Abstract
Although androgen-deprivation treatment (ADT) is the main treatment for advanced prostate cancer (PCa), it eventually fails. This failure invariably leads to castration-resistant prostate cancer (CRPC) and the development of the neuroendocrine (NE) phenotype. The molecular basis for PCa progression remains unclear. Previously, we and others have demonstrated that the sex-determining region Y-box 4 (SOX4) gene, a critical developmental transcription factor, is overexpressed and associated with poor prognosis in PCa patients. In this study, we show that SOX4 expression is associated with PCa progression and the development of the NE phenotype in androgen deprivation conditions. High-throughput microRNA profiling and bioinformatics analyses suggest that SOX4 may target the miR-17-92 cluster. SOX4 transcriptionally upregulates miR-17-92 cluster expression in PCa cells. SOX4-induced PCa cell proliferation, migration, and invasion are also mediated by miR-17-92 cluster members. Furthermore, RB1 is a target gene of miR-17-92 cluster. We found that SOX4 downregulates RB1 protein expression by upregulating the miR-17-92 expression. In addition, SOX4-knockdown restrains NE phenotype and PCa cell proliferation. Clinically, the overexpression of miR-17-92 members is shown to be positively correlated with SOX4 expression in PCa patients, whereas RB1 expression is negatively correlated with SOX4 expression in patients with the aggressive PCa phenotype. Collectively, we propose a novel model of a SOX4/miR-17-92/RB1 axis that may exist to promote PCa progression.
Introduction
With 1,276,106 new cases and 358,989 deaths estimated in 2018, prostate cancer (PCa) is rated the second most common cancer type and fifth leading cause of cancer-related deaths among men [1]. Androgen deprivation treatment (ADT) remains the standard treatment for patients with advanced PCa [2]. However, patients inevitably recur with a more aggressive castration-resistant prostate cancer (CRPC) [3]. With the use of abiraterone or enzalutamide, a subset of patients with late-stage CRPC eventually develops neuroendocrine prostate cancer (NEPC), which is associated with extremely poor overall prognosis [4], [5], [6]. The mechanisms of PCa progression, particularly pathways involved in the development of CRPC and NEPC, need to be better understood in order to develop more effective treatments.
We and others have previously reported that SOX4, an important developmental transcription factor, acts as a transforming oncogene and is overexpressed in multiple malignancies including PCa [7], [8], [9], [10]. SOX4 can bind to the promoters to regulate many genes that play significant roles in cancer progression including EGFR, EZH2, HSP70 and Tenascin C [11], [12]. Most recently, we defined an important role for SOX4 in the progression of PCa by orchestrating an epithelial-mesenchymal transition (EMT) [8], [13]. Overexpression of SOX4 has been associated with poor clinical outcome of PCa patients [8]. Additionally, we demonstrated that SOX4 is a dihydrotestosterone (DHT)-repressed androgen receptor (AR) target gene and is overexpressed in CRPC tumors as compared to the hormone-dependent PCa [14].
MicroRNAs (miRNAs) are a class of small noncoding RNA species that regulate the translation and stability of mRNAs posttranscription [15], [16]. Aberrant expression of many miRNAs has been linked to multiple human diseases including cancers [17]. Similar to classical oncogenes and tumor suppressors, miRNAs play important roles in PCa development and progression [16], [18], [19].
In this study, we hypothesized that SOX4 may promote PCa progression via the regulation of specific miRNAs. To test this, we analyzed the high-throughput miRNA expression profiling and identified the miR-17-92 cluster that is transcriptionally upregulated by SOX4. RB1 was shown to be the target of the SOX4/miR-17-92 axis. The biological role of the SOX4/miR-17-92/RB1 axis in PCa progression was further investigated. Finally, we proposed a model of SOX4/miR-17-92/RB1 axis which may provide insight as to how SOX4 could contribute to PCa progression.
Material and Methods
Cell Lines
HEK293T cell and human PCa cell lines LNCaP, VCaP, 22RV1, PC3 and DU145 were obtained from the American Type Culture Collection (Rockville, MD) and cultured following the manufacturer’s recommendations. To establish a PCa cell line with the NE phenotype, LNCaP cells were continuously cultured in a medium for 3 months with 10% FBS without steroids as previously described [20], [21], [22]. NEPC markers including CHGA, CHGB, SYP, and NCAM1 were significantly increased, and the resulting subline was designated as LNCaP-NEPC. The stable LNCaP-NEPC cells were authenticated by short tandem repeat analysis in our study.
Transient Transfection
SOX4-specific siRNAs, SOX4 cDNA expression vectors, and miR-17-92 mimics and inhibitors were designed and synthesized by Gene-Pharma (Shanghai, China). The effective sequences of siRNA were listed in Supplemental Table 1. SOX4 cDNA was subcloned into the p-ENTER eukaryotic expression vector. PCa cells and HEK293T were transiently transfected with siRNA, miRNA mimics, miRNA inhibitor, plasmids, and the corresponding control using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) following the manufacturer’s recommendations. Transfection efficiency was confirmed by real-time quantitative polymerase chain reaction (RT-qPCR) and Western blot.
Stable Cell Line Generation
Lentiviral vectors encoding siRNA of SOX4 and the empty vector control were synthesized by Gene-Pharma (Shanghai, China). For stable infection, 1×104 VCaP/LNCaP-NEPC cells were plated in six-well plates for 48 hours. Following lentiviral infection, 2 μg/ml of puromycin was used for 2 weeks to select expression-stable cell lines. Transfection efficiency was confirmed by RT-qPCR and Western blot.
RNA Isolation and RT-qPCR Assays
The TRIzol reagents (Invitrogen) were used for total RNA extraction. The mRNA was reverse-transcribed into cDNA by using the ReverTra Ace qPCR RT kit (TOYOBO, Japan). The qRT-PCR assay was carried out with FastStart Universal SYBR Green Master (Roche, USA) according to manufacturer's instructions. Mature miRNAs were detected by using the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal loading control for mRNA and pri-miRNA. Pre-U6 and U6 were used as an endogenous control for pre-miRNA and miRNA. The primers of miRNAs precursors were utilized according to previous study [23]. All primers used in this study are listed in Supplemental Table 2. The primers of mature miRNAs were designed and synthesized by Gene-Pharma (Shanghai, China).
MiRNA-seq and Bioinformatics Analysis
VCaP cells were transiently transfected with SOX4-specific siRNA or the control for 48 hours, and the total RNA was isolated using the TRIzol reagent (Invitrogen). MiRNA-seq analysis (KangCheng, Shanghai, China) was used to compare the miRNA expression profiles in VCaP-siSOX4 and VCaP-NC cells. MiRNA target analysis (http://www.targetscan.org and http://www.microrna.org) was carried out to predict the target genes.
Western Blot
Western blot was performed as previously described [24]. Antibodies used in this study are as listed: anti-SOX4 (1:500; cat. no. NBP1-50776; Novus), anti-RB1 (1:1000; cat. no. CY5661; Abways), anti-E2F1 (1:1000; cat. no.CY6580; Abways), anti-CHGA (1:1000; cat. no.CY6701, Abways), anti-NCAM1 (1:1000; cat. no. CY6683, Abways), anti-TP53 (1:1000; cat. no. AB3126, Abways), and anti-GAPDH (1:1000; cat. no. ab9385; Abcam). The signals were detected with the Rapid-Step TM ECL Reagent (Millipore, USA). Independent experiments were performed for at least three separate times. Quantifications of the band intensity of Western blot were digitally analyzed using Image J software.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) was carried out as previously described [24]. The EZ-Magna ChIP assay kit (Millipore, Billerica, MD) was used according to the manufacturer’s recommendations. In brief, 1.0×107 VCaP or LNCaP cells were cross-linked with 1% formaldehyde, immunoprecipitated, and reacted with 1-5 μg of antibodies overnight at 4°C. Then, DNA was purified, and the DNA enrichment template was analyzed by PCR and RT-qPCR with the primers specific for miR-17-92 promoter. All primers used in this study are listed in Supplemental Table 1.
Dual Luciferase Assay
A dual luciferase assay was performed as previously described [24]. In brief, VCaP or HEK293T cells were co-transfected with the indicated plasmid and siRNA or miRNA mimics/inhibitors. The cells were harvested for the luciferase reporter assay using the dual luciferase assay reporter system following the manufacturer’s instructions (Promega).
