Abstract
Background
Several 4,6-diarylpyrimidin-2-amine derivatives show anticancer properties. However, their mode of action is not fully characterized. To develop potent anticancer chemotherapeutic agents, we designed and synthesized 25 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety.
Methods
Clonogenic long-term survival assays were performed to screen anticancer compounds. To derive the structural conditions showing good cytotoxicities against cancer cells, quantitative structure-activity relationships (QSAR) were calculated. Biological activities were determined by flow cytometry for cell cycle analysis and by immunoblot analysis for the detection of Aurora kinase A (AURKA) activity. Because 2-(2-Amino-6-(2,4-dimethoxyphenyl)pyrimidin-4-yl) phenol (derivative 12) selectively inhibited AURKA activity from the kinome assay, in silico docking experiments were performed to elucidate the molecular binding mode between derivative 12 and AURKA.
Results
The pharmacophores were derived based on the QSAR calculations. Derivative 12 inhibited AURKA activity and reduced phosphorylation of AURKA at Thr283 in HCT116 human colon cancer cells. Derivative 12 caused the accumulation of the G2/M phase of the cell cycle and triggered the cleavages of caspase-3, caspase −7, and poly(ADP-ribose) polymerase. The binding energies of 30 apo-AURKA – derivative 12 complexes obtained from in silico docking ranged from −16.72 to −11.63 kcal/mol.
Conclusions
Derivative 12 is an AURKA inhibitor, which reduces clonogenicity, arrests the cell cycle at the G2/M phase, and induces caspase-mediated apoptotic cell death in HCT116 human colon cancer cells. In silico docking demonstrated that derivative 12 binds to AURKA well. The structure-activity relationship calculations showed hydrophobic substituents and 1-naphthalenyl group at the R2 position increased the activity. The existence of an H-bond acceptor at C-2 of the R1 position increased the activity, too.
Graphical abstract.
Derivative 12 inhibits Aurora kinase A activity and causes the G2/M phase arrest of the cell cycle
Electronic supplementary material
The online version of this article (10.1007/s40199-019-00272-5) contains supplementary material, which is available to authorized users.
Keywords: Apoptosis; Aurora kinase A inhibitor; Cell cycle; Clonogenicity; 4,6-Diphenylpyrimidin-2-amine; In silico docking; QSAR
Background
2-(2-Arylmethylthio-4-chloro-5-methylbenzenesulfonyl)-1-(1,3,5-triazin-2-ylamino)guanidine derivatives (Fig. 1a) show anticancer activity via targeting transmembrane tumor-associated isozymes, such as human carbonic anhydrase IX and XII [1]. Guanidinated anthrathiophenediones, such as 1-(2-((11-amino-5,10-dioxo-5,10-dihydroanthra[2,3-b]thiophen-4-yl)amino)ethyl)guanidine (Fig. 1b) show antitumor characteristics [2]. 3-Amino-2-(4-chloro-2-mercaptobenzenesulfonyl)-guanidine derivatives (Fig. 1c) show anticancer properties in three cancer cell lines, including MCF-7 breast cancer,NCI-H460 lung cancer, and SF-268 CNS cancer cells [3]. They all contain a guanidine moiety. Therefore, based on these starting points, to develop potent chemotherapeutic agents, we designed and synthesized 25 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety (Fig. 1d). Of them, several compounds were reported previously, but the biological activities of some compounds were related with antibacterial, antifungal, and anticonvulsant activities, and the synthetic methods of other compounds were reported [4–7]. Only derivative 16 was reported to show anticancer activity [8]. The structures of the derivatives synthesized here were identified using NMR spectroscopy and high-resolution mass spectrometry (HRMS). To evaluate whether the synthetic compounds exhibited anticancer effects, we selected HCT116 human colon cancer cell line. Colon cancer is the third most commonly diagnosed cancer in South Korea, as well as in the United States. Additionally, the causative factors are not clear, although one of the causes is the western dietary life [9]. However, the relationship between dietary patterns and colon cancer incidence remains unclear. If colon cancer does not spread, the first treatment is surgery. After stage II or surgery, chemotherapy is required. Furthermore, there are many drugs for chemotherapy of colon cancer, but because of side-effects or drug resistance, new chemotherapeutic agents are needed.
Fig. 1.
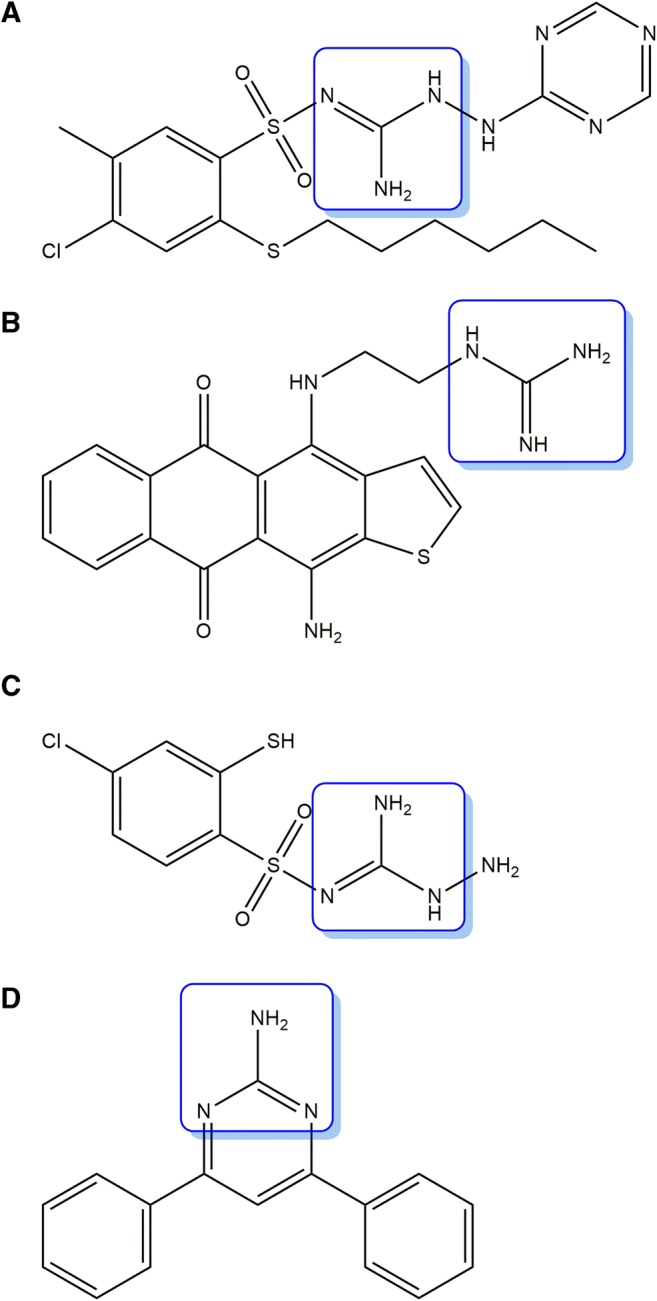
The structures of a 2-(2-Arylmethylthio-4-chloro-5-methylbenzenesulfonyl)-1-(1,3,5-triazin-2-ylamino)guanidine, b 1-(2-((11-amino-5,10-dioxo-5,10-dihydroanthra[2,3-b]thiophen-4-yl)amino)ethyl)guanidine, c 3-Amino-2-(4-chloro-2-mercaptobenzenesulfonyl)-guanidine, and d 4,6-diphenylpyrimidin-2-amine. The boxes denote the guanidine moiety
In this study, we used a cell-based clonogenic long-term survival assay (CLSA) to determine anticancer activity. To determine the mechanism by which these compounds exhibit cytotoxicities, a kinome assay was performed with the title compound exhibiting the best GI50 value in the test. Because the title compound selectively inhibited Aurora kinase A (AURKA) activity from the kinome assay, we performed in silico docking experiments to elucidate the molecular binding mode between the title compound and AURKA. To derive the structural properties showing good cell growth inhibitory effects, quantitative structure-activity relationship (QSAR) calculations were conducted using comparative molecular field analysis (CoMFA) and comparative molecular similarity index analysis (CoMSIA). The structural properties derived from the structure-activity relationships could help us design new compounds that exhibited better cancer cell growth inhibitory effects.
Materials and methods
Preparation of the synthetic compounds
Beginning with an ethanol solution of variously substituted acetophenone (I) and substituted benzaldehydes or naphthylaldehydes (II), an excess amount of 50% aqueous KOH was added, and the mixtures were stirred at room temperature for 20 h. After completion of the reaction, the reaction mixture was poured into 6 N HCl under an ice-bath to produce precipitates of chalcones (III). The solids were filtered and washed with ethanol and were used for the next reaction without further purification. The chalcone (III, 1 eq) and guanidine HCl salt (1.5 eq) was dissolved in DMF solution and was added to a solid of K2CO3 (3 eq) to make a solution. The reaction mixture was refluxed for 2 h and cooled to room temperature. The reaction mixture was poured into 3 N HCl under an ice-bath to produce precipitates of 2-aminopyrimidine products (1–25). When the solids were impure, the 2-aminopyrimidines were purified by recrystallization from ethanol. The synthetic procedure of 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety is summarized in Scheme 1. The structures and names of 25 derivatives are listed in Table 1. All NMR experiments were carried out on a Bruker Avance 400 NMR spectrometer (9.4 T, Karlsruhe, Germany). The deuterated solvents and NMR tubes were purchased from Sigma-Aldrich (St. Louis, MO, USA). The high-resolution mass spectroscopic (HRMS) data were obtained using an ultraperformance liquid chromatography-hybrid quadrupole time-of-flight mass spectrometer with a Waters Acquity UPLC system (Waters Corp., Milford, MA).
Scheme 1.
The synthetic procedures to prepare 25 4,6-diphenylpyrimidin-2-amine derivatives containing guanidine moiety.
