Abstract
Background
Many aspects of our lives are now digitized and connected to the internet. As a result, individuals are now creating and collecting more personal data than ever before. This offers an unprecedented chance for human-participant research ranging from the social sciences to precision medicine. With this potential wealth of data comes practical problems (e.g., how to merge data streams from various sources), as well as ethical problems (e.g., how best to balance risks and benefits when enabling personal data sharing by individuals).
Results
To begin to address these problems in real time, we present Open Humans, a community-based platform that enables personal data collections across data streams, giving individuals more personal data access and control of sharing authorizations, and enabling academic research as well as patient-led projects. We showcase data streams that Open Humans combines (e.g., personal genetic data, wearable activity monitors, GPS location records, and continuous glucose monitor data), along with use cases of how the data facilitate various projects.
Conclusions
Open Humans highlights how a community-centric ecosystem can be used to aggregate personal data from various sources, as well as how these data can be used by academic and citizen scientists through practical, iterative approaches to sharing that strive to balance considerations with participant autonomy, inclusion, and privacy.
Keywords: personal data, crowdsourcing, citizen science, database, open data, participatory science, peer production
Background
Research involving human participants, from biomedical and health research to social sciences studies, is experiencing rapid changes. The rise of electronic records, online platforms, and data from devices contributes to a sense that these collected data can change how research in these fields is performed [1–4].
Among the affected disciplines is precision medicine—which takes behavioral, environmental, and genetic factors into account and has become a vision for health care in the United States [5]. By taking individual parameters into account, precision medicine aims to improve health outcomes, e.g., by optimizing drugs based on a patient’s genetic makeup [6, 7].
Access to large-scale data sets, along with availability of appropriate methods to analyze these data [8, 9], is often described as a major prerequisite for the success of precision medicine [10]. Decreasing costs for large-scale, individualized analyses such as whole-genome sequencing [11] have already helped facilitate both research in precision medicine and its adoption. In addition, an increasing number of patients and healthy individuals are collecting health-related data outside traditional health care, e.g., through smartphones and wearable devices [12, 13] or through direct-to-consumer (DTC) genetic testing [14].
Indeed, ≥12–17 million individuals have taken a DTC genetic test [15, 16], while >25 million such tests have been purchased [17]. Meanwhile, it is estimated that by 2020 >2 exabytes of storage will be needed for health care data [18] alone. Furthermore, data from social network sites such as Facebook and Twitter are increasingly likely targets for medical data mining [19]. Additionally, more data are becoming available from personal medical devices, both in real time and for retrospective analyses [20].
These changes to research and medical practice bring with them a number of challenges, including the problems of data silos, ethical data sharing, and participant involvement. A participant-centered approach to personal data aggregation, sharing, and research has the potential to address these issues. To achieve this, we created "Open Humans" as a digital ecosystem designed to facilitate individual data aggregation across data sources, granular management of data sharing, and co-created research.
Data silos
To fully realize the promises of these large personal data collections, not only in precision medicine but in all fields of research, access to both big data and smaller data sources is needed, as is the ability to tap into a variety of data streams and link these data [10, 21]. Data silos can hinder the merging and reuse of data by third parties for a number of reasons: they can be incompatible due to different data licenses [22] or inaccessible due to privacy, ethical, and regulatory concerns [23–25]. For example, the US National Human Genome Research Institute’s Database of Genotypes and Phenotypes remains an underused resource because of logistical and regulatory/ethical oversight challenges for would-be users [26]. In addition to legal barriers, there are typically technical challenges in rendering data accessible, usable, and/or anonymized, and a data controller typically has incentives to seek compensation in return for these activities.
Beyond biomedical data sets, there are data from wearable devices, social media, and other data held by private companies, from which data exports are often not available. In other cases data access might be legally mandated, but the practical outcomes are mixed or in progress [27, 28], e.g., for clinical health data in the United States as mandated by the 1996 Health Insurance Portability and Accountability Act and 2009 Health Information Technology for Economic and Clinical Health Act (HIPAA and HITECH Act) and for personal data in the European Union as mandated by rights to data access and data portability in the 2016 General Data Protection Regulation (GDPR) [29, 30]. In addition, within the context of research involving human participants, data access may be recommended [31] but not legally required, and as a result is not typically provided [32]. Data portability and easy access to research data by participating individuals could empower them to steer research in directions that affect their lives and health outcomes.
Ethical data reuse
While the sharing and reuse of biomedical data can potentially transform medical care and medical research, it brings along a number of ethical considerations [33, 34]. In the field of human genetics, the ethics of sharing data has been extensively considered with respect to how research participants and patients can give informed consent for studies that carry risks of genetic discrimination, loss of privacy, and reidentification in publicly shared data [35, 36]. Owing to access and portability issues, however, research with biomedical data is rarely driven by the individuals from whom the data originated—and as a result, such research fails to give patients much power over how their data can be used [37]. For example, it is now abundantly clear that DTC genetic testing companies routinely share their customers' deidentified (but reidentifiable) data with third parties [38]. Open Humans seeks to be among the agents for change in this regard. Bottom-up research initiatives have included disease- and/or mutation-specific efforts [39, 40] and the development of platforms meant to allow participants to control data sharing at a granular level [41]. Open Humans is meant to complement such initiatives and enable the creation of multiple ”sandboxes” where both personal and biomedical data can be leveraged to help grow empirical knowledge and further downstream development of diagnostics and therapies.
Elsewhere, social media is also gaining importance in research as well as public health [42]. Differing perceptions on the sensitivity of social media data can lead to privacy concerns. For example, an analysis performed on 70,000 users of an online dating website, where private personal data were scraped by researchers and then publicly shared, caused a public outcry [43]. Such cases have sparked calls for caution in performing ”big data” research with these new forms of personal data [44, 45].
Research that interacts with social media users raises additional concerns. For example, Facebook was widely criticized for an experiment to study emotional contagion among 700,000 of its users without their consent or debriefing, prompting discussion of the ethics of unregulated human subjects research and ”A/B testing” by private entities [46–48]. And the 2018 disclosure of the Cambridge Analytica controversy, in which a private firm harvested information from 50 million Facebook users without their permission, led Facebook to tighten control over its APIs, turning it into even more of a silo that does not allow for research to be performed by outside researchers [49].
For the foreseeable future, researchers who reuse data from commercial sources will have to decide how to balance the interests of commercial data controllers, participants, and society. While there is no consensus on how research consent for existing personal data should be performed, we know that participants desire more granular abilities to manage data sharing: to decide who can and cannot see it, under what circumstances, and what can and cannot be done with it [50]. Such individual control will be especially critical in the sensitive context of precision medicine [24].
Participant involvement
"Citizen science" mostly describes the involvement of volunteers in the data collection, analysis, and interpretation phases of research projects [51], thus both supporting the research process itself and helping with public engagement. Furthermore, the Universal Declaration of Human Rights describes a broad human right to access science as a whole, implying a right to participate in all aspects of the scientific enterprise [52].
Traditionally, many participatory science projects have focused on the natural sciences, such as natural resource management, environmental monitoring/protection, and astrophysics [53–55]. In many of these examples volunteers are asked to crowd-source and support scientists in the collection of data, e.g., by field observations or through sensors [56] or by performing human computation tasks such as classifying images [57] or generating folded protein structures [58].
Analogous to the movement in other realms of citizen science, there is a growing movement toward more participant/patient involvement in research on humans, including in fields such as radiology, public health, psychology, and epidemiology [59, 60]. Patients often have a better understanding of their disease and needs than medical/research professionals [61, 62], and that patient involvement can help catalyze policy interventions [63]. Examples include the studies on amyotrophic lateral sclerosis initiated by PatientsLikeMe users [64], crowd-sourcing efforts such as American Gut [65], and a variety of other "citizen genomics" efforts [66]. It is likely that involving patients in clinical research not only can help minimize cost but can lead to drugs being brought to market sooner [67].
