Ammonia-oxidizing archaea (AOA) are the most abundant chemolithoautotrophic microorganisms in the oxygenated water column of the global ocean. Although H2O2 appears to be a universal by-product of aerobic metabolism, genes encoding the hydrogen peroxide (H2O2)-detoxifying enzyme catalase are largely absent in genomes of marine AOA. Here, we provide evidence that closely related marine AOA have different degrees of sensitivity to H2O2, which may contribute to niche differentiation between these organisms. Furthermore, our results suggest that marine AOA rely on H2O2 detoxification during periods of high metabolic activity and release organic compounds, thereby potentially attracting heterotrophic prokaryotes that provide this missing function. In summary, this report provides insights into the metabolic interactions between AOA and heterotrophic bacteria in marine environments and suggests that AOA play an important role in the biogeochemical carbon cycle by making organic carbon available for heterotrophic microorganisms.
KEYWORDS: Nitrosopumilus, ammonia-oxidizing archaea, hydrogen peroxide, metabolic interactions, oxidative stress, proteomics
ABSTRACT
Ammonia-oxidizing archaea (AOA) play an important role in the nitrogen cycle and account for a considerable fraction of the prokaryotic plankton in the ocean. Most AOA lack the hydrogen peroxide (H2O2)-detoxifying enzyme catalase, and some AOA have been shown to grow poorly under conditions of exposure to H2O2. However, differences in the degrees of H2O2 sensitivity of different AOA strains, the physiological status of AOA cells exposed to H2O2, and their molecular response to H2O2 remain poorly characterized. Further, AOA might rely on heterotrophic bacteria to detoxify H2O2, and yet the extent and variety of costs and benefits involved in these interactions remain unclear. Here, we used a proteomics approach to compare the protein profiles of three Nitrosopumilus strains grown in the presence and absence of catalase and in coculture with the heterotrophic alphaproteobacterium Oceanicaulis alexandrii. We observed that most proteins detected at a higher relative abundance in H2O2-exposed Nitrosopumilus cells had no known function in oxidative stress defense. Instead, these proteins were putatively involved in the remodeling of the extracellular matrix, which we hypothesize to be a strategy limiting the influx of H2O2 into the cells. Using RNA-stable isotope probing, we confirmed that O. alexandrii cells growing in coculture with the Nitrosopumilus strains assimilated Nitrosopumilus-derived organic carbon, suggesting that AOA could recruit H2O2-detoxifying bacteria through the release of labile organic matter. Our results contribute new insights into the response of AOA to H2O2 and highlight the potential ecological importance of their interactions with heterotrophic free-living bacteria in marine environments.
IMPORTANCE Ammonia-oxidizing archaea (AOA) are the most abundant chemolithoautotrophic microorganisms in the oxygenated water column of the global ocean. Although H2O2 appears to be a universal by-product of aerobic metabolism, genes encoding the hydrogen peroxide (H2O2)-detoxifying enzyme catalase are largely absent in genomes of marine AOA. Here, we provide evidence that closely related marine AOA have different degrees of sensitivity to H2O2, which may contribute to niche differentiation between these organisms. Furthermore, our results suggest that marine AOA rely on H2O2 detoxification during periods of high metabolic activity and release organic compounds, thereby potentially attracting heterotrophic prokaryotes that provide this missing function. In summary, this report provides insights into the metabolic interactions between AOA and heterotrophic bacteria in marine environments and suggests that AOA play an important role in the biogeochemical carbon cycle by making organic carbon available for heterotrophic microorganisms.
INTRODUCTION
Ammonia-oxidizing archaea (AOA) are a major component of marine microbial communities and represent the dominant ammonia oxidizers in the ocean, carrying out the first and rate-limiting step of nitrification (1–3). AOA are members of the phylum Thaumarchaeota (4, 5) and are particularly abundant in the mesopelagic zone of the open ocean (6) and in oxygen minimum zones (7).
Although more than a dozen strains of autotrophic AOA have been enriched from marine waters and sediments (8–14), their isolation and maintenance on a mineral medium have repeatedly proven difficult. To some extent, this difficulty may be attributed to the dependence of some AOA on the presence of alpha-ketoacids or, alternatively, that of cocultivated heterotrophic bacteria to achieve exponential growth in batch cultures (9, 15, 16). Recently, this dependency has been linked to their sensitivity to hydrogen peroxide (H2O2), which is detoxified by alpha-ketoacids and/or cocultivated heterotrophs (14). The sensitivity of AOA to H2O2 is somewhat surprising as H2O2 appears to be a universal by-product of aerobic metabolism and the vast majority of aerobic organisms encode H2O2-scavenging enzymes, including catalases and peroxidases (17, 18). The H2O2 sensitivity reported for some AOA strains isolated from marine environments correlates with the lack of genes encoding canonical catalase homologs in their genomes (14, 19).
In the ocean, H2O2 is mainly produced by the photooxidation of chromophoric dissolved organic matter (20) but is also introduced via precipitation (21) and metabolic processes (22, 23). The lack of a recognizable form of H2O2 detoxification machinery in AOA was hypothesized to result from their limited exposure to this oxidant under oligotrophic conditions (14). However, in marine surface waters, AOA might be chronically exposed to H2O2, where its concentrations can reach up to 500 nM (24). While the ammonia oxidization activity of the marine archaeon Nitrosopumilus strain DDS1 was completely inhibited after production of ∼200 nM H2O2 (14), Nitrosopumilus maritimus SCM1 showed no decrease in ammonia oxidation after additions of H2O2 (up to 1 μmol liter−1) to the culture medium (25). Differences in H2O2 sensitivity have also been reported for environmental AOA populations and were suggested previously to be defining features of distinct AOA ecotypes (19). These observations indicate that the degrees of H2O2 tolerance differ across the vast diversity of AOA species, and yet exploration of the entire spectrum of their phenotypic response to H2O2 has just started. Furthermore, it is unclear whether the inability to achieve exponential growth in the absence of an external H2O2 scavenger (i.e., alpha-ketoacids [14–16]) represents a consequence or the absence of a molecular response to H2O2.
In this study, we compared the growth levels of three strains of marine AOA in the presence and absence of commercial catalase or under conditions of growth in coculture with the heterotrophic alphaproteobacterium Oceanicaulis alexandrii and evaluated the concurrent levels of production and eventual scavenging of H2O2 in the culture medium. The investigated AOA comprised all marine axenic cultures with closed genomes which are currently available, including the first reported AOA isolate, Nitrosopumilus maritimus SCM1 (26), as well as two isolates from coastal surface waters of the Northern Adriatic Sea, Nitrosopumilus adriaticus NF5 and Nitrosopumilus piranensis D3C (27). O. alexandrii was the most persistent contaminant prior to obtaining axenic cultures of N. adriaticus NF5 and N. piranensis D3C (9). Furthermore, the molecular response of the investigated strains to H2O2 was assessed by comparing the proteomes of cells growing in the presence or in the absence of catalase and/or the alphaproteobacterium O. alexandrii. Additionally, metabolic interactions of the three Nitrosopumilus strains with O. alexandrii were explored using stable isotope probing and comparative proteome analysis.
Collectively, the results of this study provide insights into the molecular and physiological responses of AOA to H2O2 and highlight the potential ecological importance of their interactions with heterotrophic free-living bacteria in marine environments.
RESULTS AND DISCUSSION
The effect of H2O2 on the growth and ammonia oxidation activity of three Nitrosopumilus strains.
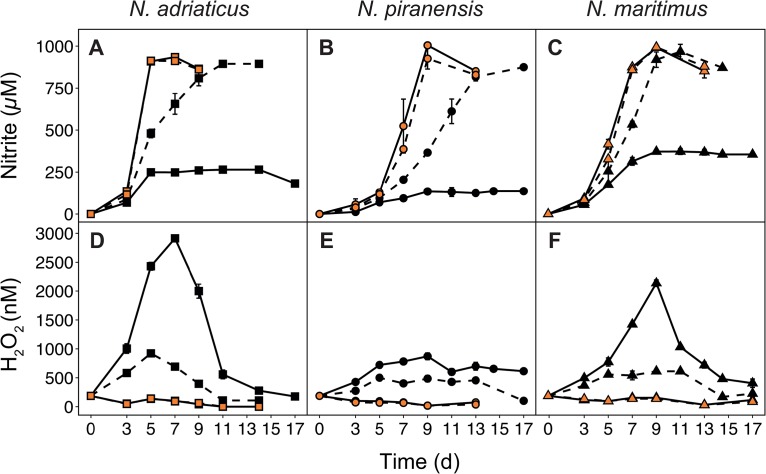
Ammonia oxidation activity and growth of three Nitrosopumilus strains (N. adriaticus NF5, N. piranensis D3C, and N. maritimus SCM1) were assessed both in the presence and absence of commercial catalase and under conditions of growth in coculture with the heterotrophic alphaproteobacterium Oceanicaulis alexandrii. When catalase was added to the cultures, all three Nitrosopumilus strains depleted 1 mM ammonium within 5 to 8 days of incubation (Fig. 1A to C), yielding nearly stoichiometric amounts of nitrite as previously described for these strains (9, 25). In cocultures with O. alexandrii, however, nitrite production was consistently slower than in axenic cultures supplemented with catalase. In cocultures, the complete conversion of ammonium to nitrite took 10 to 15 days, possibly due to the proportionally small O. alexandrii cell population size and consequently low H2O2-detoxifying capacity of O. alexandrii relative to purified catalase. Adding both catalase and O. alexandrii did not result in an increased rate of nitrite production relative to those observed for axenic AOA cultures grown in the presence of catalase.