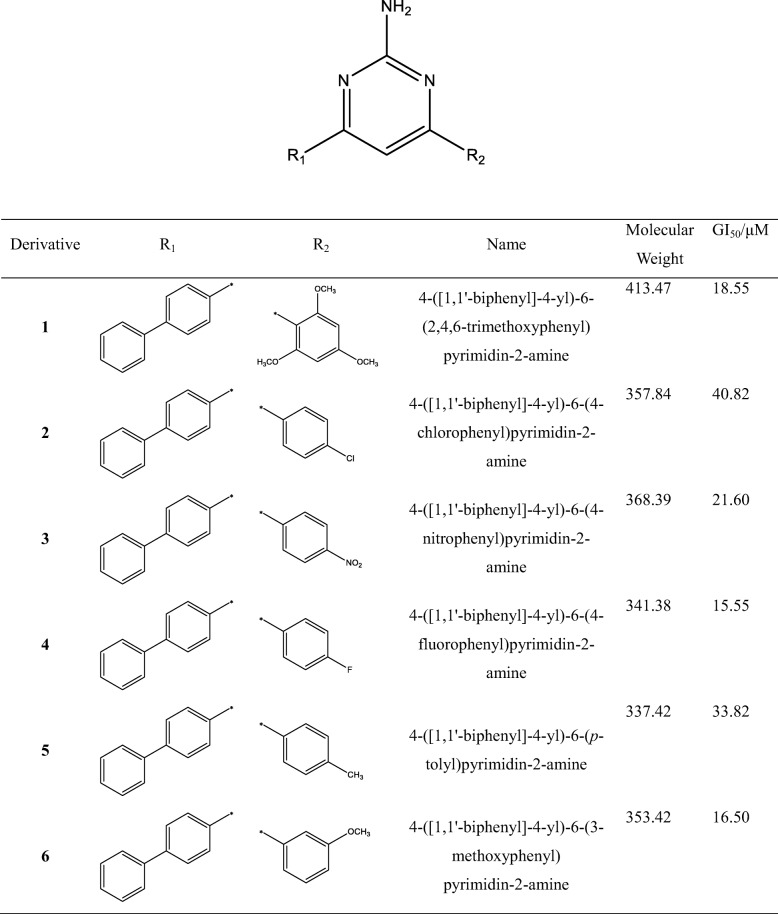
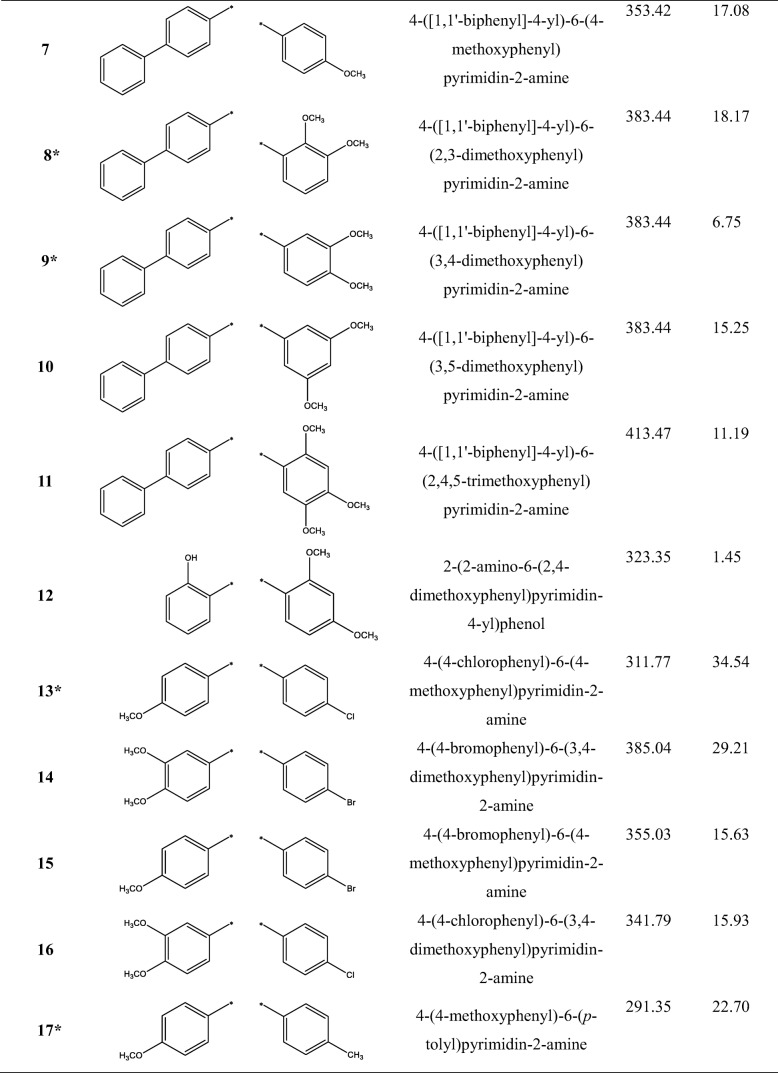
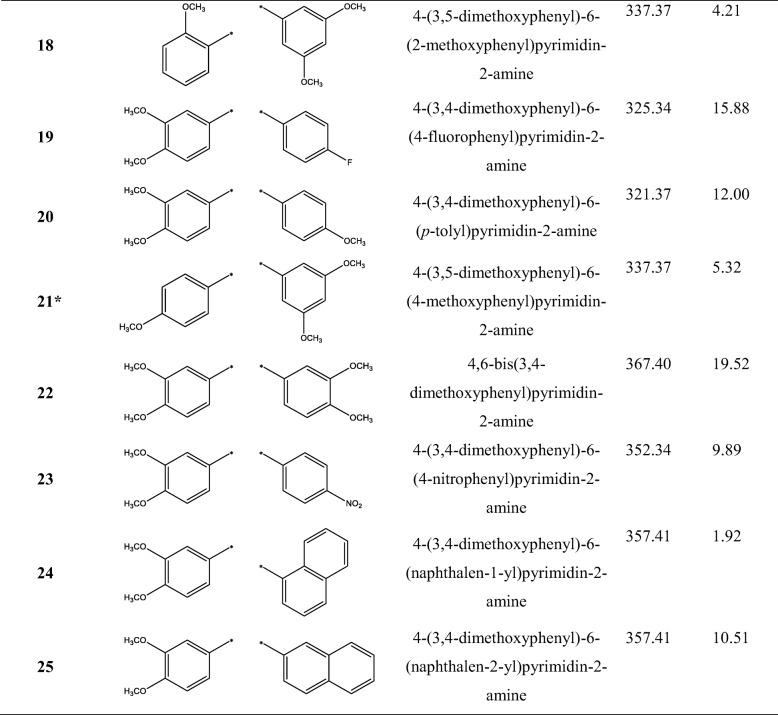
Table 1.
The structures and names of 25 4,6-diphenylpyrimidin-2-amine derivatives, and their half-maximal cell growth inhibitory concentrations (GI50), where * denotes the test set used for the QSAR calculations
4-([1,1′-biphenyl]-4-yl)-6-(2,4,6-trimethoxyphenyl)pyrimidin-2-amine (1)
1H NMR (400 MHz, DMSO-d6) δ 8.17 (ddd, 2H, H-3, H-6, J = 8.4, 2.1, 1.7 Hz), 7.78 (ddd, 2H, H-2, H-5, J = 8.4, 2.1, 1.7 Hz), 7.74 (ddd, 2H, H-2′, H-6′, J = 7.8, 1.9, 1.4 Hz), 7.49 (ddd, 2H, H-3′, H-5′, J = 7.8, 7.4, 1.5 Hz), 7.39 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.4, 1.4 Hz), 7.02 (s, 1H, py-5H), 6.60 (s, 1H, NH2), 6.32 (s, 2H, H-3″, H-5″), 3.83 (s, 1H, 4”-OCH3), 3.69 (s, 2H, 2”-OCH3, 6”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.4 (py-6), 163.9 (py-2), 162.5 (py-4), 161.1 (C-4″), 158.0 (C-2″, C-6″), 141.7 (C-1), 139.4 (C-1′), 136.3 (C-4), 129.0 (C-3′, C-5′), 127.8 (C-4′), 127.3 (C-3, C-5), 126.8 (C-2, C-6), 126.7 (C-2′, C-6′), 110.7 (C-1″), 108.3 (py-5), 90.8 (C-3″, C-5″), 55.6 (2”-OCH3, 6”-OCH3), 55.4 (4”-OCH3);HRMS (m/z): Calcd. for (M + H)+: 414.1818; Found: 414.1824.
4-([1,1′-biphenyl]-4-yl)-6-(4-chlorophenyl)pyrimidin-2-amine (2)
1H NMR (400 MHz, DMSO-d6) δ 8.33 (ddd, 2H, H-3, H-6, J = 8.4, 1.9, 1.7 Hz), 8.28 (d, 2H, H-2″, H-6″, J = 8.6 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.4, 1.9, 1.7 Hz), 7.79 (s, 1H, py-5H), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.7, 1.9, 1.5 Hz), 7.59 (d, 2H, H-3″, H-5″, J = 8.6 Hz), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.7, 7.4, 1.5 Hz), 7.40 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.5, 1.5 Hz), 6.80 (s, 1H, NH2); 13C NMR (400 MHz, DMSO-d6) δ 164.6 (py-4), 164.0 (py-2), 163.6 (py-6), 142.1 (C-1), 139.3 (C-1′), 136.2 (C-4, C-1″), 135.2 (C-4″), 129.0 (C-3′, C-5′), 128.8 (C-2″, C-6″), 128.6 (C-3″, C-5″), 127.9 (C-4′), 127.6 (C-3, C-5), 126.80 (C-2, C-6), 126.75 (C-2′, C-6′), 101.7 (py-5); HRMS (m/z): Calcd. for (M + H)+: 358.1111; Found: 358.1125.
4-([1,1′-biphenyl]-4-yl)-6-(4-nitrophenyl)pyrimidin-2-amine (3)
1H NMR (400 MHz, DMSO-d6) δ 8.50 (d, 2H, H-2″, H-6″, J = 8.7 Hz), 8.36 (d, 2H, H-3″, H-5″, J = 8.7 Hz), 8.34 (ddd, 2H, H-3, H-6, J = 8.4, 2.0, 1.9 Hz), 7.90 (s, 1H, py-5H), 7.83 (ddd, 2H, H-2, H-5, J = 8.4, 2.0, 1.9 Hz), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.9, 1.8, 1.3 Hz), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.9, 7.3, 1.4 Hz), 7.40 (dddd, 1H, H-4′, J = 7.3, 7.3, 1.3, 1.3 Hz), 6.93 (s, 1H, NH2); 13C NMR (400 MHz, DMSO-d6) δ 165.1 (py-4), 164.1 (py-2), 162.6 (py-6), 148.5 (C-4″), 143.5 (C-1″), 142.3 (C-1), 139.3 (C-1′), 135.9 (C-4), 129.0 (C-3′, C-5′), 128.2 (C-2″, C-6″), 127.9 (C-4′), 127.7 (C-3, C-5), 126.84 (C-2, C-6), 126.76 (C-2′, C-6′), 123.7 (C-3″, C-5″), 102.7 (py-5); HRMS (m/z): Calcd. for (M + H)+: 369.1352; Found: 369.1377.