Elsewhere, the "quantified self" movement, in which individuals perform self-tracking of biological, behavioral, or environmental information and design experiments with an n= 1 to learn about themselves [68], can be placed on this continuum of participant-led research [69]. By performing self-experiments and recording their own data, individuals can gain critical knowledge about themselves and the process of performing research. Analogous to the benefits of patient insights in clinical research, individuals engaged in self-tracking and personal data analysis have the potential to contribute their insights to a variety of other research areas.
A participant-centered approach to research
As shown above, substantially involving patients and participants in the research process has multiple benefits. Participants as primary data holders can help in breaking down walls among data silos and in aggregating and sharing personal data streams. Furthermore, by being involved in the research process and actively providing data, they can gain autonomy and can actively consent to their data being used, thus mitigating (but not eliminating) the likelihood of subsequent ethical concerns. Last but not least, enabling individuals to analyze and explore their own data, individually and collectively, can result in valuable feedback that helps researchers incorporate the needs, desires, and insights of participants.
In recent years a number of projects have started to explore both data donations and crowd-sourcing research with an extended involvement of participants. In genomics, both academic projects such as DNA.Land [70] and community-driven projects such as openSNP [71] are enabling crowdsourcing via personal genetic data set donations. Furthermore, the idea of "health data cooperatives" that are communally run to manage access to health data has emerged [24].
However, most of these projects limit participants’ involvement in the research process: a participant is limited, for example, to providing specific types of data for a specific data repository. Additionally, participants are rarely given an easy way to help in designing a study, let alone running their own.
To close these gaps we developed Open Humans, a community-based platform that enables its members to share a growing number of personal data types, participate in research projects and create their own, and facilitate the exploration of personal data by and for the individual member. Open Humans was initially conceived as an iteration of work with the Harvard Personal Genome Project [72]. Along with a description of the platform itself and its power and limitations, we present a set of examples of how the platform is already being used for academic and participant-led research projects.
Results
We designed Open Humans as a web platform with the goal of easily enabling connections to existing and newly created data sources and data (re-)using applications. Platform members import data about themselves from various sources into their Open Humans account. They can then explore their aggregated data and share it with projects from citizen scientists and academic researchers.
Design
In the center of the design are 3 main components: Members, Projects, and Data objects. Members can join various Projects and authorize them to read Data that are stored in their account as well as write new Data for this Member (see Figure 1 for a dataflow diagram).
Figure 1.
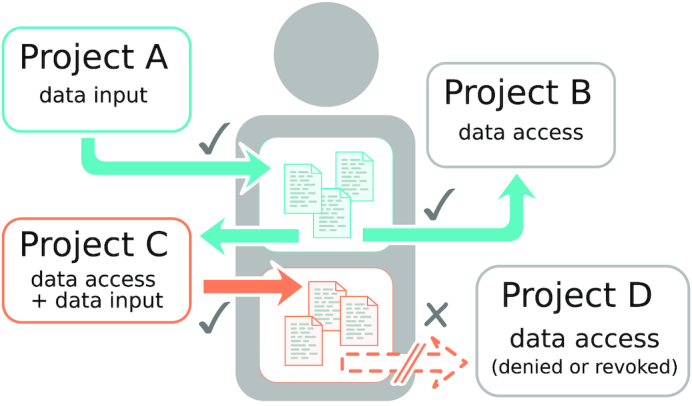
The Open Humans authorization flow. A Member (center) can join Projects and approve them to read or write Data. The Member approves Project A to deposit files (blue) into their account. They also approve Project B to read the files that Project A has deposited. Additionally, the Member approves Project C to both read the files of Project A and write new files. The Member declines to give access to their personal data to Project D.
Projects
Projects are the primary way for Members to interact with Open Humans. Projects can be created by any Member. During project creation a prospective project lead must provide a description of their project and specify the access permissions they request from Members who decide to join. These may include:
Username By default projects do not get access to a Member’s username; each Member is identified with a random, unique identifier specific to that project. This way Members can join a project pseudonymously.
Data Access A Project may ask permission to read Data that have been deposited into a Member’s account by other projects. A Project lead needs to specify to which existing projects’ data they want to have access, and only these data will be shared with the new project.
Through the permission system, Members get a clear idea of the amount of Data they are sharing by joining a given Project and whether their username will be shared and with whom.
Furthermore, all Projects have the following permissions for any Members who have joined them: (1) they can send messages to Members, which are received as emails; and (2) they can upload new Data into the Member account. Thus, in addition to acting as potential data recipients, Projects are also the avenue by which Data are added to Member accounts.
Projects can be set up in 2 different ways: As an On-site Project or as an OAuth2 Project. The OAuth2 Project format offers a standard OAuth2 user authorization process commonly used to connect across web services. Projects that implement this can connect an Open Humans user to a separate mobile or web application and can be fully automated. For Projects that do not have separate applications, the On-site Project format allows the Project to present ”consent” or ”terms of use” information within Open Humans, thereby minimizing the need for technical work on the part of a Project. Both formats have access to APIs for performing data uploads, data access, and member messaging.
Projects also need to clearly signal whether they are a research study that is subject to ethical oversight by an institutional review board (IRB) or equivalent, or whether they are not performing such research (i.e., not subject to this oversight). This allows for participant-led projects outside an academic research setting, provided Members see a notification alerting them to the absence of IRB oversight.
Thus, any Member can create a Project in the site, at any time, and all APIs work immediately. However, a Project will not be publicly listed for Members to see, and has a cap limiting the number of Members that may join. Public listing and unlimited usage is granted when a Project is marked as ”approved” following a community review process. Projects that have IRB oversight are required to provide documentation of IRB approval as part of this review process.
In summary, given the broad potential features available, a Project can cover anything from data import tools, to data processing tools, to research projects, to self-quantification projects that visualize and analyze a Member’s data.
Members
Members interact with Projects that are run on Open Humans. By joining Projects that act as data uploaders, they can add specific Data into their Open Humans accounts. This is a way to connect external services: e.g., put their genetic data or activity tracking data into their Open Humans account. Once they have connected to relevant Projects that import their own data, Members can opt in to joining additional Projects that they wish to grant access to their account’s data.
As Members are able to selectively join Projects, they can elect which projects their Data should be shared with. Members may withdraw from a Project at any time. This results in immediate revocation of Data-sharing authorization for that Project, as well as a removal of Data upload and message permissions. Projects may also support data erasure requests upon withdrawal, and any remaining Data uploaded by a project may be retained or deleted by the Member. Open Humans also allows Members to delete their entire account at any time, resulting in an immediate removal from the database, cessation of data-processing activities, and permanent deletion following the automated turnover of backup storage.
Data input and management
Data are uploaded into a Member’s account, which allows any joined Projects with requisite permissions to access these data. To be fully available to all of the possible projects that can be run on Open Humans, all data are stored in files that can be downloaded by users and Projects that have gotten permission. For any file that a Project deposits into a Member’s account, the uploading Project needs to specify at least a description and tags as metadata for the files.
Members can always review and access the Data stored in their own accounts. By default, the Data uploaded into their accounts is not shared with any projects but the one that deposited the Data, unless and until other Projects are joined and specifically authorized to access these Data. In addition to being able to share data with other Projects, Members can also opt in to making the data of individual data sources publicly available on a project-by-project basis. Data that have been publicly shared are then discoverable through the Open Humans Public Data API and are visible on a Member’s user profile.
Open Humans in practice
Using this design, the platform now features a number of projects that import data directly into Open Humans. Among data sources that can be imported and connected are 23andMe, AncestryDNA, Fitbit, Runkeeper, Withings, uBiome, and a generic VCF importer for genetic data such as whole-exome or genome sequences. Furthermore, as a special category, the Data Selfie project allows Members to add additional data files that are not supported by a specialized project yet.
