Abstract
We describe a single-color digital PCR assay that detects and quantifies cancer mutations directly from circulating DNA collected from the plasma of cancer patients. This approach relies on a double-stranded DNA intercalator dye and paired allele-specific DNA primer sets to determine an absolute count of both the mutation and wild-type–bearing DNA molecules present in the sample. The cell-free DNA assay uses an input of 1 ng of nonamplified DNA, approximately 300 genome equivalents, and has a molecular limit of detection of three mutation DNA genome-equivalent molecules per assay reaction. When using more genome equivalents as input, we demonstrated a sensitivity of 0.10% for detecting the BRAF V600E and KRAS G12D mutations. We developed several mutation assays specific to the cancer driver mutations of patients' tumors and detected these same mutations directly from the nonamplified, circulating cell-free DNA. This rapid and high-performance digital PCR assay can be configured to detect specific cancer mutations unique to an individual cancer, making it a potentially valuable method for patient-specific longitudinal monitoring.
In cancer patients, apoptotic and necrotic tumor cells release their DNA content into the blood. This is commonly referred to as circulating tumor DNA (ctDNA); one uses either DNA sequencing or molecular genotyping assays to identify and quantitatively measure those ctDNA molecules bearing somatic mutations.1, 2 Liquid biopsy specimens are used to detect specific cancer mutations in ctDNA from the plasma fraction of a standard blood draw, and detecting mutations in ctDNA has numerous applications.2 These detected mutations may indicate the type of tumor present, thus reducing the need for an invasive tissue biopsy specimen.3, 4, 5 Longitudinal, quantitative monitoring of cancer mutations in ctDNA provides information about the general tumor burden for an individual patient during treatment.4 For example, elevated levels of detectable ctDNA mutations are associated with the presence of higher tumor burden.2, 6 One can measure and quantitatively track the abundance of cancer mutations from ctDNA after surgical resection of the tumor. Furthermore, one can identify mutations that can provide insight into treatment options; an example of this would be the presence of hotspot KRAS mutations and the negative predictive response to anti–epidermal growth factor receptor therapy.5, 7, 8
Current methods of ctDNA analysis include next-generation targeted sequencing and fluorescent probe–based digital PCR assays.9, 10 Regardless of the method, several challenges exist in identifying ctDNA mutations. Blood-borne DNA is highly fragmented into low-molecular-weight species that are on average <200 bp in length.11 The ctDNA intermingles with the normal DNA present in circulation. Thus, the overall ctDNA fraction of the total circulating cell-free DNA is typically low in concentration, even in those patients with high metastatic tumor burden. Furthermore, the analysis of ctDNA is complicated by its short half-life that ranges from several minutes to hours.1, 12, 13, 14 Some next-generation targeted sequencing and digital PCR methods rely on preamplification of the cell-free DNA to increase the amount of DNA template and improve the quantitation of measurable ctDNA with mutations.15, 16 However, a preamplification step introduces polymerase-based errors and PCR bias that may obscure the accurate detection of mutations.
To address these challenges, we developed a molecular assay that uses single-color digital droplet PCR (ddPCR).17 This approach involves partitioning one or several DNA molecules into individual microvolume droplet reactions, where a specific PCR amplicon product is generated. We use single-color intercalator dye (eg, EvaGreen) that binds to double-stranded DNA, such as PCR amplicons. After droplet partitioning and thermal cycling, one measures the droplet-specific fluorescence generated from the interaction between the single-color intercalator dye and the PCR amplicon (Figure 1). The resulting single-color dye fluorescent signal has a direct relationship to the size of the amplicon product.17, 18
Figure 1.
Single-color single-nucleotide variant mutation detection of single-allele (ie, simplex) and paired-allele genotyping assays. A: Simplified representation of a patient-derived sample assay using ddPCR technology. (1) Spin whole blood to isolate plasma, (2) extract circulating cell-free DNA, and (3–5) single-color ddPCR workflow and output. B: A single-allele genotyping assay is designed with a common forward primer that targets genomic DNA paired with an allele-specific reverse primer, thus targeting one allele per ddPCR. The allele-specific base is the last 3′ base of the primer. A paired-allele genotyping assay, in the context of this article, refers to combining the wild-type allele genotyping assay with the mutation allele genotyping assay in one ddPCR. To facilitate separation of droplets based on fluorescence, a noncomplementary tail is added to one of the allele-specific primers, herein shown as the mutation.
PCR primers were designed to detect somatic single-nucleotide variants in cancer mutations. Each assay detects and quantitates a single mutation. A mutation- or wild-type–specific reverse primer is paired with a common target forward primer (Figure 1B). These primer sets are used in two categories of assays that detect either a single variant allele or paired alleles in which one quantitates both the wild-type and variant allele. For the paired-allele assay, the mutation-specific primer has a configurable extension tail that differentiates the mutation amplicons from the wild-type amplicons. The extension tail produces a different size amplicon product (Figure 1B). When the droplet fluorescence data are visualized, this difference leads to the separation of fluorescent clusters composed of either mutation or wild-type amplicons. By quantitatively measuring and counting the number of fluorescence-positive droplets, one determines the number of mutation and wild-type DNA molecules present for a specific mutation (Figure 1A).17 The primers were designed to produce small amplicons that cover genomic segments <150 bases. As a result, one can readily amplify the highly fragmented, low-molecular-weight ctDNA molecules bearing cancer mutations. As already noted, the amplicon extension tail varies the size of PCR products, thus enabling different amplicons and multiplexing to be used for single-color ddPCR. In addition, this single-color assay design avoids the extensive optimization required of a third fluorescent probe (ie, TaqMan), as is commonly used in most ctDNA genotyping assays.
In this study, we demonstrate how a series of specific features of this assay enable the high performance for detecting mutations. This method's flexible design allows for efficient configuration across different cancer mutations, high sensitivity in detecting mutations at the resolution of several DNA molecules from ctDNA, and high specificity of detection for single-nucleotide alterations. Only several nanograms of cell-free DNA are required without the need for preamplification, and one can detect as few as three mutation-bearing molecules in a single reaction. This level of sensitivity enables one to detect a 0.10% mutation fraction. By eliminating preamplification, some bias in the absolute quantification of mutation signal can be avoided. Moreover, these assays can be completed in a matter of hours (Figure 1A).
First, we demonstrate how specific features of the assay can be adjusted for improving the detection of different mutations—this is particularly critical given the small amount of ctDNA that exists in clinical samples. Second, we developed a series of mutation assays specific to the cancers of individual patients. We applied these optimized single-color ddPCR assays on clinical DNA samples, including cell-free DNA, and identified cancer driver mutations in the ctDNA specific to a patient's primary malignancy.
Materials and Methods
DNA Samples and Processing
The Institutional Review Board at Stanford University School of Medicine (Stanford, CA) approved the study, and informed consent was obtained. Each patient had undergone diagnostic cancer gene sequencing of his or her colorectal adenocarcinoma or cholangiocarcinoma and had diagnostic cancer mutation reports available at the time of enrollment into the study. For all cell-free samples, two EDTA vials (BD, Franklin Lakes, NJ) of blood were collected. Within 2 hours of the initial collection, the blood was spun for 10 minutes at 2000 × g. The plasma fraction was transferred to a fresh Eppendorf (Hamburg, Germany) LoBind microtube. The plasma was spun a second time for 10 minutes at 2000 × g. The spun plasma was subsequently transferred into Eppendorf LoBind microtubes in 1-mL aliquots and flash frozen in liquid nitrogen before being stored at −80°C until the time of extraction. The Promega (Madison, WI) Maxwell 16 Circulating DNA Plasma Kit was used to extract cell-free DNA from 0.5- to 1.0-mL aliquots of the spun plasma. To extract DNA from formalin-fixed, paraffin-embedded (FFPE) tissue, a total of 10 sections (5 μm thick) was collected from each block and the DNA was extracted using the Promega Maxwell 16 FFPE Plus LEV DNA extraction kit.
