Abstract
Peroxisome proliferator–activated receptor α (PPARα) is a key nuclear receptor involved in the control of lipid homeostasis. In rodents, PPARα is also a potent hepatic mitogen. Hepatocyte-specific disruption of PPARα inhibits agonist-induced hepatocyte proliferation; however, little is known about the exact role of PPARα in partial hepatectomy (PHx)–induced liver regeneration. Herein, using hepatocyte-specific PPARα-deficient (PparaΔHep) mice, the function of hepatocyte PPARα in PHx-induced liver regeneration was investigated. PPARα protein level and transcriptional activity were increased in the liver after PHx. Compared with the Pparafl/fl mice, PparaΔHep mice exhibited significantly reduced hepatocyte proliferation at 32 hours after PHx. Consistently, reduced Ccnd1 and Pcna mRNA and CYCD1 and proliferating cell nuclear antigen protein were observed at 32 hours after PHx in PparaΔHep mice. Furthermore, PparaΔHep mice showed increased hepatic lipid accumulation and enhanced hepatic triglyceride contents because of impaired hepatic fatty acid β-oxidation when compared with that observed in Pparafl/fl mice. These results indicate that PPARα promotes liver regeneration after PHx, at least partially via regulating the cell cycle and lipid metabolism.
The liver has a remarkable ability to regenerate. A 70% partial hepatectomy (PHx) animal model is commonly used to study liver regeneration. After PHx, the hepatocyte, the main cell type of the liver, replicates through both hypertrophic and hyperplastic mechanisms.1 Liver regeneration is a compensatory hyperplasia process involving multiple growth factors, cytokines, hormones, and nuclear receptors.2, 3, 4, 5, 6, 7, 8 Almost all the cell types in liver participate in the PHx response; hepatocytes and cholangiocytes rapidly enter into the cell cycle, whereas stellate cells, Kupffer cells, and endothelial cells follow hepatocytes in the procession of regeneration,8 which takes 5 to 7 days in mice, with cell proliferation peaking at days 2 or 3.2, 3, 4, 8 In the early stage after liver regeneration, the blood concentrations of hepatocyte growth factor, epidermal growth factor, tumor necrosis factor α (TNFα), IL-6, norepinephrine, bile acids, and serotonin were rapidly increased.2, 3, 4, 5, 6, 7 It is widely accepted that PHx causes a transient increase in lipopolysaccharide levels, which stimulates Kupffer cells to produce IL-6 and TNFα that act on hepatocytes to induce survival and proliferation.9, 10
Systemic and liver local metabolic changes are believed to trigger and participate in liver regeneration. Hypoglycemia and increased circulating insulin levels also occur soon after PHx.11, 12, 13 The liver presents transient steatosis on day 2 or 3 after PHx, which could influence liver regeneration.11, 14 An early study shows that mice rapidly develop hypoglycemia after the operation, which causes adipose tissue lipolysis and leads to periphery lipid redistribution in the regenerating liver.15 This study suggests that hepatic steatosis after PHx could promote liver regeneration.15 However, liver regeneration was impaired after PHx in hepatic steatosis mice,16 mice with genetic obesity,17 or mice fed with a high-fat diet.18 Disruption of hepatic lipid accumulation also decreases liver regeneration after PHx in mice.19 Until now, the underlying mechanisms of these results remain unclear.
Many regulators of lipid droplet formation and peripheral lipid mobilization were reported to participate in liver regeneration, such as farnesoid X receptor,20, 21 caveolin-1,22, 23 and peroxisome proliferator–activated receptors (PPARs).24, 25, 26 PPARα is a ligand-activated nuclear receptor, which plays an important role in regulating liver and skeletal muscle fatty acid metabolism and glucose homeostasis.27 Short exposure of mice and rats to peroxisome proliferator chemicals and fibrate drugs leads to hepatocyte proliferation and hepatomegaly through activation of PPARα.28, 29, 30 Chronic exposure to hypolipidemia drugs, such as WY-14643 and fenofibrate, results in prolonged PPARα activation, leading to hepatocellular carcinoma in mice and rats.31 A previous study revealed that increased PPARα signaling influenced the cell cycle regulation of late-phase liver regeneration.32 However, using PPARα whole-body knockout mice (Ppara-null mice), one report found a delayed hepatocyte proliferation after PHx, accompanied by decreased expression of Ccnd1 and Myc involved in cell cycle regulation and impaired RAS membrane association, which indicate that PPARα could promote cell proliferation after PHx.26, 31 In summary, the findings from these studies suggest that PPARα plays a key role in the liver regeneration program, but the definitive role of PPARα in liver cell proliferation after PHx remains unresolved. A recent study revealed that WY-14643–activated PPARα induces cell proliferation, which is mainly dependent on hepatocyte PPARα,33 rather than PPARα expressed in nonparenchymal cells, indicating the function of PPARα in hepatocyte proliferation is cell type specific. However, the precise effect of hepatocyte PPARα on liver regeneration after PHx remains unknown. In this study, using mice with conditional ablation of PPARα in hepatocyte (PparaΔHep mice), hepatocyte PPARα was found to promote liver regeneration after PHx via regulating cell cycle and lipid metabolism.
Materials and Methods
Animals
Male 8- to 10-week–old mice were used in the present study. Hepatocyte-specific PPARα-deficient mice (PparaΔHep mice) were generated, as described,33 and Pparafl/fl mice were used as littermate controls. These mice were kept on a standard 12-hour light/dark cycle with free access to chow diet and water. The animal studies were performed in accordance with protocols approved by the Capital Medical University Animal Care and Use Committee.
To investigate the transcriptional activity of PPARα after PHx, transgenic mice containing a transgene expressing luciferase under control of a PPARA response element repeat were used. These mice were generated, as previously described,34 and obtained from Charles River Company (Boston, MA). The animal studies were performed in the Laboratory of Metabolism, National Cancer Institute, NIH (Bethesda, MD), in accordance with protocol approved by the National Cancer Institute Animal Care and Use Committee.
Partial Hepatectomy Model and Tissue Harvesting
The two-third partial hepatectomy (PHx) surgery was performed as previously described.35, 36, 37 Mice were injected with 5-bromo-2-deoxyuridine (50 mg/kg body weight) 2 hours before sacrificing at the indicated time points. Liver tissues were formalin fixed or frozen in OCT for cryosection, whereas the remaining liver tissue was snap frozen for further analysis.
Hematoxylin-Eosin Staining and Immunohistochemistry Staining
Paraffin-embedded liver tissues were cut into sections (4 μm thick) for hematoxylin-eosin and immunohistochemistry staining. Hematoxylin-eosin staining was performed following standard methods. Immunohistochemistry analysis was performed using antibodies against 5-bromo-2-deoxyuridine (BD Bioscience, San Jose, CA), as previously described.9
Oil Red O Staining
For oil red O staining, OCT-embedded blocks were cut into sections (10 μm thick) and stained with oil red O solution, as previously described.38
Quantitative Real-Time RT-PCR
Total RNA was extracted from Pparafl/fl and PparaΔHep livers using Trizol reagent (Life Technologies, Carlsbad, CA), then reverse transcripted into cDNA with GoScript Reverse Transcriptase (Promega, Madison, WI) and used for quantitative real-time RT-PCR analysis with SYBR Green premix (TaKaRa, Nojihigashi, Kusatsu, Shiga, Japan). Quantitative real-time RT-PCR assays were performed on CFX Connect Real-Time System (Bio-Rad, Hercules, CA). Expression of target genes was normalized to that of the housekeeping gene β-actin mRNA. The primers are listed in Table 1.
Table 1.