The community around the Open Humans platform has expanded the support to additional Data sources by writing their own data importers and data connections. These include a bridge to openSNP, and importers for data from FamilyTreeDNA, Apple HealthKit, Gencove, Twitter, and the Nightscout (open source diabetes) community. Across these data importers, the platform supports data sources covering genetic and activity-tracking data as well as recorded GPS tracks, data from glucose monitors, and social media.
The platform has grown significantly since its launch in 2015: as of 30 May 2019, 6,976 members have signed up with Open Humans. Of these, 2,945 members have loaded 19,949 data sets into their accounts. In cases where external data sources support the import of historical data (e.g., Fitbit, Twitter), data sets can include data that reach back before the launch of Open Humans. Furthermore, overall there are now 30 projects that are actively running on Open Humans, with an additional 12 projects that have already finished data collection and thus have been concluded (see Table 1 for the most heavily engaged projects).
Table 1.
Open Humans projects with >250 members
| Project name | Description | Members | Data deposited | Data access requested |
|---|---|---|---|---|
| 23andMe Upload | Enables members to import their 23andMe data | 1,202 | 23andMe data | |
| Genevieve Genome Report | Matches a member’s genome against public variant data and invites them to contribute to shared notes. | 845 | 23andMe Upload, Harvard PGP, Genome/Exome Upload, Username, and public data | |
| Harvard Personal Genome Project | Enables members to import their data from the Personal Genome Project | 812 | Full genome sequencing data and survey data | |
| Twitter Archive Analyzer | Enables members to import their Twitter archives and analyzes them | 531 | Twitter archives | |
| Personal Data Notebooks | Enables personal data analyses with Jupyter Notebooks | 524 | Jupyter Notebooks | All Data |
| Keeping Pace | Seeks to study data about how we move around, to understand how seasons and local environment influence our movement patterns | 403 | Fitbit, Jawbone, Moves, Apple HealthKit, Runkeeper | |
| AncestryDNA Upload | Enables members to import their AncestryDNA data | 438 | AncestyDNA data | |
| Fitbit Connection | Connects a member’s Fitbit account to add data from their Fitbit activity trackers and other Fitbit devices | 404 | Data from a Fitbit account | |
| GenomiX Genome Exploration | A study of how people interact with their genome data using GenomiX, a visualization tool | 365 | Username and public data | |
| Circles | A research study that aims to discover the genetic basis for a mysterious and remarkable human trait: the areola | 321 | 23andMe, AncestryDNA, Data Selfies, Harvard PGP, Genome/Exome Upload | |
| Gencove | Your genome app—get your ancestry, microbiome, and more! Contribute your data to OpenHumans | 311 | Sequencing bam files | |
| openSNP | Enables members to connect their Open Humans and openSNP accounts | 308 | openSNP user details | Username and public data |
| Nightscout Data Transfer | A tool to easily enable the upload of data from individual Nightscout databases | 293 | Nightscout data |
Data were collected on 25 April 2019.
Use cases
To demonstrate the range of projects made possible through the platform and how the community improves the ecosystem that is growing around Open Humans, here we highlight some of the ongoing projects, covering participant-led research, academic research, and projects originating in the self-quantification community.
OpenAPS and Nightscout Data and Data Commons
There are a variety of open source diabetes tools and applications that have been created to aid individuals with type 1 diabetes in managing and visualizing their diabetes data from disparate devices. One such tool is Nightscout, which allows users to access continuous glucose monitoring (CGM) data. Another such example is OpenAPS, the Open Source Artificial Pancreas System, which is designed to automatically adjust an insulin pump’s insulin delivery to keep users’ blood glucose in a safe range overnight and between meals [73]. These tools enable real-time and retrospective data analysis of rich and complex diabetes data sets from the real world.
Traditionally, gathering this level of diabetes data would be time-consuming, expensive, and otherwise burdensome to the traditional researcher, and often pose a prohibitive barrier to researchers interested in getting started in the area of diabetes research and development. Using Open Humans, individuals from the diabetes community have created a data uploader tool called Nightscout Data Transfer Tool to enable individuals to share their CGM and related data with the Nightscout and/or OpenAPS Data Commons [74]. Sharing is done pseudonymously via random identifiers, enabling an individual to protect their privacy. Furthermore, sharing is facilitated because a single data upload may be used in multiple studies and projects. These 2 patient-led data commons have requirements for use that allow both traditional and citizen science (e.g., patient) researchers to use these data for research. These data commons were created with the goal of facilitating more access to diabetes data such as CGM data sets that are traditionally expensive to access. By doing so, they enable more researchers to explore innovations for people with diabetes. Additionally, OpenAPS is the first open source artificial pancreas system with hundreds of users, who are hoping such data sharing will facilitate better tools and better innovations for academic and commercial innovators in this space. To date, dozens of researchers and many community members have accessed and used data from each of these commons. Some publications and presentations have also showcased the work and the data donated by members of the community, further allowing other researchers to build on this body of work and these data sets [75, 76].
In addition to facilitating easier access to more and richer diabetes data, the Nightscout and OpenAPS communities have also been developing a series of open source tools to enable individuals to more easily work with the data sets [77].
Linking across communities: openSNP
openSNP is a database for personal genomics data that takes a different approach than Open Humans. While Open Humans focuses on granular control in terms of whom Members share their data with, openSNP focuses on maximizing reuse of data, by exclusively allowing individuals to donate raw DTC genetic test data into the public domain [71]. With >4,500 genetic data sets already, openSNP is one of the largest openly crowd-sourced genome databases. In addition to the genetic data, members of openSNP annotate their data with additional trait data. There is no integration of further data sources into openSNP.
Despite the differences between openSNP and Open Humans, there is overlap of members who use both platforms, with openSNP members having additional non-genetic public data sets in Open Humans. By linking the public data sets across both platforms, both ecosystems can be enriched and members can avoid having to upload their data twice.
The connection of accounts is performed by each platform providing links to the same member on the other platform: the openSNP project for Open Humans asks members for permission to read their Open Humans username during the authentication phase. By recording a member's Open Humans username, it becomes possible to link the public data sets on Open Humans to a given openSNP member. Furthermore, openSNP deposits a link to the public openSNP data sets in their Open Humans member account. So far >250 people have taken advantage of linking their openSNP and Open Humans accounts to each other.
Genetic data augmentation
Most DTC genetic testing companies genotype customers using single-nucleotide polymorphism (SNP) genotyping technology, which genotypes a fraction of the total available sites in a human genome. Because any 2 human genomes are >99% identical, these genotyped sites are carefully selected to capture human variation across global subpopulations. These sites (or genetic variants) can inform customers about their genetic ancestry, predict traits such as eye color, and determine susceptibility to some recessive diseases. While DTC testing may only genotype a tiny fraction of total sites available in the genome, it’s offered at a fraction of the price when compared to more comprehensive genotyping methods such as exome or genome sequencing. Until recently, individuals who wanted to know their genotypes at sites not covered by DTC testing needed to purchase a significantly more expensive genotyping test.
Genome-wide genotype imputation is an increasingly popular technique that offers a no- or low-cost alternative to comprehensive genotyping methods. In short, imputation is performed by scanning the entire genome in large intervals and using high-quality genotype calls from a large reference population to statistically determine a sample’s (or samples’) genotype likelihoods at missing sites based on shared genotypes with the reference population. Traditionally, genotype imputation has not been readily accessible to DTC customers because it entails a complex multi-step process requiring technical expertise and computing resources. Recently, the Michigan Imputation Server launched a free-to-use imputation pipeline [78]. The server was designed to be user-friendly and greatly lowered the barrier to entry for everyday DTC customers to have access to imputed genotypes.