In Vitro Proliferation, Migration and Invasion Assays
Cell-Light EdU DNA Cell Proliferation (EdU) assay (Ribobio, Guangzhou, China) and MTS assays (Promega, Madison, WI) were carried out to measure cell proliferation. Transwell assays were used to measure cell migration and invasion. These assays were performed as previously described [24], [25].
Tumor Xenografts
Male BALB/c mice (4 weeks old) were purchased from Weitonglihua Biotechnology (Beijing, China). They were housed in a specific pathogen-free environment and fed a normal chow diet. The experimental protocol was approved by the Shandong University Animal Care Committee, and all procedures were performed in compliance with the institution’s guidelines. A total of 1.0×107 VCaP cells in 100 μl of PBS and the same volume of Matrigel were injected subcutaneously into the mice (n=5/group). The tumor size was measured 6 weeks after tumor formation, and the tumor volume was calculated with the formula: tumor volume=length × (width) 2 × 1/2.
Tissue Specimens
The study consisted of 141 PCa patients who underwent radical prostatectomy between 2009 and 2016 at the Qilu Hospital of Shandong University (Jinan, China). In addition, 7 PCa cases with small cell carcinoma were included. Two tissue microarrays (TMAs) were constructed by incorporating two 1-mm cores from each representative tumor. The morphology was confirmed by two pathologists (B.H. and W.X.L.). This study was approved by the Shandong University Research Ethics Committee, and written informed consent was obtained from each patient.
Immunohistochemistry
Immunohistochemistry was conducted as previously described [8]. Slides were incubated with antibodies overnight at 4°C (with antibodies against SOX4 and RB1). The slides were then blindly evaluated by two independent pathologists (B.H. and W.X.L.). The scoring criteria for the validation of SOX4 and RB1 were described previously [8], [26]. The nuclear staining for SOX4 and RB1 were scored into four grades (0, negative; 1-3, weak; 4-6, moderate; and 8-12, strong) based on its staining intensity. The percentages of positive cells were scored into five categories: 0 (0%), 1 (1%-25%), 2 (26%-50%), 3 (51%-75%), and 4 (76%-100%) [8], [26], [27]. In this study, we combined negative and weakly SOX4 positive tumors into one group and moderately and strongly positive SOX4 into the other (SOX4 overexpression). PCa cases were considered to have lost Rb protein if any TMA spots showed loss (0+ staining) in >95% of the tumor nuclei. Positive nuclear staining in surrounding endothelial cells provided an internal control.
Statistical Analysis
All the analyses in this study were completed by Graphpad Prism 6 or the SPSS 20.0 software, with P<.05 considered statistically significant. The two-tailed unpaired t test was used to calculate statistical significance between the mean values of the two groups. Pearson’s correlation test was used to measure the correlation between the two factors.
Results
Aberrant SOX4 Expression Is Associated with PCa Progression and NE Phenotype
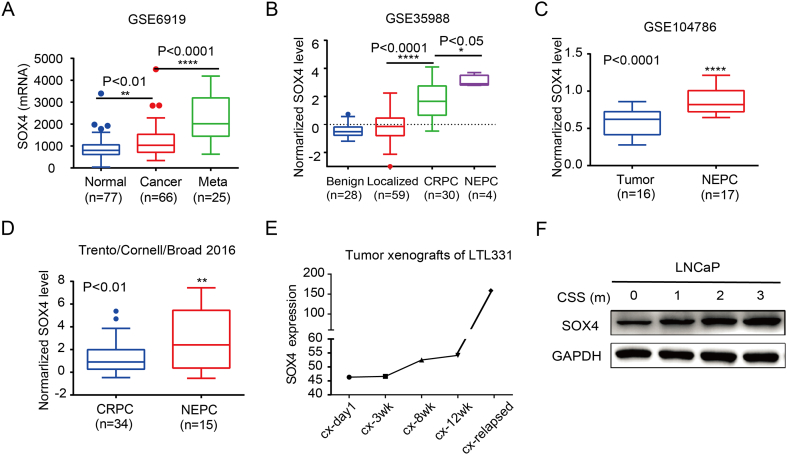
Previously, we and others have shown that SOX4 overexpression is associated with higher Gleason scores and poor prognosis in PCa patients [8], [10]. Using publicly available datasets GSE6919 and GSE35988, we demonstrated that the expression level of SOX4 increased with PCa progression (Figure 1, A and B) [28], [29], [30], [31]. In the GSE104786 and Trento/Cornell/Broad 2016 cohort, SOX4 expression is significantly higher in NEPC when compared to CRPC (Figure 1, C and D) [31], [32]. Analysis of SOX4 expression in patient-derived xenografts (tumor xenografts of LTL331) indicates that SOX4 is upregulated during transdifferentiation of PCa, especially in relapsed xenograft tumors recognized as NEPC (Figure 1E) [33]. In addition, SOX4 expression is increased during the ADT treatment of LNCaP cells (Figure 1F). These findings suggest that the overexpression of SOX4 may be associated with PCa progression and the NE phenotype.
Figure 1.
SOX4 overexpression is associated with PCa progression and NEPC. SOX4 expression is upregulated during the PCa progression in GSE6919 dataset (A) and GSE35988 dataset (B). SOX4 expression in CRPC and NEPC patients from GSE104768 dataset (C) and Trento/Cornell/Broad 2016 dataset (D). Number of samples (n) and P values (determined by a two-tailed Student’s t test) are as shown. (E) MRNA expression of SOX4 during NE transdifferentiation in the LTL331 system. (F) Western blot tested SOX4 expression in ADT treatment of LNCaP cells. LNCaP cell were continuously cultured in medium with 10% FBS depleted of steroids for 0-3 months.
MiRNA Expression Profiling in SOX4-Knockdown Expression PCa Cells
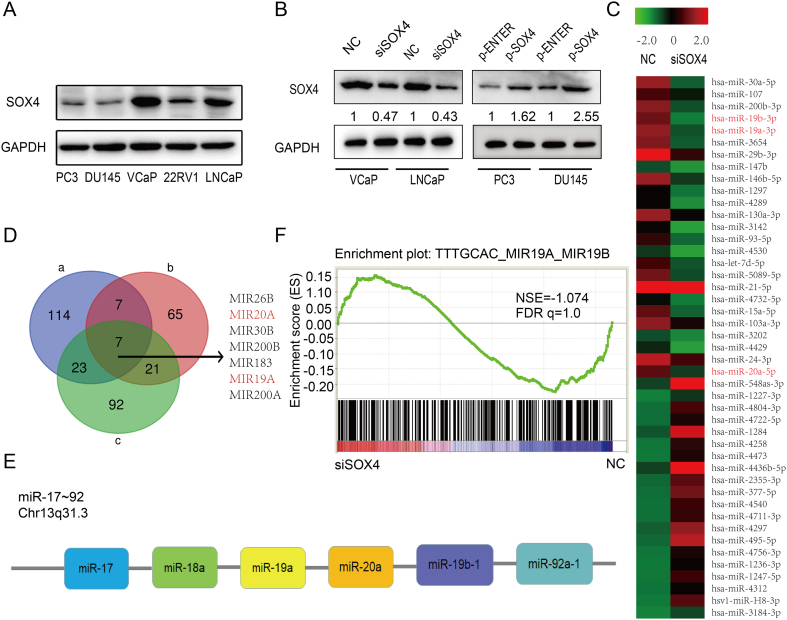
Previous reports have shown that many miRNAs are involved in promoting PCa progression including miR-17-92 [34], [35], miR-106b-25 [36], [37] and miR-424 [38]. However, it remains unclear whether or not SOX4 regulates the miRNAs expression in PCa cells. To identify the miRNAs that are regulated by SOX4, we firstly chose an appropriate PCa cell model to modulate the SOX4 expression levels. The protein expression level of SOX4 in PCa cell lines was measured by Western blot (Figure 2A). VCaP and LNCaP cells were chosen for SOX4 knockdown, while DU145 and PC3 cells were chosen for SOX4 overexpression. The transfection efficiency was determined by Western blot (Figure 2B).
Figure 2.