NA18507 wild-type DNA was obtained from the Coriell Institute (Camden, NJ). Cancer cell lines and related DNA were obtained through the following sources: ATCC (Manassas, VA), the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany), the National Institute of Biomedical Innovation's Japanese Collection of Research Bioresources Cell Bank (Asagi Saito Ibaraki-City, Osaka, Japan), and Sigma Aldrich (St. Louis, MO). Additional cancer cell line information matched to the relevant cancer mutation is provided in Table 1. DNA was extracted from the cell lines LS123 and RPMI-8226 (ATCC) using the Promega Maxwell 16 Cell DNA Purification Kit. DNA was quantified with the Qubit dsDNA BR Assay Kit (Invitrogen, Carlsbad, CA) as well as with a custom simplex single-color ddPCR assay targeting the human RPP30 gene (forward, 5′-TGCCCTCAATCAGCCCCTGG-3′; reverse, 5′-TTGCCAAGGAAAATCTAAAGG-3′). As described later, this simplex ddPCR assay was used to quantify cell-free samples.
Table 1.
DNA Samples, Sources, Type, and Mutations
| Sample name | Source | Mutation of interest | Variant allele fraction |
|---|---|---|---|
| NA18507 | Coriell Institute (Camden, NJ) | None | 0.00 |
| LS411N | ATCC (Manassas, VA) | BRAF V600E (c.1799 T>A) | 0.68 |
| RKO | ATCC | PIK3CA H1047R (c.3140 A>G) | 0.49 |
| LS123 | ATCC | TP53 R175H (c.524 G>A) | 0.99 |
| RPMI 8226 | ATCC | TP53 E285K (c.853 G>A) | 1.00 |
| KU1919 | DSMZ (Braunschweig, Germany) | AKT1 E17K (c.49 G>A) | 0.71 |
| PL-21 | DSMZ | KRAS A146V (c.437 C>T) | 0.65 |
| GP2D | Sigma Aldrich (St. Louis, MO) | KRAS G12D (c.35 G>A) | 0.43 |
| RCM-1 | JCRB (Asagi Saito Ibaraki-City, Osaka, Japan) | KRAS G12V (c.35 G>T) | 1.00 |
Included are the catalog names and sample names of all DNA used in this study. The variant allele fraction of the mutation of interest for each DNA sample was determined using the cBio website to explore Cancer Cell Line Encyclopedia data (http://www.cbioportal.org, accessed February 14, 2017).
DSMZ, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures; JCRB, National Institute of Biomedical Innovation's Japanese Collection of Research Bioresources Cell Bank.
Assay Design
Using the results from clinical diagnostic sequencing of the patients' primary colorectal cancer, two or three cancer driver mutations were chosen, specific to each patient, that lead to amino acid substitutions. The selected cancer driver genes for this study included BRAF, KRAS, TP53, AKT1, and PIK3CA. Wild-type and mutation-specific single-allele genotyping assays were designed for selected mutations from these driver genes. These assays were configured with a single common forward primer and two reverse primers that were identical except at the 3′ base (Figure 1B). The 3′ base of the reverse primers was determined based on the target. It was either the single-nucleotide variant mutation of interest or a wild-type reference base. Each primer length was generally 20 nucleotides—when needed, the length was adjusted to improve the overall melting temperature. The primers amplified between 65 and 150 bp of genomic DNA for each mutation of interest. As previously published, a bioinformatics process of selecting primer sequences that are unique in the human genome and whose melting temperatures are within a 3°C range was used.19
Designing Paired-Allele Assays
For assays that simultaneously detect DNA molecules for both the wild type and mutation, a nonspecific 5′ extension tail (ie, repeats of AAAT) was added to the allele-specific primer of one of the single-allele genotyping primer sets (Figure 1B). For a given amplicon, primers containing different tail lengths were tested, and the extension tail length that provided the best separation between the two positive cluster populations was selected (Figure 2). Because there is no universal tail length for cluster separation, this extension tail length experiment was used to identify the optimal length that provided a distinct separation between the mutation and wild-type droplet clusters for other paired-allele mutation detection assays.
Figure 2.
Varied length of 5′ artificial tail allows for mutation (Mut) and wild-type (WT) allele multiplexing. ddPCR amplitude results of the paired-allele KRAS G12V assay using a 1:1 mixture of normal diploid control DNA (NA18507) and KRAS G12V cancer cell line DNA (RCM-1) as template. Shown from left to right are the two-dimensional results of increasing the length of the 5′ artificial tail on the mutation reverse primer. A visual representation of each tail length condition is included above each plot. Populations are colorized green and blue to be representative of the distinct mutation and wild-type populations, respectively.
All primers were synthesized through Integrated DNA Technologies (Coralville, IA), Exiqon (Woburn, MA), and the Stanford Protein and Nucleic Acid Facility (Stanford, CA). Primer sequences are noted in Table 2. Primer sets were optimized using a thermal gradient, and confirmed the PCR specificity using cancer cell line DNA already known to have the mutation and normal human cell line DNA (Table 1).
Table 2.
Primer Sequences and Individual Assay Conditions for All Customized SNV Genotyping Assays
| Assay target | Primer name | Primer sequence | Amplicon length, bp | Assay format | Anneal/extend temperature and time∗ | Final primer concentration, nmol/L |
|---|---|---|---|---|---|---|
| BRAF V600E (c.1799 T>A) | BRAF_600_Forward | 5′-(GGGGGG)CATGAAGACCTCACAGTAAA-3′ | Paired allele | 58°C and 1:00 | 100 | |
| BRAF_600_WT_39T | 5′-(AAATAAATAAATAAATAAATAAATAAATAAATAAATAAA)CCACTCCATCGAGATTTCA-3′ | 104 | ||||
| BRAF_V600E_Mut_6T | 5′-(GGGGGG)CCACTCCATCGAGATTTCT-3′ | 71 | ||||
|
(LNA) BRAF V600E (c.1799 T>A) |
BRAF_600_Forward | 5′-(GGGGGG)CATGAAGACCTCACAGTAAA-3′ | Paired allele | 54°C and 2:00 | 100 | |
| BRAF_600_L_WT_39T | 5′-(AAATAAATAAATAAATAAATAAATAAATAAATAAATAAA)CCACTCCATCGAGATTTC+A-3′ | 104 | ||||
| BRAF_V600E_L_Mut_6T | 5′-(GGGGGG)CCACTCCATCGAGATTTC+T-3′ | 71 | ||||
| KRAS G12D (c.35 G>A) | KRAS_12__Forward | 5′-(GCG)TGTTGGATCATATTCGTCCACAA-3′ | Paired allele | 61°C and 2:00 | 50 | |
| KRAS_12_WT | 5′-(GCG)ACTTGTGGTAGTTGGAGCAGG-3′ | 88 | ||||
| KRAS_G12D_12T | 5′-(AAATAAATAAAT)ACTTGTGGTAGTTGGAGCAGA-3′ | 100 | ||||
| KRAS G12V (c.35 G>T) | KRAS_12_Forward | 5′-(GCG)TGTTGGATCATATTCGTCCACAA-3′ | Paired allele | 63°C and 2:00 | 50 | |
| KRAS_12_WT | 5′-(GCG)ACTTGTGGTAGTTGGAGCAGG-3′ | 88 | ||||
| KRAS_G12V_Mut_100T | 5′-25(AAAT)ACTTGTGGTAGTTGGAGCAGT-3′ | 188 | ||||
| PIK3CA H1047R (c.3140 A>G) | PIK3CA_1047_Forward | 5′-(CG)CGAAAGACCCTAGCCTTAGA-3′ | Single allele | 63.5°C and 2:00 | 50 | |
| PIK3CA_1047_WT_R | 5′-(GC)TTGTCCAGCCACCATGACGT-3′ | 93 | ||||
| PIK3CA_H1047R_Mut_R | 5′-TTGTCCAGCCACCATGACGC-3′ | 93 | ||||
| TP53 R175H (c.524 G>A) | TP53_175_Forward | 5′-GGGGCCAGACCTAAGAGCAATC-3′ | Single allele | 68°C and 1:00 | 100 | |
| TP53_175_WT_R | 5′-ATGACGGAGGTTGTGAGGCG-3′ | 144 | ||||
| TP53_R175H_Mut_R | 5′-ATGACGGAGGTTGTGAGGCA-3′ | 144 | ||||
| AKT1 E17K (c.49 G>A) | AKT1_17_Reverse | 5′-GGTCTGACGGGTAGAGTGT-3′ | Single allele | 64°C and 2:00 | 50 | |
| AKT1_17_WT_F | 5′-CGCCAGGTCTTGATGTACTC-3′ | 71 | ||||
| AKT1_E17K_Mut_F | 5′-CGCCAGGTCTTGATGTACTT-3′ | 71 | ||||
| KRAS A146V (c.437 C>T) | KRAS_146_Forward | 5′-CCTAGTATAGCATAATTGAG-3′ | Single allele | 52°C and 2:00 | 50 | |
| KRAS_146_WT_R | 5′-CCTTTTATTGAAACATCGGC-3′ | 132 | ||||
| KRAS_A146V_Mut_R | 5′-CCTTTTATTGAAACATCGGT-3′ | 132 | ||||
| TP53 E285K (c.853 G>A) | TP53_285_Forward | 5′-CGCTTAGTGCTCCCTGGGGG-3′ | Single allele | 64°C and 1:30 | 50 | |
| TP53_285_WT_R | 5′-TGGGAGAGACCGGCCCATAG-3′ | 84 | ||||
| TP53_E285K_Mut_R | 5′-TGGGAGAGACCGGCCCATAA-3′ | 84 |
Tail sequences are in parenthesis, mismatch bases are underlined, and +N locked nucleic acids are boldfaced.