List of Real-Time Quantitative PCR Primer Sequences
| Genes | NCBI accession number∗ | Forward | Reverse |
|---|---|---|---|
| Ppara | NM_001113418 | 5′-CTGCCTTCCCTGTGAACTGA-3′ | 5′-ACAGAGCGCTAAGCTGTGAT-3′ |
| Ccna2 | NM_009828 | 5′-TCGCTGCATCAGGAAGACC-3′ | 5′-CTTAAGAGGAGCAACCCGTCG-3′ |
| Ccnd1 | NM_007631 | 5′-TCAAGTGTGACCCGGACTGC-3′ | 5′-CCTTGGGGTCGACGTTCTG-3′ |
| Ccne1 | NM_007633 | 5′-ACTTCCCGTCTTGAATTGGGG-3′ | 5′-AGGATGACGCTGCAGAAAGT-3′ |
| Pcna | NM_011045 | 5′-TCGTCTCACGTCTCCTTGGT-3′ | 5′-TTTTGGACATGCTGGTGAGGT-3′ |
| Check1 | NM_007691 | 5′-CTGGCAAAGGACTGCTTGTC-3′ | 5′-GTTGAACTTCTCCATAGGCACC-3′ |
| Mcm2 | NM_008564 | 5′-CAGCATTGCACCCTCCATCT-3′ | 5′-TGCTTTCCACCTGGGTTCTT-3′ |
| Mcm5 | NM_001302540 | 5′-AACGAACCAATAGGAGCGCA-3′ | 5′-ATGACTGTACCTCAGCCCTC-3′ |
| Prkdc | NM_011159 | 5′-ACAGAGACGGTAATCACGGGT-3′ | 5′-CCAGCAGGAAAGCTGGGTT-3′ |
| Rad51 | NM_011234 | 5′-TTCACGGTTAGAGCAGTGTGG-3′ | 5′-TTCGGTGCATAAGCAACAGC-3′ |
| G6pc | NM_008061.4 | 5′-GTCTTGTCAGGCATTGCTGTG-3′ | 5′-GAATCCAAGCGCGAAACCAA-3′ |
| Pck1 | NM_011044.2 | 5′-ATGAAAGGCCGCACCATGTA-3′ | 5′-GGGCGAGTCTGTCAGTTCAA-3′ |
| Pkm | NM_001253883.1 | 5′-ATGCAGCACCTGATAGCTCG-3′ | 5′-AGGTCTGTGGAGTGACTGGA-3′ |
| Pfkm | NM_001163487.1 | 5′-GGAGAGCTAAAACTACAAGAGTGG-3′ | 5′-CTCCACCAGAGGTCAACACG-3′ |
| Gck | NM_001287386.1 | 5′-CCTCGGGAGTCAGGAACATC-3′ | 5′-ATCTGCTCTACCAGAGTCAACG-3′ |
| Hadha | NM_178878.2 | 5′-AAGAGCTTTCGTCCTCTTCTGC-3′ | 5′-AAATGCAGCCTCTGGAGCGTA-3′ |
| Hadhb | NM_001289798.1 | 5′-CGGACGTTTGTCAGTCTGGA-3′ | 5′-CTGAAATCTGCCTGTGGGGA-3′ |
| Cyp4a10 | NM_010011.3 | 5′-AGGAGCCAGGAACTGCATTG-3′ | 5′-GACCCTGGTAGGATCTGGCA-3′ |
| Cyp4a14 | NM_007822.2 | 5′-TTGCTCACGAGCACACAGAT-3′ | 5′-TCCTCCATTCTGGCAAACAAGA-3′ |
| Cpt2 | NM_009949.2 | 5′-ATCTCAGGCCCCTGGTTTGA-3′ | 5′-ATCTCAGGCCCCTGGTTTGA-3′ |
| Ehhadh | NM_023737.3 | 5′-CGGTCAATGCCATCAGTCCA-3′ | 5′-AGCACCTGCACAGAAGTTGT-3′ |
| Hmgcs2 | NM_008256.4 | 5′-AGAAATCCCTGGCTCGGTTG-3′ | 5′-AGCTTTAGACCCCTGAAGGC-3′ |
| Txnip | NM_001009935.2 | 5′-GAAGGCTTTTCTCGATCGCC-3′ | 5′-GGCAGACACTGGTGCCATTA-3′ |
| Vnn1 | NM_011704.3 | 5′-GCATGCTGTGATCCTGCCTAA-3′ | 5′-TAATGTGCGCACCCTGCT-3′ |
| Cpt1a | NM_013495.2 | 5′-TCGGTGAGCCTGGCCT-3′ | 5′-TTGAGTGGTGACCGAGTCTG-3′ |
NCBI, National Center for Biotechnology Information.
Available from https://www.ncbi.nlm.nih.gov/nuccore.
Protein Extraction and Western Blot Analysis
Whole-cell lysate was extracted using radioimmunoprecipitation assay buffer (Applygen, Beijing, China), and nuclear extraction was done with a nuclear and cytoplasmic protein extraction kit (KeyGENBioTECH, Nanjing, China). The protein concentration was measured using the bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, MA). Specific primary antibodies used were as follows: antibody against PPARα and LaminB1 from Abcam (Cambridge, UK), cyclin D1 and β-actin antibodies from Cell Signaling Technology (Boston, MA), and proliferating cell nuclear antigen (PCNA) antibody from Santa Cruz Biotechnology (Dallas, TX). The dilutions were 1:1000 in 5% bovine serum albumin. After incubating with horseradish peroxidase–conjugated secondary antibody, the immunocomplexes were visualized with FluorChem-R (ProteinSimple, San Jose, CA). Total protein levels were normalized to β-actin (ACTB), and nuclear protein levels were normalized to LaminB1.
Microarray Analysis
Microarray analysis was performed by CNKINGBIO Company (China). Total RNA was extracted and purified, and the quality was examined. Purified RNA was labeled and hybridized to an AffymetrixHuman Gene 1.1 ST array plate (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning were performed on an Affymetrix GeneTitan platform following the instruction. Kyoto Encyclopedia of Genes and Genomes is a knowledge base for systematic analysis of gene functions, linking genomic information with higher-order functional information, and now is used widely for pathway-related analysis. The hypergeometric distribution was used to calculate the pathway enrichment, and false discover rate was used to adjust the P values for multiple comparisons.
Hepatic Triglyceride Measurement
Hepatic triglycerides were extracted in 50 mmol/L Tris buffer, homogenized, and incubated at 37°C with shaking overnight. A triglyceride measurement kit (Applygen) was used following the manufacturer's instruction to measure triglyceride contents.
Serum β-Hydroxybutyrate Assay
Serum concentrations of β-hydroxybutyrate were detected by enzyme-linked immunosorbent assay (NanJingJianCheng Bioengineering Institute, Nanjing, China), strictly according to the manufacturer's instruction.
Luciferase Assay
PPARA response element repeat mice, aged 8 to 10 weeks, were subjected to PHx or sham surgery, as outlined in the original protocol. Thirty-two hours after surgery, livers were harvested and luciferase activity was measured by the Dual-Luciferase Assay System and normalized by protein concentration. PPARA response element repeat mice treated with PPARα agonist Wy-14643 were served as positive control, and nontreatment served as negative control.
Statistical Analysis
All data are presented as means ± SD. Significant difference between groups was determined using unpaired t-test. Values obtained from three or more groups were determined by one-factor analysis of variance (ANOVA) followed by Tukey's post hoc test using GraphPad Prism5.0 software (GraphPad Software, San Diego, CA). P < 0.05 was considered statistically significant.