As part of the Open Humans platform, Imputer is a participant-created project that performs genome-wide genotype imputation on one of a Member’s connected genetic data sources, such as 23andMe or AncestryDNA. First, Imputer must be authorized by a Member; once connected, the Imputer interface [79] allows the Member to select which genetic data source they would like to impute and launches the imputation pipeline in 1 click. Imputer submits the imputation job to a queue on a server where the imputation is performed. Once the job has finished, the imputed genotypes are uploaded as a .vcf file and an email is sent to the Member notifying them that their data are available. Imputer makes it easy for Members to augment their existing genetic data sources using techniques that were previously difficult to access. The Imputer imputation pipeline was built using genipe [80] and uses the 1000 Genomes Project [81] genotype data as the reference population.
Reuse of public data for understanding health behavior
A research team at the Universities of Copenhagen and Geneva, the Quality of Life (QoL) Technologies Lab, has been able to perform preliminary research using public data in Open Humans. Because physical inactivity is one of the strongest risk factors for preventable chronic conditions [82], the QoL Lab’s goal is to leverage self-quantification data to assess and subsequently enhance the well-being of individuals and possibly, in the long term, reduce the prevalence of some chronic diseases. At this stage, the QoL Lab has used the Open Humans public data sets of Fitbit and Apple HealthKit projects.
In Open Humans, individuals who donate public data uploaded from Fitbit and Apple HealthKit projects share with others the daily summaries taken with their Fitbit and Apple devices such as steps, resting heart rate (HR), and minutes asleep. The public data sets contain time-series data from ≥30 members, each of whom decides whether to provide access to the aforementioned measurements. The number of records for each variable available in the Open Humans database varies because not all the devices record the same variables and participants may choose not to share a particular measurement (see Fig. 2).
Figure 2.
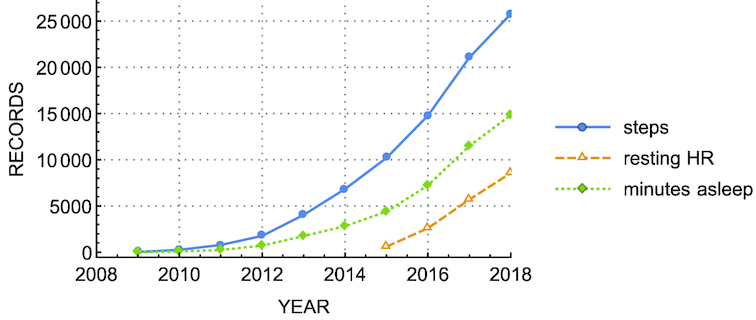
Self-quantification data from Fitbit project. Number of public records from January 2009 until October 2018 (cumulative total).
The QoL Technologies Lab team reports that access to public data has facilitated its research planning. While the number of public data sets is smaller in terms of the number of members who give this kind of access, they are very useful for running observational studies over long periods and can be used to prepare data cleaning and processing methods, which can then be applied to follow-up studies. Because running studies on Open Humans and accessing private data as part of a research institution requires approval from an IRB—a potentially lengthy process—the availability of the public data allows development and testing of methods during an earlier stage of the research process. A study is now being developed on the basis of this preliminary work. Additionally, the QoL Lab was granted ethics approval from University of Copenhagen in November 2018 (No. 504-0034/18-5000).
Data reuse in genetic data visualization research
With the increasing number of individuals engaging with their genetic data, including via DTC products, there is a need for research into how individuals interact with these data to explore and understand it. The Human-Computer Interaction for Personal Genomics (PGHCI) project at Wellesley College and New York University has focused on exploring these questions. Research was initially conducted by creating visualizations of genetic data interpretations based on public genetic data sets and associated reports. The research initially recruited participants via Amazon Mechanical Turk to evaluate a set of visualizations; this approach, however, was not based on participants’ own information, which is preferred to improve experimental validity.
Open Humans provided an opportunity to work with individuals and their data in a manner that leveraged pre-existing genetic data for reuse in new research while minimizing privacy risks. A project, GenomiX Genome Exploration, was created in Open Humans that invited Members who had publicly shared their genetic data in Open Humans to engage with a custom visualization derived from their existing public data and associated interpretations. The study found various design implications in genome data engagement, including the value of affording users the flexibility to examine the same report using multiple views [83].
Personal data exploration
Open Humans aggregates data from multiple sources connected to individual Members. This makes it a natural starting point for a Member to explore their personal data. To facilitate this, Open Humans includes the Personal Data Notebooks project.
Through a JupyterHub setup [84] that authenticates Members through their Open Humans accounts, Members can write Jupyter Notebooks [85] that get full access to their personal data in their web browser. This allows Members to explore and analyze their own data without the need to download or install specialized analysis software on their own computers. Furthermore, it allows Members to easily analyze data across the various data sources, e.g., combining data about their social media use as well as activity-tracking data from wearable devices. This allows Members to explore potential correlations such as whether a decrease in physical activity correlates with more time spent on social media.
Because the notebooks themselves do not store any of the personal data but rather the generic methods to access the data, they can be easily shared between Open Humans Members without leaking a Member’s personal data. This property facilitates not only the sharing of analysis methods but also reproducible n = 1 experiments in the spirit of self-quantification.
To make these notebooks not only interoperable and reusable but also findable and accessible [86], the sister project to the Personal Data Notebooks—the Personal Data Exploratory—was started. Members can upload notebooks right from their Personal Data Notebook interface to Open Humans and can publish them on the Personal Data Exploratory site with just a few clicks. The Exploratory publicly displays the published notebooks to the wider community and categorizes them according to the data sources used, tags, and their content.
The categorization allows other Members to easily discover notebooks of interest. Notebooks written by other Members can be launched and run on a Member’s own personal data through the Personal Data Notebooks, requiring only a single click of a button. Through the close interplay between the Personal Data Notebooks and the shared notebook library of the Personal Data Exploratory, Open Humans offers an integrated personal data analysis environment that allows personal data to be disseminated in a private and secure way, while simultaneously growing a library of data exploration tools that can be reused by other Members, as shown in Figure 3.
Figure 3.
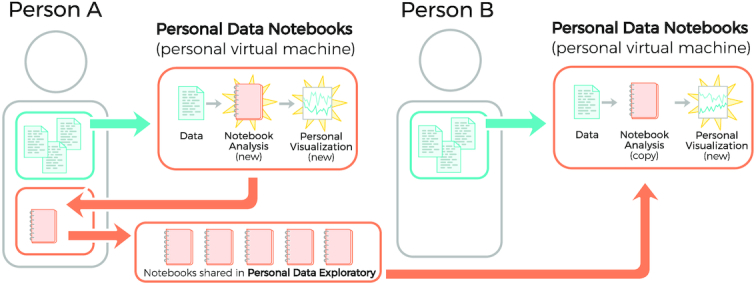
Personal Data Notebooks in Open Humans. Any Member (e.g., ”Person A,” left) can create a Notebook to explore their personal Data using the Personal Data Notebooks project. They can then choose to share a Notebook via the Personal Data Exploratory. This allows another Member (e.g., ”Person B,” right) to load a copy of the Notebook and run it, privately, to produce their own analysis.
Google Search History Analyzer and community review
The Google Search History Analyzer is a project that highlights the Open Humans community review process for Projects, demonstrating how this process can help improve not only a project that is reviewed but also the infrastructure of Open Humans. The Google Search History Analyzer invites individuals to upload their Google Search History data and analyze them in a quantitative way, through Personal Data Notebooks. Examples of analyses that users can perform through the Personal Data Notebooks include retrieving graphs of their most common search terms and their daily or weekly evolution, as well as visualizing connections among their top search terms and their co-occurrence. One goal of this project is to raise awareness on the breadth and deeply personalized content that web searches might carry. Another long-term goal is to provide social scientists who are currently using web search history data for predicting social trends, e.g., unemployment [87] or interest in medical conditions [88], with the means to have access to a pool of individuals who can provide informed consent to the use of their search history data along with additional metadata (e.g., demographic information or survey questions) that could render their Google search history terms more informative.