4-([1,1′-biphenyl]-4-yl)-6-(4-fluorophenyl)pyrimidin-2-amine (4)
1H NMR (400 MHz, DMSO-d6) δ 8.33 (ddd, 2H, H-3, H-6, J = 8.5, 2.1, 1.7 Hz), 8.31 (dd, 2H, H-2″, H-6″, J = 8.9, 5.7 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.5, 2.1, 1.7 Hz), 7.77 (s, 1H, py-5H), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.8, 1.6, 1.2 Hz), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.8, 7.4, 1.4 Hz), 7.40 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.2, 1.2 Hz), 7.35 (dd, 2H, H-3″, H-5″, J = 8.9, 8.9 Hz), 6.77 (s, 1H, NH2); 13C NMR (400 MHz, DMSO-d6) δ 164.5 (py-4), 164.0 (py-6), 163.8 (py-2), 163.6 (d, 1C, C-4″, J = 247.8 Hz), 142.0 (C-1), 139.4 (C-1′), 136.2 (C-4), 133.8 (d, 1C, C-1″, J = 2.7 Hz), 129.3 (d, 2C, C-2″, C-6″, J = 8.5 Hz), 129.0 (C-3′, C-5′), 127.9 (C-4′), 127.6 (C-3, C-5), 126.79 (C-2, C-6), 126.75 (C-2′, C-6′), 115.5 (d, 2C, C-3″, C-5″, J = 21.4 Hz), 101.6 (py-5); HRMS (m/z): Calcd. for (M + H)+: 342.1407; Found: 342.1433.
4-([1,1′-biphenyl]-4-yl)-6-(p-tolyl)pyrimidin-2-amine (5)
1H NMR (400 MHz, DMSO-d6) δ 8.32 (ddd, 2H, H-3, H-6, J = 8.4, 1.7, 1.3 Hz), 8.15 (d, 2H, H-2″, H-6″, J = 8.2 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.4, 1.7, 1.3 Hz), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.2, 2.1, 1.2 Hz), 7.73 (s, 1H, py-5H), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.6, 7.2, 1.2 Hz), 7.40 (dddd, 1H, H-4′, J = 7.6, 7.6, 1.2, 1.2 Hz), 7.33 (d, 2H, H-3″, H-5″, J = 8.2 Hz), 6.71 (s, 1H, NH2), 2.38 (s, 1H, 4”-CH3); 13C NMR (400 MHz, DMSO-d6) δ 164.8 (py-6), 164.2 (py-4), 164.0 (py-2), 141.9 (C-1), 140.2 (C-4″), 139.4 (C-1′), 136.4 (C-4), 134.6 (C-1″), 129.2 (C-3″, C-5″), 129.0 (C-3′, C-5′), 127.8 (C-4′), 127.6 (C-3, C-5), 126.9 (C-2″, C-6″), 126.8 (C-2, C-6), 126.7 (C-2′, C-6′), 101.4 (py-5), 20.9 (4”-CH3); HRMS (m/z): Calcd. for (M + H)+: 338.1657; Found: 338.1671.
4-([1,1′-biphenyl]-4-yl)-6-(3-methoxyphenyl)pyrimidin-2-amine (6)
1H NMR (400 MHz, DMSO-d6) δ 8.31 (ddd, 2H, H-3, H-6, J = 8.4, 2.0, 1.8 Hz), 7.81 (ddd, 3H, H-2, H-5, H-6″, J = 8.4, 2.0, 1.8 Hz), 7.74 (ddd, 2H, H-2′, H-6′, J = 7.9, 2.1, 1.6 Hz), 7.729 (s, 1H, py-5H), 7.728 (dd, 1H, H-2″, J = 1.0, 1.0 Hz), 7.49 (ddd, 2H, H-3′, H-5′, J = 7.9, 7.4, 1.3 Hz), 7.44 (dd, 1H, H-5″, J = 8.2, 8.2 Hz), 7.39 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.6, 1.6 Hz), 7.09 (ddd, 1H, H-4″, J = 8.2, 2.6, 1.0 Hz), 6.71 (s, 1H, NH2), 3.85 (s, 1H, 3”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.7 (py-6), 164.4 (py-4), 164.0 (py-2), 159.6 (C-3″), 142.0 (C-1), 139.4 (C-1′), 138.9 (C-1″), 136.3 (C-4), 129.7 (C-5″), 129.0 (C-3′, C-5′), 127.9 (C-4′), 127.6 (C-3, C-5), 126.80 (C-2, C-6), 126.75 (C-2′, C-6′), 119.4 (C-6″), 116.2 (C-4″), 112.2 (C-2″), 102.0 (py-5), 55.3 (3”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 354.1606; Found: 354.1623.
4-([1,1′-biphenyl]-4-yl)-6-(4-methoxyphenyl)pyrimidin-2-amine (7)
1H NMR (400 MHz, DMSO-d6) δ 8.32 (ddd, 2H, H-3, H-6, J = 8.4, 2.1, 1.8 Hz), 8.23 (d, 2H, H-2″, H-6″, J = 8.8 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.4, 2.1, 1.8 Hz), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.8, 1.7, 1.2 Hz), 7.71 (s, 1H, py-5H), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.8, 7.4, 1.6 Hz), 7.40 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.2, 1.2 Hz), 7.07 (d, 2H, H-3″, H-5″, J = 8.8 Hz), 6.67 (s, 1H, NH2), 3.84 (s, 1H, 4”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.5 (py-6), 164.1 (py-4), 163.9 (py-2), 161.3 (C-4″), 141.9 (C-1), 139.4 (C-1′), 136.5 (C-4), 129.6 (C-1″), 129.0 (C-3′, C-5′), 128.6 (C-2″, C-6″), 127.9 (C-4′), 127.5 (C-3, C-5), 126.8 (C-2, C-6), 126.7 (C-2′, C-6′), 113.9 (C-3″, C-5″), 101.0 (py-5), 55.3 (4”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 354.1606; Found: 354.1621.
4-([1,1′-biphenyl]-4-yl)-6-(2,3-dimethoxyphenyl)pyrimidin-2-amine (8)
1H NMR (400 MHz, DMSO-d6) δ 8.18 (ddd, 2H, H-3, H-6, J = 8.5, 2.1, 1.8 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.5, 2.1, 1.8 Hz), 7.74 (ddd, 2H, H-2′, H-6′, J = 7.9, 1.8, 1.5 Hz), 7.51 (s, 1H, py-5H), 7.49 (ddd, 2H, H-3′, H-5′, J = 7.9, 7.4, 1.5 Hz), 7.40 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.5, 1.5 Hz), 7.32 (dd, 1H, H-6″, J = 6.2, 2.9 Hz), 7.179 (dd, 1H, H-4″, J = 8.1, 2.9 Hz), 7.177 (dd, 1H, H-5″, J = 8.1, 6.2 Hz), 6.72 (s, 1H, NH2), 3.86 (s, 1H, 3”-OCH3), 3.76 (s, 1H, 2”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.8 (py-6), 163.9 (py-2), 163.5 (py-4), 152.9 (C-3″), 147.2 (C-2″), 142.0 (C-1), 139.4 (C-1′), 136.4 (C-4), 132.7 (C-1″), 129.1 (C-3′, C-5′), 127.9 (C-4′), 127.4 (C-3, C-5), 127.0 (C-2, C-6), 126.8 (C-2′, C-6′), 124.0 (C-5″), 121.6 (C-6″), 114.1 (C-4″), 106.0 (py-5), 60.8 (2”-OCH3), 55.9 (3”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 384.1712; Found: 384.1731.
4-([1,1′-biphenyl]-4-yl)-6-(3,4-dimethoxyphenyl)pyrimidin-2-amine (9)
1H NMR (400 MHz, pyridine-d5) δ 8.53 (ddd, 2H, H-3, H-6, J = 8.4, 2.1, 1.7 Hz), 8.23 (d, 1H, H-2″, J = 2.0 Hz), 8.10 (dd, 1H, H-6″, J = 8.4, 2.0 Hz), 7.97 (s, 1H, py-5H), 7.86 (ddd, 2H, H-2, H-5, J = 8.4, 2.1, 1.7 Hz), 7.77 (ddd, 2H, H-2′, H-6′, J = 7.1, 2.1, 1.4 Hz), 7.51 (ddd, 2H, H-3′, H-5′, J = 7.5, 7.1, 1.4 Hz), 7.41 (dddd, 1H, H-4′, J = 7.5, 7.5, 1.4, 1.4 Hz), 7.15 (d, 1H, H-5″, J = 8.4 Hz), 6.06 (s, 1H, NH2), 3.87 (s, 1H, 3”-OCH3), 3.83 (s, 1H, 4”-OCH3); 13C NMR (400 MHz, pyridine-d5) δ 166.2 (py-6), 165.9 (py-4), 165.6 (py-2), 152.8 (C-4″), 150.6 (C-3″), 143.7 (C-1), 141.1 (C-1′), 138.0 (C-4), 131.5 (C-1″), 129.9 (C-3′, C-5′), 128.8 (C-3, C-5), 128.7 (C-4′), 128.04 (C-2, C-6), 127.97 (C-2′, C-6′), 121.5 (C-6″), 112.6 (C-5″), 111.9 (C-2″), 102.9 (py-5), 56.5 (3”-OCH3), 56.4 (4”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 384.1712; Found: 384.1732.
4-([1,1′-biphenyl]-4-yl)-6-(3,5-dimethoxyphenyl)pyrimidin-2-amine (10)
1H NMR (400 MHz, DMSO-d6) δ 8.35 (ddd, 2H, H-3, H-6, J = 8.4, 2.1, 1.8 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.4, 2.1, 1.8 Hz), 7.76 (ddd, 2H, H-2′, H-6′, J = 7.8, 2.0, 1.3 Hz), 7.75 (s, 1H, py-5H), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.8, 7.4, 1.5 Hz), 7.41 (d, 2H, H-2″, H-6″, J = 2.3 Hz), 7.40 (dddd, 1H, H-4′, J = 7.4, 7.4, 1.3, 1.3 Hz), 6.76 (s, 1H, NH2), 6.67 (dd, 1H, H-4″, J = 2.3, 2.3 Hz), 3.85 (s, 2H, 3”-OCH3, 5”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.7 (py-6), 164.4 (py-54), 163.9 (py-2), 160.7 (C-3″, C-5″), 142.0 (C-1), 139.6 (C-1″), 139.4 (C-1′), 136.3 (C-4), 129.0 (C-3′, C-5′), 127.9 (C-4′), 127.7 (C-3, C-5), 126.8 (C-2, C-6, C-2′, C-6′), 105.0 (C-2″, C-6″), 102.4 (C-4″), 102.1 (py-5), 55.4 (3”-OCH3, 5”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 384.1712; Found: 384.1724.
4-([1,1′-biphenyl]-4-yl)-6-(2,4,5-trimethoxyphenyl)pyrimidin-2-amine (11)
1H NMR (400 MHz, DMSO-d6) δ 8.15 (ddd, 2H, H-3, H-6, J = 8.4, 1.9, 1.8 Hz), 7.82 (ddd, 2H, H-2, H-5, J = 8.4, 1.9, 1.8 Hz), 7.74 (ddd, 2H, H-2′, H-6′, J = 7.8, 2.0, 1.3 Hz), 7.71 (s, 1H, py-5H), 7.61 (s, 1H, H-6″), 7.50 (ddd, 2H, H-3′, H-5′, J = 7.8, 7.3, 1.6 Hz), 7.40 (dddd, 1H, H-4′, J = 7.3, 7.3, 1.3, 1.3 Hz), 6.82 (s, 1H, H-3″), 6.61 (s, 1H, NH2), 3.92 (s, 1H, 2”-OCH3), 3.88 (s, 1H, 4”-OCH3), 3.77 (s, 1H, 5”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 163.8 (py-2), 163.5 (py-6), 163.1 (py-4), 153.2 (C-2″), 151.4 (C-4″), 142.7 (C-5″), 141.7 (C-1), 139.4 (C-1′), 136.7 (C-4), 129.0 (C-3′, C-5′), 127.8 (C-4′), 127.3 (C-3, C-5), 126.9 (C-2, C-6), 126.7 (C-2′, C-6′), 117.7 (C-1″), 113.7 (C-6″), 106.1 (py-5), 98.5 (C-3″), 56.6 (2”-OCH3), 56.2 (5”-OCH3), 55.8 (4”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 414.1818; Found: 414.1826.