High-throughput miRNA expression profiling in SOX4 knockdown PCa cells. (A) The protein expression levels of SOX4 in human PCa cell lines determined by Western blot. (B) VCaP and LNCaP cells with silencing SOX4 expression as well as DU145 and PC3 cells with SOX4 overexpression were established by transiently transfection in 48 hours. Levels of SOX4 knockdown and overexpression were quantified and normalized to GAPDH levels. (C) Heatmap show the top 25 downregulated miRNAs and top 20 upregulated miRNAs by siSOX4 in VCaP cells. (D) A Venn diagram depicting seven miRNAs that meet the following properties: (a) >2.0-fold change miRNAs downregulated by siSOX4 in VCaP; (b) top 100 miRNAs Pearson correlated with SOX4 in TCGA; (c) >2.0-fold change miRNAs upregulated in castration-resistant xenograft tissue compared to androgen-dependent xenograft tissue from GSE55829. (E) miR17-92 cluster members located within the same chromosome. (F) miR-19a and miR-19b downregulated genes are enriched for upregulation upon SOX4 knockdown from GSE11914.
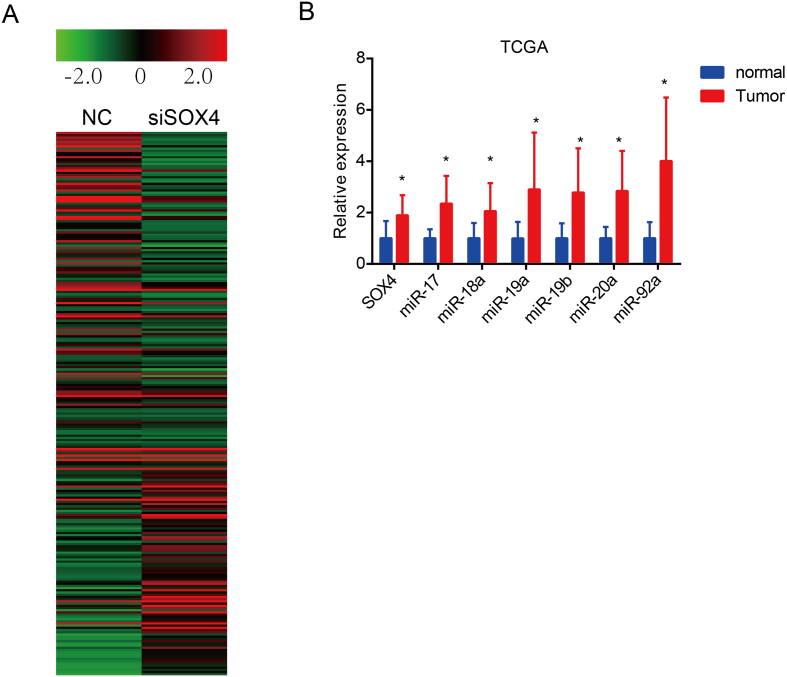
Next, we preformed high-throughput miRNA expression profiling in SOX4-knockdown VCaP cells. A total of 92 miRNAs were upregulated and 154 miRNAs were downregulated in the SOX4-knockdown VCaP cells as compared to the negative control (fold change >2.0) (Supplementary Figure 1A). Of these miRNAs, the top 25 downregulated and the top 20 upregulated miRNAs were shown in the heat maps (Figure 2C). Next, we analyzed the top 100 miRNAs that were increased in castration-resistant xenograft tissues as compared to androgen-dependent xenograft tissues from GSE55829 dataset [39]. Furthermore, the top 100 miRNAs correlated to SOX4 in The Cancer Genome Atlas (TCGA) were chosen by Pearson correlation. Cross-comparison of all clustered miRNAs allowed us to generate a “consensus” of seven miRNAs (Figure 2D). Interestingly, we found that miR-19a and miR-20a which were significantly downregulated by SOX4 knockdown (Figure 2C) belong to the miR-17-92 cluster which is located on the same chromosome (Figure 2E). In addition, we analyzed the GSE11914 dataset by Gene Set Enrichment Analysis (GSEA) [11]. This dataset is about expression data from LNCaP cells by SOX4 knockdown and overexpression using high-throughput mRNA expression profiling. As shown in Figure 2F, miR-19a and miR-19b downregulated genes were enriched for upregulation upon SOX4 knockdown. All the above findings suggest that miR-17-92 cluster is more likely to be directly regulated by SOX4. Clinically, the expression of SOX4 and miR-17-92 cluster members is increased in PCa patients as compared to normal cases, respectively (Supplementary Figure 1B).
Supplementary Figure 1.
High-throughput miRNA expression profiling in VCaP cells by siSOX4. (A) Heatmap show 92 miRNAs upregulated and 154 miRNAs downregulated in the siSOX4 PCa cells compared to negative control (fold change >2.0). (B) Expression of SOX4 and miR-17-92 cluster members is higher in PCa patients to compared normal samples, respectively, from TCGA dataset.
SOX4 Upregulates miR-17-92 Cluster Expression by Binding to Its Promoter
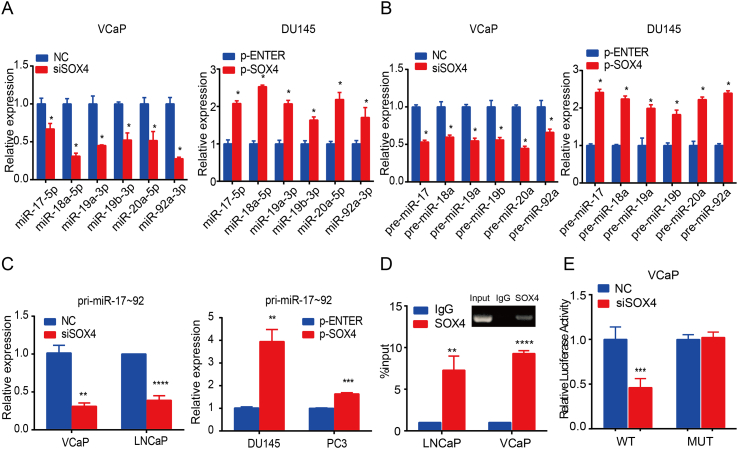
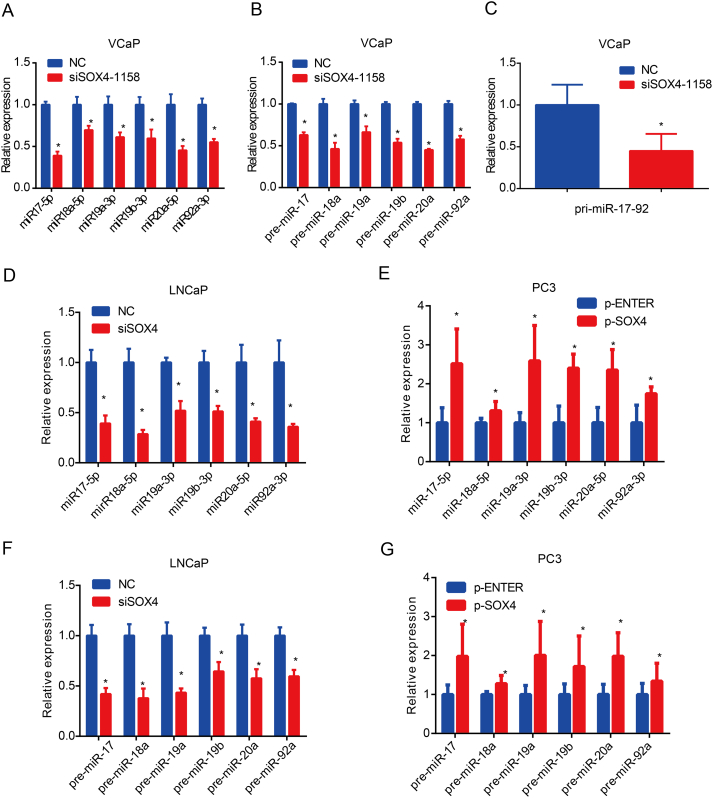
Next, we characterized how SOX4 regulates the expression of the miR-17-92 cluster in PCa in vitro. SOX4 inhibition significantly decreased miR-17-92 cluster member expression in VCaP cells, whereas overexpression of SOX4 significantly increased expression of miR-17-92 cluster members in DU145 cells (Figure 3A). Considering that SOX4 is a transcription factor, we hypothesized that SOX4 might regulate miR-17-92 cluster expression at the transcriptional level. To test this hypothesis, we investigated the expression of pre-miR-17-92 cluster members and pri-miR-17-92 by SOX4 knockdown/overexpression. As shown in Figure 3B and C, our results showed that SOX4 knockdown repressed pre-miR-17-92 members and pri-miR-17-92 expression in VCaP cells, whereas SOX4 overexpression increased pre-miR-17-92 members and pri-miR-17-92 expression. These results suggest that SOX4 transcriptionally regulates the miR-17-92 cluster. Similar results were obtained using the other siRNA-SOX4 in VCaP cells (Supplementary Figure 2, A-C). Similar results were obtained using the LNCaP and PC3 cells (Supplementary Figure 2, D-G).