SNV, single-nucleotide variant.
Time is given in minutes:seconds.
Digital PCR Assay Conditions
For both the validation experiments and patient cell-free experiments, all samples were analyzed with at least three or four replicates. Before ddPCR, the cell line and FFPE genomic DNA samples were treated with an EcoRI (New England BioLabs, Ipswich, MA) restriction digest, incubating for 8 hours at 37°C, followed by subsequent 65°C incubation for 20 minutes to terminate the reaction.17 Patient cell-free DNA did not require enzymatic fragmentation given that these samples are already highly fragmented. All ddPCRs were performed using the Bio-Rad EvaGreen QX200 ddPCR system and protocol (Bio-Rad Laboratories, Hercules, CA). Each 22 μL ddPCR used 1× EvaGreen ddPCR supermix and a final concentration of 50 to 100 nmol/L of each genotyping primer set per reaction (Table 2). The PCR master mix was partitioned and plated per Bio-Rad's QX200 droplet generation protocol. The thermal conditions were as follows, with a heated lid temperature set to 105°C: one cycle of enzyme activation at 95°C for 5 minutes; denaturation at 95°C for 30 seconds, immediately followed by annealing/extension between 54°C and 68°C for between 1 and 2 minutes, depending on the assay target, for a total of 40 cycles; a final droplet stabilization step at 4°C for 5 minutes; 90°C for 5 minutes; and an extended 4°C hold. The annealing and extension temperature and time are noted for each mutation detection assay in Table 2. After thermal cycling, the plate was transferred and read by the Bio-Rad QX200 Droplet reader. Amplitude files were generated using the Bio-Rad droplet reader software, QuantaSoft version 1.7.4.0917.
Determining Number of Mutation DNA Molecules
For reporting results, mass was converted to molarity and the molecular haploid genome equivalents were included—this number accurately reflects the molecule-based detection for these assays. Supplemental Table S1 shows a detailed conversion of nanograms to molecular genome equivalents.
To calculate the number of wild-type and cancer haploid genome equivalents per ddPCR, the following Poisson distribution formula was used:
| (1) |
We calculated the mass of one haploid human genome (m) from a single normal cell in grams using the genome size in bp (n). One haploid genome is approximately 3.3 pg.5 The number of haploid genome equivalents present for all assays was calculated with the following equation: .
The number of haploid genome equivalents calculated above was used in the following formula to determine the fraction or percentage of mutation genome equivalents detected:
| (2) |
DNA Mixtures for Mutation Quantitation
Using the cBio portal to explore Cancer Cell Line Encyclopedia data (http://www.cbioportal.org, accessed February 14, 2017), cancer cell lines that had the same driver mutations identified from diagnostic sequencing of the patient's primary tumor were identified.20, 21 After obtaining these mutation-matched cancer cell lines, the concentration of all DNA samples in terms of haploid genome equivalents was measured. To determine this genome equivalent concentration, a simplex ddPCR assay that amplifies the 110 bases of the human RPP30 gene (forward, 5′-TGCCCTCAATCAGCCCCTGG-3′; reverse, 5′-TTGCCAAGGAAAATCTAAAGG-3′) was used. The master mix and thermal cycling conditions are identical to the digital PCR assay conditions previously noted in Materials and Methods, but used a final primer concentration of 100 nmol/L and a thermal cycling annealing/extension temperature of 54°C for 1 minute. Using the calculated genome equivalent concentration and the reported mutation allele fraction from the cBio website above (Table 1), the number of mutation and wild-type bearing molecules in each of the cancer cell line DNA samples was calculated. A series of DNA mixtures containing different fractional amounts of mutation-bearing genome equivalents (cancer cell line DNA) to normal DNA genome equivalents (NA18507) was prepared. Generally, for any given ddPCR, approximately 300 genome equivalents of the mixed template DNA were used per reaction, equivalent to a final concentration of approximately 1 ng/μL.
In addition, the above RPP30 simplex DNA assay was used to measure the concentration of each patient DNA sample in terms of haploid genome equivalents. This concentration was used to adjust the overall concentration of the DNA dilution series to be equivalent to the cell-free patient sample. This DNA dilution was then used as a standard curve and was run in parallel to every patient sample.
Clustering and Quantitation of Mutation Signal
The QuantaSoft version 1.7.4.0917 (Bio-Rad) experiments were used to assign clusters for simplex assays. The R version 3.2.3 programing language (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org) was used for additional analysis and generating plots. Supplemental Figure S1 provides a simplified workflow of the automated clustering of paired-allele assays. A data file was generated that contained up to four replicates of a positive control mixture condition (ie, 50:50 mutation/wild type), which established CIs for all cluster types and served as an empirical reference model. These reference model data were used to classify droplets in all assays, including those with unknown abundances of mutation and wild-type alleles. A droplet was classified as belonging to a specific cluster type based on the distance of its fluorescent signal to a given reference cluster's CI. By running individual reactions with no template DNA, the baseline fluorescence of negative droplets was identified, and this result was used to determine the threshold of nonspecific amplification events. Significant droplet fluorescence arising from the correct amplicon products was defined to be when, on average, more than one positive droplet was identified among all replicates. Generally, the negative control replicates did not have any significant amplicon products, as indicated by the vast majority of the replicates having no positive droplets.
For each mutation assay, a series of DNA mixtures containing different fractions of mutation and wild-type molecules was generated. These DNA mixture series were used during optimization and validation of all assays. As mentioned in DNA Mixtures for Mutation Quantitation, these series were also adjusted to be the same concentration as clinical cell-free DNA samples and then used as a standard curve when testing patient ctDNA. This standard curve was used in two capacities: to generate the reference model and establish CIs, and to validate the linear range of detection of assays in line with patient mutation detection assays. If the negative control of a standard curve showed positive fluorescent signal, linear regression was performed on the in-line standard curve and was used to interpolate the fraction or percentage of mutation genome equivalent molecules for a given patient ctDNA of FFPE sample. Inclusion of a multipoint standard curve alongside patient cell-free samples provides an in-line measure of the assay function; however, in practice, multiple replicates of mixed template (ie, 50% mutation) and negative control template (ie, 0% mutation) are sufficient to generate the CIs required to automatically cluster patient data, as described in the previous paragraph.