FIG 1.
Nitrite production (A, B, and C) and hydrogen peroxide (H2O2) concentrations (D, E, and F) of three Nitrosopumilus strains grown in pure culture (solid line) or in coculture with O. alexandrii (dashed line). Orange shapes indicate the addition of catalase. Error bars represent standard deviations of measurements from triplicate cultures. d, days.
In the absence of catalase and O. alexandrii, all three Nitrosopumilus strains depleted the supplied ammonium only partially and nitrite concentrations remained constant at 150 to 300 μM after 5 to 9 days of incubation (Fig. 1A to C). The stalled production of nitrite in axenic cultures devoid of catalase correlated with a growth arrest of the cultures. Maximum cell abundances ranged between 4 × 106 and 1 × 107 cells ml−1 after 3 to 6 days of incubation, which was on average 5 to 10 times lower than in cultures containing catalase (see Table S1 in the supplemental material). Growth arrest was observed 2 to 3 days earlier than the arrest of nitrite production in all three strains, indicating that the remaining level of ammonia oxidation was insufficient to meet the energy demands of dividing cells during H2O2 exposure (Fig. 1A to C; see also Table S1).
Cell counts of Nitrospumilus and Oceanicaulis alexandrii cultures grown in different treatments (CAT, catalase; coculture; without addition of catalase or O. alexandrii). Cell counts were derived from one of three biological replicates per strain and treatment. Download Table S1, DOCX file, 0.02 MB (18.4KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the medium of cultures lacking both catalase and O. alexandrii, H2O2 concentrations reached 800 nM to 3 μM after 7 to 9 days of incubation (Fig. 1D to F), which was far above the background levels of H2O2 (∼150 nM) in the culture medium (see Fig. S1A in the supplemental material). Concentrations of H2O2 remained fairly stable in the abiotic controls (Fig. S1A), indicating that the investigated Nitrosopumilus strains produced H2O2 as a result of their metabolic activity. Although the fluorescence-based assay used in this study (see Materials and Methods) could potentially detect oxidizing agents other than H2O2, complete loss of the signal after catalase addition suggests that H2O2 was the primary oxidant measured. Importantly, the presence of the organic buffer HEPES in the culture medium did not appear to represent a dominant source of H2O2, in contrast to previous reports on phytoplankton cultures (28) (Fig. S1A). Indeed, the HEPES buffer may release H2O2 only under conditions of exposure to light whereas the three Nitrosopumilus strains were grown in the dark. After reaching their peak, H2O2 concentrations also declined in axenic cultures devoid of catalase (Fig. 1D to F). This decline in the H2O2 concentration was also observed in the abiotic controls, where H2O2 was added at ∼3.5 μM (Fig. S1B), suggesting that H2O2 was not actively scavenged by the investigated strains. In all cultures containing the purified catalase, H2O2 concentrations never increased above background levels, whereas in cocultures, H2O2 concentrations sporadically reached levels as high as 900 nM in N. adriaticus-O. alexandrii cocultures (Fig. 1D).
(A) Hydrogen peroxide (H2O2) concentrations of SCM medium controls with or without addition of catalase (5 U ml−1) incubated in the dark at 30°C. (B) Abiotic reduction of H2O2 in SCM medium over time. Error bars represent the range of the mean from measurements of duplicate cultures (sometimes too small to be visible). Download FIG S1, PDF file, 0.2 MB (229.7KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparing the three strains, complete inhibition occurred at lower H2O2 concentrations in N. piranensis (∼800 nM; Fig. 1E) than in N. adriaticus and N. maritimus (2.5 μM and 2 μM H2O2, respectively; Fig. 1D and F). Although N. maritimus produced more nitrite than N. adriaticus prior to inhibition (∼350 μM nitrite versus ∼250 μM, respectively; Fig. 1A and C), N. adriaticus exhibited a higher cell-specific net level of H2O2 production (Fig. 1; see also Table S1), suggesting that the molecular machinery responsible for H2O2 production in distinct AOA strains could have different H2O2 production yields. In contrast to N. piranensis and N. maritimus, which encode two putative superoxide dismutases, N. adriaticus encodes three superoxide dismutases, potentially explaining the higher observed level of H2O2 production by this strain.
The three strains investigated in this study appeared to tolerate higher concentrations of H2O2 than Nitrosopumilus strain DDS1, which was reported to be completely inhibited at ∼200 nM H2O2 (14). While strain DDS1 was isolated from a water depth of 200 m, N. adriaticus NF5 and N. piranensis D3C were isolated from coastal surface waters where H2O2 concentrations are typically ∼10 to 100 times higher than in deeper waters (29, 30). Both strains putatively encode cyclobutane pyrimidine dimer (CPD) photolyase (9), an enzyme activated by UV-A radiation to repair DNA damage (31), suggesting that they might be more tolerant of conditions typically found in surface waters (i.e., higher H2O2 concentrations) than strains isolated from deeper water layers. N. maritimus was previously observed to be insensitive to additions of H2O2 and was still able to oxidize 1 mM ammonia when H2O2 was added to the culture medium at 5 μmol liter−1 (25), which contrasts with the complete inhibition of N. maritimus at 2 μM H2O2 that we report here (Fig. 1F). Considering the different experimental setup and the lack of H2O2 concentration measurements in the incubations cited above (26), the explanation of the different results that we report here remains currently unclear.
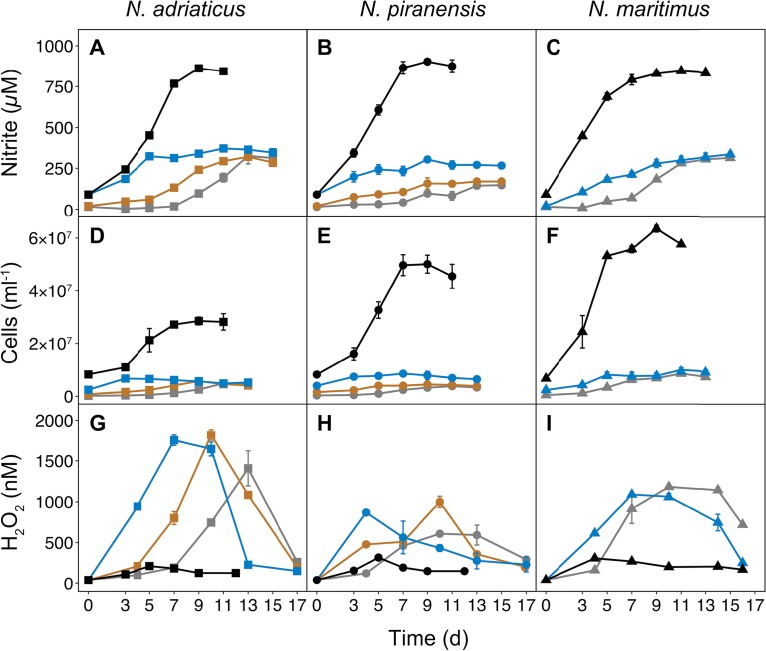
All three Nitrosopumilus strains could overcome H2O2-induced growth arrest when initial cell abundances were higher than or equal to 7 × 106 ml−1, and growth of these cultures was similar to those containing catalase (Fig. 2A to F). While H2O2 concentrations appeared to increase linearly with nitrite production in cultures with low initial cell abundances (∼2 × 105, ∼8 × 105, and ∼3 × 106 ml−1; Fig. 2A, C, and G to I), the detected H2O2 concentrations were much lower (<300 nM) in cultures with high initial cell abundances (7 × 106 ml−1; Fig. 2G to I), indicating either that less H2O2 was produced or that it was scavenged by an unknown mechanism. Similar cell abundance-dependent H2O2 sensitivity patterns have been described in axenic Prochlorococcus cultures, which grew well in concentrated but not in dilute cultures (32, 33). In the absence of H2O2 scavengers, growth and ammonia oxidation activity did not show a linear response to the size of the inoculum but rather were induced at a certain cell abundance level in all three Nitrosopumilus strains (Fig. 2A to F). Even though knowledge of the exact mechanism of this phenomenon remains elusive, cell abundance-dependent cellular responses are commonly induced by quorum sensing.
FIG 2.
Nitrite concentrations (A, B, and C), cell abundances (D, E, and F), and hydrogen peroxide (H2O2) concentrations (G, H, and I) of three Nitrosopumilus strains grown at various initial cell abundances, which are represented by different colors: ∼2 × 105 ml−1 (gray lines), ∼8 × 105 ml−1 (brown lines), ∼3 × 106 ml−1 (blue lines), ∼7 × 106 ml−1 (black lines). Error bars represent the range of the mean from measurements of duplicate cultures.
While the culture conditions in this study did not reflect the oligotrophic conditions typically found in the ocean, note that all three Nitrosopumilus strains also produced large amounts of H2O2 at environmentally relevant cell abundances of ∼2 × 105 ml−1 (Fig. 2D to F). Thus, in their natural environment, marine AOA might potentially rely on H2O2 detoxification by other microorganisms during periods of high activity and/or in nutrient-rich microniches.
