Abstract
T cells in the immune system are activated by binding to foreign peptides (from an external pathogen) or mutant peptide (derived from endogenous proteins) displayed on the surface of a diseased cell. This triggers a series of intracellular signaling pathways, which ultimately dictate the response of the T cell. The insights from computational models have greatly improved our understanding of the mechanisms that control T-cell activation. In this review, we focus on the use of ordinary differential equation–based mechanistic models to study T-cell activation. We highlight several examples that demonstrate the models’ utility in answering specific questions related to T-cell activation signaling, from antigen discrimination to the feedback mechanisms that initiate transcription factor activation. In addition, we describe other modeling approaches that can be combined with mechanistic models to bridge time scales and better understand how intracellular signaling events, which occur on the order of seconds to minutes, influence phenotypic responses of T-cell activation, which occur on the order of hours to days. Overall, through concrete examples, we emphasize how computational modeling can be used to enable the rational design and optimization of immunotherapies.
INTRODUCTION
T cells are the members of the immune system responsible for identifying and killing diseased cells, cells that are either infected with a foreign pathogen or are mutated within the host to become cancerous. T cells perform this immune response through binding of the T-cell receptor (TCR), expressed on the surface of T cells, to a major histocompatibility complex (MHC) displaying a foreign antigen on the surface of a diseased cell. This triggers a series of intracellular signaling pathways1,2 (Fig 1), which integrate to enable the T cell to make a switch-like decision to become activated or not. Considering the complexity of the positive and negative feedback loops and multiprotein interactions in these pathways, it is difficult to predict how changes to T cells (ie, modifying the TCR, expressing engineered chimeric antigen receptors [CARs], or targeting specific intracellular signaling molecules) would affect T-cell activation. Therefore, systems biology approaches combining quantitative experiments with mechanistic modeling are needed to predict T-cell dynamics. Systems biology modeling identifies how individual components influence the system as a whole.3
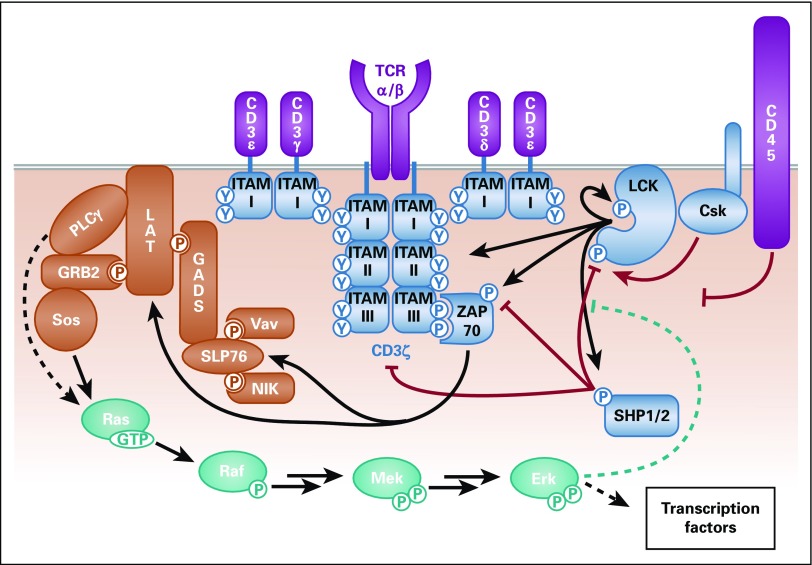
FIG 1.
Schematic of T-cell receptor (TCR) signaling. The main TCR signaling pathways incorporated into mechanistic computational models are shown here. Purple: Antigen discrimination: the TCR alpha/beta (α/β) chains bind to an antigen displayed on a major histocompatibility complex on a diseased cell; the CD3 chains are associated with the TCR. Blue: Intracellular receptor phosphorylation: various kinases, such as lymphocyte-specific protein tyrosine kinase (LCK), CSK, and ZAP-70, and phosphatases, such as SHP-1, SHP2, and CD45, influence the phosphorylation of state of the CD3 immunoreceptor tyrosine-based activation motifs (ITAMs) on the TCR. These molecules can either phosphorylate or dephosphorylate the TCR directly, influence the activity of a molecule that directly acts on the TCR, or bind to phosphorylated ITAMs to protect them from dephosphorylation. Gold: LAT signalosome: many different adaptor proteins and kinases bind together in the LAT signalosome to activate downstream signaling pathways. Teal: Mitogen-activated protein kinase (MAPK) pathway: the MAPK pathway acts as a positive feedback to prevent LCK association with the phosphatase SHP-1, as well as activate downstream transcription factors. Solid lines represent direct activity, dashed lines represent indirect activity, arrows represent phosphorylation events, and blunted lines represent dephosphorylation events. Red lines are inhibitory mechanisms, and teal lines are positive feedback mechanisms.