Statistical Analysis
Correlation analyses were performed using the Pearson correlation coefficient and associated two-tailed P values. These values and linear plots were generated using Prism 7 statistical analysis software version 7.0a (GraphPad Software, Inc., La Jolla, CA). Prism 7 was also used to perform the linear regression analysis method mentioned above to interpolate the percentage mutation. Additional plots were generated using the R programing language (R Foundation for Statistical Computing, Vienna, Austria; http://www.r-project.org). A one-sided t-test was used for determining significance among different assay conditions and background signal.
Results
For this single-color, droplet-based dPCR assay, we describe several of the critical parameters that enable this assay to detect rare mutations from ctDNA. All experiments were run in triplicate or greater per sample and used genomic or cell-free DNA directly without any preamplification steps. For assay development and performance testing, we used a series of mixed DNA dilutions of normal control and cancer cell line DNA containing known fractions of mutation to wild-type genome equivalents. Using the critical parameters that are described, we designed a series of wild-type and mutation-specific assays that were present in the primary colon adenocarcinomas of the patients in the study. The selected cancer driver genes for this study included BRAF, KRAS, TP53, AKT1, and PIK3CA. As a demonstration of this assay's application on ctDNA, we ran our single-color mutation detection assays directly on cell-free DNA. Finally, the assay was also used on degraded DNA of poor quality originating from clinical FFPE tumor samples.
Amplicon Length Affects Performance for Paired-Allele Assays
For paired-allele assays, a nonspecific 5′ extension (ie, repeats of AAAT) was added to the mutation-specific primer (Figure 1B). It was observed that different mutations showed variable characteristics in terms of the fluorescent droplet quantitation and clustering of mutation versus wild-type amplicons. We determined how different tail extension lengths influenced the separation of clustering and accurate genotype measurements (Figure 2). For these studies, primers were designed for the KRAS G12V (c.35 G>T) mutation, and tested primer with different tail extensions ranging from 12 up to 100 bp. These assays used DNA composed of a 1:1 mixture of the normal control NA18507 DNA and the cancer cell line RCM-1 DNA (Figure 2). For the KRAS G12V amplicons with 12, 24, or 39 bp length, the mutation cluster was not clearly separated from the wild-type cluster. At 60 bp or greater, the mutation droplet cluster separated without any overlap from the droplet cluster composed of wild-type amplicons. As a result, the fractional representation of the KRAS G12V (c.35 G>T) mutation versus wild-type alleles matched the reported values in the cancer cell line. Similarly, for the other mutations, we determined the appropriate tail length. The primer sequences, including tails, for all assays are noted in Table 2.
Comparing the Single- versus Paired-Allele Assays
A single-color detection assay targeting the hotspot mutation KRAS G12D (c.35 G>A) was designed. Using a total of 300 genome equivalents for each sample, a series of DNA mixtures using normal NA18507 DNA and cancer cell line GP2d DNA such that the fraction of mutation genome equivalents ranged from 43.0% to 0.0% of the total 300 (Table 1) was prepared. For these experiments, the performance of the single-allele (ie, simplex) assay was compared with the paired-allele assays using these different mixture samples. Figure 3A shows the results of this comparison as a two-dimensional clustering plot. The fractional representation of the detected mutation versus wild-type genome equivalents matched the expected values of the DNA mixtures for both the single-allele and paired-allele assay formats. Figure 3B displays a plot of the expected percentage mutation at each fraction versus the average detected percentage mutation, with the variance as determined from replicates. The Pearson correlation coefficient R2 was calculated to be 0.9971 (P = 0.0342) and 0.9999 (P < 0.0001) for the single- and paired-allele assays, respectively. Overall, these high correlation values indicated that the single- versus paired-allele assays demonstrate similar performance for this mutation.
Figure 3.
Detecting the hotspot mutation KRAS G12D. A: A ddPCR scatterplot representation of the channel 2 (x axis) and channel 1 (y axis) fluorescent amplitude for individual droplets of reactions from an NA18507 (normal control) and GP2d (KRAS G12D) DNA dilution series with mutation fractions ranging from 10.0% to 0% of the total genome equivalents (GEs). Each graph represents a single replicate of the KRAS G12D detection assay in both single-allele (top panels) and paired-allele (bottom panels) formats colorized to reflect mutation-positive droplets in green, wild-type–positive droplets in blue, and negative droplets in black, as determined by the clustering algorithm described in Materials and Methods. The targeted concentration for each sample was 300 GEs, and the number of GEs identified after clustering is printed within the plot. Mutation and wild-type GEs are written in green and blue, respectively. B: Results of the standard curve mutation detection experiments plotted as the expected percentage mutation (x axis) versus the average detected percentage mutation (y axis) for the single-allele assay (triangles) and paired-allele assay (circles). Included is a zoom view of each to visualize low fraction conditions (indicated by the red boxes). The Pearson correlation R2 and associated two-tailed P value are displayed within each plot. Error bars represent SD (B). n = 3 (B, single-allele assay); n = 4 (B, paired-allele assay).
Primer Features that Improve Detection Specificity
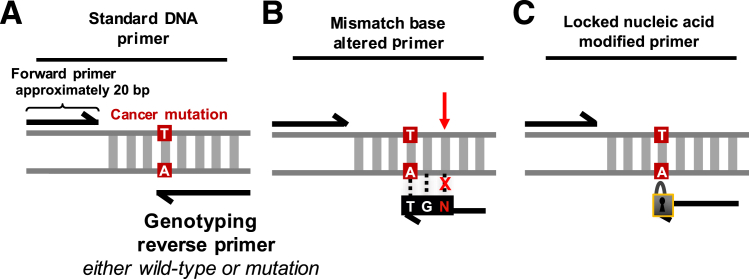
To improve the assay's base-pair specificity for mutation detection in some cases, the allele-specific primers (Figure 4A) were altered to contain a single mismatched base—this change in the primer sequence decreased the binding efficiency for nontarget alleles (Figure 4B). The approach was tested for the KRAS A146V (c.437 C>T) mutation. Both mutation and wild-type primers were tested with all possible options for the third base in from the 3′ portion of the reverse primer. All experiments were run with three or more replicates.
Figure 4.
Simplification of primer sequence design options shown as single-allele assays. A: The standard DNA primer option contains a complementary genomic sequence apart from the allele-specific 3′ base. B: Mismatch base primers are made up entirely of a complementary genomic sequence apart from the allele-specific 3′ base and the third base into the primer from the 3′ end of the primer. The identity of the base is determined experimentally. C: Locked nucleic acid (LNA)–containing primers are made up entirely of complementary DNA genomic bases apart from the allele-specific 3′ base. This 3′ base is an allele-specific LNA rather than a standard nucleotide.
Supplemental Figure S2 summarizes the mismatch optimization results for the KRAS A146V. For this experiment, normal control DNA from NA18507 was used as the template. Different mutation-specific versus wild-type primers were tested to evaluate the presence of off- versus on-target amplification. The results are visualized using a two-dimensional plot of droplet fluorescent amplitude. The mutation-specific primer without mismatch alterations demonstrated significant nonspecific amplification (Supplemental Figure S2A). After substituting the third base for all possible alternate nucleotide options (Supplemental Figure S2B), it was evident from the decrease in off-target positive droplets that the mismatched base reduced the off-target primer binding. For example, when substituting the third base for either a G or a T, no off-target molecules were amplified, as noted by the lack of positive droplets. In contrast, when substituting a C, as indicated by positive droplets, off-target molecules were identified. When comparing the wild-type clusters for every substitution condition, the primers containing G bases provided more distinct clusters in the wild-type assay than the primers containing T bases. In combination, the G base substitution provided the most specific and distinct clusters for both the mutation and the wild-type sequences. The same method was used to improve the detection specificity of primers for the PIK3CA H1047R (c.3140 A>G), KRAS G12D (c.35 G>A), and KRAS G12V (c.35 G>T) mutations, as listed in Table 2.