The proteomic response of three Nitrosopumilus strains to H2O2.
Comparative proteomics was used to investigate the molecular response of the three Nitrosopumilus strains to H2O2 exposure relative to cultures grown in the presence of commercial catalase or in coculture with the heterotrophic bacterium O. alexandrii. Additionally, we distinguished between axenic cultures that were completely inhibited in their growth (H2O2 inhibited) and axenic cultures that were grown at a high initial cell abundance (7 × 106 ml−1) and were able to grow despite the absence of an H2O2 scavenger (H2O2 noninhibited).
A total of 1,020 to 1,372 proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and accounted for ∼55% to 68% of the predicted coding DNA sequences in the genomes of N. adriaticus NF5, N. piranensis D3C, and N. maritimus SCM1 (Table S2A). Between the four treatments (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited), 1,040, 1,027, and 856 proteins were shared by N. adriaticus, N. piranensis, and N. maritimus, respectively (Table S2A). While the relative abundances of the majority of proteins remained constant between the different treatments, 56 to 109 proteins significantly changed in their relative abundances (increased or decreased; adjusted P value, <0.05) in the absence of catalase or O. alexandrii compared to the results seen with cultures grown in the presence of either catalase or O. alexandrii (Table S2B). Of these, 33 proteins were shared among all three strains. Comparing the proteome composition of cultures grown in the presence of catalase to that of cocultures grown with O. alexandrii, 12 to 20 proteins changed in their relative abundances, 5 of which were shared by all three strains (Table S2B). However, these 5 shared proteins also changed in relative abundance when Nitrosopumilus cells were exposed to H2O2, suggesting that they did not represent a specific response to the presence of the heterotrophic bacterium itself. Instead, they might have been induced as a result of the lower capacity of O. alexandrii to detoxify H2O2 than purified catalase (Fig. 1D to F).
(A) Proteins shared between proteomes of three Nitrosopumilus strains grown under conditions of different treatments (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited). (B) Significant changes in relative protein abundances of three Nitrosopumilus strains based on pairwise comparisons between different treatments. Each treatment included proteins shared by three biological replicates. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Furthermore, despite the reduced sensitivity to H2O2 in Nitrosopumilus cultures with high initial cell abundances (H2O2 noninhibited) (Fig. 2), we did not detect any changes in the proteome composition shared by all three strains relative to cultures that were completely inhibited by H2O2 (Table S2B). This suggests that the proteome composition of cells exposed to H2O2 does not directly indicate whether the cells are active, indicating the existence of further regulatory mechanisms (i.e., noncoding RNAs or posttranslational protein modifications) determining growth. Previous studies have reported that most of the abundant transcripts are relatively invariant across growth phases and environmental conditions in AOA (12, 34).
Several strain-specific changes in the proteome composition were identified across the four treatments in each of the three strains (Fig. S2A to C; see also Text S1 in the supplemental material); however, we focus in the following sections on the proteomic features shared by all three strains.
In-depth descriptions of protocols used for protein extraction, LC-MS/MS analysis, and peptide identification, RNA extraction, quantitative PCR, flow cytometry, and CARD-FISH. Discussion of additional information on extracellular matrix-associated proteins, strain-specific proteomic responses of the individual Nitrosopumilus strains to H2O2, and potential H2O2 production sites in AOA. Download Text S1, DOCX file, 0.07 MB (74.5KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map of the proteins that showed a significant change in relative abundance in three Nitrosopumilus strains under different culture conditions (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited). Columns and rows were clustered based on Euclidean distances corresponding to differences between treatments and relative protein abundances, respectively. (A) Nitrosopumilus adriaticus NF5. (B) Nitrosopumilus piranensis D3C. (C) Nitrosopumilus maritimus SCM1. Download FIG S2, PDF file, 0.8 MB (811KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(i) Thaumarchaeal homologs of proteins involved in canonical oxidative stress defense. Microbial cells use various intracellular scavenging and repair mechanisms to limit and repair damage caused by reactive oxygen species (ROS). In bacteria, the basal scavenging system for O2- is superoxide dismutase, whereas H2O2 is scavenged by catalase and peroxidases (18, 35). Genetic responses to H2O2 stress in Gram-negative and Gram-positive bacteria are controlled by the transcriptional regulons OxyR and PerR, respectively (36–38), which regulate the expression of catalase, alkyl hydroperoxide reductase (Ahp), and proteins involved in disulfide reduction, heme synthesis, iron scavenging (ferritin and related proteins), and iron import control, as well as proteins involved in divalent cation import (mostly manganese) (see reference 39 and references therein). ROS scavenging systems in archaea, although much less extensively studied, have been shown to be similar to those in bacteria (40, 41).
Although the genomes of all cultured marine AOA do not encode canonical catalase homologs, genes encoding putative catalases have recently been reported from two single-cell genomes obtained from AOA cells sampled from Antarctic surface waters (42). These genes were predicted to be horizontally acquired genes (42). We reanalyzed these single-cell genomes and observed that catalase-encoding genes are part of short contigs that harbor only genes of presumed bacterial origin (due to their similarity to known bacterial homologs), suggesting that they represent contaminating genome fragments that originated from bacterial genomes (see Data Set S2A in the supplemental material). Although there is tangible evidence of the presence of catalase-encoding genes in AOA isolated from terrestrial environments (43, 44) and from a wastewater treatment plant (45), thus far, there is no evidence suggesting that marine AOA encode catalases.
In spite of the notable absence of catalase homologs, we identified homologs of known ROS-scavenging enzymes and, more generally, of proteins involved in canonical pathways of oxidative stress defense in the genomes and proteomes of the three Nitrosopumilus strains that we investigated (Data Set S1D). Surprisingly, however, the relative abundances of most proteins assumed to play a role in oxidative stress defense did not change in response to H2O2 exposure. These also included the five putative Ahp proteins, which raises the issue of whether these putative candidates are functional H2O2-detoxifying enzymes as previously contested (14) or are generally not regulated on the gene expression level in members of the Nitrosopumilus genus. Alternatively, intracellular H2O2 concentrations might not have been high enough to induce a response.
(A) Orthologous groups constructed from the genomes of Nitrosopumilus adriaticus NF5, Nitrosopumilus piranensis D3C, and Nitrosopumilus maritimus SCM1. (B) Gene annotations of orthologous groups. (C) KEGG assignments of the H2O2 responsive proteins shared by all three strains. (D) Relative protein abundances of the proteins related to canonical oxidative stress defense mechanisms in all three Nitrosopumilus strains for each treatment. Download Data Set S1, XLSX file, 0.2 MB (157.4KB, xlsx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
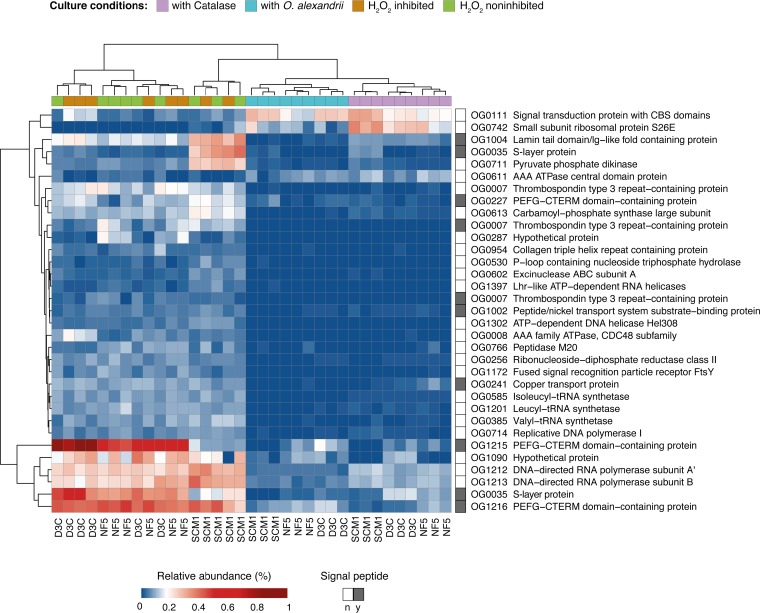
Nevertheless, more than one-third (36%) of the proteins that showed a significant response to H2O2 exposure were assigned to KEGG orthologous groups (OGs) associated with genetic information processing and nucleotide metabolism (Fig. 3; see also Data Set S1C). These included DNA polymerase I and ATP-dependent helicase, which are key enzymes of DNA replication, as well as two subunits of DNA-directed RNA polymerase, which is involved in RNA synthesis (Fig. 3). Additionally, ribonucleotide reductase, which converts ribonucleotides into deoxyribonucleotides, was identified at high relative abundance. While ribonucleotide reductase expression is necessary for DNA replication, it has also been shown to be induced in response to DNA damage and replication blocks (46, 47). Furthermore, proteins putatively involved in the repair of DNA damage, including excinuclease ABC subunit A (UvrA) and DNA helicase Hel308, were identified at high relative abundance during H2O2 exposure (Fig. 3). The latter is suggested to play a role in the repair and start of replication forks (48), whereas UvrA is part of the nucleotide excision repair mechanism (49). Nucleotide excision repair initiation involves a two-step mechanism in which UvrA initially scans the genome and locates DNA damage prior to recruiting UvrB and UvrC, which are needed for DNA damage verification, excision, and, ultimately, repair (49, 50). However, we did not detect an increase in the relative abundance of UvrBC, suggesting that H2O2 concentrations in Nitrosopumilus cells might not have been high enough to induce extensive DNA damage.