Open Humans requires Projects have an ”approval” to become visible and broadly available to Members. Prior to Google Search History Analyzer, this approval process was informal and internal; however, the sensitive data handled by this Project raised concerns regarding a need for a more formal and transparent review process because web search history terms might carry highly personalized information, such as personal interests, medical history, places a person visits, or even predictors for severe psychiatric conditions [89]. As a result, a community review process was developed for Project approvals going forward: new projects are shared with the larger community for public comments, inviting feedback from all Members. Project owners can reply to the feedback and improve their project accordingly, as well as seek help from other Open Humans Members. The community review continues until concerns, if any, have been resolved; no formal timeline for finding consensus exists—instead the process is adaptive to the levels of concern raised by the community members. If and when project approval occurs, this status is implemented by the administrators of the Open Humans platform. Project approval status can be reconsidered at any time by opening a new review process, which may be done by any community Member.
As a result of this process, the Google Search History Analyzer project was improved with added documentation, increased clarity, and additional security implementations on the project side. Furthermore, it led to the implementation of a new feature on the Open Humans platform itself, enabling a project-specific override to prevent public data sharing by Members for these data—as requested by community review—thereby reducing the risk that these sensitive data sets might be publicly released by the Members accidentally.
Discussion
Participatory/community science (also known as citizen science) is a growing field that engages people in the scientific process. But while participatory science keeps growing quickly in the environmental sciences and astronomy, its development in the humanities, social sciences, and in medical research lags behind [90], despite expectations that it will make inroads into those fields [60, 91]. Both barriers in accessing personal data that are stored in commercial entities as well as legitimate ethical concerns that surround the use of personal data contribute to this slower adoption in realms that rely on access to personal information [34, 36]. Open Humans was designed to address many of these issues; we discuss some of them in subsequent sections.
Granular and specific consent
One often suggested way to mitigate the ethical concerns around the sharing of personal data in a research framework is by giving participants granular consent options [37]. In a medical context, most patients prefer to have granular control over which medical data to share and for which purposes [92, 93], especially in the context of electronic medical records [94]. Furthermore, the GDPR requires that organizations handling personal data give the individual granular consent options for how their data are used [95].
Open Humans strongly limits the platform's use of member data to an opt-in model, implementing a form of granular consent for data sharing and data use through the use of projects that Members can opt into. On a technical level, Project organizers need to select the data sources they would like to access, and Members can give specific consent for that project’s activities. From the perspective of Open Humans this produces a format for granular consent regarding the data it manages because each potential use of data in the platform is mediated by a specific project.
Additionally, Projects on Open Humans need to adhere to the community guidelines. In addition to mandating clarity and specificity in consent, these guidelines require Projects to inform prospective participants about the level of data access they would request, how the data would be used, and what privacy and security precautions they have in place. Authorization may be withdrawn at any time, at which point Projects may no longer access de-authorized data. Furthermore, Projects may receive notification of erasure requests made by participants who withdraw, should they opt to support these.
Data portability
Much of health data is still stored in data silos managed by national institutions, sometimes further subcategorized by diseases [96]. On an individual level, the situation is not much better: while medical data are usually stored in electronic records, much of a person’s data is now held by the companies that run social media platforms, develop smartphone apps, or purvey wearable devices [97]. This fragmentation—especially when coupled with a lack of data export methods—prevents individuals from authorizing new uses of their data.
Personal information management systems (PIMS) could help individuals in re-collecting and integrating their personal data from different sources [98]. The right to data portability, as encapsulated in Article 20 of the GDPR, has the potential to boost the adoption of such systems because it guarantees individuals in the European Union a right to export the personal data they have provided to data holders in electronic and other useful formats. While Article 20 does not cover derived data, such as genetic information generated from biological samples [99], other personal data that are provided directly and thus subject to Article 20 can be highly valuable for individuals and research purposes. Additionally, Article 15 of the GDPR provides individuals with further rights to access and copies to such derived data, although without specific provisions for the format of such data. Both traditional medical research [100] as well as citizen science [101] have the potential to benefit from these data. By design, Open Humans works similar to a PIMS because it allows individuals to bundle and collect their personal data from external sources. Like other PIMS, Open Humans is likely to benefit from any increase in data export, e.g., due to the GDPR.
While the availability of data export functions is a necessary condition for making PIMS work, it alone is not sufficient. PIMS need to support the data import on their end, either by supporting the file types or by offering support for the APIs of the external services. Because file formats and APIs are not static but can change over time, especially among popular services [102], a significant amount of effort is needed to keep data import functions into PIMS up to date. This cost keeps accumulating as the number of supported data imports keeps increasing. The modular, project-based nature of Open Humans allows the distribution of the workload of keeping integrations up to date, as data importers can be provided by any third party. Existing data imports on Open Humans already demonstrate this capability: both the Nightscout and the Apple HealthKit data importers are examples of this. In the case of Nightscout, members of the diabetes community themselves built and maintain the data import into Open Humans to power their own data commons that overlays the Open Humans data storage. And the HealthKit import application was written by an individual Open Humans Member who wanted to add support for adding their own data.
Enabling individual-centric research and citizen science
Open Humans provides several benefits for citizen science efforts and individual researchers who do not work in academia. The OpenAPS and Nightscout Data Commons highlighted in the results are prime examples of how Open Humans can enable such participant-led research.
To enable research done by non-traditional researchers, the project creation workflow of Open Humans includes information for project leaders about informed consent and other key considerations. It encourages project administrators to be clear about both data management and security in a thorough community guide [103]. This guide includes best practice guidelines for data security as well as details on how to communicate to participants which data access is being requested and why. It emphasizes plain language and clarity.
To further the community’s sense of ownership in the Open Humans platform, participants are involved in the governance of the ecosystem. On a high level the community gets to elect one-third of the members of the Open Humans Foundation board of directors, enabling them to exert direct influence on the larger direction of the platform.
Furthermore, Members of Open Humans are invited to participate in the approval of new projects that want to be shared on the platform via a community review process, as illustrated by the Google Search History project use case described above. This community review process parallels efforts made elsewhere to pursue participant-centered alternatives to IRBs [104], which at present include extremely limited input from community members. Indeed, traditional policies for project approval from an ethical standpoint have been repeatedly questioned [105], and even more so for the case of participant-centric research [106], due to inconsistent levels of engagement from non-academic members [107] and lack of participant protection and autonomy [106]. Notably the review process as implemented on Open Humans is less structured than traditional approaches because it is performed by community members who choose to participate; self-selection for engagement may help maximize efficiency in a heterogeneous ecosystem. We hope this alternative design helps inform other projects seeking increased participant input in project review and oversight.
Summary
Open Humans is an active online platform for personal data aggregation and data sharing that enables citizen science and traditional academic science alike. By leaving data-sharing decisions to individual members, the platform offers a way of doing personal data-based research in an iterative, ethically sensitive way and enables individuals to engage in science as both investigators and participants.
Methods
The primary Open Humans web application, as well as data source Projects maintained directly by Open Humans, are written in Python 3 using the Django web framework. API end points, JSON and HTML data serialization, and OAuth2 authorization are managed by the Django REST Framework and Django OAuth Toolkit libraries. Web applications are deployed on Heroku and use Amazon S3 for file storage. The Personal Data Notebooks JupyterHub project is deployed via Google Cloud Platform.
Two Python packages have been developed and distributed in the Python Package Index to facilitate interactions with our API: (1) open-humans-api provides Python functions for API end points, as well as command line tools for performing many standard API operations; (2) django-open-humans provides a reusable Django module for using Open Humans OAuth2 and API features.
Open Humans complies with GDPR and provides a live records of processing activities report [108].
