Abstract
Background
Yellowhorn (Xanthoceras sorbifolium) is a species of the Sapindaceae family native to China and is an oil tree that can withstand cold and drought conditions. A pseudomolecule-level genome assembly for this species will not only contribute to understanding the evolution of its genes and chromosomes but also bring yellowhorn breeding into the genomic era.
Findings
Here, we generated 15 pseudomolecules of yellowhorn chromosomes, on which 97.04% of scaffolds were anchored, using the combined Illumina HiSeq, Pacific Biosciences Sequel, and Hi-C technologies. The length of the final yellowhorn genome assembly was 504.2 Mb with a contig N50 size of 1.04 Mb and a scaffold N50 size of 32.17 Mb. Genome annotation revealed that 68.67% of the yellowhorn genome was composed of repetitive elements. Gene modelling predicted 24,672 protein-coding genes. By comparing orthologous genes, the divergence time of yellowhorn and its close sister species longan (Dimocarpus longan) was estimated at ∼33.07 million years ago. Gene cluster and chromosome synteny analysis demonstrated that the yellowhorn genome shared a conserved genome structure with its ancestor in some chromosomes.
Conclusions
This genome assembly represents a high-quality reference genome for yellowhorn. Integrated genome annotations provide a valuable dataset for genetic and molecular research in this species. We did not detect whole-genome duplication in the genome. The yellowhorn genome carries syntenic blocks from ancient chromosomes. These data sources will enable this genome to serve as an initial platform for breeding better yellowhorn cultivars.
Keywords: Xanthoceras sorbifolium, yellowhorn, PacBio sequencing, genome assembly, Hi-C, genome annotation, conserved chromosome
Data Description
Background
Yellowhorn (Xanthoceras sorbifolium) (NCBI: txid99658) is a woody oil species [1] that belongs to the Sapindaceae family and the monotypic genus Xanthoceras. As an endemic and economically important species in northern China, it is widely used for soil and water conservation owing to its capacity to survive on arid, saline, and alkaline land and in extreme temperatures even below −40°C [2, 3]. Almost 7.5 × 105 tons of yellowhorn seeds are harvested in autumn every year [4] (Fig. 1). The oil content of its seed kernels can be as high as 67%, of which 85–93% is unsaturated fatty acid, including 37.1–46.2% linoleic acid and 28.6–37.1% oleic acid, which are essential fatty acids in the human diet [5]. Recently, as a major woody oil plant species, yellowhorn has drawn governmental and popular attention because of the shortage of vegetable oil resources in China. Notably, an essential nutrient for brain growth and maintenance—nervonic acid, which is rarely found in plants—accounts for 3.04% of the seed oil of yellowhorn [6, 7]. Recent results indicate that xanthoceraside, a novel triterpenoid saponin extracted from yellowhorn husks, has an antitumor effect and the potential to treat Alzheimer disease [8–10]. In this study, we generated a high-quality yellowhorn genome assembly and conducted annotation and genomic structure and evolution analyses. Our data provide a rich resource of genetic information for developing yellowhorn resources and understanding the special place of Xanthoceras and Sapindaceae in plant evolution.
Figure 1:
Images of yellowhorn plants. (A) Yellowhorn tree in an artificial forest. (B) Ripe fruit, which dehisce into 3 parts by the carpels. (C) A harvest scene of yellowhorn in northern China. (D) Seeds in ripe fruits, which number 18–24 in 1 fruit.
Sequenced individual and sample collection
Tender leaves were collected from an individual of X. sorbifolium cv. Zhongshi 4, which is a new variety issued by the National Forestry and Grassland Administration (variety rights No. 20180121), in Zhangwu, Liaoning, China. This tree was produced via clone of a plus tree from natural population in Tongliao, Inner Mongolia, China. The leaves were frozen in liquid nitrogen and stored at −80°C until DNA extraction.
Estimation of genome size through flow cytometry analysis
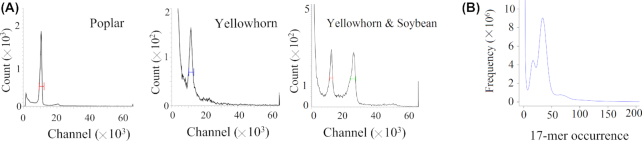
One-month-old leaves from the sequenced yellowhorn individual were subjected to flow cytometry analysis to estimate the genome size as described by Galbraith et al. [11]. Glycine max var. William 82 (2C genome size = 2.28 pg) [12, 13] and Populus trichocarpa var. Nisqually 1 (2C genome size = 0.99 pg) [14] were used as standard references. The soybean and yellowhorn samples were chopped together using a razor blade and the nuclei were stained with propidium iodide. To avoid peaks that were too close to be distinguished when run simultaneously, the poplar and yellowhorn samples were run separately. Each sample was measured 3 times on the flow cytometer. More than 3,000 nuclei were analysed per sample with a FACSAria flow cytometer (Becton, Dickinson and Company, Franklin Lakes,NJ, USA). A total of 16 samples were analysed using soybean and poplar as standard species. The software BD FACSDiva (version 8.0.1) was used for data analysis with the coefficient of variation controlled at 5%. Compared with the soybean internal standard (peak at 25,413) and poplar reference (peak at 10,363), the peak fluorescence intensity values of yellowhorn samples were 11,968 and 11,558, respectively. With the soybean genome size (1,115 Mb) and poplar genome size (485 ± 10 Mb) [13–15] taken as reference, the yellowhorn genome size was estimated to be ∼525.94 and 540.93 Mb, which were relatively close (Fig. 2A).