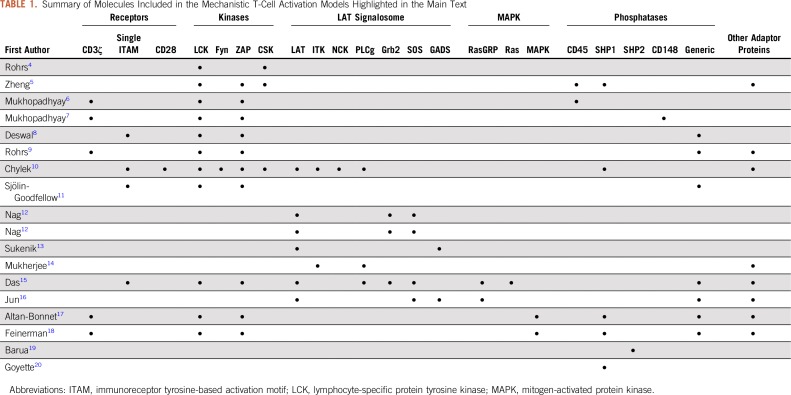
Systems biology models have been used to study T-cell activation, including mechanistic models, logic-based models, and data-driven statistical models. The latter two can identify new structural features of a pathway and bridge time scales between signal initiation and phenotypic response, but often lack mechanistic information about the system. Mechanistic models typically include known interactions between individual molecular species. Ordinary differential equation (ODE)-based deterministic models provide quantitative insights into how kinetic and physical changes to the system influence the output. Mechanistic stochastic models incorporate biologic variability and predict responses within a heterogeneous cell population. Because overactivation of a few T cells can lead to serious system effects and even death, stochastic models can identify which cell-to-cell differences significantly influence an individual cell’s response. Here, we highlight computational models of T-cell activation, with a focus on how mechanistic models (Table 1) advance our understanding so that the immune system can be engineered to better fight disease.
TABLE 1.
Summary of Molecules Included in the Mechanistic T-Cell Activation Models Highlighted in the Main Text
MECHANISTIC MODELS
Antigen Discrimination
Several theories have been proposed21-24 to explain the mechanism through which T cells become activated by as few as one or two agonist antigens, referred to as sensitivity, but not by thousands of antagonist self-antigens, even though there is only a small difference in the binding affinities of the two antigen types, referred to as specificity. The dominant hypotheses include (1) kinetic segregation, (2) kinetic proofreading, (3) receptor scanning, and (4) serial triggering, which have been reviewed previously.25,26 There is computational and experimental data to support each hypothesis, and it is likely that all of these, and more, play a role in the highly sensitive and specific initiation of TCR activation.
The theory of kinetic segregation proposes that the physical separation of the TCR from phosphatases, such as CD45, enables accumulation of phosphorylated TCR domains, which can initiate T-cell activation (Fig 1; purple).24 Experiments show that the large extracellular domains of many membrane-spanning phosphatases are excluded from the narrow contact region between a T cell and its target.27,28 This physical exclusion of phosphatases is thought to prevent aberrant signaling.29 Burroughs and Wülfing30 constructed a model of kinetic segregation to show how the size of the extracellular receptor domain, and therefore, the bond length between the T cell and antigen-presenting cell, controls the composition of the segregated regions. However, this model cannot explain T-cell specificity and sensitivity.
Kinetic proofreading and receptor scanning provide a mechanism for TCR specificity, which cannot be predicted by a simple binding-activation model. In kinetic proofreading, an antigen must remain bound for a certain amount of time to allow for enough CD3 phosphorylation to initiate downstream signaling. Several models of T-cell activation have included kinetic proofreading, each explaining complex data.25,31,32 We highlight a recent model of receptor scanning informed by new high-resolution experiments that specifically quantify antigen-receptor dwell time thresholds for CD4+ and CD8+ T cells. Stepanek et al21 created a new model, termed LCK (lymphocyte-specific protein tyrosine kinase)-come-and-stay, which predicted that the CD4 and CD8 coreceptors play an important role in setting the threshold level for T cells. Their model predicted that the difference in the LCK binding of CD4 and CD8 is sufficient to account for the activation threshold differences between CD4+ and CD8+ T cells, and they validated this prediction experimentally.
To account for the high sensitivity of T-cell activation, serial triggering33 has been proposed. In this mechanism, one peptide-MHC complex can bind and unbind from many TCRs. This allows for short dwell times to lead to strong activation such that a single antigen-MHC complex can initiate enough TCR signaling to activate downstream transcription factors. Serial triggering explains T-cell sensitivity and specificity and is also consistent with kinetic segregation and kinetic proofreading.34 Thus, multiple mechanisms of antigen discrimination work together to influence T-cell decisions.
Modeling of antigen discrimination has also been applied to CAR-engineered T cells. Harris et al35 measured T-cell activation by TCRs and CARs with the same antigen binding kinetics and showed that CARs are 10- to 100-fold less sensitive than TCRs. Using a simple model of kinetic proofreading coupled with an incoherent feedforward loop, they hypothesized that the CAR has weaker receptor activation through the proofreading step, but similar downstream activation after receptor phosphorylation.