Results
Increased PPARα Expression in the Liver after PHx
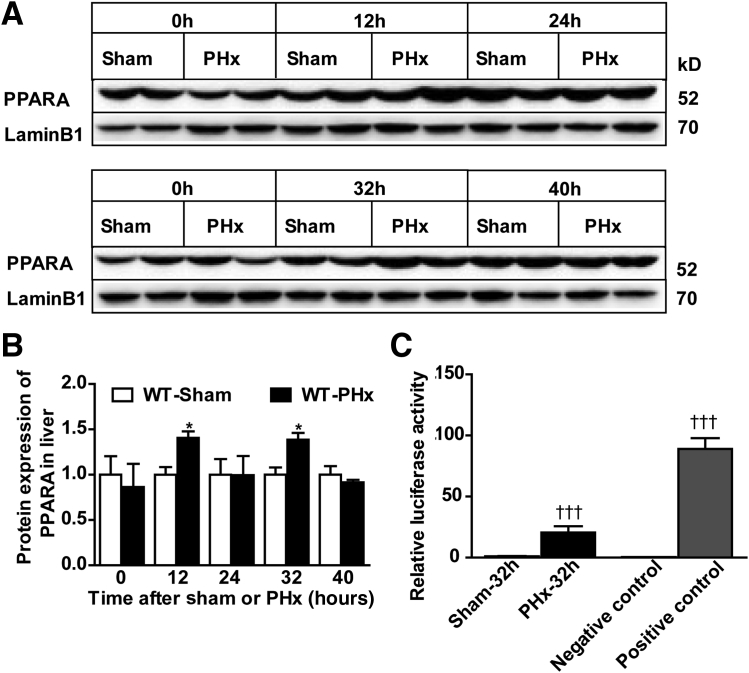
PPARα controls β-oxidation in liver and serves as a potent hepatic mitogen in rodents.39 To investigate the influence of PPARα on liver regeneration after PHx, PPARα and retinoid X receptor α protein expression was determined by Western blot analysis. PPARα protein level was increased at 12 and 32 hours after PHx (Figure 1, A and B). However, retinoid X receptor α protein levels were comparable between sham and PHx surgery (Supplemental Figure S1). To further assess the change of PPARα after PHx, in vivo luciferase assays were performed after PHx or sham at 32 hours. Luciferase activity was significantly increased after PHx surgery compared with sham-operated mice (Figure 1C). These results indicate that PPARα may be involved in PHx-induced liver regeneration.
Figure 1.
Increased PPARα activity in wild-type (WT) mouse liver after PHx. A and B: Western blot analysis of PPARα protein expression in WT livers after PHx or sham operation. C: Luciferase activity of PPARA response element repeat mice after sham or PHx, negative control, and positive control. Data are expressed as means ± SD (B and C). n = 3 (A and B); n = 5 (C). ∗P < 0.05 versus WT-sham; †††P < 0.001 versus sham or negative control.
Disruption of PPARα in Hepatocyte Inhibits Liver Regeneration after PHx
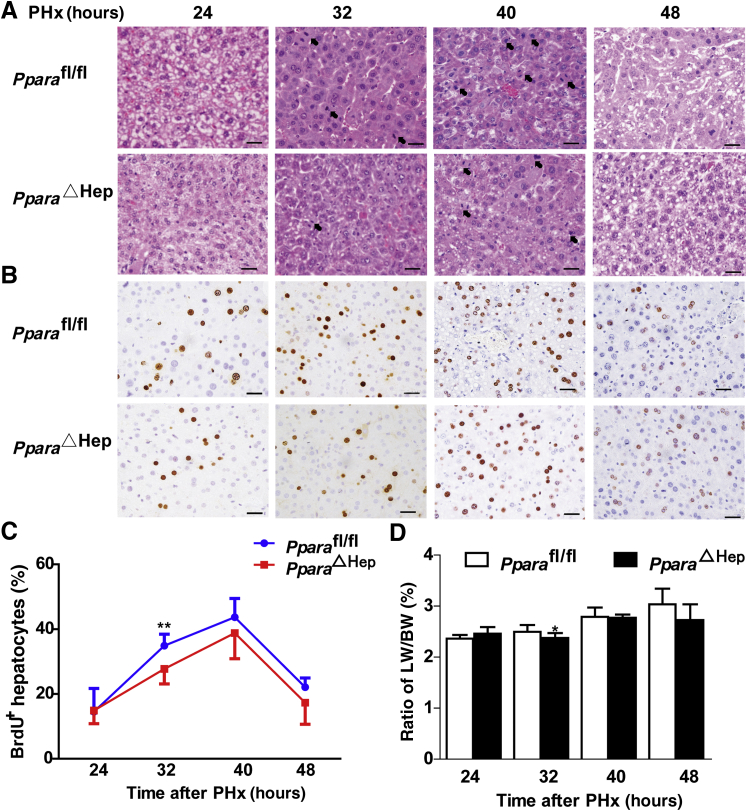
To further study the definite role of hepatocyte PPARα in PHx-induced liver regeneration, hepatocyte-specific PPARα-deficient (PparaΔHep) mice were used.33 The knockout efficiency was confirmed by real-time quantitative PCR and Western blot analysis (Supplemental Figure S2). When subjected to PHx, PparaΔHep mice exhibited significantly decreased mitosis in hepatocytes at 32 hours compared with Pparafl/fl mice (Figure 2A). BrdU incorporation showed that peak hepatocyte proliferation appears at 40 hours after PHx in both Pparafl/fl and PparaΔHep mice (Figure 2, B and C). However, PparaΔHep mice showed significantly reduced hepatocyte proliferation at 32 hours after PHx compared with Pparafl/fl mice (Figure 2C). Consistently, the liver weight/body weight ratio after PHx also decreased in PparaΔHep mice (Figure 2D). Taken together, these results suggest that liver regeneration was impaired in PparaΔHep mice after PHx compared with Pparafl/fl mice.
Figure 2.
Disruption of PPARα in hepatocyte inhibits liver regeneration after PHx. A: Representative images of hematoxylin and eosin staining of liver tissues from PparaΔHep mice and Pparafl/fl mice at 24, 32, 40, and 48 hours. The arrows refer to mitotic hepatocytes. B: Representative images of 5-bromo-2-deoxyuridine (BrdU) staining of residual liver in two groups of mice at 24, 32, 40, and 48 hours after PHx. C: BrdU-positive hepatocyte/total hepatocyte ratios per field of two groups at indicated time points. D: The ratio of liver weight/body weight (LW/BW) of the two groups at different time points. Data are expressed as means ± SD (D). n = 5 Pparafl/fl mice; n = 11 PparaΔHep mice at 24, 32, 40 and 48 hours after PHx. ∗P < 0.05, ∗∗P < 0.01 versus Pparafl/fl mice. Scale bars = 20 μm (A and B). Original magnification, ×400 (A and B).
