#). Experiments were performed at 25 ± 0.5 °C or 34–35 °C as indicated. The temperature was controlled with a control syringe heater and an inline solution heater (TC344C, SW-10/6 and SH-27B, Warner Instruments). Recordings were performed with 32 × 32 pixels (pixel size: 12.3 ± 1.72 pixel/µm) at 128 ms/frame (7.8 Hz). When we found a large drift of the position of the sample or significant photo-bleaching, we stopped the experiment and excluded from further analyses.
2pFLIM data analysis
The fluorescence lifetime of mEGFP-CaMKIIα is affected by the FRET efficiency. The change of mean fluorescence lifetime of mEGFP-CaMKIIα (τm) reflects the change of FRET efficiency and thus the binding fraction change of mEGFP-CaMKIIα to mCherry-CaM. To measure the fraction of mEGFP-CaMKIIα (donor) bound to mCherry-CaM (acceptor), the mean fluorescence lifetime of mEGFP-CaMKIIα (τm) was derived from the mean photon arrival time t as follows:
| 1 |
where F(t) is the fluorescence lifetime decay curve, t0 is offset. t0 is estimated by fitting to the fluorescence decay curve summing all pixels in all frames over a whole image session (typically 1024 frames) with a double-exponential function convolved with the Gaussian pulse response function:
| 2 |
where F0 is constant, and
| 3 |
in which PA and PAD is the fraction of free donor and donor bound with acceptor, respectively, τD is the fluorescence lifetime of the donor without any bound acceptor (τD = 2.60 ns), τAD is the fluorescence lifetime of the donor bound with acceptor τAD = 1.09 ns, τG is the width of the Gaussian pulse response function, F0 is the peak fluorescence before convolution, t0 is time offset, and erfc is the error function. τD and τAD are fixed during the curve fitting to obtain PA and PAD. For regions of interests (ROI) within a field-of-view (such as spine and dendrite), the binding fraction PAD is derived as follows:
| 4 |
Simulation of CaMKIIα kinetics scheme
We constructed a set of rate equations (elementary reaction) to describe CaMKIIα biochemical reactions based on the proposed CaMKIIα kinetics model. The law of mass action was applied to obtain non-linear ordinary differential equations (ODEs) and to solve the concentration of each species. We implemented the algorithm written in Python. To simplify the simulation, the influx of NMDA-receptor mediated Ca2+ during repetitive glutamate uncaging is modeling as:
| 5 |
where i is the number of uncaging pulses (integers, i = 0…29, 30 pulses), td is the uncaging interval (2 s), Ai is the peak [Ca2+] at ith uncaging pulse, R = 50 nM is the resting [Ca2+], and τCa = 100 ms is the Ca2+ decay time constant6,10,39. Peak Ca2+ amplitude Ai decays after each uncaging pulse6,10,39, perhaps due to desensitization of NMDARs40. We model this as:
| 6 |
where A0 = 4 µM is the peak [Ca2+] in response to the first uncaging pulse, and τn = 5 is the decay constant, P1 = 0.5 and P2 = 0.5 are constants (P1 + P2 = 1).
For spike-timing-dependent plasticity (Fig. 7), we used:
| 7 |
where A = 0.8 µM and τCa = 20 ms for back-propagating action potentials (bAPs), and when paired with a synaptic release, A = 2.4 µM was used. Because the synaptic release is simulated as a stochastic event, we repeated 20 times and averaged them for release probability <1.
Ca2+ binding to CaM was modeled using the previous scheme22. Thr286 phosphorylation occurs when two adjacent subunits are active9. We assume that the rate of phosphorylation of a subunit (k1, Fig. 6b) is proportional to the chance that the adjacent subunit is active:
| 8 |
where kphospho = 12.6 s−1 is the peak phosphorylation rate23, and F is the active CaMKII fraction:
| 9 |
where CaMKIIT = 70 µM is the total CaMKIIα subunit concentration6,22. Total CaM concentration was assumed to be 30 µM22,41. Dephosphorylation before dissociation of CaM and rebinding of CaM to Thr286 phosphorylated-CaMKIIα (P or P2, Fig. 6b and Fig. 7a) are assumed not to occur for the model in Fig. 6, following the previous models22. However, it is assumed to be 10% of the binding to non-phosphorylated CaMKIIα in the model in Fig. 7. Kinetic parameters other than k1 (k2–k5) are obtained as follows: we obtain k2 = 1/3 s−1 from the time constant of CaM dissociation (3 s) (Fig. 3c), and k3 = 1/6 s−1 and k5 = 1/60 s−1 from two time constants of CaMKII activity decay (6 s and 60 s)10. We obtain k4 from the fraction of slow CaMKII decay (25%), which can be approximated by the ratio between k3 and k4: k4 = 0.25 k3. The activity of autonomous activity was assumed to be 60% of that in the CaM bound form. All kinetic parameters are summarized in Table 1.