Intracellular Receptor Phosphorylation
Once the TCR binds to its respective agonist target, the intracellular signaling domains on the receptors become phosphorylated. The endogenous TCR contains three CD3 dimers, each bearing several pairs of tyrosine phosphorylation sites called immunoreceptor tyrosine-based activation motifs (ITAMs; Fig 1; blue).36 A variety of models attempt to describe the intracellular signaling events that lead to CD3 phosphorylation.
ODE-based mechanistic models explore the mechanisms that regulate individual proteins in the system. Rohrs et al4 investigated how LCK, the main activating kinase in T cells, is regulated. This model showed that the catalytic rates of phosphorylated LCK species preferentially phosphorylate specific tyrosine substrates, resulting in a feedback loop in which active LCK prefers to downregulate LCK activity by phosphorylating at an inhibitory phosphorylation site, whereas unphosphorylated and doubly phosphorylated LCK prefer the activating site. Other models have explored the regulation of phosphatases associated with T-cell activation signaling.5,19,20 These minimal models provide a basis to explore how various proteins influence phosphorylation of the TCR itself, including how the three ITAMs on CD3ζ amplify T-cell signaling.8,11,15,17,18 These minimal models can be expanded to incorporate new experimental data regarding the roles of CD3ζ.37
There are several models of TCR phosphorylation.34 An early model of intracellular T-cell signaling assumes that the TCR is immediately phosphorylated on TCR binding. This model by Zheng et al5 predicted strong early phosphorylation of ZAP-70 and slower dephosphorylation due to negative feedback from the phosphatases, matching experimental data. However, this model does not include the influence of activation or feedback mechanisms on the ITAM domains themselves. Activation of the CD3ζ ITAMs has been studied by Mukhopadhyay et al.6 Their first model predicted that the structure of CD3ζ, with six tyrosine phosphorylation sites, could promote a switch-like decision in the on/off state of an individual CD3ζ protein, termed ultrasensitivity.6 The model tested many possible mechanisms of phosphorylation and ZAP-70 binding, showing that ultrasensitivity requires that multiple ITAMs be phosphorylated sequentially with increasing ZAP-70 binding affinities. Interestingly, follow-up experiments using a reconstituted cellular system did not show ultrasensitivity.7 On the basis of these experiments, Mukhopadhyay et al7 created a new model that correlates the CD3ζ structure with its rate of activation. The model showed that CD3ζ phosphorylation increases with subsequent phosphorylation events because of increasing rigidity of the CD3ζ chain.
Several groups have developed detailed mechanistic models of TCR activation using phosphoproteomic mass spectrometry. Rohrs et al9 paired phosphoproteomic measurements of recombinant proteins with mechanistic modeling to quantify LCK-mediated site-specific phosphorylation of CD3ζ ITAMs with or without CD28. This work predicted that the ITAMs are phosphorylated with distinct kinetics and shed light on differences between the various ITAMs and costimulatory domains. Chylek et al10 used mass spectrometry-based phosphotyrosine measurements to create a model of the early time points (less than 1 minute) of T-cell activation. Their work specified individual pathways for different CD3 proteins, showing that CD3ε influences recruitment of the actin remodeling protein WAS, which was originally thought to be regulated by CD3ζ. Sjölin-Goodfellow et al11 modeled a minimal system of four proteins (CD3ζ, LCK, ZAP-70, and generic phosphatases), focusing on the role of ZAP-70 binding and catalytic activity. The model explained the different roles of ZAP-70 binding and catalytic activities and provided possible mechanistic explanations for the observation that the CD3 ITAMs are phosphorylated asymmetrically (where LCK binds to singly phosphorylated ITAMs before phosphorylating the second ITAM site). Taken together, these models present a picture of the subtle kinetic differences between the ITAM tyrosine site phosphorylation that may directly influence downstream signaling outputs.