Disruption of PPARα in Hepatocytes Results in Decreased Cell Cyclin and DNA Repair Gene Expression after PHx
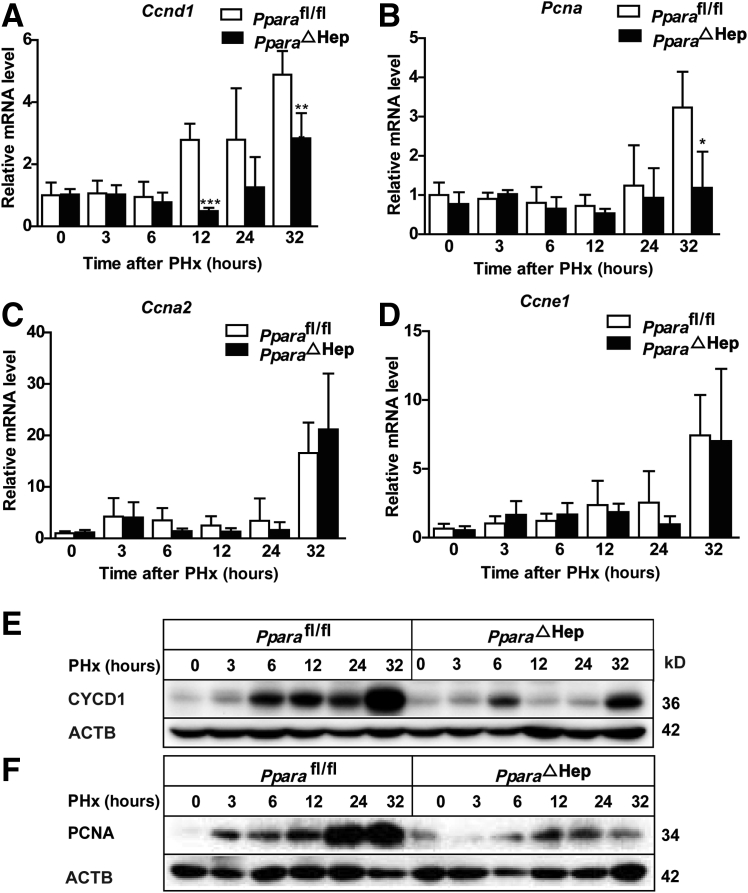
To further confirm whether hepatocyte PPARα deficiency inhibits the cell cycle after PHx, the kinetics of key cell cycle proteins, including CYCD1, CYCA2, CYCE1, and PCNA expression, were measured. Ccna2 and Ccne1 mRNA levels were comparable between Pparafl/fl and PparaΔHep mice; however, Ccnd1 mRNA, expressed during G1-S phase progression, was significantly decreased in PparaΔHep mice (Figure 3, A–C) after PHx compared with Pparafl/fl mice, and Pcna mRNA levels also decreased at 32 hours after PHx compared with Pparafl/fl mice (Figure 3D), consistent with BrdU incorporation assay (Figure 2). Finally, the CYCD1 and PCNA protein levels were determined in Pparafl/fl and PparaΔHep mice after PHx at 0, 3, 6, 12, 24, and 32 hours. PparaΔHep mice showed a decrease of CYCD1 protein expression at different time points after PHx compared with that in Pparafl/fl mice (Figure 3E). PCNA protein also decreased at 12, 24, and 32 hours after PHx in contrast with Pparafl/fl mice (Figure 3F). These results suggest that hepatocyte PPARα may regulate cell proliferation by inducing cell cycle–related gene expression, which is in agreement with previous findings.40
Figure 3.
Disruption of PPARα in hepatocytes results in decreased cell cycle gene expression after PHx. A–D: Real-time PCR analysis of cell cycle–related genes Ccna2, Ccnd1, Ccne1, and Pcna mRNA levels in PparaΔHep and Pparafl/fl mice at different time points after PHx. E and F: Western blot analysis of CYCD1 and proliferating cell nuclear antigen (PCNA) protein expression in PparaΔHep mice and Pparafl/fl mice at 0, 3, 6, 12, 24, and 32 hours after PHx. Data are expressed as means ± SD (A–D). n = 5 (A–D). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 versus Pparafl/fl. ACTB, β-actin.
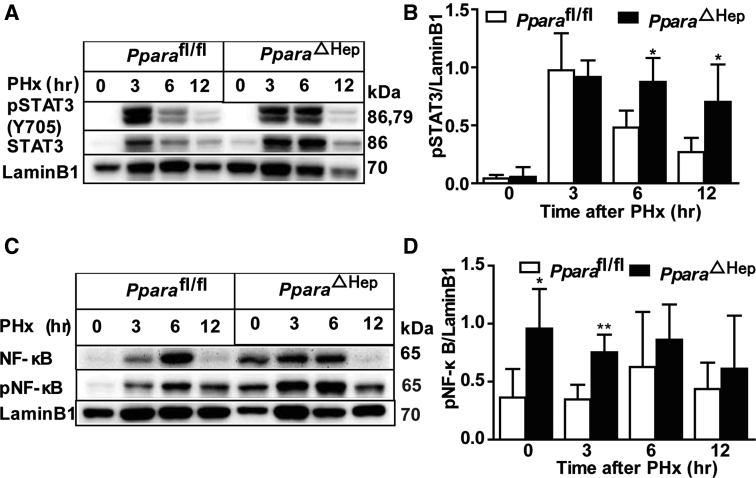
It is well established that STAT3 and NF-κB signaling pathways are activated in the early stage of liver regeneration.4 Activation of STAT3 and NF-κB was found at the early time points of 3 and 6 hours after PHx, as revealed by Western blot analyses (Supplemental Figure S3).27 Surprisingly, the phosphorylated STAT3 level was slightly higher at 6 and 12 hours after PHx in PparaΔHep mice, and phosphorylated NF-κB was also higher at 0 and 3 hours after PHx in PparaΔHep mice when compared with that in Pparafl/fl mice (Supplemental Figure S3). Activation of STAT3 and NF-κB was found to contribute to early liver regeneration after PHx.4, 9 Despite the slight increase in activation of STAT3 and NF-κB in the early stages of PHx, the decreased cell cycle–related protein expression in PparaΔHep mice suggested that the hepatocyte-specific PPARα-stimulated cell proliferation is STAT3/NF-κB independent.
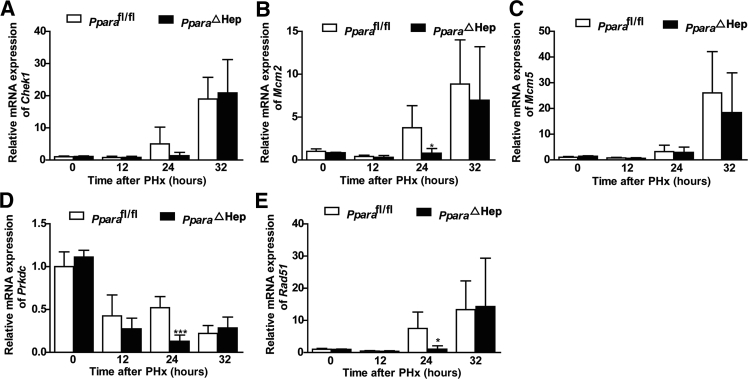
Increased reactive oxygen species are found in liver after PHx, which could damage cellular DNA.41 A previous study demonstrated that hepatic nonparenchymal cell DNA repair gene activation was dependent on PPARα.42 Accordingly, hepatocyte PPARα may play a role in regulating DNA repair. DNA repair gene mRNA Rad51, Prkdc, and Mcm2 were decreased at 24 hours in PparaΔHep mice when compared with that in Pparafl/fl mice (Figure 4).
Figure 4.
Disruption of PPARα in hepatocytes results in decreased DNA repair gene expression after PHx. A–E: Quantitative PCR analysis of DNA repair–related gene Chek1, Mcm2/5, Prkdc, and Rad51 mRNAs at different time points after PHx. Data are expressed as means ± SD (A–E). n = 5 (A–E). ∗P < 0.05, ∗∗∗P < 0.001 versus Pparafl/fl.