Availability of source code and requirements
Project name: Open Humans
Project home page: http://www.openhumans.org
Operating system(s): Platform independent
Programming language: Python3
Other requirements: full list on GitHub https://github.com/openhumans/open-humans/
License: MIT
Project name: Open Humans API
Project home page: https://open-humans-api.readthedocs.io/en/latest/
Operating system(s): Platform independent
Programming language: Python3
Other requirements: full list on GitHub https://github.com/openhumans/open-humans-api
License: MIT
Project name: Django Open Humans
Project home page: https://django-open-humans.readthedocs.io/en/latest
Operating system(s): Platform independent
Programming language: Python3
Other requirements: full list on GitHub https://github.com/OpenHumans/django-open-humans
License: MIT
Abbreviations
API: application programming interface; CGM: continuous glucose monitor; DTC: direct to consumer; GDPR: General Data Protection Regulation; GPS: Global Positioning System; HR: heart rate; HTML: Hypertext Markup Language; IRB: institutional review board; JSON: JavaScript Object Notation; OpenAPS: Open Source Artificial Pancreas System; PGHCI: Human-Computer Interaction for Personal Genomics; PIMS: personal information management systems; QoL: quality of life; SNP: single-nucleotide polymorphism; VCF: variant call format.
Competing interests
B.G.T. is supported by a fellowship from Open Humans Foundation, which operates Open Humans. M.P.B. is funded for full-time work at Open Humans Foundation as Executive Director and President. M.A. is a paid consultant to Genetic Alliance and Variant Bio.
Funding
The development and operation of Open Humans has been supported through grants from the Robert Wood Johnson Foundation, John S. and James L. Knight Foundation, and Shuttleworth Foundation. "AT is supported by the Interfaculty Research Cooperation "Decoding Sleep: From Neurons to Health & Mind“ of the University of Bern”
Authors' contributions
B.G.T.: conceptualization, data curation, investigation, methodology, project administration, software, supervision, writing—original draft, writing—review and editing. M.A.: supervision, writing—review and editing. K.A.: data curation, software, validation, writing—original draft, writing—review and editing. M.D.: software, writing—review and editing. V.E.: data curation, formal analysis, investigation, validation, visualization, writing—original draft, writing—review and editing. B.G.: data curation, resources, software, validation. T.H.: methodology, resources, software. D.L.: data curation, formal analysis, validation, writing—original draft, writing—review and editing. O.N.: investigation, validation. O.S.: investigation, validation, writing—review and editing. A.T.: data curation, software, validation, writing—original draft, writing—review and editing. J.B.: conceptualization, funding acquisition, resources, investigation, project administration, supervision. M.P.B.: conceptualization, data curation, funding acquisition, investigation, methodology, project administration, resources, software, supervision, writing—original draft, writing—review and editing.
Supplementary Material
Joon-Ho Yu -- 11/28/2018 Reviewed
Joon-Ho Yu -- 5/27/2019 Reviewed
Birgit Wouters -- 12/11/2018 Reviewed
Birgit Wouters -- 5/23/2019 Reviewed
ACKNOWLEDGEMENTS
The authors would like to thank all members of the Open Humans community for their diverse contributions to Open Humans: developing the process as well as platforms that link to Open Humans, sharing their personal data, advancing public knowledge sources, being active community members. In this spirit, this manuscript was written as a community project done by and with Open Humans members following an open call for contributions.
In particular, the authors would like to thank Rosy Gupta, Manaswini Das, Jasmine Tamak, and Tarannum Khan. They made valuable contributions as summer interns with Open Humans through the Outreachy internship program. The authors are grateful to Mike Escalante, who contributed in software development as well as mentoring for Outreachy.
The authors also would like to thank the reviewers—their input significantly improved the manuscript.
References
- 1. McCormick TH, Lee H, Cesare N, et al.. Using Twitter for demographic and social science research: tools for data collection and processing. Sociol Methods Res. 2015;46(3):390–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Özdemir V, Dove ES, Gürsoy UK, et al.. Personalized medicine beyond genomics: alternative futures in big data—proteomics, environtome and the social proteome. J Neural Transm (Vienna). 2015;124(1):25–32. [DOI] [PubMed] [Google Scholar]
- 3. Athey S. Beyond prediction: using big data for policy problems. Science. 2017;355(6324):483–5. [DOI] [PubMed] [Google Scholar]
- 4. Cappella JN. Vectors into the future of mass and interpersonal communication research: big data, social media, and computational social science. Hum Commun Res. 2017;43(4):545–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chhibber A, Kroetz DL, Tantisira KG, et al.. Genomic architecture of pharmacological efficacy and adverse events. Pharmacogenomics. 2014;15(16):2025–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kummar S, Williams PM, Lih CJ, et al.. Application of molecular profiling in clinical trials for advanced metastatic cancers. J Nati Cancer Inst. 2015;107(4), doi: 10.1093/jnci/djv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dilsizian SE, Siegel EL. Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep. 2013;16(1):441. [DOI] [PubMed] [Google Scholar]
- 9. Moon H, Ahn H, Kodell RL, et al.. Ensemble methods for classification of patients for personalized medicine with high-dimensional data. Artif Intell Med. 2007;41(3):197–207. [DOI] [PubMed] [Google Scholar]
- 10. Kohane IS. Ten things we have to do to achieve precision medicine. Science. 2015;349(6243):37–8. [DOI] [PubMed] [Google Scholar]
- 11. Wetterstrand LA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) 2018. https://www.genome.gov/sequencingcostsdata/. Accessed 19 June 2019. [Google Scholar]
- 12. Swan M. Emerging patient-driven health care models: an examination of health social networks, consumer personalized medicine and quantified self-tracking. Int J Environ Res Public Health. 2009;6(2):492–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gay V, Leijdekkers P. Bringing health and fitness data together for connected health care: mobile apps as enablers of interoperability. J Med Internet Res. 2015;17(11):e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corpas M, Valdivia-Granda W, Torres N, et al.. Crowdsourced direct-to-consumer genomic analysis of a family quartet. BMC Genomics. 2015;16:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Regalado A. 2017 was the year consumer DNA testing blew up. MIT Technology Review. 2018. https://www.technologyreview.com/s/610233/2017-was-the-year-consumer-dna-testing-blew-up/. Accessed 19 June 2019. [Google Scholar]
- 16. Khan R, Mittelman D. Consumer genomics will change your life, whether you get tested or not. Genome Biol. 2018;19(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Regalado A. More than 26 million people have taken an at-home ancestry test. MIT Technology Review. 2019. https://www.technologyreview.com/s/612880/more-than-26-million-people-have-taken-an-at-home-ancestry-test/. Accessed 19 June 2019. [Google Scholar]
- 18. EMC. The digital universe: Driving data growth in healthcare 2014. https://www.emc.com/analyst-report/digital-universe-healthcare-vertical-report-ar.pdf. Accessed 19 June 2019. [Google Scholar]
- 19. Rozenblum R, Bates DW. Patient-centred healthcare, social media and the internet: the perfect storm?. BMJ Qual Saf. 2013;22(3):183–6. [DOI] [PubMed] [Google Scholar]
- 20. DeAngelis SF. Patient monitoring, big data, and the future of healthcare. Wired. 2014. https://www.wired.com/insights/2014/08/patient-monitoring-big-data-future-healthcare/. Accessed 19 June 2019. [Google Scholar]
- 21. Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. JAMA. 2014;311(24):2479–80. [DOI] [PubMed] [Google Scholar]