Figure 3.
SOX4 upregulates miR-17-92 cluster expression by binding to its promoter. VCaP and DU145 cells were transfected with SOX4 knockdown/overexpression or its negative control. After incubation for 48 hours, RT-qPCR detected the expression levels of miR-17-92 cluster members (A) and pre-miR-17-92 cluster members (B). (C) pri-mir-17-92 expression was detected by RT-qPCR in indicated cells transfected with knockdown/overexpression SOX4 or its negative control at 48 hours. (D) SOX4 directly binds to miR-17-92 promoter. VCaP and LNCaP cells were subjected to ChIP assay with SOX4 antibodies. IgG was used as control. The y-axis represented the fold enrichment of the promoter fragments captured by the two different antibodies. IgG was used as a loading control. (E) SOX4 induced miR-17-92 promoter activity. VCaP cells were transfected with pGL3-miR-17-92-Luc-WT/Mut together with SOX4-knockdown or its negative control. *P<.05, **P<.01, ***P<.001, based on Student’s t test.
Supplementary Figure 2.
SOX4 upregulates the expression of miR-17-92 cluster. VCaP cells were transfected with siSOX4-1158 or its negative control. After incubation for 48 hours, RT-qPCR detected the expression levels of miR-17-92 cluster members (A), pre-miR-17-92 cluster members (B), and pri-miR-17-92 (C). LNCaP and PC3 cells were transfected with SOX4 knockdown/overexpression or its negative control. After incubation for 48 hours, RT-qPCR detected the expression levels of miR-17-92 cluster members (D, E) and pre-miR-17-92 cluster members (F, G). *P <.05; P values based on Student’s t test.
Next, we determined whether SOX4 could activate miR-17-92 promoter activity. For this aim, we first utilized MatInspector (http://www.genomatix.de) to identify the potential SOX4 binding site to the miR-17-92 promoter. There was one SOX4 binding site at 313-322 of the miR-17-92 promoter. We then performed ChIP with a pair of primers flanking the binding motif. ChIP assay showed that SOX4 indeed binds to this binding site in both VCaP and LNCaP cells (Figure 3D). Furthermore, SOX4 was able to induce miR-17-92 promoter activity but failed to do so in mutant miR-17-92 promoter luciferase activities in VCaP cells (Figure 3E). All these findings suggest that SOX4 induces miR-17-92 expression by binding to its promoter.
SOX4-Induced PCa Cell Proliferation, Migration, and Invasion Are Mediated by miR-17-92 Cluster Members
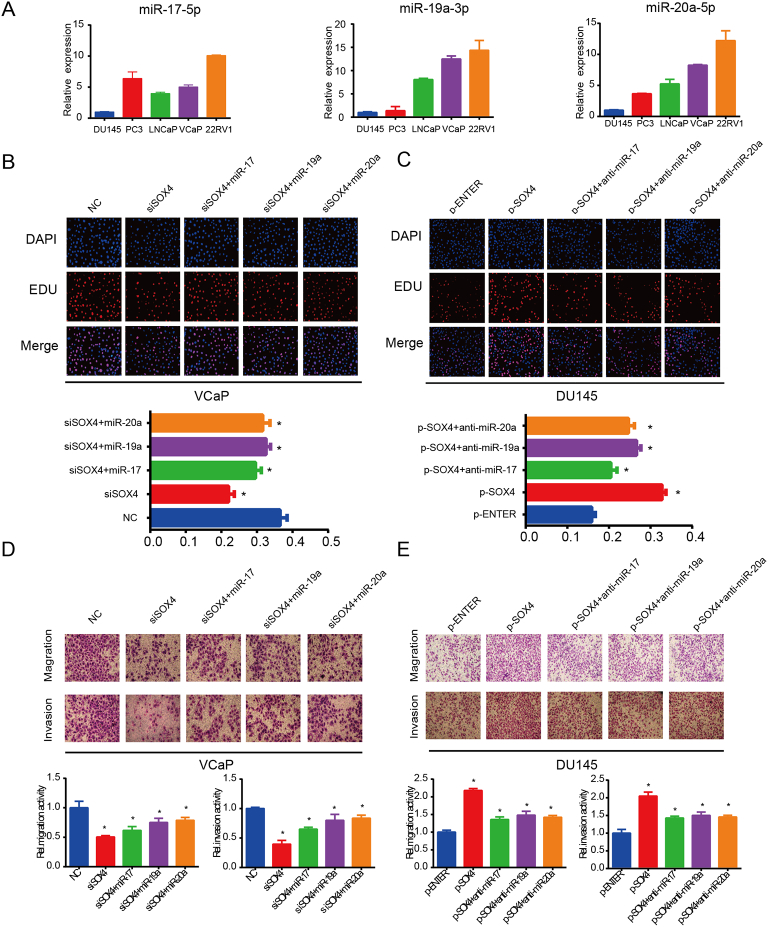
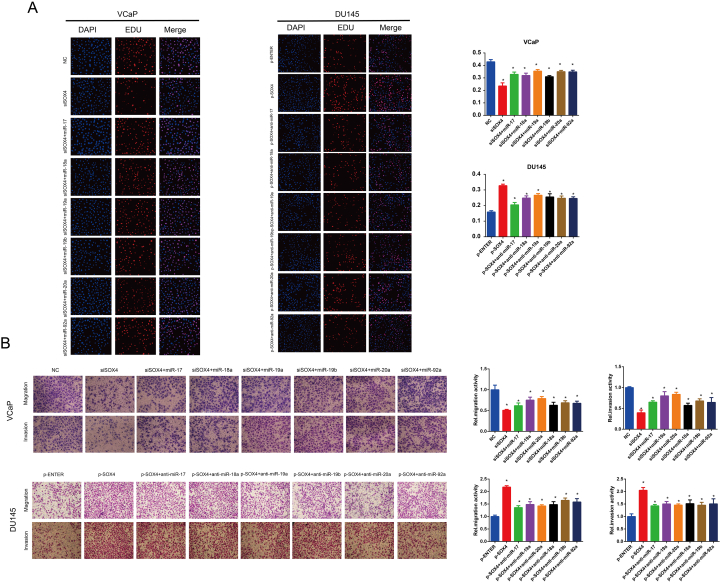
Since SOX4 upregulates miR-17-92 cluster expression, we next investigated whether modulation of miR-17-92 cluster could mediate SOX4 oncogenic activity. Expression levels of miR-17-5p, miR-19a-3p, and miR-20a-5p are shown in Figure 4A. Next, we co-transfected SOX4 knockdown and respective miR-17-92 cluster member mimics in VCaP cells, while we co-transfected SOX4 overexpression and respective miR-17-92 cluster member inhibitors in DU145 cells. Consistent with our previous reports, knockdown of SOX4 inhibited PCa cell proliferation, migration, and invasion, whereas reconstitution of miR-17-92 cluster members partially blocked the repressed effects due to SOX4-knockdown. Additionally, inhibitors of miR-17-92 cluster members partially blocked SOX4-overexpression–induced cell proliferation, migration, and invasion (Supplementary Figure 3). Results of miR-17-5p, miR-19a-3p, miR-20a-5p, and their anti-mimics were shown in the Figure 4, B-E. Collectively, these findings suggest that SOX4-induced PCa cell proliferation, migration, and invasion are mediated by miR-17-92 cluster members.
Figure 4.