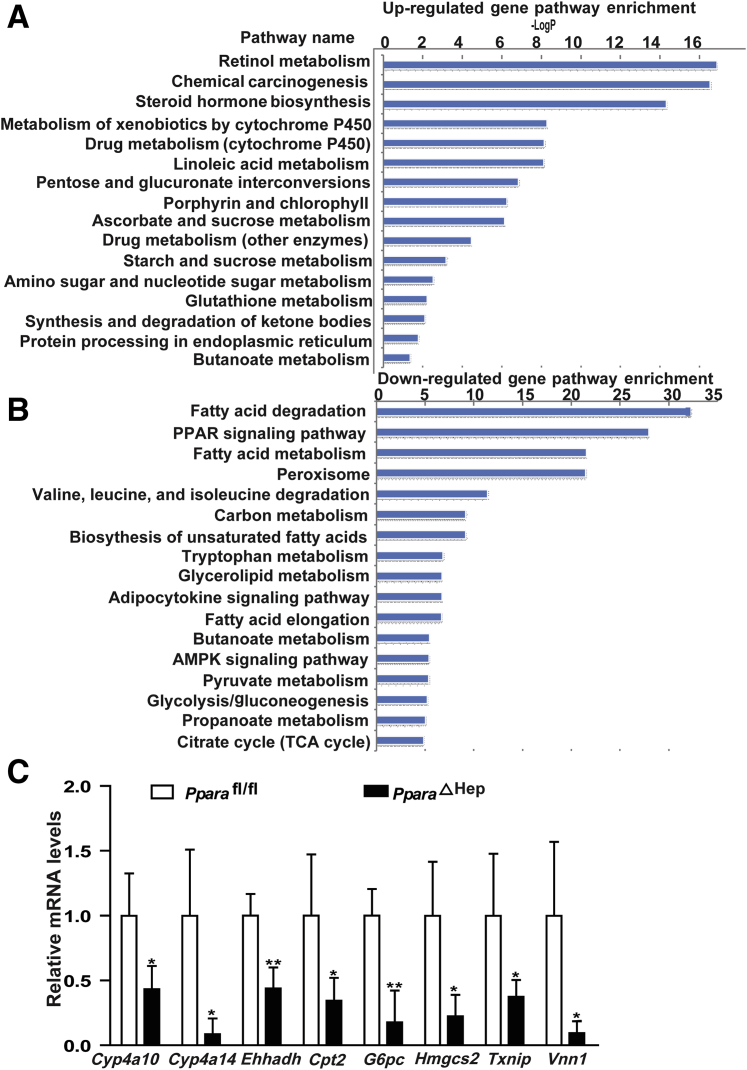
Microarray Analysis for Hepatic Gene Profile Changes from Hepatocyte PPARα-Deficient Mice at 12 Hours after PHx
PPARα acts as the master of fatty acid oxidation in liver; however, the changes of its target genes after PHx and the consequent effects on regenerative genes were unknown. At 12 to 24 hours after PHx, the mice develop marked steatosis.11 Therefore, the time point of 12 hours after PHx was chosen to perform microarray analysis. Hepatic mRNA profiles were examined in PparaΔHep and Pparafl/fl mice liver at 12 hours after PHx (Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo; accession number GSE114223). Pathway enrichment analysis showed that compared with control mice, the up-regulated genes in PparaΔHep mice are mainly involved in drug metabolism and detoxification and the cytochrome P450 family (Figure 5A). Because there were many down-regulated genes, only the top 20 pathway types were shown (Figure 5B). These genes were mainly involved in hepatic fatty acid metabolism, PPARα signaling, amino acid metabolism, glycolysis, and gluconeogenesis. The most obvious change was lipid metabolism (Figure 5B). To confirm this result, some of the mRNAs were quantitated, including Cyp4a10/14, Cpt2, Ehhadh, G6pc, Hmgcs2, Txnip, and Vnn1, revealing that they were lower, consistent with the microarray analysis (Figure 5C).
Figure 5.
Microarray analyses for hepatic gene profiles from hepatocyte PPARα-deficient mice at 12 hours after PHx. A: Up-regulated gene pathways in PparaΔHep mice compared with control mice at 12 hours after PHx. B: Down-regulated gene pathways in PparaΔHep mice compared with Pparafl/fl mice at 12 hours after PHx, as determined by microarray analysis. C: Quantification of Cyp4a10, Cyp4a14, Cpt2, Ehhadh, G6pc, Hmgcs2, Txnip, and Vnn1 mRNAs in PparaΔHep and Pparafl/fl mice. Data are expressed as means ± SD (C). n = 4 (A–C). ∗P < 0.05, ∗∗P < 0.01 versus Pparafl/fl. AMPK, adenosine 5’-monophosphate-activated protein kinase; TCA, tricarboxylic acid cycle.
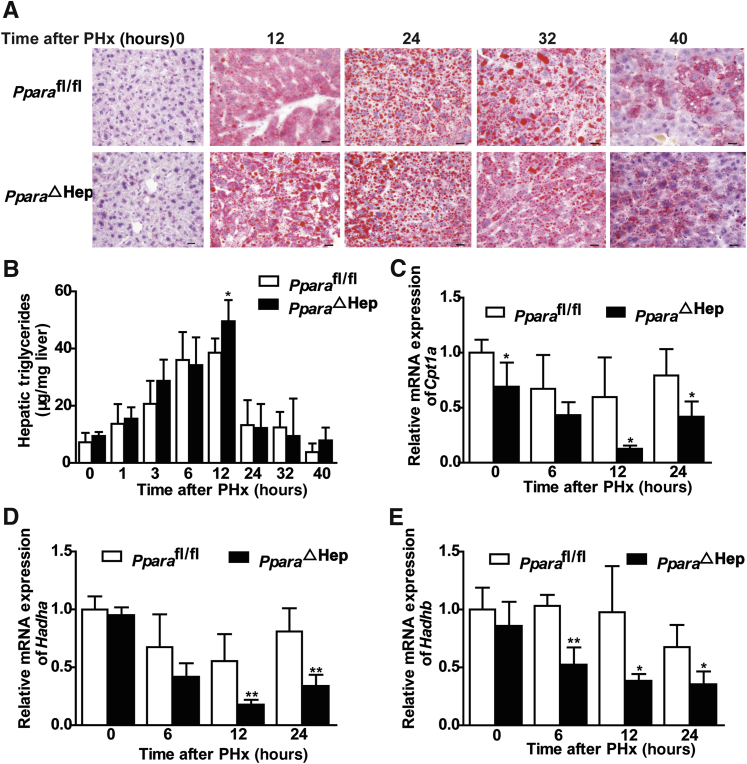
Hepatocyte PPARα Deficiency Promotes Hepatic Lipid Accumulation during Liver Regeneration
PPARα plays an important role in regulating fatty acid oxidation.43 Microarray analysis results indicate that hepatic PPARα may promote liver regeneration after PHx by regulating the lipid metabolism to meet the energy requirement. To study the hepatic lipid metabolism changes after PHx, oil red O staining was used to measure the hepatic lipid accumulation. PparaΔHep mice showed more hepatic lipid accumulation and increased triglyceride contents than those in Pparafl/fl mice at 12 hours after PHx (Figure 6, A and B). In accordance with this result, mRNAs encoded by genes involved in hepatic fatty acid β-oxidation, such as Cpt1a and Hadha/b, all decreased at 12 and 24 hours after PHx in PparaΔHep mice when compared with Pparafl/fl mice (Figure 6, C–E). These results indicate that hepatic PPARα could promote liver regeneration by increasing fatty acid oxidation and lipid accumulation.
Figure 6.
Hepatic PPARα deficiency promotes hepatic lipid accumulation during liver regeneration. A: Representative images for oil red O staining of liver tissues from PparaΔHep mice and Pparafl/fl mice at 0, 12, 24, 32, and 40 hours. B: Hepatic triglyceride content in liver tissues. C–E: Fatty acid β-oxidation related Cpt1a and Hadha/b mRNAs. Data are expressed as means ± SD (B–E). n = 5 (B–E). ∗P < 0.05, ∗∗P < 0.01 versus Pparafl/fl. Scale bars = 20 μm (A). Original magnification, ×400 (A).