Comparison of Different Primer Sets to Detect the BRAF V600E
We determined whether modified nucleotides improve the performance of single-color ddPCR genotyping, particularly for the BRAF V600E (c.1799T>A) mutation. As a test, a locked nucleic acid nucleotide (LNA) was introduced in the allele-specific position of the BRAF V600E detecting primer (Figure 4C). LNAs are nucleotide analogs that have a locked conformational structure because of an additional O2′ C4′ methylene linkage—this feature provides an improved affinity for binding target sequences in DNA.22, 23, 24 To test the performance, DNA mixtures originating from the cancer cell line LS411N DNA and the normal standard NA18507 DNA were used (Table 1). We prepared the mixtures so that the fraction of mutation genome equivalents ranged from 68.0% to 0% of the total genome equivalents assayed per reaction. The quantitative signal from the decreasing fractional abundances of mutation and wild-type molecules was measured using the BRAF assay. All experiments were run with three or more replicates.
Figure 5 shows the performance of three different BRAF V600E assays: i) single-allele DNA primer, ii) single-allele LNA primer, and iii) paired-allele DNA primer assays. The simplex DNA assay did not accurately quantify the low-fraction components of the DNA mixture series, as denoted by a low correlation coefficient R2 value of 0.0749. In contrast, when using the simplex LNA assay, the fractional representation of the detected mutation versus wild-type genome equivalents matched the expected values of the DNA mixtures. Compared with the nonmodified oligonucleotide simplex assay, the LNA assay's R2 correlation coefficient was significantly improved at 0.9985 (P = 0.0006), suggesting a linear detection range. For the LNA assay, the negative controls did not show any significant signal. Thus, the addition of the LNA base led to a significant improvement in detection sensitivity compared with the non-LNA simplex assay.
Figure 5.
Locked nucleic acids (LNAs) improve the specificity of the simplex BRAF V600E assay. The results of mutation quantification of a DNA dilution series composed of mixed NA18507 (normal control) and LS411N (BRAF V600E) DNA with three separate BRAF V600E assays. The dilutions points are 10.0%, 1.0%, and 0% mutation for the simplex DNA assay, and 68.0%, 10.0%, 1.0%, and 0% mutation for the single-allele LNA and paired-allele DNA assays. The results for each are displayed as a plot of the expected percentage mutation (x axis) versus the average detected percentage mutation (y axis) for three replicates. The Pearson correlation R2 and associated two-tailed P value are displayed within each plot. Error bars represent SD. n = 3 (simplex DNA assay); n = 4 (single-allele LNA and paired-allele DNA assays).
Finally, the paired-allele BRAF V600E assay was tested with DNA primers that lacked the LNA nucleotides. It was determined that the paired-allele assay had a correlation coefficient R2 of 0.9988 (P = 0.0006), indicating that detection behaved in a highly linear manner. The negative controls did not show any significant signal. Overall, the paired-allele assay provided mutation fraction results that corresponded to the range of mutation allelic fractions expected from the DNA mixtures. When comparing the performance of the simplex LNA versus the paired-allele DNA assays, both had similar detection performance.
Detection Specificity for Different KRAS Mutations
To determine the specificity of the genotyping detection and quantitation, we designed and tested two separate paired-allele assays for either the KRAS G12D (c.35 G>A) or KRAS G12V (c.35 G>T) mutation; these mutations differ by only a single nucleotide at the same codon position. Ideally, a paired-allele KRAS G12D genotyping assay should not amplify the KRAS G12V mutation and vice versa.
We identified two colorectal cancer cell lines that had different KRAS mutations (eg, GP2d for KRAS G12D and RCM-1 for KRAS G12V). Using each cell line DNA, we prepared two separate series of DNA mixtures with normal control NA18507 DNA to a final concentration of 300 genome equivalents per sample reaction. For both KRAS G12D and G12V mutations, we prepared two mixed DNA samples such that the fraction of mutation genome equivalents represented either 10.0% or 1.0% of the total 300 genome equivalents. Normal DNA without cancer cell line DNA (NA18507) was included as a negative control. Three assay types were tested: i) single-color, paired-allele KRAS G12D; ii) single-color, paired-allele KRAS G12V; and iii) commercially available PrimePCR paired-allele KRAS G12D assay (Bio-Rad) with the KRAS G12V 10.0% mutation condition. All samples were run with three or more replicates.
Both single-color KRAS mutation assays had base-pair specificity, only amplifying their specific mutation target when using the matched cancer cell line DNA (Table 3). For example, when the RCM-1 cell line having the KRAS G12V mutation was tested, the G12D assay did not detect either a 10.0% (P = 0.0003) or a 1.0% fraction (P = 0.0233) compared with the G12V assay. Likewise, when testing the GP2d cell line DNA for the KRAS G12D mutation, the G12V assay did not detect the 10.0% (P = 0.0014) or 1.0% (P = 0.0475) fraction compared with the G12D assay.
Table 3.
Base-Pair Sensitivity of KRAS G12V and G12D Assays
| Cell line | Mutation | Single-color paired alleles (%) |
Significance (P value) | |
|---|---|---|---|---|
| G12V assay | G12D assay | |||
| RCM-1 | ||||
| 10.0% Mutant | G12V | 10.9 ± 2.3 | 0.3 ± 0.4 | <0.001 |
| 1.0% Mutant | G12V | 1.0 ± 0.5 | 0.5 ± 0.8 | <0.05 |
| GP2D | ||||
| 10.0% Mutant | G12D | 0.4 ± 0.6 | 9.6 ± 2.4 | <0.01 |
| 1.0% Mutant | G12D | 0.2 ± 0.4 | 1.5 ± 0.1 | <0.05 |
| Normal diploid control | ||||
| 0.0% Mutant | Wild type | 0.2 ± 0.4 | 0.0 ± 0.0 | |
DNA mixtures of normal control NA18507 and the cell line specified were prepared to be composed of 10.0% or 1.0% mutation-containing template. The percentage mutation detected for each assay with the given template is denoted in the center column. A one-sided t-test was used to determine the significance between assays for each given DNA mixture condition.
A commercially available paired-allele TaqMan probe KRAS G12D (c.35 G>A) assay using the GP2d cell line DNA was tested for comparison (Table 3). Running five replicates, we analyzed GP2d and identified a 49.7% ± 0.6% mutation fraction. A 10.0% KRAS G12V DNA mixture was generated using the RCM-1 cell line. When we ran the TaqMan KRAS G12D assay on this sample, three clusters were identified: a negative cluster, a wild-type cluster, and a non–wild-type cluster that represented 7.9% ± 1.8% of the total positive droplet population. This cluster represents the G12V mutation. Thus, the TaqMan probe G12D assay demonstrated a nonspecific signal from the different mutation. These results agree with a previous report in which it was observed that the TaqMan-based KRAS G12D assay did not discriminate among different KRAS mutations.25 Overall, these results suggest that the single-color mutation detection assay provides quantitative single-nucleotide specificity for single-nucleotide variant mutations, unlike the TaqMan probe assay.
Mutation Limit of Detection at 0.1%
We sought to determine whether the single-color assay could detect a 0.1% allelic fraction for the BRAF V600E (c.1799 T>A) and KRAS G12D (c.35 G>A) mutations. A series of DNA mixtures with an increasing number of genome equivalents being assayed per sample reaction for both mutations was prepared. The lowest fraction was 0.1%. Each DNA sample had between 2000 and 3000 genome equivalents per reaction. For the BRAF V600 assay mixture series, DNA from the cancer cell line LS411N and normal control DNA from NA18507 were used (Table 1). For the KRAS G12D mixture series, DNA from the cancer cell line GP2d and normal control DNA from NA18507 were used (Table 1).
For this experiment, the BRAF V600E and KRAS G12D paired-allele assay used standard oligonucleotides and had no LNAs. More than five replicates were run for each sample, and average values of the mutation fractions with SDs are reported for each sample. These results are summarized in Table 4. Using the paired-allele assays, we detected on average 0.7% ± 0.3% and 1.0% ± 0.3% mutation in the 1.0% mutation condition for the BRAF V600E and KRAS G12D assays, respectively, both of which were significantly greater than the negative control DNA condition (P < 0.0001 for both assays). This result is additionally supported seeing that no signal from mutation-bearing molecules was detected across wild-type DNA control replicates. Converting from the fraction mutation to the number of mutation genome equivalent molecules detected, we observed an average of 15.2 ± 6.5 BRAF V600E mutation genome equivalents and 20.8 ± 6.3 KRAS G12D mutation genome equivalents.