The LAT Signalosome
The models summarized in the Antigen Discrimination and Intracellular Receptor Phopshorylation sections describe T-cell activation up to the point of ZAP-70 binding to the activated TCR. Once bound, ZAP-70 is phosphorylated at several sites and is able to phosphorylate downstream proteins in the LAT signalosome (Fig 1; gold). LAT is a membrane-bound intracellular scaffolding protein with several phosphorylation sites, where the four membrane distal tyrosine residues strongly regulate T-cell activation.12 The three C-terminal tyrosine sites bind to the SH2 domains of adaptor proteins GRB2, GADS, and Grap,38 which bind different proteins required for T-cell activation (ie, PLCγ, SOS, SLP76, ITK, NCK, and Vav). Multiple different combinations of LAT signalosomes are necessary for T-cell activation.39-41
Computational tools can aid in modeling the large number of multiprotein interactions in LAT signalosomes. One tool is BioNetGen, in which rules define possible site-specific interactions, such as binding or phosphorylation, regardless of the state at other sites on the interacting proteins.42 BioNetGen can generate a set of ODEs; however, for the case of branching LAT complexes, it enables a network-free simulation that can enumerate the evolution of these protein complexes.43 Nag et al 12,44 used BioNetGen to construct a model focusing on the aggregation of LAT into clusters containing GRB2 and SOS. These are high valence proteins that aggregate in branched structures to help colocalize downstream signaling molecules and increase signal strength. Nag et al12 predicted the amounts of monovalent, bivalent, and trivalent LAT in the clusters and proposed that LAT bound to adaptor proteins besides Grb2 could account for the monovalent and bivalent forms. Sukenik et al13 showed experimentally that GADS can dimerize through interactions in the SH2 domain, and this homodimerization promotes LAT binding. They developed a model on the basis of the experimental measurements and predicted that cooperative binding of GADS dimers to phosphorylated LAT could increase the sensitivity of TCR signaling. Mukherjee et al14 used BioNetGen to simplify the multiprotein interactions involved in LAT binding and investigate ITK kinase activity, a functional output. Rather than including binding events that lead to LAT signalosome recruitment of ITK, they assumed that ITK and its substrate PLCγ are directly recruited to phospho-LAT. The model predicted that cooperative allosteric regulation of ITK by IP4, a signaling modulator, is important in controlling ITK activity.
The Mitogen-Activated Protein Kinase Pathway
Many of the molecules in the LAT signalosome lead to pathways that activate transcription factors. The ERK/mitogen-activated protein kinase (MAPK) pathway (Fig 1; teal) exhibits rapid and robust activation, controls TCR activation via feedback,45 and influences T-cell functional response.46 This pathway displays ultrasensitivity in T cells and is thought to help make an on/off activation decision from the graded upstream inputs after TCR binding.15 Several models have explored the ERK/MAPK pathway and its role in T-cell activation.
ERK response in T cells is typically digital, resulting in a bimodal population as the stimulus level increases above threshold. Das et al15 used computational modeling to better understand how RasGRP and SOS coordinate MAPK activation. They found that RasGRP activation leads to a graded response, whereas SOS is subject to positive feedback that results in a digital response. These results were validated by Jun et al,16 who expanded the model to include another MAPK pathway leading to p38 activation. They found that p38 activation requires SOS binding, without allosteric regulation.
ERK also provides feedback upstream of the MAPK pathway to LCK. ERK phosphorylates LCK at a protection site, preventing it from being dephosphorylated by the phosphatase SHP-1, maintaining LCK’s catalytic activity. Altan-Bonnet and Germain17 modeled these competing feedback loops with kinetic proofreading and demonstrated that the feedback loops determine the threshold for antigen discrimination, while keeping a rapid and sensitive response. Altan-Bonnet and Germain17 also showed that the cellular response is highly dependent on SHP-1 concentration. Feinerman et al47 further tested the effects of protein expression heterogeneity on T-cell signaling using a stochastic model implementation. The model predicted that CD8 controls the half-maximal concentration of antigen required for cells to respond, and SHP-1 controls the maximal percent of cells that respond. Coregulation of these two proteins was predicted to limit population-level variability in T-cell response, which they validated experimentally.
Cytosolic Messengers
Cytosolic messengers are activated downstream of the LAT signalosome. One of the most important messengers in T-cell activation is calcium. In T-cell activation, PLCγ in the LAT signalosome promotes formation of IP3, which releases calcium from intracellular vesicles. This calcium promotes signaling to activate the transcription factor NFAT. Perley et al48 modeled TCR-mediated activation of calcium and downstream cytokines. Their model showed the importance of positive feedback from ERK to kinases such as LCK to promote calcium release.
Several groups have used computational modeling to explore how messengers influence kinetic proofreading.49-51 These studies incorporate the temporal summation model, in which the messenger accumulates downstream if TCRs are serially triggered.49 Once this messenger reaches a certain level, it activates downstream signaling without waiting for kinetic proofreading at the receptor level, a phenomenon known as kinetic proofreading escape. Calcium has been shown to exhibit these characteristics, but more work is needed to better understand how other messengers influence T-cell activation.
Phenotypic Responses
Extending models of T-cell signaling beyond activation of transcription factors requires a change in time scale. All of the intracellular signaling events described previously occur on a time scale of seconds to minutes after antigen binds to the TCR; however, gene regulation and protein synthesis take much longer (hours to days). Although some mechanistic models attempt to explain these events over time, the exact mechanisms and parameters that govern these events are not well defined.52 Therefore, more coarse-grained mechanistic models are used to explain phenotypic responses of T-cell activation. For example, Locasale53 used an ODE-based model to explore how memory can develop in long-term interrupted T-cell stimulation. Their model simplified signaling such that T-cell antigen binding directly stimulates transcription factor activation. The model identified possible gene regulation mechanisms that allow T cells to integrate activation signals over longer time scales; however, their work lumps many of the intracellular signaling pathways described earlier into a single intermediate step.