FIG 3.
Heat map of proteins that showed a significant change in relative abundance in three Nitrosopumilus strains (N. adriaticus NF5, N. piranensis D3C, and N. maritimus SCM1) comparing different culture conditions (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited). Columns and rows were clustered based on Euclidean distances corresponding to differences between treatments and relative protein abundances, respectively. The presence of a signal peptide is indicated in dark gray.
The concentration of H2O2 inside cells is dependent on the rate of its endogenous formation and influx, balanced against the rate of H2O2 scavenging and efflux (51). A few studies have suggested previously that extent of passage of H2O2 through biological membranes is limited (51–53). In Escherichia coli, endogenous H2O2 is rapidly scavenged by Ahp (and to only a lower extent by catalase) to levels below 20 nM and does not persist long enough to penetrate the membrane, with less than 10% escaping the cell (51, 54). Hence, there is typically no measurable accumulation of H2O2 in the culture medium (54), in contrast to the range of H2O2 concentrations of ∼1 to 2.5 μM measured in the Nitrosopumilus culture medium in this study. Additionally, putative Ahp proteins have been identified in the proteomes of all three Nitrosopumilus strains at similar abundance levels in all treatments as mentioned above. On the basis of these observations, we hypothesize that production of H2O2 in AOA possibly takes place at the outer side of the membrane (e.g., in the pseudoperiplasmic space).
(ii) Increase of relative abundances of membrane-associated proteins in response to H2O2. Among the proteins that showed a significant change in relative abundance in AOA cultures exposed to H2O2 (H2O2 inhibited and noninhibited treatments), ∼30% contained a signal peptide indicating secretion and/or localization on the outer side of the cytoplasmic membrane (Fig. 3). Concomitantly, tRNA synthetases for branched-chain amino acids, essential components of membrane spanning helices and thus of membrane bound/surface proteins (55), were identified at higher relative abundances (Fig. 3). Moreover, the high level of representation of the signal recognition particle (SRP) receptor protein FtsY, which likely mediates the delivery of SRP-nascent chain complexes to the cell membrane (56), suggests an increased rate of transport of proteins to the membrane.
Both putative membrane-bound S-layer proteins were among the 10 to 50 most abundant proteins detected in axenic cultures devoid of catalase (Fig. 3). The putative S-layer proteins were on average 4 to 30 times more abundant in the proteomes of axenic cultures grown in the absence of catalase than in those of cultures grown in the presence of catalase or in coculture with O. alexandrii, potentially indicating an increased renewal or restructuring of the S-layer coat under conditions of H2O2 exposure. In addition, three PEFG-CTERM domain-containing proteins were detected at 4 to 10 times higher relative abundance in axenic cultures without addition of catalase than cultures grown with catalase or in coculture with O. alexandrii. The PEFG-CTERM motif resembles the PEP-CTERM domain typically present in glycoproteins which are transported and anchored into the plasma membrane, such as S-layer proteins in Haloferax volcanii (57). Furthermore, two thrombospondin type 3-like repeat (TT3R)-containing proteins were on average 15 to 40 times more abundant in cultures grown under conditions of H2O2 exposure than in cultures containing catalase or O. alexandrii (Fig. 3). Proteins containing TT3R motifs in AOA show sequence similarity to hypothetical proteins of the myxobacterial thrombospondin-like gene cluster, which has been suggested to play a role in the construction of the cell surface matrix (58). However, besides the well-known calcium-binding capacities of TT3R motifs, knowledge of their exact functions in prokaryotes remains elusive (59). Nevertheless, we detected putative structural proteins (OG1004 and OG0954) which share homology with proteins known to interact with thrombospondins in eukaryotes at high relative abundance levels in all three Nitrosopumilus strains under conditions of exposure to H2O2 (Fig. 3) (further discussed in Text S1 in the supplemental material).
The increase in the relative abundance of putative components of the extracellular matrix of AOA is reminiscent of the protective barrier formed by the eukaryote Saccharomyces cerevisiae to limit the influx of H2O2 into its cells (60). Furthermore, cell aggregation, exopolysaccharide (EPS) production, and, ultimately, biofilm formation represent common physiological responses of bacteria exposed to H2O2 and may promote survival (61, 62). AOA species, including members of the Nitrosopumilus genus, contain the genomic repertoire for exopolysaccharide production and cell surface modifications (63). Formation of some small aggregates was observed in H2O2-exposed cultures (Fig. S3A and B), indicating that members of the Nitrosopumilus genus potentially remodel their membrane and/or extracellular matrix, including cell-to-cell attachment properties, in response to H2O2. However, the response of secreted/membrane-bound proteins observed for the three AOA strains investigated here could also represent a result of the renewal of damaged proteins in close proximity to the H2O2 production site (further discussed in Text S1 in the supplemental material).
(A and B) Cell aggregation of Nitrosopumilus adriaticus NF5 observed during growth in medium without hydrogen peroxide (H2O2) scavengers. (C and D) CARD-FISH images of Nitrosopumilus adriaticus NF5 (DAPI [4′,6-diamidino-2-phenylindole]; blue) grown in coculture with Oceanicaulis alexandrii (bacterial EUBI probe; green). Download FIG S3, PDF file, 2.7 MB (2.7MB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Furthermore, a putative membrane-bound copper transport protein was detected at significantly higher relative abundance in the proteomes of the three Nitrosopumilus strains grown under conditions of exposure to H2O2 than in the proteomes derived from cultures containing catalase or O. alexandrii (Fig. 3). The N-terminal side of this protein exhibits homology with CopC family proteins, which are periplasmic copper binding proteins suggested to primarily play a role in bacterial copper homeostasis (64). E. coli cells containing excess copper were shown to be less sensitive to H2O2-induced DNA damage (65). Furthermore, reactions of Cu(I) and Cu(II) with H2O2 have recently been suggested to be involved in the formation of Cu(III) and O2−, respectively, instead of OH· (66). Hence, while classical Fenton reactions might induce oxidative damage and inactivation of iron-containing enzymes (67), copper import could represent a strategy to reduce or even prevent damage to macromolecules induced by OH·. Nevertheless, the function of this protein in AOA remains to be confirmed and requires further investigations.
Metabolic interactions between Nitrosopumilus and Oceanicaulis alexandrii.
The peak concentrations of H2O2 in cocultures of the heterotrophic alphaproteobacterium Oceanicaulis alexandrii with Nitrosopumilus strains were on average 2 to 3 times lower than those measured in axenic Nitrosopumilus cultures (Fig. 1D to F), suggesting that O. alexandrii is capable of reducing the H2O2 concentration in AOA cultures. The initial description of O. alexandrii noted that strains of this bacterium are catalase positive (68), and, accordingly, the genome of the type strain O. alexandrii DSM 11625T encodes two homologs of heme-containing catalase peroxidases belonging to the class I catalases (69). Amino acid residues forming the catalytic site of biochemically characterized heme-containing catalases are conserved in catalase homologs of O. alexandrii (Data Set S2B), suggesting that these homologs are bona fide catalases. One of these two homologs is characterized by an N-terminal signal peptide (Data Set S2B), suggesting that it is addressed to the periplasm and might be further secreted into the culture medium. Additionally, we detected proteotypic peptides of these catalase homologs in protein extracts prepared from cocultures of O. alexandrii and Nitrosopumilus spp., further suggesting that reduced concentrations of H2O2 in cocultures of O. alexandrii and AOA could result from O. alexandrii catalase activity.
(A) Catalase-containing contigs of thaumarchaeal single-cell genomes AB-661-L21 and AB-661-M19 from Luo et al. (42) (catalases are marked in yellow). (B) Multiple-sequence alignment of class I catalase peroxidases, including sequences from Oceanicaulis alexandrii (blue) and sequences with known three-dimensional (3D) structures obtained from PeroxiBase. Color scheme: black = 100% sequence similarity; gray = 90% to 99% sequence similarity. Signal peptides are indicated in red. Essential residues involved in catalysis are labeled with an asterisk. Download Data Set S2, DOCX file, 0.2 MB (236.9KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
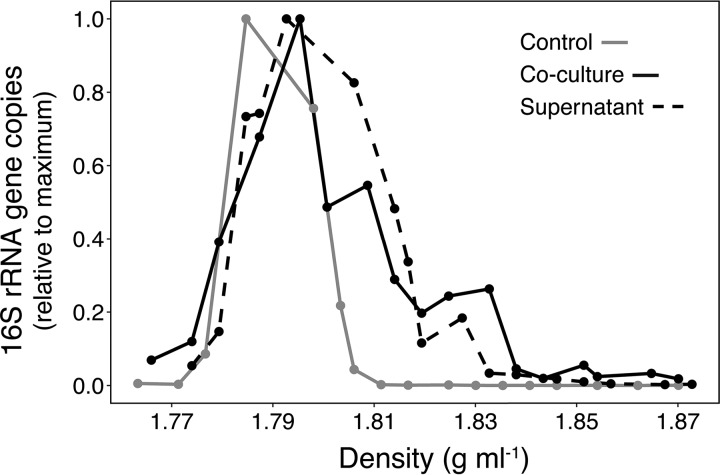
O. alexandrii was able to grow in coculture with all three Nitrosopumilus strains, as well as in the supernatant of Nitrosopumilus cultures (Fig. S4A; see also Table S1). RNA stable isotope probing (RNA-SIP) was used to directly confirm the transfer of organic carbon from autotrophic Nitrosopumilus cells to O. alexandrii. After two consecutive passages in medium containing 13C-labeled bicarbonate, 16S rRNA sequences of N. piranensis showed a clear enrichment in the “heavy” (13C) fraction (approximately 25% of the RNA was labeled) relative to control incubations (Fig. S5). When O. alexandrii was subsequently grown in coculture with N. piranensis cells or in N. piranensis culture supernatant, 16S rRNA sequences of O. alexandrii showed an enrichment in the “heavy” (13C) fraction (approximately 17% in coculture and 8% on supernatant) compared to the control incubations (Fig. 4). Furthermore, no incorporation of 13C directly from bicarbonate via anaplerotic reactions was observed in the control treatment (i.e., O. alexandrii cells growing in yeast extract-peptone medium supplemented with 13C-labeled bicarbonate) (Fig. 4).