SOX4-induced PCa cell proliferation, migration, and invasion are mediated by miR-17-5p, miR-19a-3p, and miR-20a-5p. (A) RT-qPCR analysis of miR-17-5p, miR-19a-3p, and miR-20a-5p expression in PCa cell lines. VCaP cells were transiently transfected with indicated siRNA and mimics (B, D), and DU145 cells were transiently transfected with indicated plasmid and anti-mimics (C, E). After incubation for 48 hours, cell proliferation was examined by EDU assay, and migration invasion ability of cells was examined by Transwell assay. *P<.05, P values based on Student’s t test.
Supplementary Figure 3.
SOX4-induced PCa cell proliferation, migration, and invasion are mediated by miR-17-92 cluster members. VCaP cells were transiently transfected with indicated siRNA and mimics, and DU145 cells were transiently transfected with indicated plasmid and anti-mimics. After incubation for 48 hours, cell proliferation was examined by EDU assay (A), and migration and invasion ability of cells was examined by transwell assay (B). *P <0.05; P values based on Student’s t test.
RB1 Protein Is Downregulated by the SOX4/miR-17-92 Axis
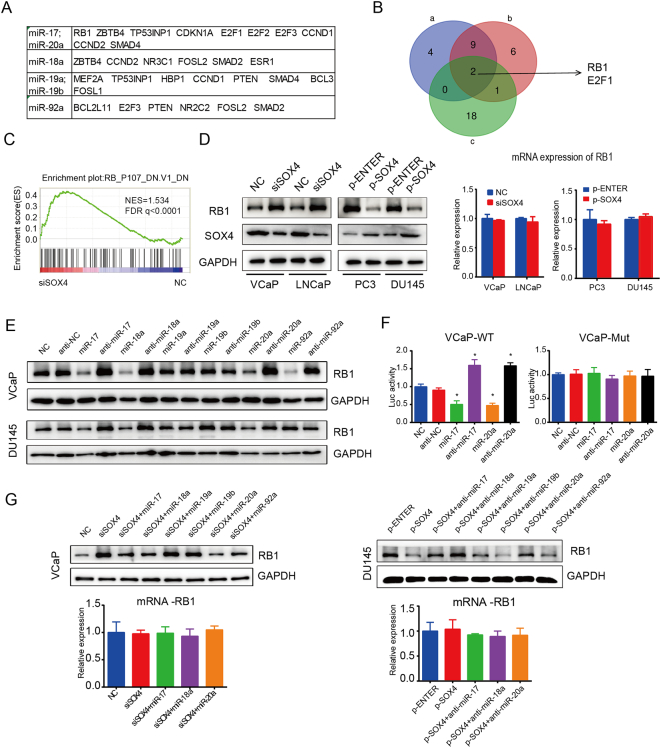
In order to better understand the network of SOX4/miR-17-92 in PCa cells, we searched for potential targets of SOX4 and miR-17-92. Using GSEA, we analyzed the genes that are potentially regulated by SOX4 from GSE11914 dataset [11]. Reported and important target genes of miR-17-92 were shown in Figure 5A [40]. Cross-comparison of all clustered genes allowed us to generate a “consensus” of two genes, namely, RB1 and E2F1 (Figure 5B). RB1 is a classical tumor suppressor gene, and RB1 dysfunction has been shown to promote PCa development and progression [27], [41], [42], [43], [44]. RB1-upregulated gene signatures were significantly enriched after SOX4 knockdown (Figure 5C). Next, we confirmed that RB1 is a target of the SOX4/miR-17-92 axis. We found that SOX4 downregulated the RB1 protein expression but failed at mRNA level at 48 hours (Figure 5D). So our results suggest that SOX4 represses the RB1 protein at the posttranscriptional level. Interestingly, miR-17-5p, miR-18a-5p, and miR-20a-5p repressed RB1 protein expression, and their respective inhibitors increased RB1 protein expression. No significant alteration of RB1 protein was found in the miR-19a-3p, miR-19b-3p, and miR-92a-3p mimics and inhibitors (Figure 5E). RB1 has been reported to be a direct target gene of miR-17 and miR-20a. The relative luciferase activity of the wild-type RB1 3′-UTR was inhibited by miR-17-5p and miR-20a-5p mimics but increased by their respective inhibitors. However, such effects were not observed with the mutant RB1 3′-UTR (Figure 5F).
Figure 5.
SOX4 represses RB1 protein expression via upregulating miR17-92 cluster. (A) Table of miR-17-92 cluster members downstream target genes. (B) A Venn diagram depicting two genes that meet the following properties: (a) Statistically significant genes of GSEA analysis of siSOX4 vs. NC from GSE11914. (b) Statistically significant genes of GSEA analysis of SOX4 overexpression vs. NC from GSE11914. (c) miR-17-92 downstream target genes shown in A. (C) RB1 knockout downregulated gene signatures were enriched for upregulation upon SOX4 knockdown from GSE11914. (D) The RB1 protein expression detected by Western blot and RB-mRNA analyzed by RT-qPCR after knockdown or overexpression of SOX4 in PCa cells. (E) Indicated mimics and anti-mimics were transiently transfected into VCaP and DU145 cells. After incubation for 48 hours, RB1 protein expression was detected by Western blot. (F) RB1 is a direct target of miR-17-5p and miR-20a-5p. Overexpression/knockdown of miR-17 and miR-20a suppresses/increases RB1 3′-UTR but not mutant 3′-UTR luciferase activities in VCaP cells. (G) MiR-17-5p and miR-20a-5p were able to override the downregulation of RB1 protein by SOX4. We co-transfected VCaP/DU145 cells with miR-17-92 cluster members mimics/inhibitors and SOX4 knockdown/overexpression vector. After incubation for 48 hours, Western blot and RT-qPCR were carried out to detect the expression of RB1. *P<.05; P values based on Student’s t test.
To further explore whether miR-17-92 is required for the downregulation of RB1 protein by SOX4, we co-transfected SOX4 knockdown or overexpression and miR-17-5p, miR-18a-5p, and miR-20a-5p mimics/inhibitors in VCaP/DU145 cells. As shown in Figure 5G, SOX4 knockdown significantly induced RB1 protein expression, whereas miR-17-5p, miR-18a-5p, and miR-20a-5p mimics significantly blocked this increase in VCaP cells. Accordingly, miR-17-5p, miR-18a-5p, and miR-20a-5p inhibitors significantly restored RB1 expression in DU145 cells. No significant changes were found at mRNA level of RB1. Taken together, our data support that SOX4 represses RB1 protein expression via upregulating miR-17-92 cluster expression.