FUTURE DIRECTIONS
Mechanistic models are useful for predicting the dynamics of intracellular signaling. However, more work can be done to improve their predictive ability. These models can be expanded to account for additional signaling mechanisms, such as binding of adaptor proteins to CD3ζ,54 differences between the individual ITAMs,55 and T-cell costimulatory domains.56 More detailed models are also needed to describe how the LAT signalosome triggers downstream pathways that activate transcription factors associated with T-cell activation and differentiation.57
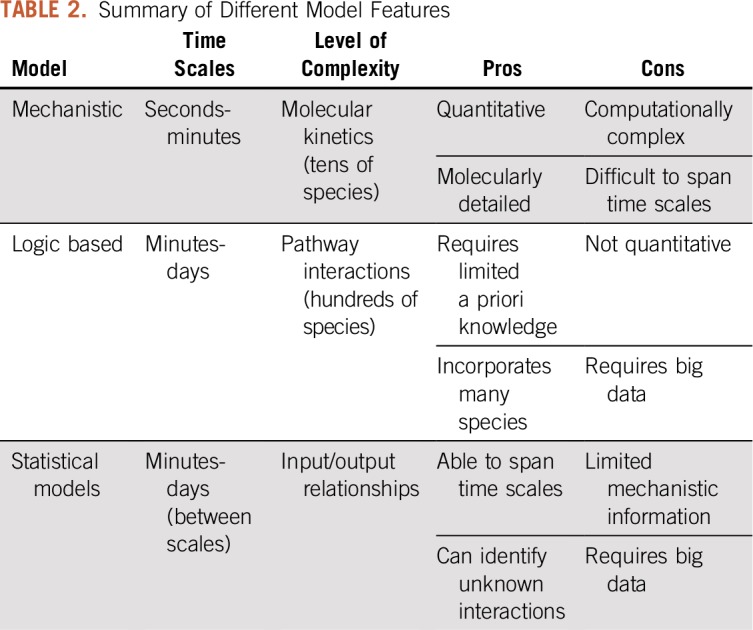
In addition, mechanistic models can be complemented by other modeling approaches (Table 2). Logic-based models help identify new interactions between proteins in a pathway.58,59 Logic-based models have the ability to incorporate hundreds of proteins, compared with tens of proteins, which are typically included in mechanistic ODE-based models. Although logic-based models cannot provide quantitative information about the signaling reactions, they can predict how various signaling events influence the phenotypic responses of T cells. For example, Kaufman et al60 used a Boolean formalism to explore how kinases remain active, even after the antigen unbinds from the TCR, linking the timing of antigen binding and intracellular signal initiation to longer time scales of T-cell response. Miskov-Zivanov et al61 created a Boolean model that predicted T-cell differentiation into regulator or helper cells on the basis of the strength and timing of antigen presentation. Zhang et al62 compared how activation of cancerous T cells differs from healthy T-cell activation. Thus, logical models can provide new insights and predict functional responses.
TABLE 2.
Summary of Different Model Features
Data-driven statistical modeling can be used to bridge the time scales between the initial signaling events and T cells’ phenotypic responses. Kemp et al63 created a multivariable regression model that relates the levels of intermediates in the TCR activation pathway to production of interleukin-2, a cytokine that mediates the cells’ immune response. They measured levels of 11 proteins downstream of the TCR and CD28 after T-cell activation with different antigen peptides. They then used partial least squares regression to identify the combination of signaling events that could explain the interleukin-2 production. The model also predicted the effects of altering individual pathways. This work showed that T-cell activation is regulated by a combination of multiple signaling nodes downstream of the TCR. More recently, data-driven models have benefited from mass cytometry techniques (CyTOF), which can measure more than 40 proteins at once.64 Such models of T cells65 and NK cells66 provide new insight into the complexity of signaling dynamics.
It is increasingly evident that the spatial arrangement of molecules plays an important role in T-cell activation.49,67 New experimental data are rapidly emerging to indicate the importance of molecular movement and arrangement in T-cell activation.68-70 Some models of T-cell activation account for the molecules’ spatial distribution,8 and software such as Simmune, developed by the National Institutes of Health/National Institute of Allergy and Infectious Diseases, has opened new opportunities to explore these spatial features.71
With the renewed focus on immunotherapy, it is important to understand how T cells are activated so that they can be engineered to fight diseases.72,73 Computational models can identify promising methods for optimizing activation through the TCR74 and CARs,75 reducing the time and money spent on guess-and-check preclinical experiments. Accurately predicting how modifying signaling mechanisms affects T-cell phenotypic responses requires a mechanistic model with downstream signaling outputs that are detailed and precise enough to be used as inputs to course-grained logic or statistical models that predict phenotypic responses. Ultimately, by combining various computational modeling approaches with robust experimental data, we can gain insight needed to optimize new immunotherapies.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Jennifer A. Rohrs, Pin Wang
Administrative support: Pin Wang
Data analysis and interpretation: Pin Wang, Stacey D. Finley
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Pin Wang
Stock and Other Ownership Interests: Immune Design
Consulting or Advisory Role: TCRCure Biopharma
Patents, Royalties, Other Intellectual Property: A technology for engineering dendritic cells for targeted vaccination. A series of patents was awarded to California Institute of Technology and University of Southern California. Immune Design has licensed these patents, in which certain royalty fees can be awarded to inventors such as myself.