FIG 4.
Proportion of O. alexandrii 16S rRNA gene copies recovered from RNA-SIP gradient fractions. O. alexandrii was grown in coculture with N. piranensis in medium containing 13C-labeled bicarbonate (solid black line) and on N. piranensis supernatant (dashed black line). Growth in medium containing unlabeled yeast extract-peptone and 13C-labeled bicarbonate served as a control (solid gray line).
(A) Growth curves of Oceanicaulis alexandrii and Nitrosopumilus adriaticus NF5 in coculture with addition of catalase. (B) Growth of O. alexandrii on culture supernatant of three Nitrosopumilus strains (N. adriaticus NF5, N. piranensis D3C, and N. maritimus SCM1) and in medium without the addition of any carbon source (Control). Error bars represent standard deviations of measurements from triplicate cultures. Download FIG S4, PDF file, 0.3 MB (259.2KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proportion of N. piranensis D3C 16S rRNA gene copies recovered from RNA-SIP gradient fractions under conditions of growth in medium containing unlabeled bicarbonate (gray line) and after two passages into medium containing 13C-labeled bicarbonate (black line). Download FIG S5, PDF file, 0.2 MB (206.7KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While the 13C enrichment clearly confirms the incorporation of Nitrosopumilus-derived organic carbon into the biomass of O. alexandrii, these results do not clarify the identity of organic carbon compounds that mediate the metabolic interaction between AOA and O. alexandrii. The alphaproteobacterium generally showed the highest growth rates during exponential growth of the AOA strains (Table S1; see also Fig. S4B), indicating growth on compounds released by active Nitrosopumilus cells rather than subsistence on dead cell material. We showed in another study that all investigated strains indeed released organic matter, including labile compounds such as amino acids and thymidine (70). However, the possibility of proteolytic growth on Nitrosopumilus cells cannot be completely excluded. Nevertheless, growth on released, soluble substances is also supported by the ability of O. alexandrii to grow on Nitrosopumilus culture supernatant, suggesting a rather unspecific interaction. And yet, O. alexandrii cells often appeared to be attached to Nitrosopumilus cells during growth in coculture (Fig. S3C and D). The highest growth yields of O. alexandrii were observed in Nitrosopumilus cocultures with added catalase (Table S1), suggesting either a higher level of release of organic matter by Nitrosopumilus under optimal conditions or growth of O. alexandrii on the purified catalase itself. Alternatively, the presence of a purified catalase could provide a growth advantage to O. alexandrii by reducing the amount of catalase it needs to produce. Furthermore, O. alexandrii achieved higher cell abundances in coculture with N. piranensis (composing up to 25% of the cells during the incubation period, Table S1) than were seen with N. adriaticus or N. maritimus. Hence, the levels of quantity and/or quality of organic carbon released by different Nitrosopumilus strains potentially differ under the same culture conditions.
Conclusions.
Our results, combined with the results of previous studies, suggest that sensitivity to H2O2 is common among different members of the Nitrosopumilus genus and contribute to the understanding of the physiological and molecular responses of AOA to H2O2. The extent of the sensitivities of marine AOA to H2O2 appears to differ between strains, which may lead to niche differentiation.
The absence and/or loss of a specific function (i.e., H2O2 detoxification) has been suggested to provide a selective advantage by conserving an organism’s limiting resources (71). The ocean’s most abundant free-living prokaryotes, including Prochlorococcus, “Candidatus Pelagibacter” (SAR11 clade), and AOA, can grow axenically only when such missing metabolic functions are provided (14, 33, 72). Our results suggest that marine AOA rely on H2O2 detoxification during periods of high activity and release organic compounds, thereby attracting heterotrophic prokaryotes that provide the missing catalase function.
Interactions between Nitrosopumilus spp. and the alphaproteobacterium O. alexandrii are reminiscent of interactions between heterotrophic bacteria and phytoplankton cells (i.e., within the “phycosphere”), and the importance of these microscale interactions for aquatic ecosystems is widely acknowledged (73). Similarly, metabolic interactions within the immediate surroundings of AOA cells might represent a successful ecological strategy for heterotrophic bacteria, especially in locations below the euphotic layer of the ocean. Microbial radiocarbon signatures indicate that chemolithoautotrophic production can supply up to 95% of the organic carbon incorporated by free-living microbial communities in mesopelagic waters (74). AOA are the most abundant chemolithoautotrophic microbes in the global ocean, suggesting that they could play a crucial role in the production of reduced carbon compounds from inorganic carbon and therefore in the provision of labile organic matter for heterotrophic prokaryotes.
MATERIALS AND METHODS
Cultivation procedures and H2O2 sensitivity experiments.
Axenic cultures of Nitrosopumilus adriaticus NF5, Nitrosopumilus piranensis D3C, and Nitrosopumilus maritimus SCM1 were routinely grown in synthetic Crenarchaeota medium (SCM) in the dark as previously described (27, 75) with the addition of catalase (Sigma catalog no. C1345) (5 units ml−1 final concentration). Cultures were maintained in 30-ml polypropylene plastic bottles, and growth was monitored via flow cytometry (described in Text S1 in the supplemental material) and by measuring nitrite production levels (76).
Prior to establishing different culture treatments, Nitrosopumilus cultures were grown without the addition of catalase for one passage (initial cell abundances in the preculture were ≥7 × 106 ml−1) to ensure exclusion of the remaining catalase activity and catalase carryover to the culture medium. Subsequently, each AOA strain was grown under four distinct sets of conditions and triplicate cultures were prepared for each growth condition and strain. The tested growth conditions were as follows: (i) no H2O2 scavenger, (ii) supplementation with catalase, (iii) inoculation with Oceanicaulis alexandrii, and (iiv) both inoculation with O. alexandrii and supplementation with catalase. To establish cocultures, O. alexandrii was grown in SCM medium with 0.01% yeast extract-peptone and cells were harvested via centrifugation (10,000 × g, 10°C, 15 min) after 3 days, washed three times with SCM culture medium, and added to freshly inoculated Nitrosopumilus cultures (5% O. alexandrii and 95% Nitrosopumilus spp. [based on cell abundance measurements]). Catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) was performed on cocultures to differentiate between bacterial cell abundance and archaeal cell abundance (described in Text S1 in the supplemental material).
Furthermore, the effect of the AOA inoculum size on their cellular response to H2O2 was tested by establishing duplicate cultures of Nitrosopumilus spp. with various initial cell abundances (∼2 × 105, ∼8 × 105, ∼3 × 106, and 7 × 106 cells ml−1) without catalase addition. H2O2 concentrations were measured with a fluorescence-based assay (Sigma-Aldrich, catalog no. MAK165) according to the manufacturer’s protocol.
Proteomics and differential protein expression analysis.
Triplicate cultures of Nitrosopumilus spp. were grown in 250-ml Schott bottles for each of the following treatments: with addition of catalase, in coculture with Oceanicaulis alexandrii, without catalase and O. alexandrii (H2O2 inhibited), and without catalase and O. alexandrii at a high (7 × 106 cells ml−1) initial cell abundance (H2O2 noninhibited). Culture conditions were established as described in the section above. Cells were harvested during exponential growth via centrifugation (18,500 × g, 4°C, 1.5 h), and cell pellets were immediately frozen at −80°C until whole-cell protein extraction was performed.