2-(2-amino-6-(2,4-dimethoxyphenyl)pyrimidin-4-yl)phenol (12)
1H NMR (400 MHz, DMSO-d6) δ 7.90 (d, 1H, H-6″, J = 8.6 Hz), 7.88 (dd, 1H, H-6, J = 8.7, 1.6 Hz), 7.71 (s, 1H, py-5H), 7.34 (ddd, 1H, H-4, J = 8.0, 7.1, 1.6 Hz), 7.04 (s, 1H, NH2), 6.92 (dd, 1H, H-5, J = 8.7, 7.1 Hz), 6.91 (d, 1H, H-3, J = 8.0 Hz), 6.70 (d, 1H, H-3″, J = 2.3 Hz), 6.67 (dd, 1H, H-5″, J = 8.6, 2.3 Hz), 3.91 (s, 1H, 4”-OCH3), 3.84 (s, 1H, 2”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.3 (py-4), 163.7 (py-6), 162.3 (C-2″), 161.1 (py-2), 160.3 (C-2), 159.3 (C-4″), 132.4 (C-4), 131.5 (C-6″), 127.2 (C-6), 118.9 (C-5), 118.8 (C-1″), 118.1 (C-3), 117.7 (C-1), 105.6 (C-5″), 103.9 (py-5), 98.0 (C-3″), 55.9 (4”-OCH3), 55.5 (2”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 324.1348; Found: 324.1357.
4-(4-chlorophenyl)-6-(4-methoxyphenyl)pyrimidin-2-amine (13)
1H NMR (400 MHz, DMSO-d6) δ 8.24 (d, 2H, H-2″, H-6″, J = 8.6 Hz), 8.20 (d, 2H, H-2, H-6, J = 8.9 Hz), 7.67 (s, 1H, py-5H), 7.57 (d, 2H, H-3″, H-5″, J = 8.6 Hz), 7.06 (d, 2H, H-3, H-5, J = 8.9 Hz), 6.69 (s, 1H, NH2), 3.83 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.7 (py-4), 163.9 (py-2), 163.2 (py-6), 161.3 (C-4), 136.3 (C-1″), 135.1 (C-4″), 129.5 (C-1), 128.7 (C-2″, C-6″), 128.6 (C-2, C-6, C-3″, C-5″), 113.9 (C-3, C-5), 101.0 (py-5), 55.3 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 312.0904; Found: 312.0911.
4-(4-bromophenyl)-6-(3,4-dimethoxyphenyl)pyrimidin-2-amine (14)
1H NMR (400 MHz, DMSO-d6) δ 8.18 (d, 2H, H-2″, H-6″, J = 8.6 Hz), 7.87 (dd, 1H, H-6, J = 8.6, 2.0 Hz), 7.80 (d, 1H, H-2, J = 2.0 Hz), 7.72 (s, 1H, py-5H), 7.72 (d, 2H, H-3″, H-5″, J = 8.6 Hz), 7.08 (d, 1H, H-5, J = 8.6 Hz), 7.04 (s, 1H, NH2), 3.88 (s, 1H, 3-OCH3), 3.84 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.7 (py-4), 163.6 (py-6), 163.2 (py-2), 151.5 (C-4), 148.9 (C-3), 136.3 (C-1″), 131.8 (C-3″, C-5″), 129.3 (C-2″, C-6″), 129.2 (C-1), 124.5 (C-4″), 120.7 (C-6), 111.6 (C-5), 110.4 (C-2), 101.5 (py-5), 55.9 (3-OCH3), 55.8 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 386.0504; Found: 386.0515.
4-(4-bromophenyl)-6-(4-methoxyphenyl)pyrimidin-2-amine (15)
1H NMR (400 MHz, DMSO-d6) δ 8.20 (d, 2H, H-2, H-6, J = 8.9 Hz), 8.17 (d, 2H, H-2″, H-6″, J = 8.6 Hz), 7.71 (d, 2H, H-3″, H-5″, J = 8.6 Hz), 7.67 (s, 1H, py-5H), 7.06 (d, 2H, H-3, H-5, J = 8.9 Hz), 6.70 (s, 1H, NH2), 3.83 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.7 (py-4), 163.9 (py-2), 163.3 (py-6), 161.3 (C-4), 136.7 (C-1″), 131.6 (C-3″, C-5″), 129.5 (C-1), 129.0 (C-2″, C-6″), 128.6 (C-2, C-6), 124.0 (C-4″), 114.0 (C-3, C-5), 101.0 (py-5), 55.3 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 356.0398; Found: 356.0431.
4-(4-chlorophenyl)-6-(3,4-dimethoxyphenyl)pyrimidin-2-amine (16)
1H NMR (400 MHz, DMSO-d6) δ 8.24 (d, 2H, H-2″, H-6″, J = 8.7 Hz), 7.84 (dd, 1H, H-6, J = 8.4, 1.9 Hz), 7.78 (d, 1H, H-2, J = 1.9 Hz), 7.67 (s, 1H, py-5H), 7.57 (d, 2H, H-3″, H-5″, J = 8.7 Hz), 7.07 (d, 1H, H-5, J = 8.4 Hz), 6.67 (s, 1H, NH2), 3.87 (s, 1H, 3-OCH3), 3.83 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.9 (py-4), 163.9 (py-2), 163.3 (py-6), 151.1 (C-4), 148.8 (C-3), 136.3 (C-1″), 135.2 (C-4″), 129.7 (C-1), 128.8 (C-2″, C-6″), 128.7 (C-3″, C-5″), 120.4 (C-6), 111.5 (C-5), 110.3 (C-2), 101.3 (py-5), 55.7 (3-OCH3), 55.6 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 342.1009; Found: 342.1026.
4-(4-methoxyphenyl)-6-(p-tolyl)pyrimidin-2-amine (17)
1H NMR (400 MHz, DMSO-d6) δ 8.19 (d, 2H, H-2, H-6, J = 8.9 Hz), 8.11 (d, 2H, H-2″, H-6″, J = 8.2 Hz), 7.61 (s, 1H, py-5H), 7.31 (d, 2H, H-3″, H-5″, J = 8.2 Hz), 7.05 (d, 2H, H-3, H-5, J = 8.9 Hz), 6.59 (s, 1H, NH2), 3.83 (s, 1H, 4-OCH3), 2.37 (s, 1H, 4”-CH3); 13C NMR (400 MHz, DMSO-d6) δ 164.5 (py-6), 164.3 (py-4), 163.9 (py-2), 161.2 (C-4), 140.1 (C-4″), 134.7 (C-1″), 129.7 (C-1), 129.2 (C-3″, C-5″), 128.5 (C-2, C-6), 126.9 (C-2″, C-6″), 114.0 (C-3, C-5), 100.8 (py-5), 55.3 (4-OCH3), 21.0 (4”-CH3); HRMS (m/z): Calcd. for (M + H)+: 292.1450; Found: 292.1440.
4-(3,5-dimethoxyphenyl)-6-(2-methoxyphenyl)pyrimidin-2-amine (18)
1H NMR (400 MHz, DMSO-d6) δ 7.76 (dd, 1H, H-6, J = 7.5, 1.7 Hz), 7.49 (s, 1H, py-5H), 7.45 (ddd, 1H, H-4, J = 8.4, 7.5, 1.7 Hz), 7.18 (d, 2H, H-2″, H-6″, J = 2.2 Hz), 7.16 (d, 1H, H-3, J = 8.4 Hz), 7.06 (dd, 1H, H-5, J = 7.5, 7.5 Hz), 6.65 (s, 1H, NH2), 6.64 (dd, 1H, H-4″, J = 2.2, 2.2 Hz), 3.85 (s, 1H, 2-OCH3), 3.82 (s, 2H, 3”-OCH3, 5”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.5 (py-4), 163.8 (py-2), 163.4 (py-6), 160.7 (C-3″, C-5″), 157.4 (C-2), 139.8 (C-1″), 131.0 (C-4), 130.2 (C-6), 127.0 (C-1), 120.4 (C-5), 112.0 (C-3), 106.9 (py-5), 104.7 (C-2″, C-6″), 102.1 (C-4″), 55.7 (2-OCH3), 55.3 (3”-OCH3, 5”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 338.1505; Found: 338.1519.
4-(3,4-dimethoxyphenyl)-6-(4-fluorophenyl)pyrimidin-2-amine (19)
1H NMR (400 MHz, DMSO-d6) δ 8.38 (dd, 2H, H-2″, H-6″, J = 8.9, 5.4 Hz), 7.98 (dd, 1H, H-6, J = 8.6, 2.2 Hz), 7.90 (s, 1H, py-5H), 7.87 (d, 1H, H-2, J = 2.2 Hz), 7.45 (dd, 2H, H-3″, H-5″, J = 8.9, 8.9 Hz), 7.16 (d, 1H, H-5, J = 8.6 Hz), 3.91 (s, 1H, 3-OCH3), 3.87 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 165.1 (d, 1C, C-4″, J = 250.7 Hz), 163.8 (py-2), 163.5 (py-6), 159.4 (py-4), 153.0 (C-4), 149.5 (C-3), 131.3 (d, 1C, C-1″, J = 2.9 Hz), 131.0 (d, 2C, C-2″, C-6″, J = 9.1 Hz), 126.3 (C-1), 122.4 (C-6), 116.6 (d, 2C, C-3″, C-5″, J = 21.7 Hz), 111.7 (C-5), 110.8 (C-2), 102.1 (py-5), 56.4 (3-OCH3), 56.3 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 326.1305; Found: 326.1320.
4-(3,4-dimethoxyphenyl)-6-(p-tolyl)pyrimidin-2-amine (20)
1H NMR (400 MHz, DMSO-d6) δ 8.12 (d, 2H, H-2″, H-6″, J = 8.2 Hz), 7.83 (dd, 1H, H-6, J = 8.3, 2.0 Hz), 7.78 (d, 1H, H-2, J = 2.0 Hz), 7.63 (s, 1H, py-5H), 7.32 (d, 2H, H-3″, H-5″, J = 8.2 Hz), 7.07 (d, 1H, H-5, J = 8.3 Hz), 6.60 (s, 1H, NH2), 3.88 (s, 1H, 3-OCH3), 3.83 (s, 1H, 4-OCH3), 2.37 (s, 1H, 4”-CH3); 13C NMR (400 MHz, DMSO-d6) δ 164.5 (py-6), 164.4 (py-4), 163.8 (py-2), 150.9 (C-4), 148.8 (C-3), 140.1 (C-4″), 134.7 (C-1″), 129.9 (C-1), 129.2 (C-3″, C-5″), 126.9 (C-2″, C-6″), 120.2 (C-6), 111.4 (C-5), 110.2 (C-2), 101.0 (py-5), 55.7 (3-OCH3), 55.6 (4-OCH3), 21.0 (4”-CH3); HRMS (m/z): Calcd. for (M + H)+: 322.1556; Found: 322.1562.