No other potential conflicts of interest were reported.
REFERENCES
- 1.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: Branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 2.Mustelin T, Abraham RT, Rudd CE, et al. Protein tyrosine phosphorylation in T cell signaling. Front Biosci. 2002;7:d918–d969. doi: 10.2741/A821. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty AK, Das J. Pairing computation with experimentation: A powerful coupling for understanding T cell signalling. Nat Rev Immunol. 2010;10:59–71. doi: 10.1038/nri2688. [DOI] [PubMed] [Google Scholar]
- 4.Rohrs JA, Wang P, Finley SD. Predictive model of lymphocyte-specific protein tyrosine kinase (LCK) autoregulation. Cell Mol Bioeng. 2016;9:351–367. doi: 10.1007/s12195-016-0438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Balakrishnan V, Buzzard G, et al. Modeling and analysis of early events in T-lymphocyte antigen-activated intracellular-signaling pathways. J Comput Appl Math. 2005;184:320–341. [Google Scholar]
- 6.Mukhopadhyay H, Cordoba S-P, Maini PK, et al. Systems model of T cell receptor proximal signaling reveals emergent ultrasensitivity. PLOS Comput Biol. 2013;9:e1003004. doi: 10.1371/journal.pcbi.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukhopadhyay H, de Wet B, Clemens L, et al. Multisite phosphorylation of the T cell receptor ζ-chain modulates potency but not the switch-like response. Biophys J. 2016;110:1896–1906. doi: 10.1016/j.bpj.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deswal S, Schulze AK, Höfer T, et al. Quantitative analysis of protein phosphorylations and interactions by multi-colour IP-FCM as an input for kinetic modelling of signalling networks. PLoS One. 2011;6:e22928. doi: 10.1371/journal.pone.0022928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohrs JA, Zheng D, Graham NA, et al. Computational model of chimeric antigen receptors explains site-specific phosphorylation kinetics. Biophys J. 2018;115:1116–1129. doi: 10.1016/j.bpj.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chylek LA, Akimov V, Dengjel J, et al. Phosphorylation site dynamics of early T-cell receptor signaling. PLoS One. 2014;9:e104240. doi: 10.1371/journal.pone.0104240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjölin-Goodfellow H, Frushicheva MP, Ji Q, et al. The catalytic activity of the kinase ZAP-70 mediates basal signaling and negative feedback of the T cell receptor pathway. Sci Signal. 2015;8:1–14. doi: 10.1126/scisignal.2005596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nag A, Monine M, Perelson AS, et al. Modeling and simulation of aggregation of membrane protein LAT with molecular variability in the number of binding sites for cytosolic Grb2-SOS1-Grb2. PLoS One. 2012;7:e28758. doi: 10.1371/journal.pone.0028758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukenik S, Frushicheva MP, Waknin-Lellouche C, et al. Dimerization of the adaptor Gads facilitates antigen receptor signaling by promoting the cooperative binding of Gads to the adaptor LAT. Sci Signal. 2017;10:eaal1482. doi: 10.1126/scisignal.aal1482. [DOI] [PubMed] [Google Scholar]
- 14.Mukherjee S, Rigaud S, Seok SC, et al. In silico modeling of Itk activation kinetics in thymocytes suggests competing positive and negative IP4 mediated feedbacks increase robustness. PLoS One. 2013;8:e73937. doi: 10.1371/journal.pone.0073937. [Erratum: PLoS One 9:10.1371/annotation/285bb79f-ef1a-467e-9fd3-4e518d65acd3, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das J, Ho M, Zikherman J, et al. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun JE, Yang M, Chen H, et al. Activation of extracellular signal-regulated kinase but not of p38 mitogen-activated protein kinase pathways in lymphocytes requires allosteric activation of SOS. Mol Cell Biol. 2013;33:2470–2484. doi: 10.1128/MCB.01593-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinerman O, Jentsch G, Tkach KE, et al. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Mol Syst Biol. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barua D, Faeder JR, Haugh JM. Structure-based kinetic models of modular signaling protein function: Focus on Shp2. Biophys J. 2007;92:2290–2300. doi: 10.1529/biophysj.106.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyette J, Salas CS, Coker-Gordon N, et al. Biophysical assay for tethered signaling reactions reveals tether-controlled activity for the phosphatase SHP-1. Sci Adv. 2017;3:e1601692. doi: 10.1126/sciadv.1601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stepanek O, Prabhakar AS, Osswald C, et al. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell. 2014;159:333–345. doi: 10.1016/j.cell.2014.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valitutti S, Müller S, Cella M, et al. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 24.Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. 2006;7:803–809. doi: 10.1038/ni1369. [DOI] [PubMed] [Google Scholar]
- 25.Coombs D, Goldstein B. T cell activation: Kinetic proofreading, serial engagement and cell adhesion. J Comput Appl Math. 2005;184:121–139. [Google Scholar]