Table 4.
High-Resolution Molecular Sensitivity
| Expected mutation fraction (%) | BRAF V600E (c.1799T>A) (%) | KRAS G12D (c.35 G>A) (%) |
|---|---|---|
| 68.00 | 67.28 ± 0.98 | NA |
| 43.00 | NA | 47.74 ± 1.57 |
| 1.00 | 0.67 ± 0.27 | 1.00 ± 0.27 |
| 0.10 | 0.11 ± 0.08 | 0.12 ± 0.10 |
| 0.00 | 0.03 ± 0.05 | 0.03 ± 0.05 |
| Pearson correlation R2 | 1.00∗ | 1.00∗ |
Average number of wild-type and mutation-bearing molecules detected per reaction for 2000 and 3000 genome equivalent standard curves containing fractional mixtures of wild-type and mutation molecules. BRAF V600E mixtures were generated using NA18507 and cancer cell line DNA from LS411. KRAS G12D mixtures were generated using NA18507 and cancer cell line DNA from GP2D. N = 8 for each condition. Included are the Pearson correlation R2, two-tailed P value, and line equation with associated P value.
∗P < 0.0001.
NA, not available.
For the 0.10% mutation fraction, we detected 0.11% ± 0.08% and 0.12% ± 0.10% for the BRAF V600E and KRAS G12D assays, respectively. These values were also determined to be significantly greater than the negative control using the wild-type DNA (P = 0.0004 for BRAF V600E, and P = 0.0016 for KRAS G12D). The negative controls with either no DNA or wild-type DNA alone demonstrated no mutation PCR product signal. The correlation coefficient R2 of the detected percentage mutation with respect to the expected percentage mutation was 1.00 (P < 0.0001) for the BRAF V600E assay and 1.00 (P < 0.0001) for the KRAS G12D assay, thus indicating linear detection. These results demonstrated that the single-color, paired-allele BRAF V600E and KRAS G12D assays accurately detected a 0.10% mutation fraction. The linear range of detection requires higher genome equivalent sample inputs to resolve lower allelic fraction mutations.
Identifying Different Cancer Mutations from ctDNA
Based on the diagnostic primary tumor sequencing and genotyping results, we identified several driver mutations in the primary tumors of six individual cancer patients; Patients 5297, 5606, 5790, 5792, and 5397 were all diagnosed with colorectal cancer, and Patient 5788 was diagnosed with cholangiocarcinoma. Three patients were not receiving active treatment at the time of sample collection (Patients 5297, 5606, and 5788), whereas three were receiving ongoing chemotherapy (Patients 5790, 5792, and 5793). A series of assays was designed that matched the mutations from these patients (Table 5).
Table 5.
Patient-Specific Clinical and Mutation Information Summary
| Patient no. | Diagnosis | Mutation | Identified in ctDNA |
|---|---|---|---|
| 5297 | CRC | TP53 R175H (c.524 G>A) | Yes |
| AKT1 E17K (c.49 G>A) | Yes | ||
| KRAS A146V (c.437 C>T) | Yes | ||
| 5606 | CRC | KRAS G12V (c.35 G>T) | Yes |
| PIK3CA H1047R (c.3140 A>G) | No | ||
| 5788 | CCA | TP53 E285K (c.853 G>A) | Yes |
| 5790 | CRC | PIK3CA H1047R (c.3140 A>G) | No |
| 5792 | CRC | KRAS G12D (c.35 G>A) | No |
| 5793 | CRC | KRAS G12V (c.35 G>T) | No |
CCA, cholangiocarcinoma; CRC, colorectal adenocarcinoma; ctDNA, circulating tumor DNA.
To enable validation of each assay's performance, different cancer cell lines that had the same mutations found among the primary tumors were selected (Table 1). For the TP53 R175H (c.524 G>A), TP53 E285K (c.853 G>A), AKT1 E17K (c.49 G>A), KRAS A146V (c.437 C>T), KRAS G12V (c.35 G>T), and PIK3CA H1047R (c.3140 A>G) mutations, we used the cancer cell lines LS123, RPMI-8226, PL-21, KU1919, RCM-1, and RKO, respectively. The sensitivity and specificity of each assay were measured by generating a series of DNA mixtures that contained known ratios of normal control DNA from NA18507 and cancer cell line DNA (described in Materials and Methods). Each reaction contained approximately 300 genome equivalents, and mixtures were prepared such that the fraction of mutation genome equivalents to normal wild-type genome equivalents ranged from 50.0% to 1.0% of the total genome equivalents assayed per reaction. A normal control DNA reaction (NA18507) was also included as a negative control for all assays.
The expected percentage mutation versus the average detected percentage mutation for each assay was plotted individually in Supplemental Figure S3; the KRAS G12D plot can be found in Figure 3B. Across all assays, we observed high Pearson correlation values, with R2 > 0.98 (P < 0.01), suggesting all of the assays had a linear detection performance. All mutations did not show any significant signal in the negative controls. The only exception was for the AKT1 E17K assay that had had a raw background droplet count of 3.0 ± 4.0 positive droplets in the negative control.
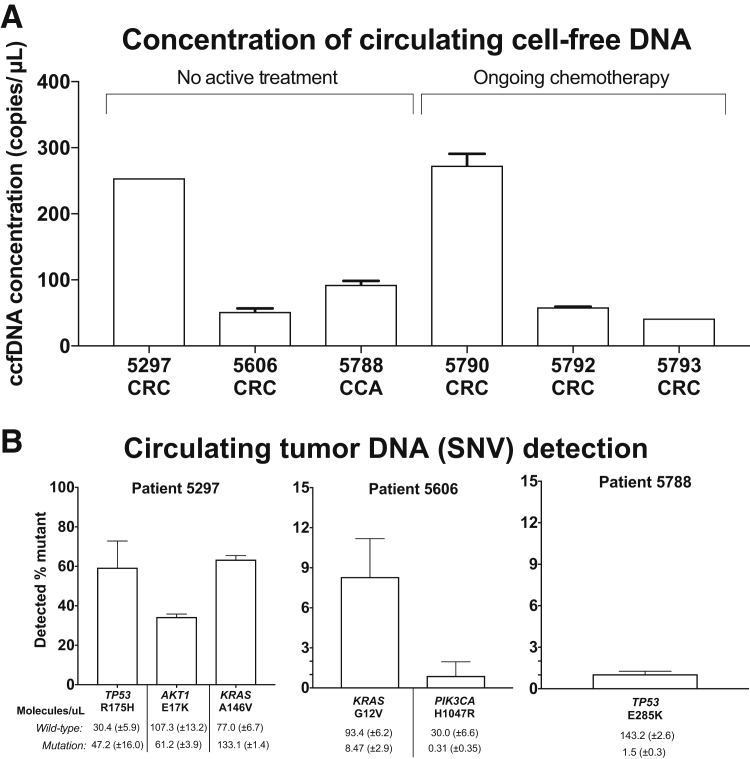
For testing the cell-free DNA, we used the cell line DNA dilution samples to provide standard curve points. Namely, the highest fraction of mutation genome equivalents provided the result for CIs that were then used to cluster each patient sample reaction (described in Materials and Methods). This step was important given that our procedure ensures that we can detect a low allelic fraction reproducibly. The droplet counts were converted to haploid genome equivalents to determine the number of mutation-bearing molecules present in the sample and then determined the mutation allelic fraction and percentage mutation.