Proteins were extracted from cell pellets and subjected to denaturing polyacrylamide gel electrophoresis (SDS-PAGE) followed by overnight trypsin in-gel digestion (described in detail in Text S1 in the supplemental material). Desalted peptides were resuspended in an aqueous solution containing 2% acetonitrile and 0.1% formic acid to a concentration of 0.2 μg μl−1 (1 μg total) prior to loading onto an Easy-spray column (Thermo Fisher Scientific PepMap RSCL) (C18; 500 mm by 75 μm; pore size of 2.0 μm). Peptides were separated during a 270-min gradient step using a flow rate of 300 nl min−1 and a one-dimensional (1D) nano-LC instrument (Dionex UltiMate 3000; Thermo Fisher Scientific) coupled to an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) (see Text S1 in the supplemental material). Each of the 36 protein extracts was analyzed twice (resulting in a total of 72 proteomic profiles), and the two technical replicates were combined for bioinformatic analysis (resulting in 36 combined proteomes). Acquired MS/MS spectra were analyzed using the SEQUEST-HT algorithm implemented in Proteome Discoverer 2.2 software (Thermo Fisher Scientific), and spectra were searched against the entire set of translated coding sequences of Nitrosopumilus adriaticus NF5 (2627854092), Nitrosopumilus piranensis D3C (2627853696), and Nitrosopumilus maritimus SCM1 (641228499), downloaded from the Integrated Microbial Genomes (IMG) database (77). Protein matches were accepted if they were identified by at least one unique peptide and with high confidence (details can be found in Text S1 in the supplemental material), and proteins were quantified using the normalized spectral abundance factor (NSAF) approach (78) as follows:
where the total number of spectral counts for the matching peptides from protein k (PSM) was divided by the protein length (L) and then divided by the sum of PSM/L for all N proteins.
In order to adequately match genes shared by the three Nitrosopumilus strains and subsequently compare their individual proteomic responses, orthologous groups (OGs) were constructed on the basis of their entire set of coding sequences using OrthoFinder (version 1.0.8) with standard settings (79). The complete list of assigned OGs and their annotations can be found in Data Set S1A and B in the supplemental material. Differential levels of expression of proteins recovered for each strain and treatment were tested with the DESeq2 Bioconductor package (version 1.20.0) (80) in the R software environment (version 3.5.0) using default parameters and spectral counts as input data based on the recommendations of Langley and Mayr (81). All possible (i.e., all six) pairwise comparisons between the four different treatments, (i.e., with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited) were performed separately for each Nitrosopumilus strain. Each test included three biological replicates per treatment, with the exception of the N. maritimus SCM1 “H2O2 inhibited” treatment, where one biological replicate was excluded from all analyses because of apparent problems during MS analyses that resulted in poor identification of the proteins. Probability values (P values) were adjusted using the Benjamini-Hochberg correction method as previously described (80, 81). The following filter criteria were applied in DESeq2: adjusted P value, <0.05; log 2-fold difference between treatments: greater than or equal to 2 and less than or equal to −2; mean of normalized counts, ≥3. Proteins that showed significant pairwise correlations were visualized with the pheatmap package (version 1.0.12) (82) in the R software environment (83). Columns and rows were clustered based on Euclidean distances corresponding to differences between treatments and relative protein abundances, respectively, as implemented in the pheatmap package. Curation of the annotations of proteins showing significant changes in relative abundance was performed by sequence similarity searches using BLAST (84) and the RefSeq (release 92) and UniprotKB/Swissprot (release 2019_01) databases (85, 86), and protein domain searches were performed using InterProScan (release 72.0) (87). Signal peptides were identified with PRED-Signal (88) and SignalP5.0 (89) to determine if proteins were potentially addressed to the membrane and/or released to the (pseudo)periplasmic space, and additional homology modeling of proteins and functional predictions were carried out with Phyre2 (90).
13C-RNA-stable isotope probing (13C-RNA-SIP).
The three AOA strains investigated in this study were grown in SCM medium containing 13C-labeled bicarbonate (2 mM final concentration) for two consecutive passages, each lasting for 5 to 7 days. Subsequently, O. alexandrii was grown with each AOA in separate cocultures and axenically on the culture supernatant of the three AOA strains. Cocultures were established as described above in the cultivation procedure section. Culture supernatants were obtained via centrifugation (10,000 × g, 10°C, 30 min) and gentle serial filtration through 0.2-μm-pore-size filters (Durapore, Millipore; 47 mm) and 0.1-μm-pore-size filters (Durapore, Millipore; 33 mm). Cells of O. alexandrii grown in yeast extract-peptone medium containing 13C-labeled bicarbonate (2 mM) served as a control to evaluate potential labeling of O. alexandrii rRNA via anaplerotic reactions. After 5 days of incubation, cultures were harvested by filtration through 0.2-μm-pore-size polycarbonate filters (Millipore; 47 mm) which were immediately frozen at −80°C. RNA was extracted according to the protocol of Angel (91) with some modifications for the use of filters (described in detail in Text S1 in the supplemental material), and samples of late-exponential-phase cultures of N. piranensis were selected for tracing the incorporation of AOA-derived organic matter into O. alexandrii 16S rRNA.
Subsequently, heavy (13C-labeled) RNA was separated from light (“natural” 13C/12C isotope ratio) RNA by isopycnic centrifugation. Approximately 300 ng of RNA was mixed with cesium trifluoroacetate (CsTFA; GE Healthcare), HiDi formamide (Thermo Fisher Scientific), and gradient buffer (0.1 M Tris-HCl [pH 8.0], 0.1 M KCl, 1 mM EDTA) as described previously (92). Samples were centrifuged at 130,000 × g at 20°C for at least 65 h in an Optima L-100 XP ultracentrifuge with a VTi 90 rotor (Beckman Coulter), and the resulting CsTFA density gradients were fractionated into 20 equal (250-μl) fractions. Fractions accounting for densities ranging between 1.760 and 1.875 g ml−1 were used for downstream analysis. RNA was precipitated at −80°C after addition of 2.5 volumes of 100% ethanol, 0.5 volumes of 5 M NH4-acetate, and 2 μl glycogen (molecular biology grade; Thermo Fisher Scientific) and pelleted by centrifugation at 20,000 × g for 30 min at 4°C. The RNA pellets were washed with ice-cold 75% ethanol, air-dried, and subsequently resuspended in 10 μl RNA storage solution (Ambion). cDNA was synthesized using SuperScript III reverse transcriptase and random hexamer primers (both from Thermo Fisher Scientific) according to the manufacturer’s protocol. 16S rRNA copies from individual SIP fraction were quantified by quantitative PCR (qPCR) (Bio-Rad) (see Text S1 in the supplemental material), and results are expressed as a proportion of the total number of 16S rRNA copies from all SIP fractions.
Data availability.
All acquired raw spectrum files and proteomic result files, including identified peptides, relative protein abundances, and DESeq outputs, are available on MassIVE (https://massive.ucsd.edu) under accession number MSV000083517 (ftp://massive.ucsd.edu/MSV000083517).
ACKNOWLEDGMENTS
We thank Martin Brenner for technical assistance with nano-LC-Orbitrap-MS, Christian Baranyi for technical assistance with cultivations, Roey Angel for helpful advice on and assistance with stable isotope probing, and Dragoslava Sibinovic for technical assistance with CARD-FISH. We also thank Christopher Sedlacek, Petra Pjevac, Christian Winter, and Melina Kerou for helpful discussions.
The experimental work was supported by the Austrian Science Fund (FWF) project (P28781-B21 to G.J.H.). B.B. was supported by the Uni:docs fellowship of the University of Vienna and the Austrian Science Fund (FWF) DK+ project “Microbial Nitrogen Cycling” (W1257-B20 to G.J.H.). M.K. was financially supported by the DFG Heisenberg program (KO 3651/3-1).
We declare that we have no conflict of interest.