Statistical analysis
Error bars shown in the figures represent standard error of the mean (sem). sem of time constants is obtained by bootstrapping. The number of samples is indicated as the number of neurons/dendritic spines. Most of the slices have only one neuron.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Dr. Giese for Camk2aT286A mice. We thank Dr. Murakoshi for His-tagged mCherry-Syn1. We thank members of the Yasuda lab for discussion, and Dr. Colgan and Dr. Raghavachari for the critical reading of the manuscript. We also thank M. Hu and J. Richards for preparing cultured slices and D. Kloetzer for laboratory management. This study was funded by Japan Society for the Promotion of Science (YN), NIH (R01MH111486, R01MH080047, and 1DP1NS096787), and the Brain Research Foundation.
Author contributions
J.Y.C. and R.Y. designed the experiments. J.Y.C. performed most of the imaging experiments, Y.H. performed biochemical experiments, and Y.N. performed fluorescence lifetime measurements in HeLa cells. J.Y.C and R.Y. constructed the simulation, analyzed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
Data availability
Time courses of all experiments and raw Western blot data are available in Data Source in Excel format. Original FLIM images will be available upon request.
Code availability
Python code for CaMKIIα simulation is available on GitHub. Matlab code for FLIM data acquisition and analysis is available on GitHub. C# code for FLIM data acquisition and analysis is available on GitHub.
Competing interests
R.Y. is a founder of Florida Lifetime Imaging LLC, a company that helps people set up FLIM. The remaining authors declare no competing interests.
Footnotes
Peer review information: Nature Communications thanks Peter Giese, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at 10.1038/s41467-019-10694-z.
References
- 1.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123:849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 2.Elgersma Y, et al. Inhibitory autophosphorylation of CaMKII controls PSD association, plasticity, and learning. Neuron. 2002;36:493–505. doi: 10.1016/S0896-6273(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 3.Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- 4.Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in alpha-calcium-calmodulin kinase II mutant mice. Science. 1992;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer T, Hanson PI, Stryer L, Schulman H. Calmodulin trapping by calcium-calmodulin-dependent protein kinase. Science. 1992;256:1199–1202. doi: 10.1126/science.256.5060.1199. [DOI] [PubMed] [Google Scholar]
- 8.Miller SG, Kennedy MB. Regulation of brain type II Ca2 + /calmodulin-dependent protein kinase by autophosphorylation: a Ca2 + -triggered molecular switch. Cell. 1986;44:861–870. doi: 10.1016/0092-8674(86)90008-5. [DOI] [PubMed] [Google Scholar]
- 9.Hanson PI, Meyer T, Stryer L, Schulman H. Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron. 1994;12:943–956. doi: 10.1016/0896-6273(94)90306-9. [DOI] [PubMed] [Google Scholar]
- 10.Chang JY, et al. CaMKII autophosphorylation is necessary for optimal integration of Ca2 + signals during LTP induction, but not maintenance. Neuron. 2017;94:800–808 e804. doi: 10.1016/j.neuron.2017.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 12.Singla SI, Hudmon A, Goldberg JM, Smith JL, Schulman H. Molecular characterization of calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 2001;276:29353–29360. doi: 10.1074/jbc.M101744200. [DOI] [PubMed] [Google Scholar]
- 13.Hanson PI, Kapiloff MS, Lou LL, Rosenfeld MG, Schulman H. Expression of a multifunctional Ca2+/calmodulin-dependent protein kinase and mutational analysis of its autoregulation. Neuron. 1989;3:59–70. doi: 10.1016/0896-6273(89)90115-3. [DOI] [PubMed] [Google Scholar]
- 14.Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat. Rev. Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colbran RJ. Inactivation of Ca2+/calmodulin-dependent protein kinase II by basal autophosphorylation. J. Biol. Chem. 1993;268:7163–7170. [PubMed] [Google Scholar]
- 16.Hashimoto Y, Schworer CM, Colbran RJ, Soderling TR. Autophosphorylation of Ca2+/calmodulin-dependent protein kinase II. Effects on total and Ca2+-independent activities and kinetic parameters. J. Biol. Chem. 1987;262:8051–8055. [PubMed] [Google Scholar]
- 17.Takao K, et al. Visualization of synaptic Ca2+ /calmodulin-dependent protein kinase II activity in living neurons. J. Neurosci. 2005;25:3107–3112. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguchi J, Matsuzaki M, Ellis-Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2 + signaling in dendrites. Neuron. 2005;46:609–622. doi: 10.1016/j.neuron.2005.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 20.Pi HJ, Otmakhov N, Lemelin D, De Koninck P, Lisman J. Autonomous CaMKII can promote either long-term potentiation or long-term depression, depending on the state of T305/T306 phosphorylation. J. Neurosci. 2010;30:8704–8709. doi: 10.1523/JNEUROSCI.0133-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colbran RJ, Soderling TR. Calcium/calmodulin-independent autophosphorylation sites of calcium/calmodulin-dependent protein kinase II. Studies on the effect of phosphorylation of threonine 305/306 and serine 314 on calmodulin binding using synthetic peptides. J. Biol. Chem. 1990;265:11213–11219. [PubMed] [Google Scholar]