The DNA concentration of the cell-free samples was measured as previously described in Materials and Methods, and all were within a 10-fold concentration range (Figure 6A). All six patient samples were tested with their respective mutation detection assays (Table 5), and identified tumor-derived ctDNA molecules from three of the six patients. Patient 5297's cancer had TP53 R175H (c.524 G>A), AKT1 E17K (c.49 G>A), and KRAS A146V (c.437 C>T) mutations. All three mutations from ctDNA were identified as well (Figure 6B). The mutation allele fractions for the TP53 R175H, AKT1 E17K, and KRAS A146V mutations were 59.3% ± 13.5%, 34.3% ± 1.5%, and 63.4% ± 2.1% mutation, respectively. On average, 47.2 ± 16.0, 61.2 ± 3.9, and 133.1 ± 1.4 mutation-bearing molecules per microliter were identified for TP53 R175H, AKT1 E17K, and KRAS A146V, respectively.
Figure 6.
Detecting cancer mutations from patient cell-free DNA. A: Bars represent the concentration of circulating cell-free DNA (ccfDNA) extracted from cancer patients using the RPP30 ddPCR assay (described in Materials and Methods). One to three replicates were run per sample, depending on sample availability. The diagnosis of each patient is abbreviated below the number, as colorectal adenocarcinoma (CRC) or cholangiocarcinoma (CCA). B: Bars represent the average percentage mutation detected for each target single-nucleotide variant (SNV) listed from patient-derived cell-free DNA. Below bars are the average number of wild-type and mutation genome equivalent molecules detected for each patient sample, represented as molecules per microliter. Three to four replicates were run per sample. Error bars represent the SD (B).
Patient 5606's primary tumor had KRAS G12V (c.35 G>T) and PIK3CA H1047R (c.3140 A>G) mutations. Analysis of the ctDNA identified the KRAS G12V mutation at 8.3% ± 2.9%; this is equivalent to 8.5 ± 2.9 mutation-bearing molecules per microliter (Figure 6B). The PIK3CA mutation was identified at 0.9% ± 1.1% mutation allelic fraction. However, after calculating the number of mutation-bearing molecules, the average number of positive mutation-bearing molecules was only 0.3 ± 0.4 molecules per microliter. This did not constitute significant signal over that of the background signal. Compared with the KRAS mutation, the lower levels of the PIK3CA H1047R mutation may be an indication that this driver event may occur in only a fraction of the tumor metastatic sites.
Finally, Patient 5788's tumor had a TP53 E285K (c.853 G>A) mutation. From the cell-free sample collected, we identified TP53 E285K at 1.1% ± 0.2% mutation (Figure 6B). This was equivalent to 1.5 ± 0.3 mutation-bearing molecules per microliter, on average.
For Patients 5790, 5792, and 5793, no detectable ctDNA molecules were observed for their mutations (PIK3CA H1047R, KRAS G12D, and KRAS G12V, respectively). The wild-type sequences were identified in all three of these cases. As the positive and negative controls showed the expected result, we concluded that our assay did not detect these mutations above our limits of detection. These patients were undergoing active treatment at the time of collection.
Precision Assays for Analysis of Patient FFPE DNA
Previous diagnostic genotyping (DNA Samples and Processing) showed that Patient 1820's primary tumor had a KRAS G12D (c.35 G>A) mutation. The KRAS G12D assay was tested on the DNA extracted from FFPE colorectal adenocarcinoma. FFPE-extracted DNA is subject to degradation and chemical cross-links that compromise the analyte quality and adversely affect the performance of molecular assays. Thus, this test provided information about the single-color ddPCR assay's performance on low-quality DNA.
For Patient 1820, we obtained matched FFPE tissue samples that included colon tissue adjacent to the tumor, dysplastic tissue without invasive carcinoma, and fully invasive carcinoma from the primary cancer. All three samples were tested in triplicate using our wild-type mutation pair KRAS G12D assay. We determined the average fraction of mutation versus wild-type genome equivalents as well as the associated SD. For the KRAS G12D mutation, the adjacent colon tissue, dysplastic, and malignant samples had fractional measurements of 1.6% ± 1.1%, 12.5% ± 4.9%, and 10.4% ± 1.5%, respectively. Converting the fraction to the number of detected mutation-bearing molecules, this represented an average of 3.2 ± 2.0, 26.1 ± 9.8, and 20.3 ± 2.9 molecules from the adjacent colon tissue, dysplastic tissue, and tumor samples, respectively. Generally, the three negative control replicates did not have any significant amplicon products, as defined by having, on average, more than one positive droplet among all replicates—most negative control replicates had no positive droplets. The presence of the KRAS mutation in the colon tissue sample suggests that the KRAS mutation was an early cancer driver event before the development of invasive carcinoma and present in adjacent colon tissue as a subclone.
Discussion
With the emergence of precision cancer medicine, there is an increasing need for new tests to evaluate patients at the time of diagnosis, during treatment, and for routine longitudinal clinical monitoring. Liquid biopsy specimens are traditionally used to analyze circulating cell-free DNA molecules and enable longitudinal monitoring for precision cancer medicine.1, 2 To address the need for rapid, accurate methods for analyzing ctDNA, we developed a single-color ddPCR assay for detecting cancer mutations from ctDNA at single-molecule resolution. This study demonstrates that single-color ddPCR assays can be successfully applied to ctDNA from clinical samples.
The single-color assay has several advantages compared with traditional fluorescent probes, including the following: i) ease of design and testing, ii) no requirement for specialized probe reagents, and iii) robust detection performance for resolving point mutations. We validated the sensitivity and specificity of this assay using control DNA mixtures composed of decreasing fractional abundances of mutation-bearing cell line DNA genome equivalents. To provide highly accurate mutation-bearing molecule quantification, we used a standard curve in parallel when analyzing each patient cell-free DNA sample. We demonstrated the sensitivity and specificity at 1.0% fraction using 300 genome equivalent, or approximately 1 ng per assay, thus minimizing the blood and DNA sample requirements for analysis. By increasing the number of genome equivalents assayed per reaction, one can also improve the sensitivity of detection—we demonstrated robust detection of the BRAF V600E and KRAS G12D at a 0.10% fraction. By avoiding the preamplification of patient circulating cell-free DNA, we avoid some of the complexity and issues of preamplification steps. In addition to the high sensitivity, the assay provides single bp specificity for the intended target mutation, as shown when using KRAS mutation assays that only differ by a single nucleotide.
In summary, our single-color ddPCR assay can be customized for a variety of different mutations and thus designed for an individual patient. For future development, we are seeking to improve the scalability of designing assays with highly parallel evaluation of primer candidates across tens of thousands of cancer mutations. If successful, this scalability would improve our ability to generate customized mutation primers.
Acknowledgments
We thank Laura Miotke for technical support; John Bell for advice on primer design; and Li Xia for statistical consultation.
Footnotes
Supported by NIH National Human Genome Research Institute grant P01HG000205 (C.W.-B., B.T.L., C.M.H., and H.P.J.) and National Cancer Institute grant R33CA174575 (H.P.J.), the American Cancer Society Research Scholar grant RSG-13-297-01-TBG (H.P.J.), and the Clayville Foundation and the Seiler Family Foundation (H.P.J. and C.W.-B.).
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2017.05.003.
Supplemental Data
Simplified workflow of steps of paired-allele assay clustering. Combine the amplitude files for replicates of the highest percentage mutation condition (1) to use for establishing CIs (2). After model generation, model file and all independent wells of the plate are passed through the clustering step (3). Herein, all wells of the plate are clustered using the model-generated CIs, and individual points are assigned to specific clusters. The data generated as output are plots, droplet counts for individual clusters, and a new amplitude file with the results of the cluster assignment for individual droplets.
Mismatch base experiment to improve primer binding specificity. Visual representation of the results from a series of single-allele assays performed when designing primers for the KRAS A146V detection assay using the normal diploid control genome NA18507 as plots for the channel 1 fluorescent amplitude (x axis) and the channel 2 fluorescent amplitude (y axis) for all droplets. Each droplet is colorized to be black (negative), blue (wild type), or green (mutation) based on manual clustering by the QuantaSoft tools. A: Fluorescent amplitude plot of the nonmodified wild-type and mutation single-allele KRAS A146V assays. B: Fluorescent amplitude plot of the mismatch modified wild-type and mutation single-allele targeting KRAS A146V assays. Mismatch sequence context displayed for each condition above the amplitude plot. Plots are arranged in order of highest primer binding specificity for both wild-type and mutation simplex assays.