- 26.Courtney AH, Lo WL, Weiss A. TCR signaling: Mechanisms of initiation and propagation. Trends Biochem Sci. 2018;43:108–123. doi: 10.1016/j.tibs.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, Weiss A. The tyrosine phosphatase CD148 is excluded from the immunologic synapse and down-regulates prolonged T cell signaling. J Cell Biol. 2003;162:673–682. doi: 10.1083/jcb.200303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leupin O, Zaru R, Laroche T, et al. Exclusion of CD45 from the T-cell receptor signaling area in antigen-stimulated T lymphocytes. Curr Biol. 2000;10:277–280. doi: 10.1016/s0960-9822(00)00362-6. [DOI] [PubMed] [Google Scholar]
- 29.Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J Cell Biol. 1999;147:447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burroughs NJ, Wülfing C. Differential segregation in a cell-cell contact interface: The dynamics of the immunological synapse. Biophys J. 2002;83:1784–1796. doi: 10.1016/S0006-3495(02)73944-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKeithan TW. Kinetic proofreading in T-cell receptor signal transduction. Proc Natl Acad Sci USA. 1995;92:5042–5046. doi: 10.1073/pnas.92.11.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lever M. Architecture of a minimal signaling pathway explains the T-cell response to a 1 million-fold variation in antigen affinity and dose. Proc Natl Acad Sci USA. 2016;113:E6630–E6638. doi: 10.1073/pnas.1608820113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sousa J, Carneiro J. A mathematical analysis of TCR serial triggering and down-regulation. Eur J Immunol. 2000;30:3219–3227. doi: 10.1002/1521-4141(200011)30:11<3219::AID-IMMU3219>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling . Nat Immunol. 2014;15:798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris DT, Hager M V, Smith SN, et al. Comparison of T cell activities mediated by human TCRs and CARs that use the same recognition domains. J Immunol. 2018;200:1088–1100. doi: 10.4049/jimmunol.1700236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love PE, Hayes SM. ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harb Perspect Biol. 2010;2:a002485. doi: 10.1101/cshperspect.a002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James JR. Tuning ITAM multiplicity on T cell receptors can control potency and selectivity to ligand density. Sci Signal. 2018;11:eaan1088. doi: 10.1126/scisignal.aan1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balagopalan L, Coussens NP, Sherman E, et al. The LAT story: A tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barr VA, Sherman E, Yi J, et al. Development of nanoscale structure in LAT-based signaling complexes. J Cell Sci. 2016;129:4548–4562. doi: 10.1242/jcs.194886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman E, Barr V, Manley S, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su AX, Ditlev JA, Hui E, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction . Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 43.Sneddon MW, Faeder JR, Emonet T. Efficient modeling, simulation and coarse-graining of biological complexity with NFsim. Nat Methods. 2011;8:177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
- 44.Nag A, Monine MI, Faeder JR, et al. Aggregation of membrane proteins by cytosolic cross-linkers: Theory and simulation of the LAT-Grb2-SOS1 system. Biophys J. 2009;96:2604–2623. doi: 10.1016/j.bpj.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stefanová I, Hemmer B, Vergelli M, et al. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat Immunol. 2003;4:248–254. doi: 10.1038/ni895. [DOI] [PubMed] [Google Scholar]
- 46.Alberola-Ila J, Forbush KA, Seger R, et al. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 47.Feinerman O, Veiga J, Dorfman JR, et al. Variability and robustness in T cell activation from regulated heterogeneity in protein levels1081. Science. 2008;32:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perley JP, Mikolajczak J, Buzzard GT, et al. Resolving early signaling events in T-cell activation leading to IL-2 and FOXP3 transcription. Processes (Basel) 2014;2:867–900. http://www.mdpi.com/2227-9717/2/4/867 [Google Scholar]
- 49.Rachmilewitz J, Lanzavecchia A. A temporal and spatial summation model for T-cell activation: Signal integration and antigen decoding. Trends Immunol. 2002;23:592–595. doi: 10.1016/s1471-4906(02)02342-6. [DOI] [PubMed] [Google Scholar]
- 50.Rosette C, Werlen G, Daniels MA, et al. The impact of duration versus extent of TCR occupancy on T cell activation: A revision of the kinetic proofreading model. Immunity. 2001;15:59–70. doi: 10.1016/s1074-7613(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 51.Hlavacek WS, Redondo A, Metzger H, et al. Kinetic proofreading models for cell signaling predict ways to escape kinetic proofreading. Proc Natl Acad Sci USA. 2001;98:7295–7300. doi: 10.1073/pnas.121172298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hancock EJ, Stan GB, Arpino JAJ, et al. Simplified mechanistic models of gene regulation for analysis and design. J R Soc Interface. 2015;12:20150312. doi: 10.1098/rsif.2015.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locasale JW. Computational investigations into the origins of short-term biochemical memory in T cell activation. PLoS One. 2007;2:e627. doi: 10.1371/journal.pone.0000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osman N, Turner H, Lucas S, et al. The protein interactions of the immunoglobulin receptor family tyrosine-based activation motifs present in the T cell receptor zeta subunits and the CD3 gamma, delta and epsilon chains. Eur J Immunol. 1996;26:1063–1068. doi: 10.1002/eji.1830260516. [DOI] [PubMed] [Google Scholar]