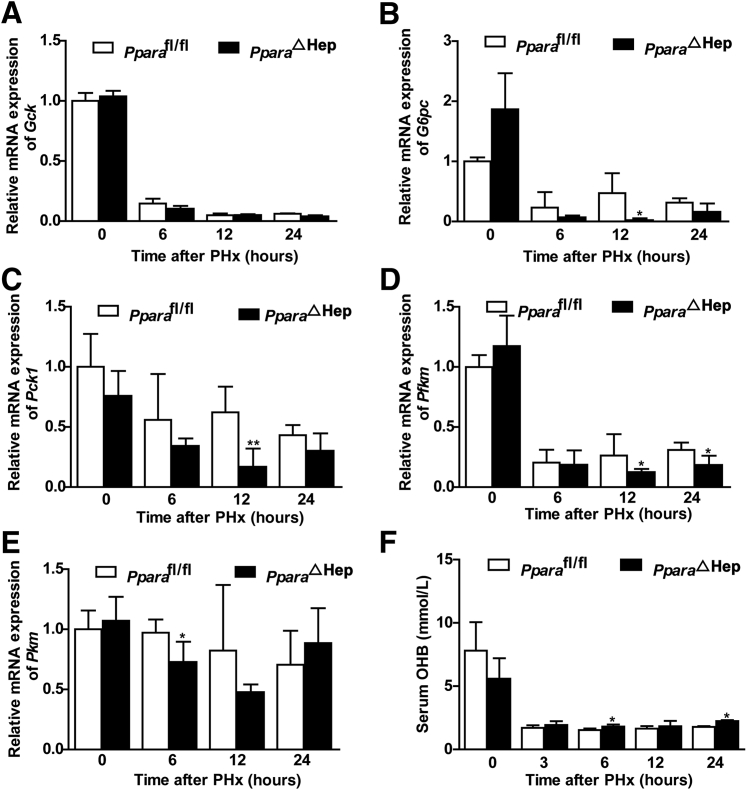
Hepatocyte PPARα Deficiency Reduces Hepatic Glucose Metabolism Gene Expression during Liver Regeneration
Previous studies showed that PPARα plays an essential role in maintaining glucose, lipid, and cholesterol homeostasis.44 Moreover, glucose metabolism is a source of energy for liver regeneration.11 Therefore, glycolysis and gluconeogenesis gene expression in liver after PHx were measured. G6pc, Pck1, Pkm, and Pfkm mRNA levels were decreased in PparaΔHep mice when compared with those in Pparafl/fl mice (Figure 7, A–E). These results show that PPARα may supply energy for liver regeneration by regulating glucose metabolism.
Figure 7.
Hepatic PPARα deficiency reduces hepatic glucose metabolism gene expression during liver regeneration. A–E: mRNA expression of gluconeogenesis genes G6pc and Pck1 (A and B) and glycolysis genes Gck, Pkm, and Pfkm (C–E). F: Serum hydroxybutyrate levels after PHx at 0, 3, 6, 12, and 24 hours in PparaΔHep and Pparafl/fl mice. Data are expressed as means ± SD (A–F). n = 5 (A–F). ∗P < 0.05, ∗∗P < 0.01 versus Pparafl/fl.
β-Hydroxybutyrate is a ketone body mainly produced in the liver from fatty acids during fasting, prolonged exercise, or absence of dietary carbohydrates, when the glucose supply is too low to meet the energy needs. β-Hydroxybutyrate is then distributed via the circulation to metabolically active tissues as a glucose-sparing energy source.45, 46 β-Hydroxybutyrate can promote histone hyperacetylation and reduce lipolysis and metabolic rates.45 Hepatocyte-specific disruption of PPARα increased serum β-hydroxybutyrate and slightly decreased the metabolic rate at 6 and 24 hours after PHx (Figure 7F). Therefore, the energy supply was not enough for normal liver regeneration compared with Pparafl/fl mice.
Discussion
In this study, hepatocyte PPARα was found to promote liver regeneration through regulating cell cycle and lipid/glucose metabolism. In addition to whole body PPARα's role in liver regeneration after PHx,26, 31, 47 the current study revealed that PPARα expressed in the hepatocyte controls energy supplies to ensure normal liver cell cycle progression. A previous study showed that PPARα agonist induced hepatocyte proliferation mainly dependent on hepatocyte PPARα but not PPARα expressed in nonparenchymal cell in the liver.33 Herein, more evidence is provided supporting a role for PPARα in hepatocyte in regulating cell proliferation in the PHx model.
PPARα is a ligand-activated nuclear receptor that participates in multiple pathophysiological processes, including lipid metabolism, glucose homeostasis, cell proliferation, and inflammatory response.27 PPARα is mainly expressed in liver, heart, and kidney and to a lesser degree in other organs. Mice with global knockout of PPARα develop transiently impaired liver regeneration after PHx, which is associated with changes in expression of cell cycle control genes, cytokine signaling, and fat metabolism.26 In the present study, conditional disruption of PPARα in hepatocytes resulted in a decreased liver regeneration after PHx. Moreover, in contrast with Pparafl/fl mice, a decrease of Ccnd1 mRNA and CYCD1 protein levels was noted at 12, 24, and 32 hours in PparaΔHep mice after PHx. CYCD1 is a key cell cycle protein that forms complexes with cyclin-dependent kinases 4 and 6, which promote cell cycle progression from the G1 phase to the S phase.48 Consistently, PparaΔHep mice showed decreased Pcna mRNA and PCNA protein expression at 32 hours after PHx. Furthermore, the present data indicate that the decreased CYCD1 and PCNA at the early stage of PHx in PparaΔHep mice is likely not because of activation of the STAT3/NF-κB pathway. These data suggest that hepatocyte PPARα promotes cell proliferation by regulating CYCD1 and PCNA expression.
Transient liver steatosis is found 12 to 24 hours after PHx and is critical for normal liver regeneration, but excess hepatic steatosis and disruption of hepatic lipid accumulation before PHx impair liver regeneration in mice.19 A previous study reported that Ppara whole body knockout mice showed transiently reduced liver regeneration with decreased fatty acid β-oxidation.26 To fully understand the relationship between liver regeneration and lipid metabolism in mice, microarray analysis was performed in PparaΔHep and Pparafl/fl control mice after PHx, revealing decreased expression of mRNAs encoding hepatic fatty acid metabolism enzymes, PPARα signaling, amino acid metabolism, glycolysis, and gluconeogenesis gene expression in PparaΔHep mice. In support of these findings, hepatic lipid and triglyceride contents were increased, whereas fatty acid β-oxidation gene mRNAs were decreased in PparaΔHep mice compared with Pparafl/fl mice at 12 hours after PHx. On the other hand, gluconeogenesis G6pc, Pck1, Pkm, and Pfkm mRNA levels were decreased in PparaΔHep mice compared with Pparafl/fl mice, and serum hydroxybutyrate levels increased. These results suggest that hepatic PPARα promotes fatty acid β-oxidation, lipid metabolism, and glucose metabolism to meet the large energy requirement needed for liver regeneration.
In conclusion, the present study found that hepatocyte PPARα promotes liver regeneration after PHx at least partially via regulating cell cycle–related gene expression and promoting fatty acid β-oxidation and glucose metabolism.
Footnotes
Supported by the National Natural Science Foundation of China grants 81100311 (S.Y.), 81470879 (S.Y.), 81770588 (H.W.), 81522009 (H.W.), 81670400 (A.Q.), and 91739120 (A.Q.); and Beijing Municipal Institutions Importation and Development of High-Caliber Talents Project CIT&TCD20150325 (A.Q.).
G.X. and S.Y. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.10.009.
Contributor Information
Hua Wang, Email: wanghua@ahmu.edu.cn.
Aijuan Qu, Email: aijuanqu@ccmu.edu.cn.