Co-Expression of SOX4, miR-17-92 Cluster, and RB1 In Vivo
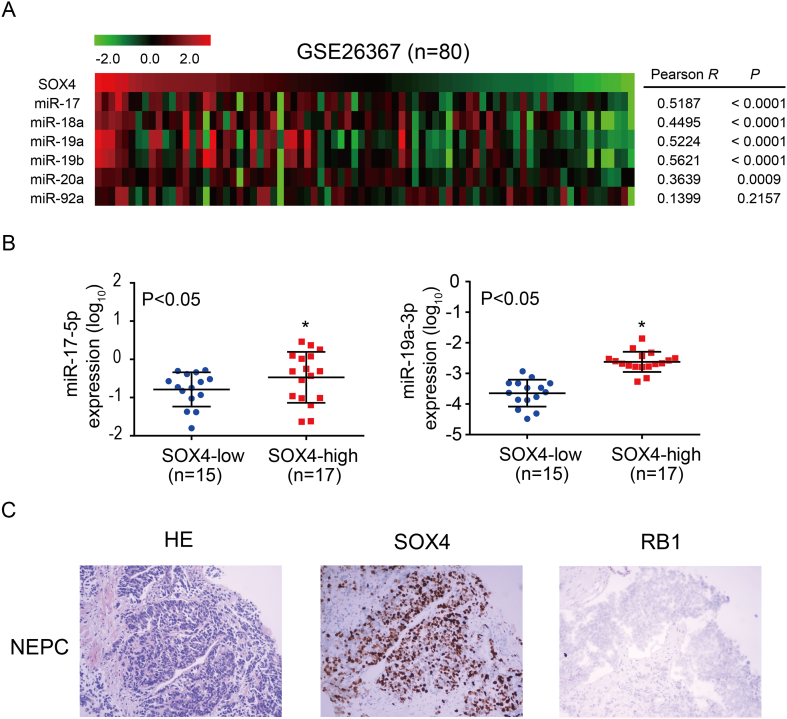
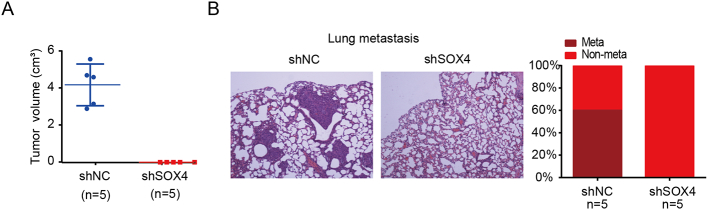
To identify the correlation between SOX4 and miR-17-92 cluster in vivo, we first analyzed public datasets. Our result showed that the expression of miR-17-92 cluster members was positively correlated with SOX4 expression in PCa patients from the GSE26367 dataset (Figure 6A) [45]. Furthermore, we examined whether SOX4 regulation of miR-17-92 cluster is physiologically relevant in PCa clinical specimens. We found that the expression of miR-17-5p and miR-19a-3p was higher in SOX4-positive patients as compared to SOX4-negative patients (Figure 6B). Using TMA and IHC, we found that SOX4 was overexpressed in 23.0% (32/140) of Chinese PCa patients. SOX4 overexpression was significantly associated with high Gleason scores and the presence of distant metastasis. IHC showed that approximately 8% of PCa patients in our cohort demonstrated RB1 protein loss. Of the seven PCa cases with small cell carcinoma, IHC also revealed that 71% (5/7) showed overexpression of SOX4 and 57.1% (4/7) demonstrated RB1 loss. Notably, IHC revealed that RB1 protein expression was negatively correlated with SOX4 expression in PCa patients with small cell carcinoma (Figure 6C). In addition, SOX4 knockdown significantly inhibited tumor formation and metastasis (Supplementary Figure 4). Collectively, our data suggest that there is a significant relationship between the co-expression of SOX4, miR-17-92, and RB1, which indicates that the SOX4/miR-17-92/RB1 axis may promote PCa progression.
Figure 6.
Expression of miR-17-92 cluster members, RB1, and SOX4 in human PCa patients. (A) MiR-17-92 cluster expression is positively correlated to SOX4 mRNA expression in PCa patients from the GSE26367 dataset; the number of PCa patients, P values, and r values are as shown. (B) Respective miR-17-5p and miR-19a-3p expression was significantly positive correlated to SOX4 protein expression in PCa patients. Number of samples (n) and P values (determined by a two-tailed Student’s t test) are as shown. (C) Representative images of H&E, SOX4, and RB1 protein expression in a PCa case with small cell carcinoma.
Supplementary Figure 4.
Knockdown of SOX4 inhibits tumor formation and lung metastasis. (A) Tumor endpoint volumes, at 6 weeks postinjection in male BALB/c mice, of VCaP cells transduced with either shNC or shSOX4 (n= 5 per group). (B) Knockdown of SOX4 inhibits lung metastasis. Representative H&E images and the percentage of lung metastasis are shown.
SOX4 Knockdown Restrains NE Phenotype and PCa Cell Proliferation
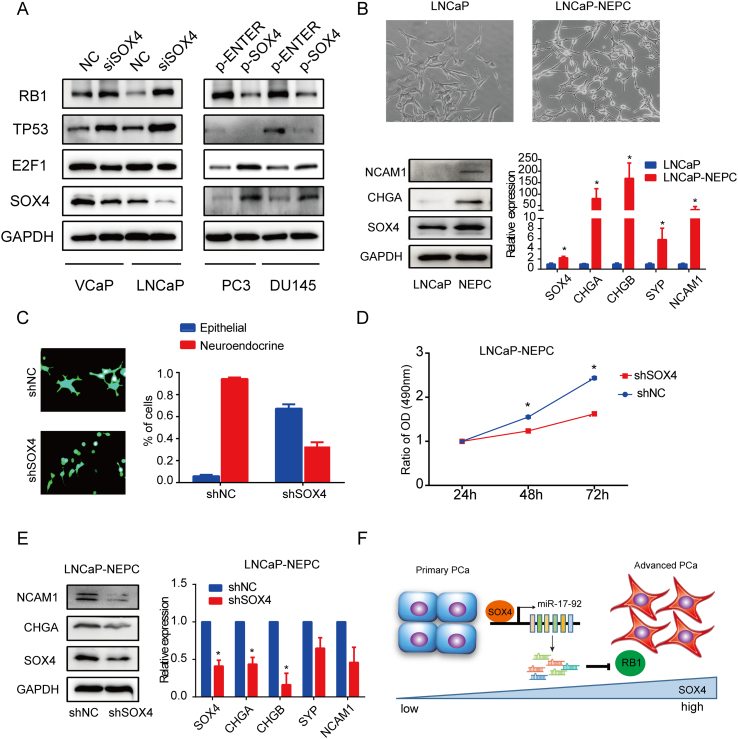
Previous studies report that RB1 loss is one characteristic of NEPC and that RB1 dysfunction promotes PCa metastasis and CRPC progression [26], [41], [43], [46]. To identify if the SOX4/miR-17-92/RB1 axis plays an important role in PCa progression, we first confirmed that SOX4 causes RB1 and TP53 downregulation but increases E2F1 protein expression (Figure 7A). We hypothesized that SOX4 knockdown may prevent NE differentiation in PCa cells.
Figure 7.
SOX4 knockdown restrains neuroendocrine differentiation and proliferation of LNCaP-NEPC cells. (A) Analysis of the protein expression of RB1, TP53, and E2F1 in SOX4 knockdown/overexpression PCa cells detected by Western blot. (B) The morphology of LNCaP and LNCaP-NEPC cells was shown in the top. LNCaP-NEPC was established by continuously culturing the LNCaP cells in medium with 10% FBS depleted of steroids for 3 months. As shown in bottom, NEPC markers and SOX4 expression were detected by Western blot and RT- qPCR. (C) LNCaP-NEPC cells were infected with shNC or shSOX4 lentivirus and selected for stable cells. The morphology of the control and SOX4 knockdown cells was shown on the left. Shown on the right is the percentage of cells with epithelial and neuroendocrine-like phenotype. (D) Cell proliferation was examined by MTS. *P<.05. P values based on Student’s t test. (E) The expression of NEPC markers and SOX4 in shNC and shSOX4 LNCaP-NEPC cells was measured by Western blot and RT-qPCR. (F) A model depicting SOX4, downregulating RB1 protein via upregulating miR-17-92 cluster, promotes PCa progression.
Next, LNCaP-NEPC cells with clear morphological changes toward an NE phenotype were chosen for further studies. Moreover, SOX4 and NEPC markers were significantly increased in LNCaP-NEPC cells as compared to LNCaP cells (Figure 7B). We then established the stable knockdown of SOX4 in LNCaP-NEPC cells. The results showed that SOX4 knockdown was able to convert LNCaP-NEPC cells with the NE phenotype to the rounded shape that is typical of epithelial cell clusters. The percentages of cells with epithelial and NE phenotype are shown in Figure 7C. Furthermore, SOX4 knockdown is shown to reduce LNCaP-NEPC cells' proliferation (Figure 7D) and NEPC markers' expression (Figure 7E) as compared to the control. Therefore, our data suggest that SOX4 knockdown restrains the NE phenotype and SOX4 may promote PCa progression.
Discussion
As a developmental transcription factor, SOX4 is overexpressed in multiple human malignancies and has been recognized as one of the 64 “cancer signature” genes, suggesting a fundamental role in tumor development and progression [9], [47]. Previously, we and others have suggested that SOX4 overexpression is correlated with high Gleason scores, abnormal cell proliferation, as well as tumor progression through the induction of EMT and metastasis in PCa [8], [10], [13], [48]. In this study, we are the first to suggest that SOX4 expression might also be associated with development of the NE phenotype in PCa. All of the above findings suggest that SOX4 promotes PCa progression.