- 22.Pepke S, Kinzer-Ursem T, Mihalas S, Kennedy MB. A dynamic model of interactions of Ca2+, calmodulin, and catalytic subunits of Ca2+/calmodulin-dependent protein kinase II. PLoS Comput Biol. 2010;6:e1000675. doi: 10.1371/journal.pcbi.1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucic V, Greif GJ, Kennedy MB. Detailed state model of CaMKII activation and autophosphorylation. Eur. Biophys. J. 2008;38:83–98. doi: 10.1007/s00249-008-0362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otmakhov N, Regmi S, Lisman JE. Fast decay of CaMKII FRET sensor signal in spines after LTP induction is not due to its dephosphorylation. PLoS One. 2015;10:e0130457. doi: 10.1371/journal.pone.0130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coultrap SJ, Buard I, Kulbe JR, Dell’Acqua ML, Bayer KU. CaMKII autonomy is substrate-dependent and further stimulated by Ca2+/calmodulin. J. Biol. Chem. 2010;285:17930–17937. doi: 10.1074/jbc.M109.069351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dan Y, Poo MM. Spike timing-dependent plasticity of neural circuits. Neuron. 2004;44:23–30. doi: 10.1016/j.neuron.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Sabatini BL, Oertner TG, Svoboda K. The life cycle of Ca2+ ions in dendritic spines. Neuron. 2002;33:439–452. doi: 10.1016/S0896-6273(02)00573-1. [DOI] [PubMed] [Google Scholar]
- 28.Yasuda R, et al. Imaging calcium concentration dynamics in small neuronal compartments. Sci. STKE. 2004;2004:pl5. doi: 10.1126/stke.2192004pl5. [DOI] [PubMed] [Google Scholar]
- 29.Koester HJ, Sakmann B. Calcium dynamics in single spines during coincident pre- and postsynaptic activity depend on relative timing of back-propagating action potentials and subthreshold excitatory postsynaptic potentials. Proc. Natl Acad. Sci. USA. 1998;95:9596–9601. doi: 10.1073/pnas.95.16.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strack S, Barban MA, Wadzinski BE, Colbran RJ. Differential inactivation of postsynaptic density-associated and soluble Ca2+/calmodulin-dependent protein kinase II by protein phosphatases 1 and 2A. J. Neurochem. 1997;68:2119–2128. doi: 10.1046/j.1471-4159.1997.68052119.x. [DOI] [PubMed] [Google Scholar]
- 31.Colbran RJ. Protein phosphatases and calcium/calmodulin-dependent protein kinase II-dependent synaptic plasticity. J. Neurosci. 2004;24:8404–8409. doi: 10.1523/JNEUROSCI.3602-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhabotinsky AM, Camp RN, Epstein IR, Lisman JE. Role of the neurogranin concentrated in spines in the induction of long-term potentiation. J. Neurosci. 2006;26:7337–7347. doi: 10.1523/JNEUROSCI.0729-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao LH, et al. Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation. Nat. Struct. Mol. Biol. 2010;17:264–272. doi: 10.1038/nsmb.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao LH, et al. A mechanism for tunable autoinhibition in the structure of a human Ca2 + /calmodulin- dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers JB, et al. The CaMKII holoenzyme structure in activation-competent conformations. Nat. Commun. 2017;8:15742. doi: 10.1038/ncomms15742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-M. [DOI] [PubMed] [Google Scholar]
- 37.Murakoshi H, et al. Kinetics of endogenous CaMKII required for synaptic plasticity revealed by optogenetic kinase inhibitor. Neuron. 2017;94:37–47 e35. doi: 10.1016/j.neuron.2017.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans Paul R., Parra-Bueno Paula, Smirnov Michael S., Lustberg Daniel J., Dudek Serena M., Hepler John R., Yasuda Ryohei. RGS14 Restricts Plasticity in Hippocampal CA2 by Limiting Postsynaptic Calcium Signaling. eneuro. 2018;5(3):ENEURO.0353-17.2018. doi: 10.1523/ENEURO.0353-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobczyk A, Svoboda K. Activity-dependent plasticity of the NMDA-receptor fractional Ca2 + current. Neuron. 2007;53:17–24. doi: 10.1016/j.neuron.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Kakiuchi S, et al. Quantitative determinations of calmodulin in the supernatant and particulate fractions of mammalian tissues. J. Biochem. 1982;92:1041–1048. doi: 10.1093/oxfordjournals.jbchem.a134019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Time courses of all experiments and raw Western blot data are available in Data Source in Excel format. Original FLIM images will be available upon request.
Python code for CaMKIIα simulation is available on GitHub. Matlab code for FLIM data acquisition and analysis is available on GitHub. C# code for FLIM data acquisition and analysis is available on GitHub.