Supplemental Data
Supplemental Figure S1.
Retinoid X receptor α protein level is comparable after PHx or sham surgery in wild-type (WT) mice. Western blot analysis of retinoid X receptor α protein expression in WT mice livers after PHx or sham operation. Data are expressed as means ± SD.
Supplemental Figure S2.
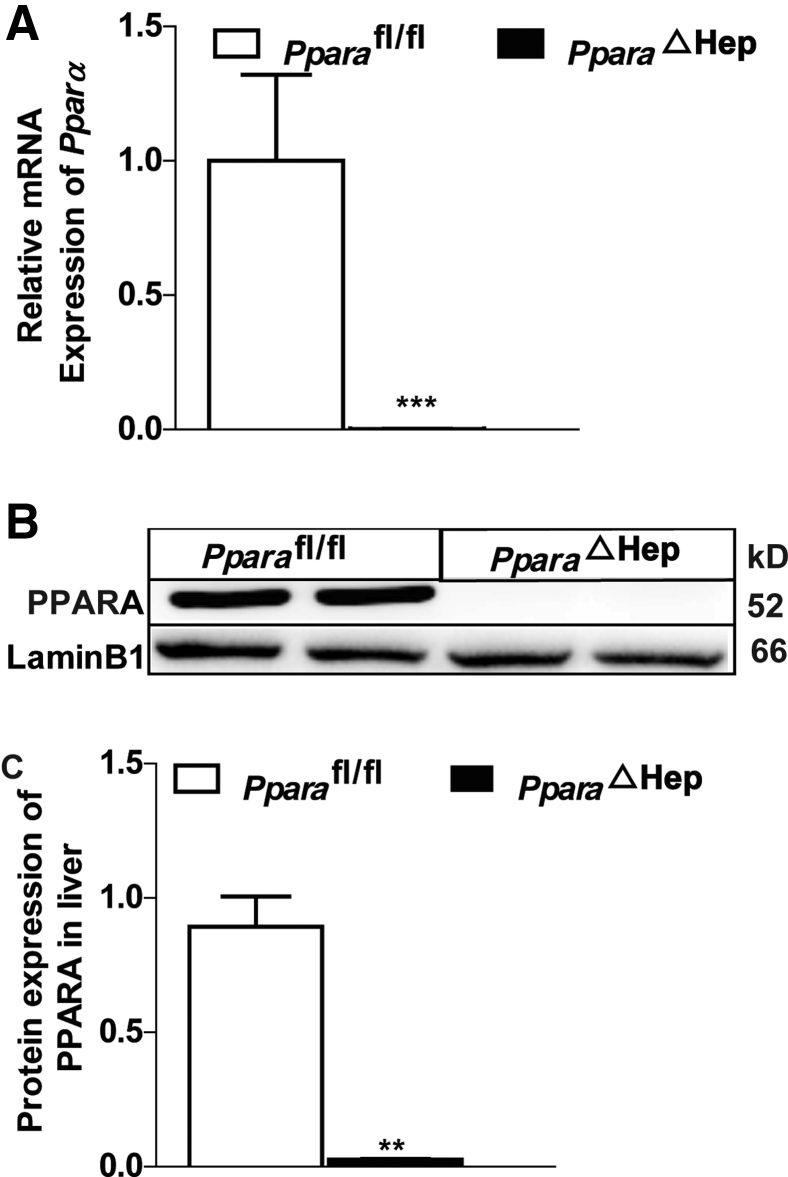
Validation of hepatocyte PPARα knockout efficiency in mice. A: Real-time PCR analysis of hepatic Ppara mRNA levels in PparaΔHep mice and Pparafl/fl mice. B and C: Western blot analysis of heptic PPARα protein expression in PparaΔHep mice and Pparafl/fl mice and statistical graph. Data are expressed as means ± SD (A and C). n = 5 (A); n = 2 (B and C). ∗∗P < 0.01, ∗∗∗P < 0.001 versus Pparafl/fl.
Supplemental Figure S3.
Hepatocyte-specific deletion of PPARα increases STAT3 and NF-κB activation. Western blot analysis of phosphorylated STAT3 (pSTAT3), STAT3, phosphorylated NF-κB (pNF-κB), and NF-κB protein expression in PparaΔHep and Pparafl/fl mice livers after PHx and statistical graph of A and B. Data are expressed as means ± SD (B and D). n = 3 (A–D). ∗P < 0.05, ∗∗P < 0.01 versus Pparafl/fl.
References
- 1.Miyaoka Y., Miyajima A. To divide or not to divide: revisiting liver regeneration. Cell Div. 2013;8:8. doi: 10.1186/1747-1028-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 3.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 4.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos G.K. Principles of liver regeneration and growth homeostasis. Compr Physiol. 2013;3:485–513. doi: 10.1002/cphy.c120014. [DOI] [PubMed] [Google Scholar]
- 6.Michalopoulos G.K. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalopoulos G.K. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalopoulos G.K. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 9.Wang H., Park O., Lafdil F., Shen K., Horiguchi N., Yin S., Fu X.Y., Kunos G., Gao B. Interplay of hepatic and myeloid signal transducer and activator of transcription 3 in facilitating liver regeneration via tempering innate immunity. Hepatology. 2010;51:1354–1362. doi: 10.1002/hep.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornell R.P. Gut-derived endotoxin elicits hepatotrophic factor secretion for liver regeneration. Am J Physiol. 1985;249:R551–R562. doi: 10.1152/ajpregu.1985.249.5.R551. [DOI] [PubMed] [Google Scholar]
- 11.Huang J., Rudnick D.A. Elucidating the metabolic regulation of liver regeneration. Am J Pathol. 2014;184:309–321. doi: 10.1016/j.ajpath.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau F., Seyfritz E., Toti F., Sigrist S., Bietigier W., Pinget M., Kessler L. Early effects of liver regeneration on endocrine pancreas: in vivo change in islet morphology and in vitro assessment of systemic effects on beta-cell function and viability in the rat model of two-thirds hepatectomy. Horm Metab Res. 2014;46:921–926. doi: 10.1055/s-0034-1389995. [DOI] [PubMed] [Google Scholar]
- 13.Huang J., Schriefer A.E., Cliften P.F., Dietzen D., Kulkarni S., Sing S., Monga S.P., Rudnick D.A. Postponing the hypoglycemic response to partial hepatectomy delays mouse liver regeneration. Am J Pathol. 2016;186:587–599. doi: 10.1016/j.ajpath.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siu J., McCall J., Connor S. Systematic review of pathophysiological changes following hepatic resection. HPB (Oxford) 2014;16:407–421. doi: 10.1111/hpb.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudnick D.A., Davidson N.O. Functional relationships between lipid metabolism and liver regeneration. Int J Hepatol. 2012;2012:549241. doi: 10.1155/2012/549241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang S.Q., Lin H.Z., Mandal A.K., Huang J., Diehl A.M. Disrupted signaling and inhibited regeneration in obese mice with fatty livers: implications for nonalcoholic fatty liver disease pathophysiology. Hepatology. 2001;34:694–706. doi: 10.1053/jhep.2001.28054. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi H., Uetsuka K., Okada T., Nakayama H., Doi K. Impaired liver regeneration after partial hepatectomy in db/db mice. Exp Toxicol Pathol. 2003;54:281–286. doi: 10.1078/0940-2993-00265. [DOI] [PubMed] [Google Scholar]
- 18.DeAngelis R.A., Markiewski M.M., Taub R., Lambris J.D. A high-fat diet impairs liver regeneration in C57BL/6 mice through overexpression of the NF-kappaB inhibitor, IkappaBalpha. Hepatology. 2005;42:1148–1157. doi: 10.1002/hep.20879. [DOI] [PubMed] [Google Scholar]
- 19.Shteyer E., Liao Y., Muglia L.J., Hruz P.W., Rudnick D.A. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology. 2004;40:1322–1332. doi: 10.1002/hep.20462. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Wang Y.D., Chen W.D., Wang X., Lou G., Liu N., Lin M., Forman B.M., Huang W. Promotion of liver regeneration/repair by farnesoid X receptor in both liver and intestine in mice. Hepatology. 2012;56:2336–2343. doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borude P., Edwards G., Walesky C., Li F., Ma X., Kong B., Guo G.L., Apte U. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology. 2012;56:2344–2352. doi: 10.1002/hep.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez M.A., Albor C., Ingelmo-Torres M., Nixon S.J., Ferguson C., Kurzchalia T., Tebar F., Enrich C., Parton R.G., Pol A. Caveolin-1 is essential for liver regeneration. Science. 2006;313:1628–1632. doi: 10.1126/science.1130773. [DOI] [PubMed] [Google Scholar]
- 23.Ren G., Wang Z.C., Cui Y.Y. [Role of caveolin-1 in hepatocyte proliferation and liver regeneration] Sheng Li Ke Xue Jin Zhan. 2009;40:341–344. Chinese. [PubMed] [Google Scholar]