- 55.Zenner G, Vorherr T, Mustelin T, et al. Differential and multiple binding of signal transducing molecules to the ITAMs of the TCR-zeta chain. J Cell Biochem. 1996;63:94–103. doi: 10.1002/(sici)1097-4644(199610)63:1<94::aid-jcb8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 56.Kohlmeier JE, Benedict SH. Alternate costimulatory molecules in T cell activation: Differential mechanisms for directing the immune response. Histol Histopathol. 2003;18:1195–1204. doi: 10.14670/HH-18.1195. [DOI] [PubMed] [Google Scholar]
- 57.Huse M. The T-cell-receptor signaling network. J Cell Sci. 2009;122:1269–1273. doi: 10.1242/jcs.042762. [DOI] [PubMed] [Google Scholar]
- 58.Saez-Rodriguez J, Simeoni L, Lindquist JA, et al. A logical model provides insights into T cell receptor signaling. PLOS Comput Biol. 2007;3:e163. doi: 10.1371/journal.pcbi.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oyeyemi OJ, Davies O, Robertson DL, et al. A logical model of HIV-1 interactions with the T-cell activation signalling pathway. Bioinformatics. 2015;31:1075–1083. doi: 10.1093/bioinformatics/btu787. [DOI] [PubMed] [Google Scholar]
- 60.Kaufman M, Andris F, Leo O. A logical analysis of T cell activation and anergy. Proc Natl Acad Sci USA. 1999;96:3894–3899. doi: 10.1073/pnas.96.7.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miskov-Zivanov N, Turner MS, Kane LP, et al. The duration of T cell stimulation is a critical determinant of cell fate and plasticity. Sci Signal. 2013;6:ra97. doi: 10.1126/scisignal.2004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang R, Shah MV, Yang J, et al. Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci USA. 2008;105:16308–16313. doi: 10.1073/pnas.0806447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kemp ML, Wille L, Lewis CL, et al. Quantitative network signal combinations downstream of TCR activation can predict IL-2 production response. J Immunol. 2007;178:4984–4992. doi: 10.4049/jimmunol.178.8.4984. [DOI] [PubMed] [Google Scholar]
- 64.Mingueneau M, Krishnaswamy S, Spitzer MH, et al. Single-cell mass cytometry of TCR signaling: Amplification of small initial differences results in low ERK activation in NOD mice. Proc Natl Acad Sci USA. 2014;111:16466–16471. doi: 10.1073/pnas.1419337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krishnaswamy S, Spitzer MH, Mingueneau M, et al. Conditional density-based analysis of T cell signaling in single-cell data . Science. 2014;346:346. doi: 10.1126/science.1250689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S, Jensen H, Stewart W, et al. In silico modeling identifies CD45 as a regulator of IL-2 synergy in the NKG2D-mediated activation of immature human NK cells. Sci Signal. 2017;10:eaai9062. doi: 10.1126/scisignal.aai9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valitutti S, Coombs D, Dupré L. The space and time frames of T cell activation at the immunological synapse. FEBS Lett. 2010;584:4851–4857. doi: 10.1016/j.febslet.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Cai H, Muller J, Depoil D, et al. Full control of ligand positioning reveals spatial thresholds for T cell receptor triggering. Nat Nanotechnol. 2018;13:610–617. doi: 10.1038/s41565-018-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jankowska KI, Williamson EK, Roy NH, et al. Integrins modulate T cell receptor signaling by constraining actin flow at the immunological synapse. Front Immunol. 2018;9:25. doi: 10.3389/fimmu.2018.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dutta D, Barr VA, Akpan I, et al. Recruitment of calcineurin to the TCR positively regulates T cell activation. Nat Immunol. 2017;18:196–204. doi: 10.1038/ni.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Germain RN. Computational analysis of T cell receptor signaling and ligand discrimination--past, present, and future. FEBS Lett. 2010;584:4814–4822. doi: 10.1016/j.febslet.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fesnak AD, June CH, Levine BL. Engineered T cells: The promise and challenges of cancer immunotherapy. Nat Rev Cancer. 2016;16:566–581. doi: 10.1038/nrc.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flemming A. Autoimmune diseases: CAR-T cells take aim at autoimmunity. Nat Rev Drug Discov. 2016;15:603. doi: 10.1038/nrd.2016.180. [DOI] [PubMed] [Google Scholar]
- 74.Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: Current status and future directions. Protein Cell. 2018;9:254–266. doi: 10.1007/s13238-016-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadelain M, Brentjens R, Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]