4-(3,5-dimethoxyphenyl)-6-(4-methoxyphenyl)pyrimidin-2-amine (21)
1H NMR (400 MHz, DMSO-d6) δ 8.22 (d, 2H, H-2, H-6, J = 8.9 Hz), 7.63 (s, 1H, py-5H), 7.36 (d, 2H, H-2″, H-6″, J = 2.3 Hz), 7.06 (d, 2H, H-3, H-5, J = 8.9 Hz), 6.64 (s, 1H, NH2), 6.64 (dd, 1H, H-4″, J = 2.3, 2.3 Hz), 3.841 (s, 2H, 3”-OCH3, 5”-OCH3), 3.835 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.4 (py-4), 164.3 (py-6), 163.8 (py-2), 161.2 (C-4), 160.7 (C-3″, C-5″), 139.7 (C-1″), 129.6 (C-1), 128.6 (C-2, C-6), 113.9 (C-3, C-5), 104.9 (C-2″, C-6″), 102.3 (C-4″), 101.4 (py-5), 55.4 (3”-OCH3, 5”-OCH3), 55.3 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 338.1505; Found: 338.1517.
4,6-bis(3,4-dimethoxyphenyl)pyrimidin-2-amine (22)
1H NMR (400 MHz, DMSO-d6) δ 7.81 (dd, 2H, H-6, H-6″, J = 8.6, 2.0 Hz), 7.74 (d, 2H, H-2, H-2″, J = 2.0 Hz), 7.61 (s, 1H, py-5H), 7.07 (d, 2H, H-5, H-5″, J = 8.6 Hz), 6.99 (s, 1H, NH2), 3.85 (s, 2H, 3-OCH3, 3”-OCH3), 3.82 (s, 2H, 4-OCH3, 4”-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 163.8 (py-4, py-6), 162.2 (py-2), 151.4 (C-4, C-4″), 148.8 (C-3, C-3″), 128.8 (C-1, C-1″), 120.8 (C-6, C-6″), 111.5 (C-5, C-5″), 110.4 (C-2, C-2″), 101.0 (py-5), 55.8 (3-OCH3, 3”-OCH3), 55.7 (4-OCH3, 4”-OCH3); HRMS (m/z): Calcd. for (M + H)+: 368.1610; Found: 368.1622.
4-(3,4-dimethoxyphenyl)-6-(4-nitrophenyl)pyrimidin-2-amine (23)
1H NMR (400 MHz, DMSO-d6) δ 8.49 (d, 2H, H-2″, H-6″, J = 9.0 Hz), 8.37 (d, 2H, H-3″, H-5″, J = 9.0 Hz), 7.94 (dd, 1H, H-6, J = 8.5, 2.0 Hz), 7.91 (s, 1H, py-5H), 7.85 (d, 1H, H-2, J = 2.0 Hz), 7.13 (d, 1H, H-5, J = 8.5 Hz), 3.90 (s, 1H, 3-OCH3), 3.86 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 164.8 (py-4), 163.0 (py-2), 162.4 (py-6), 152.3 (C-4), 149.3 (C-4″), 149.2 (C-3), 142.7 (C-1″), 129.1 (C-2″, C-6″), 128.2 (C-1), 124.3 (C-3″, C-5″), 121.6 (C-6), 112.0 (C-5), 110.6 (C-2), 103.3 (py-5), 56.24 (3-OCH3), 56.18 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 353.1250; Found: 353.1272.
4-(3,4-dimethoxyphenyl)-6-(naphthalen-1-yl)pyrimidin-2-amine (24)
1H NMR (400 MHz, DMSO-d6) δ 8.21 (dd, 1H, H-8″, J = 7.2, 2.3 Hz), 8.02 (dd, 1H, H-4″, J = 7.4, 1.5 Hz), 8.01 (dd, 1H, H-5″, J = 7.2, 2.2 Hz), 7.78 (dd, 1H, H-6, J = 8.4, 1.9 Hz), 7.76 (d, 1H, H-2, J = 1.9 Hz), 7.70 (dd, 1H, H-2″, J = 7.2, 1.5 Hz), 7.62 (dd, 1H, H-3″, J = 7.4, 7.2 Hz), 7.55 (ddd, 1H, H-7″, J = 7.2, 7.2, 2.2 Hz), 7.54 (ddd, 1H, H-6″, J = 7.2, 7.2, 2.3 Hz), 7.37 (s, 1H, py-5H), 7.06 (d, 1H, H-5, J = 8.4 Hz), 6.75 (s, 1H, NH2), 3.85 (s, 1H, 3-OCH3), 3.83 (s, 1H, 4-OCH3); 13C NMR (400 MHz, DMSO-d6) δ 167.5 (py-6), 164.1 (py-4), 163.6 (py-2), 151.0 (C-4), 148.8 (C-3), 137.2 (C-1″), 133.3 (C-10″), 130.2 (C-9″), 129.7 (C-1), 129.1 (C-4″), 128.3 (C-5″), 126.9 (C-2″), 126.5 (C-7″), 126.0 (C-6″), 125.7 (C-3″), 125.6 (C-8″), 120.1 (C-6), 111.5 (C-5), 110.1 (C-2), 106.1 (py-5), 55.61 (4-OCH3), 55.57 (3-OCH3); HRMS (m/z): Calcd. for (M + H)+: 358.1556; Found: 358.1567.
4-(3,4-dimethoxyphenyl)-6-(naphthalen-2-yl)pyrimidin-2-amine (25)
1H NMR (400 MHz, pyridine-d5) δ 9.06 (d, 1H, H-1″, J = 1.4 Hz), 8.57 (dd, 1H, H-3″, J = 8.6, 1.4 Hz), 8.25 (d, 1H, H-2, J = 2.0 Hz), 8.13 (dd, 1H, H-6, J = 8.4, 2.0 Hz), 8.10 (s, 1H, py-5H), 8.06 (dd, 1H, H-8″, J = 6.3, 1.0 Hz), 8.05 (d, 1H, H-4″, J = 8.6 Hz), 7.96 (dd, 1H, H-5″, J = 6.3, 1.0 Hz), 7.57 (ddd, 1H, H-6″, J = 6.3, 6.3, 1.0 Hz), 7.54 (ddd, 1H, H-7″, J = 6.3, 6.3, 1.0 Hz), 7.15 (d, 1H, H-5, J = 8.4 Hz), 6.56 (s, 1H, NH2), 3.88 (s, 1H, 3-OCH3), 3.83 (s, 1H, 4-OCH3); 13C NMR (400 MHz, pyridine-d5) δ 166.2 (py-6), 166.1 (py-4), 165.5 (py-2), 152.9 (C-4), 150.6 (C-3), 135.5 (C-10″), 134.6 (C-9″), 131.4 (C-1), 129.9 (C-8″), 129.6 (C-2″), 129.2 (C-4″), 128.7 (C-5″), 128.4 (C-1″), 128.1 (C-6″), 127.5 (C-7″), 125.7 (C-3″), 121.7 (C-6), 112.6 (C-5), 112.0 (C-2), 103.4 (py-5), 56.6 (3-OCH3), 56.5 (4-OCH3); HRMS (m/z): Calcd. for (M + H)+: 358.1556; Found: 358.1577.
Materials
HCT116 human colon cancer cells were obtained from the American Type Culture Collection (Rockville, MD, USA). Antibodies against phospho-pan Aurora kinase A (T288)/Aurora kinase B (T232)/Aurora kinase C (T198), caspase-3, cleaved caspase-7 (Asp198), and poly(ADP-ribose) polymerase (PARP) were obtained from Cell Signaling Technology (Beverly, MA, USA). An antibody specific to GAPDH was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Other chemicals were from Sigma-Aldrich (St. Louis, MO, USA). Enhanced chemiluminescence detection system was purchased from GE Healthcare (Piscataway, NJ, USA).
Clonogenic long-term survival assay
The clonogenic long-term survival assay (CLSA) of HCT116 human colon cancer cells followed the methods reported previously [10]. HCT116 cell lines were treated with 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety at different concentrations (0, 1, 5, 10, and 20 μM). After 7 days, cell colonies were fixed with 6% glutaraldehyde and stained with 0.1% crystal violet. Quantitation of the viable cell colonies was determined using densitometry (MultiGuage, Fujifilm, Japan). Their half-maximal cell growth inhibitory concentrations (GI50) were determined using SigmaPlot software (SYSTAT, Chicago, IL) [11].
In vitro kinases assay
The kinase assay for the title compound was performed using the EMD Millipore KinaseProfiler service assay protocol (MilliporeSigma, Burlington, MA, USA) [12]. Cancer-related kinases were selected as follows: Aurora kinase (A, B), 5’-AMP-activated protein kinase (α1,β1,γ1), RAF proto-oncogene serine/threonine-protein kinase, epidermal growth factor receptor, glycogen synthase kinase 3β, c-Jun N-terminal kinase (1a, 2a, 3), kinase insert domain receptor, mitogen-activated protein kinase (1, 2), mammalian target of rapamycin, cyclin-dependent kinase 2/cyclin E, cyclin-dependent kinase 5/p25, cyclin-dependent kinase 6/cyclin D3, insulin-like growth factor 1 receptor, protein kinase (A, Bβ, Cγ), phosphatidylinositol-4,5-bisphosphate 3-kinase, Abelson murine leukemia viral oncogene homolog 1, and apoptosis signal-regulating kinase 1. Aurora A kinase was supplied by EMD Millipore Corp. The substrate for phosphorylation, LeuArgArgAlaSerLeuGly, was 200 μM and the concentration of ATP was 10 μM [13]. All experiments were repeated three times at 10 μM.
Immunoblot analysis
HCT116 cells were treated with derivative 12 for the indicated times. The cells were lysed in an extraction buffer containing 20 mM HEPES (pH 7.2), 150 mM NaCl, 1% Triton X-100, 10% glycerol, 10 μg/mL leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Lysates (20 μg) were electrophoresed on a 10% SDS-polyacrylamide gel, transferred onto nitrocellulose membrane (Bio-Ras, Richmond, CA, USA). After blocking in TBST buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween 20) containing 10% non-fat milk for 1 h at room temperature, primary antibodies were incubated for 4 h, followed by washing in TBST and incubation with horseradish peroxidase-conjugated secondary antibodies for 4 h as described previously [14]. Antibody-reactive protein bands were visualized using an enhanced chemiluminescence detection system.