Molecular sensitivity study results for customized mutation detection assays. Each panel represents a dilution series standard curve containing fractional mixtures of normal control DNA and a different mutation-bearing cancer cell line DNA. Expected percentage mutation was determined using an off-target control gene (RPP30) for template quantification and allelic fraction as determined by sequencing. All standard curves were prepared to contain a total of 300 genome equivalents per reaction at various fractional mixtures. The results are organized as plots of the expected and detected percentage mutation for each mutation assay standard curve for four replicates of each standard curve point. Three to four replicates were run per sample. The SD for each point is represented through error bars. The Pearson correlation R2 value and the associated two-tailed P value were calculated for each data set using Prism 7 statistical analysis software version 7.0a (GraphPad Software, Inc., La Jolla, CA) and displayed on each plot.
References
- 1.Anker P., Mulcahy H., Qi Chen X., Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro B., Chakrabarty M., Cohn E.M., Leon S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::aid-cncr2820511127>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 3.Castells A., Puig P., Móra J., Boadas J., Boix L., Urgell E., Solé M., Capellà G., Lluís F., Fernández-Cruz L., Navarro S., Farré A. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: diagnostic utility and prognostic significance. J Clin Oncol. 1999;17:578–584. doi: 10.1200/JCO.1999.17.2.578. [DOI] [PubMed] [Google Scholar]
- 4.Ryan B.M., Lefort F., McManus R., Daly J., Keeling P.W.N., Weir D.G., Kelleher D. A prospective study of circulating mutant KRAS2 in the serum of patients with colorectal neoplasia: strong prognostic indicator in postoperative follow up. Mol Pathol. 2003;56:172–179. doi: 10.1136/gut.52.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A., Kinzler K.W., Vogelstein B., Diaz L.A. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pao W., Wang T.Y., Riely G.J., Miller V.A., Pan Q., Ladanyi M., Zakowski M.F., Heelan R.T., Kris M.G., Varmus H.E. KRAS mutations and primary resistance of lung adenocarcinomas to Gefitinib or Erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz L.A., Williams R., Wu J., Kinde I., Hecht J.R., Berlin J., Allen B., Bozic I., Reiter J.G., Nowak M.A., Kinzler K.W., Oliner K.S., Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson A.L., Iglehart J.D. BEAMing up personalized medicine: mutation detection in blood. Clin Cancer Res. 2012;18:3209–3211. doi: 10.1158/1078-0432.CCR-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C.W., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., Shrager J.B., Loo B.W., Jr., Alizadeh A.A., Diehn M. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahr S., Hentze H., Englisch S., Hardt D., Fackelmayer F.O., Hesch R.D., Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 12.Gray E.S., Rizos H., Reid A.L., Boyd S.C., Pereira M.R., Lo J., Tembe V., Freeman J., Lee J.H., Scolyer R.A., Siew K., Lomma C., Cooper A., Khattak M.A., Meniawy T.M., Long G.V., Carlino M.S., Millward M., Ziman M. Circulating tumor DNA to monitor treatment response and detect acquired resistance in patients with metastatic melanoma. Oncotarget. 2015;6:42008–42018. doi: 10.18632/oncotarget.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang G.A., Tadepalli J.S., Shao Y., Zhang Y., Weiss S., Robinson E., Spittle C., Furtado M., Shelton D.N., Karlin-Neumann G., Pavlick A., Osman I., Polsky D. Sensitivity of plasma BRAFmutant and NRASmutant cell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol Oncol. 2016;10:157–165. doi: 10.1016/j.molonc.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alix-Panabières C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6:479. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 15.Borsu L., Intrieri J., Thampi L., Yu H., Riely G., Nafa K., Chandramohan R., Ladanyi M., Arcila M.E. Clinical application of picodroplet digital PCR technology for rapid detection of EGFR T790M in next-generation sequencing libraries and DNA from limited tumor samples. J Mol Diagn. 2016;18:903–911. doi: 10.1016/j.jmoldx.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler J., Eder J., Pratscher B., Brandt S., Schneller D., Mullegger R., Vogl C., Trautinger F., Brem G., Burgstaller J.P. SNPase-ARMS qPCR: ultrasensitive mutation-based detection of cell-free tumor DNA in melanoma patients. PLoS One. 2015;10:e0142273. doi: 10.1371/journal.pone.0142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miotke L., Lau B.T., Rumma R.T., Ji H.P. High sensitivity detection and quantitation of DNA copy number and single nucleotide variants with single color droplet digital PCR. Anal Chem. 2014;86:2618–2624. doi: 10.1021/ac403843j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopmans E.S., Natsoulis G., Bell J.M., Grimes S.M., Sieh W., Ji H.P. A programmable method for massively parallel targeted sequencing. Nucleic Acids Res. 2014;42:e88. doi: 10.1093/nar/gku282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen M., Nielsen C.B., Nielsen K.E., Jensen G.A., Bondensgaard K., Singh S.K., Rajwanshi V.K., Koshkin A.A., Dahl B.M., Wengel J., Jacobsen J.P. The conformations of locked nucleic acids (LNA) J Mol Recognit. 2000;13:44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 23.Obika S., Nanbu D., Hari Y., Andoh J.-I., Morio K.-I., Doi T., Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 24.Jepsen J.S., Sorensen M.D., Wengel J. Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides. 2004;14:130–146. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 25.Pender A., Garcia-Murillas I., Rana S., Cutts R.J., Kelly G., Fenwick K., Kozarewa I., Gonzalez de Castro D., Bhosle J., O'Brien M., Turner N.C., Popat S., Downward J. Efficient genotyping of KRAS mutant non-small cell lung cancer using a multiplexed droplet digital PCR approach. PLoS One. 2015;10:e0139074. doi: 10.1371/journal.pone.0139074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Simplified workflow of steps of paired-allele assay clustering. Combine the amplitude files for replicates of the highest percentage mutation condition (1) to use for establishing CIs (2). After model generation, model file and all independent wells of the plate are passed through the clustering step (3). Herein, all wells of the plate are clustered using the model-generated CIs, and individual points are assigned to specific clusters. The data generated as output are plots, droplet counts for individual clusters, and a new amplitude file with the results of the cluster assignment for individual droplets.
Mismatch base experiment to improve primer binding specificity. Visual representation of the results from a series of single-allele assays performed when designing primers for the KRAS A146V detection assay using the normal diploid control genome NA18507 as plots for the channel 1 fluorescent amplitude (x axis) and the channel 2 fluorescent amplitude (y axis) for all droplets. Each droplet is colorized to be black (negative), blue (wild type), or green (mutation) based on manual clustering by the QuantaSoft tools. A: Fluorescent amplitude plot of the nonmodified wild-type and mutation single-allele KRAS A146V assays. B: Fluorescent amplitude plot of the mismatch modified wild-type and mutation single-allele targeting KRAS A146V assays. Mismatch sequence context displayed for each condition above the amplitude plot. Plots are arranged in order of highest primer binding specificity for both wild-type and mutation simplex assays.
Molecular sensitivity study results for customized mutation detection assays. Each panel represents a dilution series standard curve containing fractional mixtures of normal control DNA and a different mutation-bearing cancer cell line DNA. Expected percentage mutation was determined using an off-target control gene (RPP30) for template quantification and allelic fraction as determined by sequencing. All standard curves were prepared to contain a total of 300 genome equivalents per reaction at various fractional mixtures. The results are organized as plots of the expected and detected percentage mutation for each mutation assay standard curve for four replicates of each standard curve point. Three to four replicates were run per sample. The SD for each point is represented through error bars. The Pearson correlation R2 value and the associated two-tailed P value were calculated for each data set using Prism 7 statistical analysis software version 7.0a (GraphPad Software, Inc., La Jolla, CA) and displayed on each plot.