REFERENCES
- 1.Wuchter C, Schouten S, Boschker HTS, Sinninghe Damsté JS. 2003. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett 219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 2.Ward BB. 2011. Nitrification in the ocean, p 325–346. In Ward BB, Arp DJ, Klotz MG (ed), Nitrification. ASM Press, Washington, DC. [Google Scholar]
- 3.Santoro AE, Casciotti KL, Francis CA. 2010. Activity, abundance and diversity of nitrifying archaea. Environ Microbiol 12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- 4.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol 6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 5.Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, Streit W, Stahl DA, Wagner M, Schleper C. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Karner MB, DeLong EF, Karl DM. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 7.Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. 2012. Microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci U S A 109:15996–16003. doi: 10.1073/pnas.1205009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR, Yang Y, Orsi WD, Moran DM, Saito MA. 2015. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci U S A 112:1173–1178. doi: 10.1073/pnas.1416223112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayer B, Vojvoda J, Offre P, Alves RJE, Elisabeth NH, Garcia JAL, Volland JM, Srivastava A, Schleper C, Herndl GJ. 2016. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10:1051–1063. doi: 10.1038/ismej.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahlgren NA, Chen Y, Needham DM, Parada AE, Sachdeva R, Trinh V, Chen T, Fuhrman JA. 2017. Genome and epigenome of a novel marine Thaumarchaeota strain suggest viral infection, phosphorothioation DNA modification and multiple restriction systems. Environ Microbiol 19:2434–2452. doi: 10.1111/1462-2920.13768. [DOI] [PubMed] [Google Scholar]
- 11.Park S-J, Ghai R, Martin-Cuadrado AB, Rodriguez-Valera F, Chung W-H, Kwon K, Lee J, Madsen EL, Rhee S-K. 2014. Genomes of two new ammonia-oxidizing archaea enriched from deep marine sediments. PLoS One 9:e96449. doi: 10.1371/journal.pone.0096449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carini P, Dupont CL, Santoro AE. 2018. Patterns of thaumarchaeal gene expression in culture and diverse marine environments. Environ Microbiol doi: 10.1111/1462-2920.14107. [DOI] [PubMed] [Google Scholar]
- 13.Qin W, Heal KR, Ramdasi R, Kobelt JN, Martens-Habbena W, Bertagnolli AD, Amin SA, Walker CB, Urakawa H, Könneke M, Devol AH, Moffett JW, Armbrust EV, Jensen GJ, Ingalls AE, Stahl DA. 2017. Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int J Syst Evol Microbiol 67:5067–5079. doi: 10.1099/ijsem.0.002416. [DOI] [PubMed] [Google Scholar]
- 14.Kim J-G, Park S-J, Damsté JSS, Schouten S, Rijpstra WIC, Jung M-Y, Kim S-J, Gwak J-H, Hong H, Si O-J, Lee S, Madsen EL, Rhee S-K. 2016. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci U S A 113:7888–7893. doi: 10.1073/pnas.1605501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH, Ingalls AE, Moffett JW, Armbrust EV, Stahl DA. 2014. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci U S A 111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamocky M, Furtmüller PG, Obinger C. 2008. Evolution of catalases from bacteria to humans. Antioxid Redox Signal 10:1527–1548. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabiscol E, De Lleida U, De Lleida U, Ros J, De Lleida U. 2000. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8. [PubMed] [Google Scholar]
- 19.Tolar BB, Powers LC, Miller WL, Wallsgrove NJ, Popp BN, Hollibaugh JT. 2016. Ammonia oxidation in the ocean can be inhibited by nanomolar concentrations of hydrogen peroxide. Front Mar Sci 3:237. doi: 10.3389/fmars.2016.00237. [DOI] [Google Scholar]
- 20.Cooper WJ, Shao C, Lean DRS, Gordon AS, Scully FE. 1994. Factors affecting the distribution of H2O2 in surface waters. Env Chem Lakes Reserv 237:391–422. doi: 10.1021/ba-1994-0237. [DOI] [Google Scholar]
- 21.Cooper WJ, Saltzman ES, Zika RG. 1987. The contribution of rainwater to variability in surface ocean hydrogen peroxide. J Geophys Res 92:2970–2980. doi: 10.1029/JC092iC03p02970. [DOI] [Google Scholar]
- 22.Palenik B, Zafiriou OC, Morel F. 1987. Hydrogen peroxide production by a marine phytoplankter’. Limnol Oceanogr 32:1365–1369. doi: 10.4319/lo.1987.32.6.1365. [DOI] [Google Scholar]
- 23.Roe KL, Schneider RJ, Hansel CM, Voelker BM. 2016. Measurement of dark, particle-generated superoxide and hydrogen peroxide production and decay in the subtropical and temperate North Pacific Ocean. Deep Res I 107:59–69. doi: 10.1016/j.dsr.2015.10.012. [DOI] [Google Scholar]
- 24.Kieber DJ, Peake BM, Scully NM. 2003. Reactive oxygen species in aquatic ecosystems, p 251–290. In Helbling EW, Zagarese H (ed), UV effects in aquatic organisms and ecosystems. The Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 25.Qin W, Meinhardt KA, Moffett JW, Devol AH, Armbrust EV, Ingalls AE, Stahl DA. 2017. Influence of oxygen availability on the activities of ammonia-oxidizing archaea. Environ Microbiol Rep 9:250–256. doi: 10.1111/1758-2229.12525. [DOI] [PubMed] [Google Scholar]
- 26.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 27.Bayer B, Vojvoda J, Reinthaler T, Reyes C, Pinto M, Herndl GJ. 2019. Nitrosopumilus adriaticus sp. nov. and Nitrosopumilus piranensis sp. nov., two ammonia-oxidizing archaea from the Adriatic Sea and members of the class Nitrososphaeria. Int J Syst Evol Microbiol doi: 10.1099/ijsem.0.003360. [DOI] [PubMed] [Google Scholar]
- 28.Morris JJ, Zinser ER. 2013. Continous hydrogen peroxide production by organic buffers in phytoplankton culture media. J Phycol 49:1223–1228. doi: 10.1111/jpy.12123. [DOI] [PubMed] [Google Scholar]
- 29.Zika RG, Moffett JW, Petasne RG, Cooper WJ, Saltzman ES. 1985. Spatial and temporal variations of hydrogen peroxide in Gulf of Mexico waters. Geochim Cosmochim Acta 49:1173–1184. doi: 10.1016/0016-7037(85)90008-0. [DOI] [Google Scholar]
- 30.Yuan J, Shiller AM. 2004. Hydrogen peroxide in deep waters of the North Pacific Ocean. Geophys Res Lett 31:L01310. [Google Scholar]
- 31.Heelis PF, Kim ST, Okamura T, Sancar A. 1993. The photo-repair of pyrimidine dimers by DNA photolyase and model systems. J Photochem Photobiol B 17:219–228. doi: 10.1016/1011-1344(93)80019-6. [DOI] [PubMed] [Google Scholar]
- 32.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa T, Stahl DA. 2013. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol 79:6911–6916. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng M, Aslund F, Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 37.Christman MF, Morgan RW, Jacobson FS, Ames BN. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762. doi: 10.1016/S0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 38.Lee J-W, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 39.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maaty WS, Wiedenheft B, Tarlykov P, Schaff N, Heinemann J, Robison-Cox J, Valenzuela J, Dougherty A, Blum P, Lawrence CM, Douglas T, Young MJ, Bothner B. 2009. Something old, something new, something borrowed; how the thermoacidophilic archaeon Sulfolobus solfataricus responds to oxidative stress. PLoS One 4:e6964. doi: 10.1371/journal.pone.0006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaur A, Van PT, Busch CR, Robinson CK, Pan M, Pang WL, Reiss DJ, Diruggiero J, Baliga NS. 2010. Coordination of frontline defense mechanisms under severe oxidative stress. Mol Syst Biol 6:393. doi: 10.1038/msb.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo H, Tolar BB, Swan BK, Zhang CL, Stepanauskas R, Moran MA, Hollibaugh JT. 2014. Single-cell genomics shedding light on marine Thaumarchaeota diversification. ISME J 8:732–736. doi: 10.1038/ismej.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhalnina KV, Dias R, Leonard MT, De Quadros PD, Camargo FAO, Drew JC, Farmerie WG, Daroub SH, Triplett EW. 2014. Genome sequence of Candidatus Nitrososphaera evergladensis from group I.1b enriched from Everglades soil reveals novel genomic features of the ammonia-oxidizing archaea. PLoS One 9:e101648. doi: 10.1371/journal.pone.0101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jung MY, Kim JG, Sinninghe Damsté JS, Rijpstra WIC, Madsen EL, Kim SJ, Hong H, Si OJ, Kerou M, Schleper C, Rhee SK. 2016. A hydrophobic ammonia-oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar-contaminated sediment. Environ Microbiol Rep 8:983–992. doi: 10.1111/1758-2229.12477. [DOI] [PubMed] [Google Scholar]
- 45.Sauder LA, Albertsen M, Engel K, Schwarz J, Nielsen PH, Wagner M, Neufeld JD. 2017. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J 11:1142–1157. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrick J, Sclavi B. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol Microbiol 63:22–34. doi: 10.1111/j.1365-2958.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 47.Elledge SJ, Zhou Z, Allen JB, Navas TA. 1993. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 48.Guy CP, Bolt EL. 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res 33:3678–3690. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Houten B, Kad N. 2014. Investigation of bacterial nucleotide excision repair using single-molecule techniques. DNA Repair (Amst) 20:41–48. doi: 10.1016/j.dnarep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stracy M, Jaciuk M, Uphoff S, Kapanidis AN, Nowotny M, Sherratt DJ, Zawadzki P. 2016. Single-molecule imaging of UvrA and UvrB recruitment to DNA lesions in living Escherichia coli. Nat Commun 7:1–9. doi: 10.1038/ncomms12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seaver LC, Imlay JA. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol 183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Branco MR, Marinho HS, Cyrne L, Antunes F. 2004. Decrease of H2O2 plasma membrane permeability during adaptation to H2O2 in Saccharomyces cerevisiae. J Biol Chem 279:6501–6506. doi: 10.1074/jbc.M311818200. [DOI] [PubMed] [Google Scholar]
- 53.Antunes F, Cadenas E. 2000. Estimation of H2O2 gradients across biomembranes. FEBS Lett 475:121–126. doi: 10.1016/S0014-5793(00)01638-0. [DOI] [PubMed] [Google Scholar]
- 54.Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hildebrand PW, Preissner R, Frömmel C. 2004. Structural features of transmembrane helices. FEBS Lett 559:145–151. doi: 10.1016/S0014-5793(04)00061-4. [DOI] [PubMed] [Google Scholar]
- 56.Zwieb C, Eichler J. 2002. Getting on target: the archaeal signal recognition particle. Archaea 1:27–34. doi: 10.1155/2002/729649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdul Halim MF, Pfeiffer F, Zou J, Frisch A, Haft D, Wu S, Tolić N, Brewer H, Payne SH, Paša-Tolić L, Pohlschroder M. 2013. Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol Microbiol 88:1164–1175. doi: 10.1111/mmi.12248. [DOI] [PubMed] [Google Scholar]