To better understand of the mechanism of how SOX4 promote PCa progression, we focused on the relationship between SOX4 and miRNAs. MiRNAs play oncogenic or suppressive roles in PCa progression [15], [16], [18]. This is the first study that characterizes the miRNAs regulated by SOX4 in PCa cells. Previously, Scharer et al. have identified the direct transcriptional targets of SOX4 using a ChIP-seq approach in PCa cells, which includes the many genes involved in transcriptional regulation, developmental pathways, growth factor signaling, and tumor metastasis [11]. An increasing body of evidences has demonstrated that SOX4 directly regulates a number of genes important for PCa progression and metastasis including EGFR, tenascin C, Rac1, ADAM10, Frizzled-5, and EZH2 [9], [11], [12]. Interestingly, the SOX4 transcriptional network impacts the Notch, Wnt, and PI3K pathways; participates in modulating stem cell activation by interacting with Oct-4; and upregulates SOX2 expression [9], [49]. Although preliminary data suggested that SOX4 regulates components of the miRNA pathway such as Dicer and Argonaute 1[9], [11], it has not been reported whether SOX4 regulates the miRNAs expression in PCa cells. Using high-throughput miRNAs profiling analyses, our study discovered series of miRNAs that are potentially regulated by SOX4, including miR-106-25 cluster, miR-let7, miR-221/222, and miR-200b-3p. We also demonstrate a direct link between SOX4 and the miR-17-92 cluster. SOX4 transcriptionally upregulates the miR-17-92 cluster expression. Furthermore, SOX4-induced PCa cell proliferation, migration, and invasion are mediated by the miR-17-92 cluster.
It has been well documented that the miR-17-92 cluster plays an oncogenic role in PCa [34], [35]. Reported genes such as PTEN, BIM, RB1, Trp53inp1, and SMAD4 are well-known targets of miR-17-92 [50], [51]. Phosphatase and tensin homolog deleted on chromosome ten (PTEN) functions as a negative regulator of the PI3K/AKT signaling; however, SOX4 promotes PI3K/AKT signaling and is indispensable for prostate tumorigenesis initiated by PTEN ablation [52], [53], [54]. Additionally, both SOX4 and SMAD4 participate in EMT when induced by TGF-β [55]. Therefore, PTEN and SMAD4 maybe targets of the SOX4/miR-17-92 axis.
Among the target genes, RB1 is a key cell cycle inhibitor and tumor suppressor. RB1 interacts with E2F1 and represses its transcription activity, which leads to cell cycle arrest [56]. RB1 is often cited as a “gatekeeper,” whose inactivation, direct or indirect, is a rate-limiting step for tumor initiation [42]. Recent reports have demonstrated that RB1 dysfunction causes early-stage prostate cancer [41] and promotes PCa progression [57]. Moreover, RB1 dysfunction promotes metastasis and induces uncontrolled AR activity to promote CRPC development [27], [46]. Deficiency of RB1 protein has many causes. Among them, RB1 gene loss is the most common mechanism by which this occurs [41], [46]. However, there are still other mechanisms that have been reported to explain decreased RB1 expression. Previous studies have shown that RB1 is a target protein of PI3K/Akt signaling pathway [58] and AGE/RAGE/Akt pathway downregulates RB1 protein to promote PCa progression [59]. Furthermore, reduced or increased RB1-interacted protein also contributes to decrease RB1 protein levels [57], [60]. Additionally, RB loss is infrequently detected in primary PCa (~7%) but is predominantly associated with transition to the incurable CRPC and NEPC [26], [46].
Here, we proposed a model for a SOX4/miR-17-92/RB1 axis which gives insight into how RB1 protein is reduced (besides RB1 loss). Mechanically, SOX4 downregulates RB1 protein expression by upregulating the miR-17-92 cluster expression. Since RB1 loss is one characteristic of NEPC and promotes PCa metastasis [26], [27], we hypothesized that the SOX4/miR-17-92/RB1 axis might take part in NE phenotype development (Figure 7F). It is important to note that our data show that SOX4 knockdown represses the expression of NEPC, restraining the NE phenotype and cell proliferation. Here, we proposed a model of the SOX4/miR-17-92/RB1 axis which may be involved in PCa progression.
Conclusions
In conclusion, SOX4 downregulates RB1 by transcriptionally upregulating miR-17-92 cluster expression in PCa. The SOX4/miR-17-92/RB1 axis may promote PCa progression and development of the NE phenotype.
The following are the supplementary data related to this article.
SiRNAs Used in This Study
Primers Used in This Study
Acknowledgements
This work was supported by National Natural Science Foundation of China (grant nos. 81472417, 81672554, and 81528015 to B.H.).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zong Y, Goldstein AS. Adaptation or selection—mechanisms of castration-resistant prostate cancer. Nat Rev Urol. 2013;10:90–98. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 4.Alanee S, Moore A, Nutt M, Holland B, Dynda D, El-Zawahry A, McVary KT. Contemporary incidence and mortality rates of neuroendocrine prostate cancer. Anticancer Res. 2015;35:4145–4150. [PubMed] [Google Scholar]
- 5.Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene. 2015;34:1745–1757. doi: 10.1038/onc.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang HT, Yao YH, Li BG, Tang Y, Chang JW, Zhang J. Neuroendocrine prostate cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis—a systematic review and pooled analysis. J Clin Oncol. 2014;32:3383–3390. doi: 10.1200/JCO.2013.54.3553. [DOI] [PubMed] [Google Scholar]
- 7.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Zhang J, Yang X, Chang YW, Qi M, Zhou Z, Zhang J, Han B. SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis. 2013;16:301–307. doi: 10.1038/pcan.2013.25. [DOI] [PubMed] [Google Scholar]
- 9.Moreno CS. The sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression: crosstalk with the Wnt, Notch, and PI3K pathways. Am J Pathol. 2010;176:518–527. doi: 10.2353/ajpath.2010.090657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 11.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schubeler D, van Nimwegen E, Christofori G. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell. 2013;23:768–783. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Li Y, Yang X, Yuan H, Li X, Qi M, Chang YW, Wang C, Fu W, Yang M. ERG-SOX4 interaction promotes epithelial-mesenchymal transition in prostate cancer cells. Prostate. 2014;74:647–658. doi: 10.1002/pros.22783. [DOI] [PubMed] [Google Scholar]
- 14.Yang M, Wang J, Wang L, Shen C, Su B, Qi M, Hu J, Gao W, Tan W, Han B. Estrogen induces androgen-repressed SOX4 expression to promote progression of prostate cancer cells. Prostate. 2015;75:1363–1375. doi: 10.1002/pros.23017. [DOI] [PubMed] [Google Scholar]
- 15.Deng JH, Deng Q, Kuo CH, Delaney SW, Ying SY. MiRNA targets of prostate cancer. Methods Mol Biol. 2013;936:357–369. doi: 10.1007/978-1-62703-083-0_27. [DOI] [PubMed] [Google Scholar]
- 16.Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2018;21(2):126–144. doi: 10.1007/s12094-018-1910-8. [DOI] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang Q, Li L, Xie H, He D, Chen J, Song W, Chang LS, Chang HC, Yeh S, Chang C. Anti-androgen enzalutamide enhances prostate cancer neuroendocrine (NE) differentiation via altering the infiltrated mast cells --> androgen receptor (AR) --> miRNA32 signals. Mol Oncol. 2015;9:1241–1251. doi: 10.1016/j.molonc.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia L, Gui B, Zheng D, Decker KF, Tinay I, Tan M, Wang X, Kibel AS. Androgen receptor-regulated miRNA-193a-3p targets AJUBA to promote prostate cancer cell migration. Prostate. 2017;77:1000–1011. doi: 10.1002/pros.23356. [DOI] [PubMed] [Google Scholar]
- 20.Shi XB, Ma AH, Tepper CG, Xia L, Gregg JP, Gandour-Edwards R, Mack PC, Kung HJ, deVere White RW. Molecular alterations associated with LNCaP cell progression to androgen independence. Prostate. 2004;60:257–271. doi: 10.1002/pros.20039. [DOI] [PubMed] [Google Scholar]