Cell cycle analysis
HCT116 cells were treated with either a vehicle (0.1% DMSO) or 20 μM derivative 12 for 24 h, fixed in 70% ethanol, washed twice with phosphate-buffered saline, and then stained with 50 μg/ml propidium iodide, as previously described [15]. Analysis of the cell cyle phase was measured using a NucleoCounter NC-3000 (ChemoMetec, Allerød, Denmark).
Quantitative structure-activity relationships
Quantitative structure-activity relationships (QSAR) calculations were conducted on an Intel Core 2 Quad Q6600 (2.4 GHz) Linux PC with Sybyl 7.3 (Tripos, St. Louis, MO, USA) [16]. As the three-dimensional (3D) QSAR calculations, CoMFA and CoMSIA were adopted. Because the 3D X-ray crystallographic structure of derivative 18, 4-(3,5-dimethoxyphenyl)-6-(2-methoxyphenyl)pyrimidin-2-amine was determined by the authors [17], the 3D structures of all derivatives were determined based on the modification of derivative 18 using the Sybyl program. For all derivatives, a conformational search was performed using the grid search method with a rotation of the selected bond in 15° increments. The energy minimization process was followed using the Tripos force field and Gasteiger–Huckel charges, and ceased at the convergence criteria of the total energy (0.05 kcal/mol Å). The most stable structures were used for the QSAR calculations. The detailed experimental procedures followed to the methods previously reported [18].
In silico docking
In silico docking to elucidate the molecular binding mode between the title compound, 2-(2-amino-6-(2,4-dimethoxyphenyl)pyrimidin-4-yl)phenol, and aurora kinase A (AURKA) was performed on an Intel Core 2 Quad Q6600 (2.4 GHz) Linux PC with Sybyl 7.3 (Tripos). The 3D structure of AURKA was adopted from the X-ray crystallographic structure deposited in the protein data bank as 3uod.pdb [19]. Although it is not the 3D structure containing the greatest number of residues (2j4z.pdb) [20], because the ligand contained in this 3D structure is similar to 2-(2-amino-6-(2,4-dimethoxyphenyl)pyrimidin-4-yl)phenol, 3uod.pdb was selected. The detailed experimental procedures followed to the methods previously reported [21].
Results and discussion
Synthesis
Of 25 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety, the synthetic methods of derivatives 13 and 17 were known, but their biological activities were not reported [22]. Two derivatives, 16 and 20, have been reported to show anticancer activity [8]. Several derivatives were known to show other biological activities: 2, 3, 4, 7 (antibacterial), 14 (nucleoside binding affinity), 19 (anticonvulsant), 23 (antifungal), and 25 (antimicrobial) [4–7].
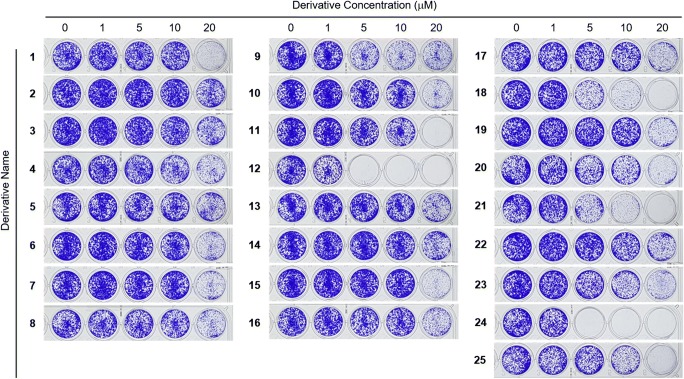
Clonogenic long-term survival assay
Of 25 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety, two (16 and 20) exhibited anticancer activity [8]; however, other derivatives remained unknown for their anticancer activity. We thus tested the effects of 25 derivatives on the inhibition of clonogenicity using a CLSA, which allowed a sensitive cell-based assay for distinguishing the growth inhibitory effect of compounds with similar structural features. HCT116 human colon cancer cells were treated with increasing concentrations of each derivative (0, 1, 5, 10, and 20 μM) for 7 days, and live cell colonies were stained with crystal violet (Fig. 2). The half-maximal cell growth inhibitory concentrations (GI50) of 25 derivatives were calculated and are listed in Table 1. They ranged between 1.45 μM (derivative 12) and 40.82 μM (derivative 2). A graph of GI50 values with error bars is plotted in Suppl. Fig. 1.
Fig. 2.
Clonogenic long-term survival assays performed at five different concentrations (0, 1, 5, 10, and 20 μM). 0.1% DMSO was used a positive control
In vitro kinases assay
We selected derivative 12 as the best inhibitor for the clonogenicity of HCT116 cells and further evaluated whether it could inhibit protein kinase activity. In vitro kinase assay shows that derivative 12 was highly specific to AURKA among the 25 cancer-associated kinases used in this test. The 95% inhibition of the AURKA kinase activity was compared to control without any compound.
Immunoblot analysis
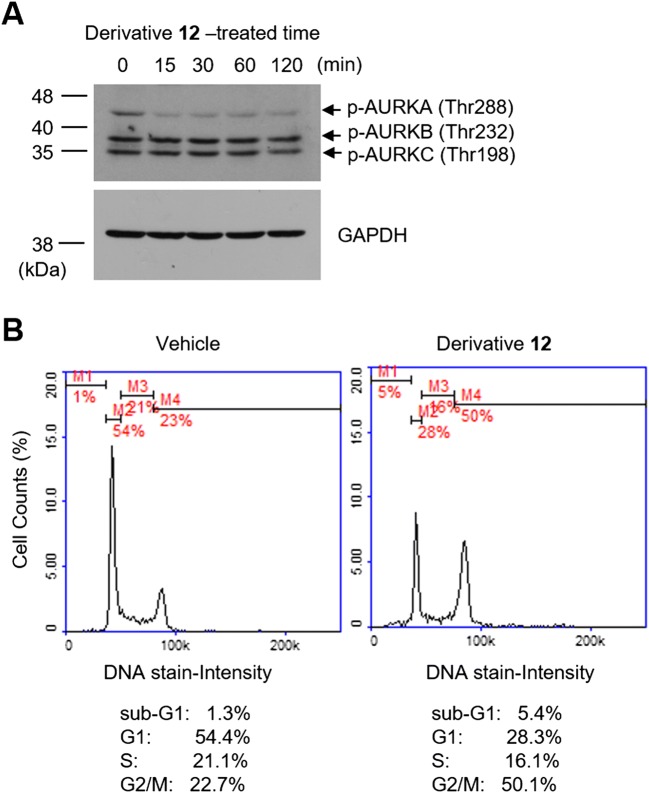
There are three subtypes of Aurora kinases: Aurora kinase A (AURKA), Aurora kinase B (AURKB), and Aurora kinase C (AURKC). To determine the specificity of derivative 12 against other Aurora kinase subtypes, we performed immunoblot analysis with a phospho-specific pan-Aurora kinase antibody. HCT116 cells were treated with 50 μM derivative 12 for different periods (0, 15, 30, 60, and 120 min), and the phosphorylation status of the three subtypes of Aurora kinases was examined. We found that only AURKA phosphorylation at Thr288 was substantially decreased, but not AURKB at Thr232 and AURKC at Thr198, in response to derivative 12 treatment (Fig. 3a).
Fig. 3.
Effect of derivative 12 on the inhibition of Aurora kinase A (AURKA) and cell cycle progression. a HCT116 cells were treated with derivative 12 for various periods (0, 15, 30, 60, and 120 min) and immunoblot analysis was performed using a phospho-specific pan-Aurora kinase antibody. GAPDH was used as an internal control. Bars, molecular weight markers. b HCT116 cells were treated with derivative 12 for 24 h, fixed with ethanol, and stained with propidium iodide. Cell cycle profiles were analyzed using a NucleoCounter NC-3000. Vehicle, 0.1% DMSO; M1, sub-G1 phase; M2, G1 phase; M3, S phase; M4, G2/M phase
Cell cycle analysis
AURKA functions during prophase of mitosis and regulates centrosome maturation and the formation of the mitotic spindle [23]. Accumulating evidence has demonstrated that inhibition of AURKA induces an abnormal mitotic spindle, cell cycle arrest at the G2/M phase, and apoptotic cell death [24]. To determine whether derivative 12 affects cell cycle progression, we carried out a flow cytometric analysis. At 24 h after derivative 12 treatment, G0/G1 phase cells decreased from 54.4% in the vehicle (DMSO)-treated control to 28.3% with a concomitant increase in the G2/M phase cell population from 22.7 to 50.1% (Fig. 3b). In general, sub-G1 phase cells appeared at the early stage of apoptosis. Derivative 12 also triggered the accumulation of sub-G1 cells from 1.3 to 5.4%. These results suggest that derivative 12 inhibition of AURKA is linked functionally to the inhibition of the cell cycle progression at the G2/M phase and the induction of apoptosis.
Activation of the caspase cascade
Caspases are a family of proteases that play essential roles in apoptosis progression [25]. There are two types of caspases involved in apoptosis; initiators (caspase-2, −8, −9, and − 10) and effectors (caspase-3 and -7). Both types of caspases are activated by proteolytic cleavages. To determine whether caspases were activated by derivative 12, HCT116 cells were treated with derivative 12 for various periods, and the cleavages of effector caspases were examined by immunoblot analysis. Both caspase-3 and caspase-7 cleavages began to be detected at 12 h after derivative 12 treatment (Fig. 4a). Also, poly(ADP-ribose) polymerase (PARP), a substrate protein of caspase-3 and caspase-7, was also proteolytically cleaved. The quantitative densitometric analysis showed that derivative 12-induced cleavages of caspase-3, caspase-7, and PARP after 12 h of treatment were all significant (Fig. 4b). These results suggested that derivative 12 could induce apoptosis through the activation of caspase cascade.
Fig. 4.