- 58.Zhang CY, Cai K, Liu H, Zhang Y, Pan HW, Wang B, Wu ZH, Hu W, Li YZ. 2007. New locus important for Myxococcus social motility and development. J Bacteriol 189:7937–7941. doi: 10.1128/JB.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai S, Sun C, Tan K, Ye S, Zhang R. 2017. Structure of thrombospondin type 3 repeats in bacterial outer membrane protein A reveals its intra-repeat disulfide bond-dependent calcium-binding capability. Cell Calcium 66:78–89. doi: 10.1016/j.ceca.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 60.Sousa-Lopes A, Antunes F, Cyrne L, Marinho HS. 2004. Decreased cellular permeability to H2O2 protects Saccharomyces cerevisiae cells in stationary phase against oxidative stress. FEBS Lett 578:152–156. doi: 10.1016/j.febslet.2004.10.090. [DOI] [PubMed] [Google Scholar]
- 61.Lehman AP, Long SR. 2013. Exopolysaccharides from Sinorhizobium meliloti can protect against H2O2-dependent damage. J Bacteriol 195:5362–5369. doi: 10.1128/JB.00681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang IA, Kim J, Park W. 2016. Endogenous hydrogen peroxide increases biofilm formation by inducing exopolysaccharide production in Acinetobacter oleivorans DR1. Sci Rep 6:21121. doi: 10.1038/srep21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kerou M, Offre P, Valledor L, Abby SS, Melcher M, Nagler M, Weckwerth W, Schleper C. 2016. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci U S A 113:E7937–E7946. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lawton TJ, Kenney GE, Hurley JD, Rosenzweig AC. 2016. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry 55:2278–2290. doi: 10.1021/acs.biochem.6b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pham AN, Xing G, Miller CJ, Waite TD. 2013. Fenton-like copper redox chemistry revisited: hydrogen peroxide and superoxide mediation of copper-catalyzed oxidant production. J Catal 301:54–64. doi: 10.1016/j.jcat.2013.01.025. [DOI] [Google Scholar]
- 67.Anjem A, Imlay JA. 2012. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem 287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strömpl C, Hold GL, Lünsdorf H, Graham J, Gallacher S, Abraham WR, Moore ERB, Timmis KN. 2003. Oceanicaulis alexandrii gen. nov., sp. nov., a novel stalked bacterium isolated from a culture of the dinoflagellate Alexandrium tamarense (Lebour) Balech. Int J Syst Evol Microbiol 53:1901–1906. doi: 10.1099/ijs.0.02635-0. [DOI] [PubMed] [Google Scholar]
- 69.Zámocký M, Gasselhuber B, Furtmüller PG, Obinger C. 2014. Turning points in the evolution of peroxidase-catalase superfamily: molecular phylogeny of hybrid heme peroxidases. Cell Mol Life Sci 71:4681–4696. doi: 10.1007/s00018-014-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bayer B, Hansman RL, Bittner MJ, Noriega BE, Niggemann J, Dittmar T, Herndl GJ. 2019. Ammonia-oxidizing archaea release a suite of organic compounds potentially fueling prokaryotic heterotrophy in the ocean. bioRxiv doi: 10.1101/558726. [DOI] [PMC free article] [PubMed]
- 71.Morris JJ, Lenski RE, Zinser ER. 2012. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. 2008. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 73.Seymour JR, Amin SA, Raina JB, Stocker R. 2017. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol 2:17065. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 74.Hansman RL, Griffin S, Watson JT, Druffel ERM, Ingalls AE, Pearson A, Aluwihare LI. 2009. The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci U S A 106:6513–6518. doi: 10.1073/pnas.0810871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA, Torre J, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 76.Griess P. 1879. Bemerkungen zu der Abhandlung der H. H. Weselsky und Benedikt “Über einige Azoverbindungen.” Ber Dtsch Chem Ges 12:426–428. doi: 10.1002/cber.187901201117. [DOI] [Google Scholar]
- 77.Markowitz VM, Chen I-M, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. 2006. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res 5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 79.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langley SR, Mayr M. 2015. Comparative analysis of statistical methods used for detecting differential expression in label-free mass spectrometry proteomics. J Proteomics 129:83–92. doi: 10.1016/j.jprot.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 82.Kolde R. 2015. pheatmap: Pretty Heatmaps. R package version 1.0.8. https://CRAN.R-project.org/package=pheatmap.
- 83.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 84.Mount DW. 2007. Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc 2007:pdb.top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 85.O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, et al. . 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. 2007. UniProtKB/Swiss-Prot. Methods Mol Biol 406:89–112. [DOI] [PubMed] [Google Scholar]
- 87.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ. 2009. Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35. doi: 10.1093/protein/gzn064. [DOI] [PubMed] [Google Scholar]
- 89.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 90.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 Web portal for protein modelling, prediction, and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Angel R. 2012. Total nucleic acid extraction from soil. Protoc Exch doi: 10.1038/protex.2012.046. [DOI] [Google Scholar]
- 92.Dumont MG, Pommerenke B, Casper P, Conrad R. 2011. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol 13:1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell counts of Nitrospumilus and Oceanicaulis alexandrii cultures grown in different treatments (CAT, catalase; coculture; without addition of catalase or O. alexandrii). Cell counts were derived from one of three biological replicates per strain and treatment. Download Table S1, DOCX file, 0.02 MB (18.4KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Hydrogen peroxide (H2O2) concentrations of SCM medium controls with or without addition of catalase (5 U ml−1) incubated in the dark at 30°C. (B) Abiotic reduction of H2O2 in SCM medium over time. Error bars represent the range of the mean from measurements of duplicate cultures (sometimes too small to be visible). Download FIG S1, PDF file, 0.2 MB (229.7KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Proteins shared between proteomes of three Nitrosopumilus strains grown under conditions of different treatments (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited). (B) Significant changes in relative protein abundances of three Nitrosopumilus strains based on pairwise comparisons between different treatments. Each treatment included proteins shared by three biological replicates. Download Table S2, DOCX file, 0.01 MB (15.1KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In-depth descriptions of protocols used for protein extraction, LC-MS/MS analysis, and peptide identification, RNA extraction, quantitative PCR, flow cytometry, and CARD-FISH. Discussion of additional information on extracellular matrix-associated proteins, strain-specific proteomic responses of the individual Nitrosopumilus strains to H2O2, and potential H2O2 production sites in AOA. Download Text S1, DOCX file, 0.07 MB (74.5KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heat map of the proteins that showed a significant change in relative abundance in three Nitrosopumilus strains under different culture conditions (with catalase, with O. alexandrii, H2O2 inhibited, and H2O2 noninhibited). Columns and rows were clustered based on Euclidean distances corresponding to differences between treatments and relative protein abundances, respectively. (A) Nitrosopumilus adriaticus NF5. (B) Nitrosopumilus piranensis D3C. (C) Nitrosopumilus maritimus SCM1. Download FIG S2, PDF file, 0.8 MB (811KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Orthologous groups constructed from the genomes of Nitrosopumilus adriaticus NF5, Nitrosopumilus piranensis D3C, and Nitrosopumilus maritimus SCM1. (B) Gene annotations of orthologous groups. (C) KEGG assignments of the H2O2 responsive proteins shared by all three strains. (D) Relative protein abundances of the proteins related to canonical oxidative stress defense mechanisms in all three Nitrosopumilus strains for each treatment. Download Data Set S1, XLSX file, 0.2 MB (157.4KB, xlsx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A and B) Cell aggregation of Nitrosopumilus adriaticus NF5 observed during growth in medium without hydrogen peroxide (H2O2) scavengers. (C and D) CARD-FISH images of Nitrosopumilus adriaticus NF5 (DAPI [4′,6-diamidino-2-phenylindole]; blue) grown in coculture with Oceanicaulis alexandrii (bacterial EUBI probe; green). Download FIG S3, PDF file, 2.7 MB (2.7MB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Catalase-containing contigs of thaumarchaeal single-cell genomes AB-661-L21 and AB-661-M19 from Luo et al. (42) (catalases are marked in yellow). (B) Multiple-sequence alignment of class I catalase peroxidases, including sequences from Oceanicaulis alexandrii (blue) and sequences with known three-dimensional (3D) structures obtained from PeroxiBase. Color scheme: black = 100% sequence similarity; gray = 90% to 99% sequence similarity. Signal peptides are indicated in red. Essential residues involved in catalysis are labeled with an asterisk. Download Data Set S2, DOCX file, 0.2 MB (236.9KB, docx) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Growth curves of Oceanicaulis alexandrii and Nitrosopumilus adriaticus NF5 in coculture with addition of catalase. (B) Growth of O. alexandrii on culture supernatant of three Nitrosopumilus strains (N. adriaticus NF5, N. piranensis D3C, and N. maritimus SCM1) and in medium without the addition of any carbon source (Control). Error bars represent standard deviations of measurements from triplicate cultures. Download FIG S4, PDF file, 0.3 MB (259.2KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proportion of N. piranensis D3C 16S rRNA gene copies recovered from RNA-SIP gradient fractions under conditions of growth in medium containing unlabeled bicarbonate (gray line) and after two passages into medium containing 13C-labeled bicarbonate (black line). Download FIG S5, PDF file, 0.2 MB (206.7KB, pdf) .
Copyright © 2019 Bayer et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All acquired raw spectrum files and proteomic result files, including identified peptides, relative protein abundances, and DESeq outputs, are available on MassIVE (https://massive.ucsd.edu) under accession number MSV000083517 (ftp://massive.ucsd.edu/MSV000083517).