- 22. Carbon S, Champieux R, McMurry J, et al.. A measure of open data: a metric and analysis of reusable data practices in biomedical data resources. bioRxiv. 2018, doi: 10.1101/282830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blasimme A, Fadda M, Schneider M, et al.. Data sharing for precision medicine: policy lessons and future directions. Health Aff (Millwood). 2018;37(5):702–9. [DOI] [PubMed] [Google Scholar]
- 24. Kossmann D, Brand A, Hafen E. Health data cooperatives – citizen empowerment. Methods Inf Med. 2014;53(2):82–6. [DOI] [PubMed] [Google Scholar]
- 25. Tenopir C, Allard S, Douglass K, et al.. Data sharing by scientists: practices and perceptions. PLoS One. 2011;6(6):e21101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simpson C, Goldenberg A, Culverhouse R, et al.. Practical barriers and ethical challenges in genetic data sharing. Int J Environ Res Public Health. 2014;11(8):8383–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lye CT, Forman HP, Gao R, et al.. Assessment of US hospital compliance with regulations for patients’ requests for medical records. JAMA Netw Open. 2018;1(6):e183014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong J, Henderson T. How portable is portable? Exercising the GDPR’s right to data portability. In: Proceedings of the 2018 ACM International Joint Conference and 2018 International Symposium on Pervasive and Ubiquitous Computing and Wearable Computers, Singapore. New York, NY: ACM; 2018:911–20. [Google Scholar]
- 29. Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363(6):501–4. [DOI] [PubMed] [Google Scholar]
- 30. Hert PD, Papakonstantinou V, Malgieri G, et al.. The right to data portability in the GDPR: towards user-centric interoperability of digital services. Comput Law Secur Rev. 2018;34(2):193–203. [Google Scholar]
- 31. Recommendations: Attachment B: Return of Individual Research Results Office for Human Research Protections, U.S. Department of Health & Human Services; 2016. https://www.hhs.gov/ohrp/sachrp-committee/recommendations/attachment-b-return-individual-research-results/index.html, Accessed 19 June 2019. [Google Scholar]
- 32. Wong CA, Hernandez AF, Califf RM. Return of research results to study participants. JAMA. 2018;320(5):435. [DOI] [PubMed] [Google Scholar]
- 33. Mason PH. The ethics of biomedical big data. J Bioeth Inq. 2017;14(4):571–4. [DOI] [PubMed] [Google Scholar]
- 34. Ross MW, Iguchi MY, Panicker S. Ethical aspects of data sharing and research participant protections. Am Psychol. 2018;73(2):138–45. [DOI] [PubMed] [Google Scholar]
- 35. Haeusermann T, Greshake B, Blasimme A. Open sharing of genomic data: who does it and why?. PLoS One. 2017;12(5):e0177158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S, Jiang X, Singh S, et al.. Genome privacy: challenges, technical approaches to mitigate risk, and ethical considerations in the United States. Ann N Y Acad Sci. 2016;1387(1):73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evans BJ. Power to the people: data citizens in the age of precision medicine. Vanderbilt J Entertain Technol Law. 2017;19(2):243–65. [PMC free article] [PubMed] [Google Scholar]
- 38. Hart K. A new data scandal: how ancestry DNA firms share your most intimate secrets. Axios. 2019. https://www.axios.com/dna-test-results-privacy-genetic-data-sharing-4687b1a0-f527-425c-ac51-b5288b0c0293.html. Accessed 19 June 2019. [Google Scholar]
- 39. Might M, Might CC. What happens when N = 1 and you want plus 1?. Prenat Diagn. 2016;37(1):70–2. [DOI] [PubMed] [Google Scholar]
- 40. Stacchiotti S, Gronchi A, Fossati P, et al.. Best practices for the management of local-regional recurrent chordoma: a position paper by the Chordoma Global Consensus Group. Ann Oncol. 2017;28(6):1230–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. PEER is transforming health systems. Genetic Alliance, Inc. 2015. https://www.peerplatform.org/idea/. Accessed 19 June 2019. [Google Scholar]
- 42. Samerski S. Individuals on alert: digital epidemiology and the individualization of surveillance. Life Sci Soc Policy. 2018;14(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cox J. 70,000 OkCupid users just had their data published. Vice 2016. http://motherboard.vice.com/read/70000-okcupid-users-just-had-their-data-published. Accessed 19 June 2019. [Google Scholar]
- 44. Zimmer M. "But the data is already public”: on the ethics of research in Facebook. Ethics Inf Technol. 2010;12(4):313–25. [Google Scholar]
- 45. Zook M, Barocas S, Danah B, et al.. Ten simple rules for responsible big data research. PLoS Comput Biol. 2017;13(3):e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jouhki J, Lauk E, Penttinen M, et al.. Facebook’s emotional contagion experiment as a challenge to research ethics. Media Commun. 2016;4(4):75. [Google Scholar]
- 47. Hunter D, Evans N. Facebook emotional contagion experiment controversy. Res Ethics. 2016;12(1):2–3. [Google Scholar]
- 48. Flick C. Informed consent and the Facebook emotional manipulation study. Res Ethics. 2015;12(1):14–28. [Google Scholar]
- 49. Bruns A. Facebook shuts the gate after the horse has bolted, hurts real research in the process. Medium 2018. https://medium.com/@Snurb/facebook-research-data-18662cf2cacb. Accessed 19 June 2019. [Google Scholar]
- 50. Golder S, Ahmed S, Norman G, et al.. Attitudes toward the ethics of research using social media: a systematic review. J Med Internet Res. 2017;19(6):e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pocock MJO, Tweddle JC, Savage J, et al.. The diversity and evolution of ecological and environmental citizen science. PLoS One. 2017;12(4):e0172579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vayena E, Tasioulas J. "We the scientists”: a human right to citizen science. Philos Technol. 2015;28(3):479–85. [Google Scholar]
- 53. McKinley DC, Miller-Rushing AJ, Ballard HL, et al.. Citizen science can improve conservation science, natural resource management, and environmental protection. Biol Conserv. 2017;208:15–28. [Google Scholar]
- 54. Conrad CC, Hilchey KG. A review of citizen science and community-based environmental monitoring: issues and opportunities. Environ Monit Assess. 2010;176(1-4):273–91. [DOI] [PubMed] [Google Scholar]
- 55. Zevin M, Coughlin S, Bahaadini S, et al.. Gravity Spy: integrating advanced LIGO detector characterization, machine learning, and citizen science. Class Quantum Gravity. 2017;34(6):064003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haklay M. Citizen science and volunteered geographic information: overview and typology of participation. In: Crowdsourcing Geographic Knowledge. Dordrecht, Netherlands: Springer; 2012:105–22. [Google Scholar]
- 57. Dickinson H, Fortson L, Lintott C, et al.. Galaxy Zoo: morphological classification of galaxy images from the Illustris simulation. Astrophys J. 2018;853(2):194. [Google Scholar]
- 58. Khatib F, Cooper S, Tyka MD, et al.. Algorithm discovery by protein folding game players. Proc Natl Acad Sci U S A. 2011;108(47):18949–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ranard BL, Ha YP, Meisel ZF, et al.. Crowdsourcing—harnessing the masses to advance health and medicine, a systematic review. J Gen Intern Med. 2013;29(1):187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rowbotham S, McKinnon M, Leach J, et al.. Does citizen science have the capacity to transform population health science?. Crit Public Health. 2017;29(1):118–28. [Google Scholar]
- 61. Mader LB, Harris T, Kläger S, et al.. Inverting the patient involvement paradigm: defining patient led research. Res Involv Engagem. 2018;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Vayena E, Brownsword R, Edwards SJ, et al.. Research led by participants: a new social contract for a new kind of research. J Med Ethics. 2015;42(4):216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Katapally TR, Bhawra J, Leatherdale ST, et al.. The SMART Study, a mobile health and citizen science methodological platform for active living surveillance, integrated knowledge translation, and policy interventions: longitudinal study. JMIR Public Health Surveill. 2018;4(1):e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wicks P, Vaughan TE, Massagli MP, et al.. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol. 2011;29(5):411–4. [DOI] [PubMed] [Google Scholar]