Effects of derivative 12 on the activation of caspases. a HCT116 cells were treated with derivative 12 for various periods (0, 6, 12, and 24 h) and immunoblot analysis was performed using caspase-3, caspase-7, and PARP antibodies. GAPDH was used as an internal control. Bars, molecular weight markers. b The band intensities of cleaved caspase-3, caspase-7, and PARP relative to GAPDH level were measured using ImageJ software. The data are presented as means ± SD (n = 3). NS, not significant. Statistical analysis was carried out using one-way analysis of variance (ANOVA) followed by Dunnet’s multiple comparisons tests using GraphPad Prism version 7.04 software
QSAR
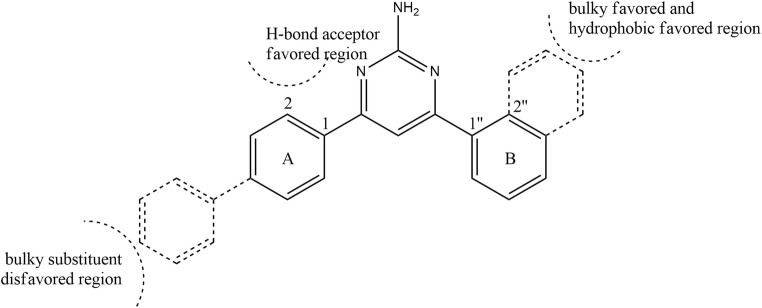
The negative logarithmic scales of GI50 values (pGI50) were adopted as the biological data for the 3D-QSAR calculations. Twenty percent of 25 derivatives was chosen arbitrarily for the test set to validate the QSAR model: 8, 9, 13, 17, and 21. They were analyzed using hierarchical cluster analysis. As shown in Suppl. Fig. 2, they belonged to separate clusters. For QSAR calculations, first, CoMFA was performed. To achieve the goal of deriving the relationships between biological activities and the 3D structures, all molecules participating in the calculations should be superimposed. All derivatives were aligned using the Sybyl/DATABASE Alignment module. As shown in Suppl. Fig. 3, because all derivatives included 4,6-diphenylpyrimidin-2-amine containing a guanidine moiety (Fig. 1d), they superimposed well. All derivatives were placed in a 2 Å spacing lattice, and their steric energy based on the Lennard-Jones potential and electrostatic energy were determined by the sp3-hybridized carbon atom with +1 charge at grid points. The detailed procedures for CoMFA followed the manufacturer’s manual. To generate the CoMFA model, a cross-validated partial least-squares (PLS) analysis was performed. Until finding a value over 0.5 for the cross-validated correlation coefficient q2, indicating the quality of the prediction, PLS analysis was iterated. To enhance the contribution of the lattice points in the CoMFA region, region focusing was performed at the same time. When the weight by discriminant power and grid spacing were 1 and 1, respectively, q2 was 0.679, which was the best cross-validation correlation coefficient generated in these CoMFA calculations. This CoMFA model provided non-cross-validated correlation coefficient (r2) of 0.922, standard error of prediction of 0.112, the number of components used in the regression was 3, and the F-test value was 62.797. Here, steric and electrostatic field descriptors contributed 75.4% and 24.6%, respectively. The pGI50 values were predicted using this CoMFA model and compared with those obtained from the clonogenic long-term survival assays performed in this research (Suppl. Table 1). A graph showing this comparison was built as shown in Suppl. Fig. 4. The differences between the pGI50 values predicted by the CoMFA model and the experimental pGI50 values ranged from 0.30 to 10.77%. To validate whether this CoMFA model wassss reliable, the pGI50 values contained in the test set were predicted using this model. As listed in Suppl. Table 1, the residuals between the experimental data and the predicted values ranged from 7.20 to 22.01%. Therefore, this CoMFA model was considered to be reliable. To visualize the steric and electrostatic field descriptors, the contour maps were generated using Sybyl software. The steric field descriptors favoring and disfavoring bulky groups were 83% and 17%, respectively (Suppl. Fig. 5). While the average GI50 value of derivatives 1–11 with the biphenyl group was 19.57 μM, that of derivatives 12–25 was 14.19 μM. The bulky group near the guanidyl group favored the activity. Derivative 24 with the 1-naphthyl group at the R2 position of the figure contained in Table 1 and derivative 12 with the 2,4-dimethoxy group showed good GI50 values of 1.92 and 1.46 μM, respectively. The contribution of the electrostatic field descriptors was not large. In addition, as mentioned later, CoMSIA did not include the electrostatic field descriptors. The visualization of the electrostatic field descriptors was ignored.
CoMSIA provided steric and electrostatic field descriptors, as well as hydrophobic, hydrogen-bond (H-bond) acceptor, and donor filed descriptors. Of several CoMSIA models generated based on the same procedures as CoMFA, a model showing an q2 of 0.575 was chosen, which included steric, hydrophobic, and H-bond donor filed descriptors. Their contributions were 21.6, 55.0, and 23.4%, respectively. In this CoMSIA model, the non-cross-validated correlation coefficient (r2), standard error of prediction, the number of components used in the regression, and F-test value were 0.962, 0.087, 6, and 54.157, respectively. The pGI50 values predicted using this CoMSIA model were compared with those obtained from the clonogenic long-term survival assay as listed in Suppl. Table 2. The experimental data was plotted against the predicted values as shown in Suppl. Fig. 6. The residuals between pGI50 values of the training set predicted by the CoMSIA model and the experimental pGI50 values ranged from 0.06 to 10.05%. For the test set, the residuals ranged between 6.98 and 27.20%. Therefore, this CoMSIA model was considered to be reliable. To visualize the results predicted using CoMSIA, contour maps were generated (Suppl. Fig. 7). The steric field descriptor of CoMSIA showed a similar result as CoMFA. Hydrophobic substituents at the R2 position of the figure contained in Table 1 increased the activity. The 1-naphthalenyl group at the R2 position showed good activity. The existence of an H-bond acceptor at C-2 of the R1 position, such as the hydroxyl or methoxy group, increased the activity: derivatives 12 and 18. The structural conditions derived from CoMFA and CoMSIA analysis can be summarized in Fig. 5.
Fig. 5.
Structural conditions to show good cancer cell growth inhibitory effects
In silico docking
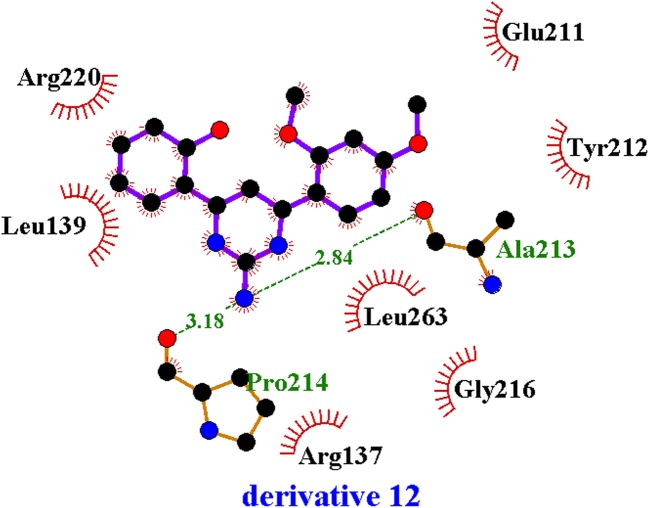
As the X-ray crystallographic structure of AURKA, 3uod.pdb was selected from the protein databank based on the reasons mentioned above. Its apo-protein was prepared for in silico docking experiments by removing its ligand, which co-crystalized with, 4-[(4-{[2-(trifluoromethyl)phenyl]amino}pyrimidin-2-yl)amino]benzoic acid (named as 0c3) (Suppl. Fig. 8). All hydrogen atoms were added and the structure was subsequently submitted to energy minimization using the conjugate gradient algorithm. Tripos force field and Gasteiger-Hückell charges were used for the energy minimization. A 3D structure of derivative 12 was obtained using the X-ray crystallographic structure of derivative 18 as mentioned above. The same procedures as apo-protein were carried out for the 3D structure of derivative 12. The binding pocket of AURKA was analyzed using the LigPlot software as previously reported [26], which consists of 13 residues; Arg137, Leu139, Gly140, Val147, Ala160, Glu211, Tyr212, Ala213, Gly216, Thr217, Arg220, Glu 260, and Leu263. These residues were used for the flexible docking experiments between AURKA and derivative 12. To confirm parameters for the docking experiments, the original ligand, 0c3 in the 3uod.pdb structure was docked into the apo-protein of AURKA (named as apo-AURKA). Because the flexible docking procedure was iterated 30 times, 30 protein-ligand complexes were generated. Their binding energies varied from −32.39 to −23.53 kcal/mol. The most poses with low binding energy were very similar to the pose of the original structure. Likewise, flexible docking was performed between apo-AURKA and derivative 12. The binding energies of 30 apo-AURKA – derivative 12 complexes ranged from −16.72 to −11.63 kcal/mol. The fourth complex was selected based on the pose of the ligand and binding energy. Its binding mode was analyzed using LigPlot. As shown in Fig. 6, there were seven hydrophobic interactions (Arg137, Leu139, Glu211, Tyr212, Gly216, Arg220, and Leu263) and two hydrogen bonds (Ala213 and Pro214) for the binding of derivative 12. Based on the 3D structure analysis using the PyMol software (PyMOL Molecular Graphics System, version 1.0r1, Schrödinger, LLC, Portland, OR, USA) [27], an additional hydrogen bond between Leu139 and 1-methoxy phenyl group was observed (Fig. 7). It was reported that only three residues (Leu215, Thr217, and Arg220) in the active site of AURKA were different from AURKB or AURKC [28]. Because the 3D structure of AURKB was known, we tried to perform in silico docking between AURKB and derivative 12. However, we could not get the protein-ligand complex, even if structures of Aurora kinases were highly conserved. Based on the current docking results, Arg220 may participate in the binding of derivative 12 through hydrophobic interactions. This phenomenon can explain the selectivity of derivative 12 for AURKA.
Fig. 6.
Residues residing in the binding site of the apo-AURKA – derivative 12 complex obtained by the LigPlot analysis
Fig. 7.
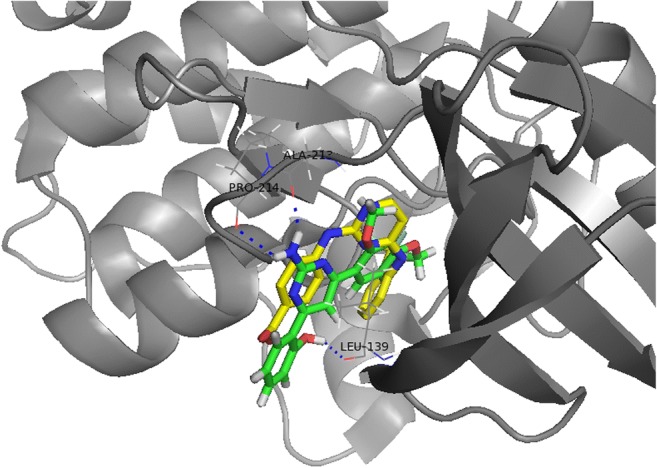
The image of the binding pocket of the apo-AURKA – derivative 12 complex visualized using the PyMol program
Conclusion
In this research, 4,6-diphenylpyrimidin-2-amine derivatives containing a guanidine moiety were designed and synthesized. As expected, they showed inhibitory effects on the cancer cell growth. An inhibitor of AURKA and AURKB, 4-(2-amino-4-methyl-5-thiazolyl)-N-[4-(4-morpholinyl)phenyl]-2-pyrimidinamine contains guanidine moiety [29]. Another inhibitor of AURKA, cyclopropanecarboxylic acid-(3-(4-(3-trifluoromethyl-phenylamino)-pyrimidin-2-ylamino)-phenyl)-amide includes guanidine moiety, too [30]. Therefore, it was considered that 2-(2-Amino-6-(2,4-dimethoxyphenyl)pyrimidin-4-yl)phenol (derivative 12) with guanidine moiety may show the inhibitory effect on AURKA. The current results demonstrated that it acts as an AURKA inhibitor, and arrests the cell cycle at the G2/M phase, and induces caspase-mediated apoptotic cell death against HCT116 human colon cancer cells. At the present time, we could not get the evidence for the title compound to bind to AURKA directly, we carried out in silico docking to elucidate the binding mode between derivative 12 and AURKA, which demonstrated that derivative 12 binds to AURKA well. The structure-activity relationship calculations showed hydrophobic substituents and 1-naphthalenyl group at the R2 position increased the activity. The existence of an H-bond acceptor at C-2 of the R1 position increased the activity, too. These results could be used to design new AURKA inhibitors exhibiting higher inhibitory effects.