- 21.Shen R, Dorai T, Szaboles M, Katz AE, Olsson CA, Buttyan R. Transdifferentiation of cultured human prostate cancer cells to a neuroendocrine cell phenotype in a hormone-depleted medium. Urol Oncol. 1997;3:67–75. doi: 10.1016/s1078-1439(97)00039-2. [DOI] [PubMed] [Google Scholar]
- 22.Zelivianski S, Verni M, Moore C, Kondrikov D, Taylor R, Lin MF. Multipathways for transdifferentiation of human prostate cancer cells into neuroendocrine-like phenotype. Biochim Biophys Acta. 2001;1539:28–43. doi: 10.1016/s0167-4889(01)00087-8. [DOI] [PubMed] [Google Scholar]
- 23.Liu HT, Gao P. The roles of microRNAs related with progression and metastasis in human cancers. Tumour Biol. 2016;37(12):15383–15397. doi: 10.1007/s13277-016-5436-9. [DOI] [PubMed] [Google Scholar]
- 24.Qi M, Yang X, Zhang F, Lin T, Sun X, Li Y, Yuan H, Ren Y, Zhang J, Qin X. ERG rearrangement is associated with prostate cancer-related death in Chinese prostate cancer patients. PLoS One. 2014;9 doi: 10.1371/journal.pone.0084959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu HT, Xing AY, Chen X, Ma RR, Wang YW, Shi DB, Zhang H, Li P, Chen HF, Li YH. MicroRNA-27b, microRNA-101 and microRNA-128 inhibit angiogenesis by down-regulating vascular endothelial growth factor C expression in gastric cancers. Oncotarget. 2015;6:37458–37470. doi: 10.18632/oncotarget.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, Mosier S, Gocke CD, Epstein JI, Netto GJ. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res. 2014;20:890–903. doi: 10.1158/1078-0432.CCR-13-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thangavel C, Boopathi E, Liu Y, Haber A, Ertel A, Bhardwaj A, Addya S, Williams N, Ciment SJ, Cotzia P. RB loss promotes prostate cancer metastasis. Cancer Res. 2017;77:982–995. doi: 10.1158/0008-5472.CAN-16-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 30.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai HK, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan TL. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer. 2017;17:759. doi: 10.1186/s12885-017-3729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akamatsu S, Wyatt AW, Lin D, Lysakowski S, Zhang F, Kim S, Tse C, Wang K, Mo F, Haegert A. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 2015;12:922–936. doi: 10.1016/j.celrep.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Zhou P, Ma L, Zhou J, Jiang M, Rao E, Zhao Y, Guo F. miR-17-92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int J Oncol. 2016;48:1737–1748. doi: 10.3892/ijo.2016.3392. [DOI] [PubMed] [Google Scholar]
- 35.Feng S, Qian X, Li H, Zhang X. Combinations of elevated tissue miRNA-17-92 cluster expression and serum prostate-specific antigen as potential diagnostic biomarkers for prostate cancer. Oncol Lett. 2017;14:6943–6949. doi: 10.3892/ol.2017.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi N, Park J, Lee JS, Yoe J, Park GY, Kim E, Jeon H, Cho YM, Roh TY, Lee Y. miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget. 2015;6:23533–23547. doi: 10.18632/oncotarget.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudson RS, Yi M, Esposito D, Glynn SA, Starks AM, Yang Y, Schetter AJ, Watkins SK, Hurwitz AA, Dorsey TH. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene. 2013;32:4139–4147. doi: 10.1038/onc.2012.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dallavalle C, Albino D, Civenni G, Merulla J, Ostano P, Mello-Grand M, Rossi S, Losa M, D'Ambrosio G, Sessa F. MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J Clin Invest. 2016;126:4585–4602. doi: 10.1172/JCI86505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Qin T, Hu W, Chen B, Dai M, Xu G. Genome-wide methylation patterns in androgen-independent prostate cancer cells: a comprehensive analysis combining MeDIP-bisulfite, RNA, and microRNA sequencing data. Genes (Basel) 2018;9 doi: 10.3390/genes9010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Li Y, Qi P, Ma Z. Biology of MiR-17-92 cluster and its progress in lung cancer. Int J Med Sci. 2018;15:1443–1448. doi: 10.7150/ijms.27341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maddison LA, Sutherland BW, Barrios RJ, Greenberg NM. Conditional deletion of Rb causes early stage prostate cancer. Cancer Res. 2004;64:6018–6025. doi: 10.1158/0008-5472.CAN-03-2509. [DOI] [PubMed] [Google Scholar]
- 42.Macleod KF. The RB tumor suppressor: a gatekeeper to hormone independence in prostate cancer? J Clin Invest. 2010;120:4179–4182. doi: 10.1172/JCI45406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355:78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He F, Mo L, Zheng XY, Hu C, Lepor H, Lee EY, Sun TT, Wu XR. Deficiency of pRb family proteins and p53 in invasive urothelial tumorigenesis. Cancer Res. 2009;69:9413–9421. doi: 10.1158/0008-5472.CAN-09-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Long Q, Johnson BA, Osunkoya AO, Lai YH, Zhou W, Abramovitz M, Xia M, Bouzyk MB, Nam RK, Sugar L. Protein-coding and microRNA biomarkers of recurrence of prostate cancer following radical prostatectomy. Am J Pathol. 2011;179:46–54. doi: 10.1016/j.ajpath.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 2010;120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Jiang H, Shao J, Mao R, Liu J, Ma Y, Fang X, Zhao N, Zheng S, Lin B. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt-p53 axis. BMC Neurol. 2014;14:207. doi: 10.1186/s12883-014-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 50.Grillari J, Hackl M, Grillari-Voglauer R. miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology. 2010;11:501–506. doi: 10.1007/s10522-010-9272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dal Bo M, Bomben R, Hernandez L, Gattei V. The MYC/miR-17-92 axis in lymphoproliferative disorders: a common pathway with therapeutic potential. Oncotarget. 2015;6:19381–19392. doi: 10.18632/oncotarget.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bilir B, Osunkoya AO, WGt Wiles, Sannigrahi S, Lefebvre V, Metzger D, Spyropoulos DD, Martin WD, Moreno CS. SOX4 is essential for prostate tumorigenesis initiated by PTEN ablation. Cancer Res. 2016;76:1112–1121. doi: 10.1158/0008-5472.CAN-15-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehta GA, Parker JS, Silva GO, Hoadley KA, Perou CM, Gatza ML. Amplification of SOX4 promotes PI3K/Akt signaling in human breast cancer. Breast Cancer Res Treat. 2017;162:439–450. doi: 10.1007/s10549-017-4139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonneau D, Longy M. Mutations of the human PTEN gene. Hum Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 55.David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, Iacobuzio-Donahue CA, Massague J. TGF-beta tumor suppression through a lethal EMT. Cell. 2016;164:1015–1030. doi: 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Padmanabhan B, Adachi N, Kataoka K, Horikoshi M. Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6. J Biol Chem. 2004;279:1546–1552. doi: 10.1074/jbc.M310266200. [DOI] [PubMed] [Google Scholar]
- 57.Qiu X, Pascal LE, Song Q, Zang Y, Ai J, O'Malley KJ, Nelson JB, Wang Z. Physical and functional interactions between ELL2 and RB in the suppression of prostate cancer cell proliferation, migration, and invasion. Neoplasia. 2017;19:207–215. doi: 10.1016/j.neo.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z, Gao N, He H, Huang C, Luo J, Shi X. Vanadate activated Akt and promoted S phase entry. Mol Cell Biochem. 2004;255:227–237. doi: 10.1023/b:mcbi.0000007278.27936.8b. [DOI] [PubMed] [Google Scholar]
- 59.Bao JM, He MY, Liu YW, Lu YJ, Hong YQ, Luo HH, Ren ZL, Zhao SC, Jiang Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am J Cancer Res. 2015;5:1741–1750. [PMC free article] [PubMed] [Google Scholar]
- 60.Fan S, Yuan R, Ma YX, Xiong J, Meng Q, Erdos M, Zhao JN, Goldberg ID, Pestell RG, Rosen EM. Disruption of BRCA1 LXCXE motif alters BRCA1 functional activity and regulation of RB family but not RB protein binding. Oncogene. 2001;20:4827–4841. doi: 10.1038/sj.onc.1204666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SiRNAs Used in This Study
Primers Used in This Study