- 24.Liu H.X., Fang Y., Hu Y., Gonzalez F.J., Fang J., Wan Y.J. PPARbeta regulates liver regeneration by modulating Akt and E2f signaling. PLoS One. 2013;8:e65644. doi: 10.1371/journal.pone.0065644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gazit V., Huang J., Weymann A., Rudnick D.A. Analysis of the role of hepatic PPARgamma expression during mouse liver regeneration. Hepatology. 2012;56:1489–1498. doi: 10.1002/hep.25880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson S.P., Yoon L., Richard E.B., Dunn C.S., Cattley R.C., Corton J.C. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36:544–554. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 27.Lefebvre P., Chinetti G., Fruchart J.C., Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corton J.C., Anderson S.P., Stauber A. Central role of peroxisome proliferator-activated receptors in the actions of peroxisome proliferators. Annu Rev Pharmacol Toxicol. 2000;40:491–518. doi: 10.1146/annurev.pharmtox.40.1.491. [DOI] [PubMed] [Google Scholar]
- 29.Peters J.M., Cattley R.C., Gonzalez F.J. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029–2033. doi: 10.1093/carcin/18.11.2029. [DOI] [PubMed] [Google Scholar]
- 30.Tien E.S., Gray J.P., Peters J.M., Vanden Heuvel J.P. Comprehensive gene expression analysis of peroxisome proliferator-treated immortalized hepatocytes: identification of peroxisome proliferator-activated receptor alpha-dependent growth regulatory genes. Cancer Res. 2003;63:5767–5780. [PubMed] [Google Scholar]
- 31.Wheeler M.D., Smutney O.M., Check J.F., Rusyn I., Schulte-Hermann R., Thurman R.G. Impaired Ras membrane association and activation in PPARalpha knockout mice after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol. 2003;284:G302–G312. doi: 10.1152/ajpgi.00175.2002. [DOI] [PubMed] [Google Scholar]
- 32.Yuan X., Yan S., Zhao J., Shi D., Yuan B., Dai W., Jiao B., Zhang W., Miao M. Lipid metabolism and peroxisome proliferator-activated receptor signaling pathways participate in late-phase liver regeneration. J Proteome Res. 2011;10:1179–1190. doi: 10.1021/pr100960h. [DOI] [PubMed] [Google Scholar]
- 33.Brocker C.N., Yue J., Kim D., Qu A., Bonzo J.A., Gonzalez F.J. Hepatocyte-specific PPARA expression exclusively promotes agonist-induced cell proliferation without influence from nonparenchymal cells. Am J Physiol Gastrointest Liver Physiol. 2017;312:G283–G299. doi: 10.1152/ajpgi.00205.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Jamal N., Dubuquoy L., Auwerx J., Bertin B., Desreumaux P. In vivo imaging reveals selective PPAR activity in the skin of peroxisome proliferator-activated receptor responsive element-luciferase reporter mice. Exp Dermatol. 2013;22:137–140. doi: 10.1111/exd.12082. [DOI] [PubMed] [Google Scholar]
- 35.Wang H., Feng D., Park O., Yin S., Gao B. Invariant NKT cell activation induces neutrophil accumulation and hepatitis: opposite regulation by IL-4 and IFN-gamma. Hepatology. 2013;58:1474–1485. doi: 10.1002/hep.26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell C., Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell C., Willenbring H. Addendum: a reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2014;9:1532. doi: 10.1038/nprot.2014.122. [DOI] [PubMed] [Google Scholar]
- 38.Qu A., Taylor M., Xue X., Matsubara T., Metzger D., Chambon P., Gonzalez F.J., Shah Y.M. Hypoxia-inducible transcription factor 2alpha promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 2011;54:472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q., Ito S., Gonzalez F.J. Hepatocyte-restricted constitutive activation of PPAR alpha induces hepatoproliferation but not hepatocarcinogenesis. Carcinogenesis. 2007;28:1171–1177. doi: 10.1093/carcin/bgm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters J.M., Aoyama T., Cattley R.C., Nobumitsu U., Hashimoto T., Gonzalez F.J. Role of peroxisome proliferator-activated receptor alpha in altered cell cycle regulation in mouse liver. Carcinogenesis. 1998;19:1989–1994. doi: 10.1093/carcin/19.11.1989. [DOI] [PubMed] [Google Scholar]
- 41.Tachibana S., Zhang X., Ito K., Ota Y., Cameron A.M., Williams G.M., Sun Z. Interleukin-6 is required for cell cycle arrest and activation of DNA repair enzymes after partial hepatectomy in mice. Cell Biosci. 2014;4:6. doi: 10.1186/2045-3701-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu A., Shah Y.M., Matsubara T., Yang Q., Gonzalez F.J. PPARalpha-dependent activation of cell cycle control and DNA repair genes in hepatic nonparenchymal cells. Toxicol Sci. 2010;118:404–410. doi: 10.1093/toxsci/kfq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferre P. The biology of peroxisome proliferator-activated receptors: relationship with lipid metabolism and insulin sensitivity. Diabetes. 2004;53(Suppl 1):S43–S50. doi: 10.2337/diabetes.53.2007.s43. [DOI] [PubMed] [Google Scholar]
- 44.Chakravarthy M.V., Pan Z., Zhu Y., Tordjman K., Schneider J.G., Coleman T., Turk J., Semenkovich C.F. “New” hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab. 2005;1:309–322. doi: 10.1016/j.cmet.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Newman J.C., Verdin E. Ketone bodies as signaling metabolites. Trends Endocrinol Metab. 2014;25:42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman J.C. Verdin E: beta-hydroxybutyrate: a signaling metabolite. Annu Rev Nutr. 2017;37:51–76. doi: 10.1146/annurev-nutr-071816-064916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao M.S., Peters J.M., Gonzalez F.J., Reddy J.K. Hepatic regeneration in peroxisome proliferator-activated receptor alpha-null mice after partial hepatectomy. Hepatol Res. 2002;22:52–57. doi: 10.1016/s1386-6346(01)00119-x. [DOI] [PubMed] [Google Scholar]
- 48.Kamarajugadda S., Becker J.R., Hanse E.A., Mashek D.G., Mashek M.T., Hendrickson A.M., Mullany L.K., Albrecht J.H. Cyclin D1 represses peroxisome proliferator-activated receptor alpha and inhibits fatty acid oxidation. Oncotarget. 2016;7:47674–47686. doi: 10.18632/oncotarget.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]