Electronic supplementary material
(DOC 4625 kb)
(PDF 19878 kb)
Acknowledgments
This work was supported by the Konkuk University Research Support Program (YHL).
Abbreviations
- AURKA
Aurora kinase A
- AURKB
Aurora kinase B
- AURKC
Aurora kinase C
- CLSA
Clonogenic long-term survival assay
- CoMFA
Comparative molecular field analysis
- CoMSIA
Comparative molecular similarity indices analysis
- GI50
Half-maximal growth inhibitory concentrations
- HR/MS
High-resolution mass spectrometry
- PARP
Poly(ADP-ribose) polymerase
- QSAR
Quantitative structure-activity relationships
Authors’ contribution
YHL: designed the experiments and wrote the manuscript. JP, YLee, JL, SYS: conducted the experiments. SA: synthesized chemicals. DK and YLim: supervised the study, analyzed the data, and edited the manuscript. All authors read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dongsoo Koh, Email: dskoh@dongduk.ac.kr.
Yoongho Lim, Email: yoongho@konkuk.ac.kr.
References
- 1.Żołnowska B, Sławiński J, Szafrański K, Angeli A, Supuran CT, Kawiak A, Wieczór M, Zielińska J, Bączek T, Bartoszewska S. Novel 2-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)-1-(1,3,5-triazin-2-ylamino)guanidine derivatives: inhibition of human carbonic anhydrase cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII, anticancer activity, and molecular modeling studies. Eur J Med Chem. 2018;143:1931–1941. doi: 10.1016/j.ejmech.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Nakkala JR, Mata R, Gupta AK, Sadras SR. Biological activities of green silver nanoparticles synthesized with Acorous Calamus rhizome extract. Eur J Med Chem. 2014;85:784–794. doi: 10.1016/j.ejmech.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 3.Brzozowski Z, Saczewski F, Sławiński J. Synthesis of novel 3-amino-2-(4-chloro-2-mercaptobenzenesulfonyl)-guanidine derivatives as potential antitumor agents. Eur J Med Chem. 2007;42(9):1218–1225. doi: 10.1016/j.ejmech.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Balasankar T, Nagarajan S. Synthesis and antibacterial activities of some 2-amino-4,6- diarylpyrimidines. Heterocycl Commun. 2004;10:451–456. doi: 10.1515/HC.2004.10.6.451. [DOI] [Google Scholar]
- 5.Alam O, Mullick P, Verma SP, Gilani SJ, Khan SA, Siddiqui N, Ahsan W. Synthesis, anticonvulsant and toxicity screening of newer pyrimidine semicarbazone derivatives. Eur J Med Chem. 2010;45(6):2467–2472. doi: 10.1016/j.ejmech.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 6.Rathwa SK, Bhoi MN, Borad MA, Patel KD, Rajani DP, Rajani SD, Patel HD. Microwave assisted ZrSiO2 mediated one-pot synthesis of spiro[chromene- 4,3′-indoline] derivatives. Curr Microw Chem. 2016;3(3):187–193. doi: 10.2174/2213335602666150728205457. [DOI] [Google Scholar]
- 7.Rong L, Ji H, Xia S, Yin S, Shi Y, Tu S. An efficient synthesis of 4-naphthylpyrimidin-2-amine derivatives under solvent-free conditions. J Heterocyclic Chem. 2012;49:696–699. doi: 10.1002/jhet.839. [DOI] [Google Scholar]
- 8.Kumar B, Sharma P, Gupta VP, Khullar M, Singh S, Dogra N, Kumar V. Synthesis and biological evaluation of pyrimidine bridged combretastatin derivatives as potential anticancer agents and mechanistic studies. Bioorg Chem. 2018;78:130–140. doi: 10.1016/j.bioorg.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 9.Satia JA, Tseng M, Galanko JA, Martin C, Sandler RS. Dietary patterns and colon cancer risk in whites and African Americans in the North Carolina colon cancer study. Nutr Cancer. 2009;61(2):179–193. doi: 10.1080/01635580802419806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 11.Kim BS, Shin SY, Ahn S, Koh D, Lee YH, Lim Y. Biological evaluation of 2-pyrazolinyl-1-carbothioamide derivatives against HCT116 human colon cancer cell lines and elucidation on QSAR and molecular binding modes. Bioorg Med Chem. 2017;25(20):5423–5432. doi: 10.1016/j.bmc.2017.07.062. [DOI] [PubMed] [Google Scholar]
- 12.Sharma SK, Kumar P, Narasimhan B, Ramasamy K, Mani V, Mishra RK, Majeed AB. Synthesis, antimicrobial, anticancer evaluation and QSAR studies of 6-methyl-4-[1-(2-substituted-phenylamino-acetyl)-1H-indol-3-yl]-2-oxo/thioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid ethyl esters. Eur J Med Chem. 2012;48:16–25. doi: 10.1016/j.ejmech.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 13.Shin SY, Yoon H, Hwang D, Ahn S, Kim DW, Koh D, Lee YH, Lim Y. Benzochalcones bearing pyrazoline moieties show anti-colorectal cancer activities and selective inhibitory effects on aurora kinases. Bioorg Med Chem. 2013;21(22):7018–7024. doi: 10.1016/j.bmc.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y, Kim BS, Ahn S, Koh D, Lee YH, Shin SY, Lim Y. Anticancer and structure-activity relationship evaluation of 3-(naphthalen-2-yl)-N,5-diphenyl-pyrazoline-1-carbothioamide analogs of chalcone. Bioorg Chem. 2016;68:166–176. doi: 10.1016/j.bioorg.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Shin SY, Ahn S, Yoon H, Jung H, Jung Y, Koh D, Lee YH, Lim Y. Colorectal anticancer activities of polymethoxylated 3-naphthyl-5-phenylpyrazoline-carbothioamides. Bioorg Med Chem Lett. 2016;26(17):4301–4309. doi: 10.1016/j.bmcl.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 16.Jung Y, Shin SY, Yong Y, Jung H, Ahn S, Lee YH, Lim Y. Plant-derived flavones as inhibitors of aurora B kinase and their quantitative structure-activity relationships. Chem Biol Drug Des. 2015;85(5):574–585. doi: 10.1111/cbdd.12445. [DOI] [PubMed] [Google Scholar]
- 17.Koh D, Lee JH. 4-(3,5-Dimethoxyphenyl)-6-(2-methoxyphenyl)pyrimidin-2-amine. IUCrData. 2018;3:x180796. doi: 10.1107/S2414314618007964. [DOI] [Google Scholar]
- 18.Jung H, Shin SY, Jung Y, Tran TA, Lee HO, Jung KY, Koh D, Cho SK, Lim Y. Quantitative relationships between the cytotoxicity of flavonoids on the human breast cancer stem-like cells MCF7-SC and their structural properties. Chem Biol Drug Des. 2015;86(4):496–508. doi: 10.1111/cbdd.12512. [DOI] [PubMed] [Google Scholar]
- 19.Martin MP, Zhu JY, Lawrence HR, Pireddu R, Luo Y, Alam R, Ozcan S, Sebti SM, Lawrence NJ, Schönbrunn E. A novel mechanism by which small molecule inhibitors induce the DFG flip in Aurora a. ACS Chem Biol. 2012;7(4):698–706. doi: 10.1021/cb200508b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fancelli D, Moll J, Varasi M, Bravo R, Artico R, Berta D, Bindi S, Cameron A, Candiani I, Cappella P, Carpinelli P, Croci W, Forte B, Giorgini ML, Klapwijk J, Marsiglio A, Pesenti E, Rocchetti M, Roletto F, Severino D, Soncini C, Storici P, Tonani R, Zugnoni P, Vianello P. 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile. J Med Chem. 2006;49(24):7247–7251. doi: 10.1021/jm060897w. [DOI] [PubMed] [Google Scholar]
- 21.Shin SY, Lee Y, Kim BS, Lee J, Ahn S, Koh D, Lim Y, Lee YH. Inhibitory effect of synthetic flavone derivatives on pan-aurora kinases: induction of g2/m cell-cycle arrest and apoptosis in hct116 human colon cancer cells. Int J Mol Sci. 2018;19(12):4086. doi: 10.3390/ijms19124086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jawale DV, Pratap UR, Bhosale MR, Mane RA. One-pot three-component synthesis of 2-amino pyrimidines in aqueous peg-400 at ambient temperature. J Heterocyclic Chem. 2016;53:1626–1630. doi: 10.1002/jhet.673. [DOI] [Google Scholar]
- 23.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora-a kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278(51):51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 24.Dar AA, Goff LW, Majid S, Berlin J, El-Rifai W. Aurora kinase inhibitors--rising stars in cancer therapeutics? Mol Cancer Ther. 2010;9(2):268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–219. doi: 10.1016/S0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 26.Jung KY, Park J, Han YS, Lee YH, Shin SY, Lim Y. Synthesis and biological evaluation of hesperetin derivatives as agents inducing apoptosis. Bioorg Med Chem. 2017;25(1):397–407. doi: 10.1016/j.bmc.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Shin SY, Jung H, Ahn S, Hwang D, Yoon H, Hyun J, Yong Y, Cho HJ, Koh D, Lee YH, Lim Y. Polyphenols bearing cinnamaldehyde scaffold showing cell growth inhibitory effects on the cisplatin-resistant A2780/Cis ovarian cancer cells. Bioorg Med Chem. 2014;22(6):1809–1820. doi: 10.1016/j.bmc.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 28.Brown JR, Koretke KK, Birkeland ML, Sanseau P, Patrick DR. Evolutionary relationships of Aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol Biol. 2004;4:39. doi: 10.1186/1471-2148-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y, Deng YQ, Wang J, Long ZJ, Tu ZC, Peng W, Zhang JQ, Liu Q, Lu G. Design, synthesis and bioevaluation of N-trisubstituted pyrimidine derivatives as potent aurora a kinase inhibitors. Eur J Med Chem. 2014;78:65–71. doi: 10.1016/j.ejmech.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Tari LW, Hoffman ID, Bensen DC, Hunter MJ, Nix J, Nelson KJ, McRee DE, Swanson RV. Structural basis for the inhibition of Aurora a kinase by a novel class of high affinity disubstituted pyrimidine inhibitors. Bioorg Med Chem Lett. 2007;17(3):688–691. doi: 10.1016/j.bmcl.2006.10.086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 4625 kb)
(PDF 19878 kb)