- 65. McDonald D, Hyde E, Debelius JW, et al.. American Gut: an open platform for citizen science microbiome research. mSystems. 2018;3(3), doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McGowan ML, Choudhury S, Juengst ET, et al.. “Let’s pull these technologies out of the ivory tower”: the politics, ethos, and ironies of participant-driven genomic research. BioSocieties. 2017;12(4):494–519. [Google Scholar]
- 67. Levitan B, Getz K, Eisenstein EL, et al.. Assessing the financial value of patient engagement. Ther Innov Regul Sci. 2017;52(2):220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Swan M. The quantified self: fundamental disruption in big data science and biological discovery. Big Data. 2013;1(2):85–99. [DOI] [PubMed] [Google Scholar]
- 69. Swan M. Health 2050: the realization of personalized medicine through crowdsourcing, the quantified self, and the participatory biocitizen. J Pers Med. 2012;2(3):93–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yuan J, Gordon A, Speyer D, et al.. DNA.Land is a framework to collect genomes and phenomes in the era of abundant genetic information. Nat Genet. 2018;50(2):160–5. [DOI] [PubMed] [Google Scholar]
- 71. Greshake B, Bayer PE, Rausch H, et al.. openSNP–A crowdsourced web resource for personal genomics. PLoS One. 2014;9(3):e89204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ball MP, Thakuria JV, Zaranek AW, et al.. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci U S A. 2012;109(30):11920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lewis D, Leibrand S, OpenAPS Community. Real-world use of open source artificial pancreas systems. J Diabetes Sci Technol. 2016;10(6):1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lewis DM, Ball MP. OpenAPS Data Commons on Open Humans. Poster presented at the 2017 Sage Assembly Bionetworks Assembly, Seattle, WA 2017, doi: 10.6084/m9.figshare.5428498.v1. [DOI] [Google Scholar]
- 75. Lewis DM, Leibrand S, Street TJ, et al.. Detecting insulin sensitivity changes for individuals with type 1 diabetes. Diabetes. 2018;67(Suppl 1):79–LB. [Google Scholar]
- 76. OpenAPS Outcomes. https://openaps.org/outcomes/. Accessed 19 June 2019. [Google Scholar]
- 77. Tools to work with data downloaded from Open Humans research platform. https://github.com/danamlewis/OpenHumansDataTools. Accessed 19 June 2019. [Google Scholar]
- 78. Das S, Forer L, Schönherr S, et al.. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Imputer. https://openimpute.com. [Google Scholar]
- 80. Lemieux Perreault LP, Legault MA, Asselin G, et al.. genipe: an automated genome-wide imputation pipeline with automatic reporting and statistical tools. Bioinformatics. 2016;32(23):3661–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dietz WH, Douglas CE, Brownson RC. Chronic disease prevention: tobacco avoidance, physical activity, and nutrition for a healthy start. JAMA. 2016;316(16):1645–6. [DOI] [PubMed] [Google Scholar]
- 83. Westendorf L, Shaer O, Pollalis C, et al.. Exploring genetic data across individuals: design and evaluation of a novel comparative report tool. J Med Internet Res. 2018;20(9):e10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Documentation for JupyterHub. https://jupyterhub.readthedocs.io. Accessed 19 June 2019. [Google Scholar]
- 85. Kluyver T, Ragan-Kelley B, Pérez F, et al.. Jupyter Notebooks – a publishing format for reproducible computational workflows. In: Loizides F, Schmidt B, eds. Positioning and Power in Academic Publishing: Players, Agents and Agendas. IOS Press;2016:87–90. [Google Scholar]
- 86. Wilkinson MD, Dumontier M, Aalbersberg IJ, et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. D’Amuri F, Marcucci J. The predictive power of Google searches in forecasting US unemployment. Int J Forecast. 2017;33(4):801–16. [Google Scholar]
- 88. Brigo F, Trinka E. Google search behavior for status epilepticus. Epilepsy Behav. 2015;49:146–49. [DOI] [PubMed] [Google Scholar]
- 89. III JFG, Lester D. Using Google searches on the internet to monitor suicidal behavior. J Affect Disord. 2013;148(2-3):411–12. [DOI] [PubMed] [Google Scholar]
- 90. Kullenberg C, Kasperowski D. What is citizen science? – a scientometric meta-analysis. PLoS One. 2016;11(1):e0147152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hoffmann-Longtin K. Power to the patients: co-design of community-based research. PLoS Blogs 2018. http://blogs.plos.org/blog/2018/08/09/power-to-the-patients-co-design-of-community-based-research/. Accessed 19 June 2019. [Google Scholar]
- 92. Schwartz PH, Caine K, Alpert SA, et al.. Patient preferences in controlling access to their electronic health records: a prospective cohort study in primary care. J Gen Intern Med. 2014;30(S1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Grando MA, Murcko A, Mahankali S, et al.. A study to elicit behavioral health patients' and providers' opinions on health records consent. J Law Med Ethics. 2017;45(2):238–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Caine K, Hanania R. Patients want granular privacy control over health information in electronic medical records. J Am Med Inform Assoc. 2013;20(1):7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nati M, Mayer S, Capossele A, et al.. Toward trusted open data and services. Internet Technol Lett. 2018;2(1):e69. [Google Scholar]
- 96. The Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352(6291):1278–80. [DOI] [PubMed] [Google Scholar]
- 97. Althoff T. Population-scale pervasive health. IEEE Pervasive Comput. 2017;16(4):75–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Allard T, Bouadi T, Duguépéroux J, et al.. From self-data to self-preferences: towards preference elicitation in personal information management systems. In: Guidotti R, Monreale A, Pedreschi D et al., et al., eds. Personal Analytics and Privacy. An Individual and Collective Perspective. Cham: Springer; 2017:10–6. [Google Scholar]
- 99. Taylor MJ, Wallace SE, Prictor M. United Kingdom: transfers of genomic data to third countries. Human Genet. 2018;137(8):637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Rumbold JMM, Pierscionek B. The effect of the General Data Protection Regulation on medical research. J Med Internet Res. 2017;19(2):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Quinn P. Is the GDPR and its right to data portability a major enabler of citizen science?. Global Jurist. 2018;18(2), doi: 10.1515/gj-2018-0021. [DOI] [Google Scholar]
- 102. Xavier L, Brito A, Hora A, et al.. Historical and impact analysis of API breaking changes: a large-scale study. In: 2017 IEEE 24th International Conference on Software Analysis, Evolution and Reengineering (SANER), Klagenfurt, Austria. IEEE; 2017, doi: 10.1109/saner.2017.7884616. [DOI] [Google Scholar]
- 103. Open Humans Project Guidelines. https://www.openhumans.org/community-guidelines/#project. Accessed 19 June 2019. [Google Scholar]
- 104. Grant AD, Wolf GI, Nebeker C. Approaches to governance of participant-led research: a qualitative case study. BMJ Open. 2019;9(4):e025633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mhaskar R, Pathak E, Wieten S, et al.. Those responsible for approving research studies have poor knowledge of research study design: a knowledge assessment of institutional review board members. Acta Inform Med. 2015;23(4):196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wilson E, Kenny A, Dickson-Swift V. Ethical challenges of community based participatory research: exploring researchers’ experience. Int J Soc Res Methodol. 2017;21(1):7–24. [Google Scholar]
- 107. Klitzman R. Institutional review board community members. Acad Med. 2012;87(7):975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Open Humans: Records of Personal Data Processing Activities. https://www.openhumans.org/data-processing-activities/. Accessed 19 June 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Joon-Ho Yu -- 11/28/2018 Reviewed
Joon-Ho Yu -- 5/27/2019 Reviewed
Birgit Wouters -- 12/11/2018 Reviewed
Birgit Wouters -- 5/23/2019 Reviewed