Figure 2:
Estimation of genome size. (A) Test results of yellowhorn, poplar, and yellowhorn + soybean samples using flow cytometry. (B) Distribution of 17-mer frequency. The x-axis and y-axis indicate the 17-mer frequency and number, respectively. The leftmost truncated peak at a low occurrence frequency (1–2) was mainly due to random base errors in the raw sequencing reads.
Illumina short-read sequencing and heterozygosity analysis
DNA was extracted from the leaves of the same individual using a DNA Secure Plant Kit (Tiangen, China). The DNA concentration and quality were assessed by 1% agarose gel electrophoresis and with a 2.0 Fluorometer (Life Technologies, CA, USA). One shotgun library with an insert size of 350 bp was prepared using a NEB Next® Ultra DNA Library Prep Kit (NEB, USA). A total of 34.51 Gb raw sequencing data were generated by the Illumina HiSeq X Ten sequencing platform. Primary data analysis was carried out using the standard Illumina pipeline [16]. Short reads were processed with Trimmomatic version 0.33 (Trimmomatic, RRID:SCR_011848) [17, 18] and Cutadapt (version 1.13) [19] to remove adapters, leading and trailing bases with a quality score <20, and reads with an average per-base quality of 20 over a 4-bp sliding window. Trimmed reads <70 nucleotides long were discarded. Finally, 34.40-Gb clean reads were used for the following analysis and error correction of Pacific Biosciences (PacBio) reads.
A k-mer analysis was performed to estimate the genomic characteristics as mentioned by Marçais and Kingsford [20]. After low-quality, duplicate, and contaminating reads were filtered out from 34.4 Gb Illumina sequencing data, 21.17-Gb high-quality clean reads were used to generate a k-mer (k = 17) depth distribution curve using Jellyfish (v2.1.1) (with the parameters -m 17 -t 10 -s 550M) and GCE v1.0.0 [21]. The frequency of 17-mer occurrence (17-mer depth) and the frequency of those 17-mers’ species at a given sequencing depth were counted and drawn distribution curves of k-mer frequency (Fig. 2B). On the basis of the flow cytometry results and computational method [21, 22], the middle peak (∼34×) was homogzygous peak. The left peak of 17× was heterozygous peak, and the right tiny peak (66×) observed in Fig. 2B was caused by repeat sequences. Depending on the formula reported by Liu et al. [21], the heterozygosity was estimated at ∼0.75%.
PacBio SMRT sequencing
Genomic DNA was extracted following the ∼40 kb SMRTbell™ Libraries Protocol [23]. The DNA was purified with a Mobio PowerClean® Pro DNA Clean-Up Kit, and its quality was assessed by standard agarose gel electrophoresis and Thermo Fisher Scientific Qubit Fluorometry. The genomic DNA was sheared to a size range of ∼40 kb using g-TUBE (Covaris) and 0.45× AMPure beads were used to enrich and purify large fragments of DNA. Damaged DNA and ends were enzymatically repaired as recommended by PacBio. Following this procedure, hairpin adapters were ligated using a blunt-end ligation reaction. The remaining damaged DNA fragments and fragments without adapters at both ends were digested using exonuclease. Subsequently, the resulting SMRTbell templates were purified by Blue Pippin electrophoresis (Sage Sciences) and sequenced on a PacBio RS II instrument using P6-C4 sequencing chemistry. A primary filtering analysis was performed on the sequencer, and the secondary analysis was performed utilizing the Single-Molecule Real-Time (SMRT) analysis pipeline version 2.1.0 (PacBio). In total, we generated 66.44 Gb (∼122.83-fold coverage of the yellowhorn genome) of single-molecule sequencing data (6,105,692 PacBio post-filtered reads), with a mean read length of 10,882 bp (Fig. S1; Table S1).
Genome assembly
After stringent filtering and correction steps using k-mer frequency-based methods [24], we assembled contigs using the PacBio reads. Preliminary assembly with the assembler Falcon v0.7 (Falcon, RRID:SCR_016089) [25] (falcon_sense_option = –output_multi –min_idt 0.70 –min_cov 4 –max_n_read 300 –n_core 8 overlap_filtering_setting = –max_diff 100 –max_cov 100 –min_cov 2 –n_core 12 –bestn 10) generated a total length of 598.65 Mb of contigs with an N50 length of 1.11 Mb, using the 66.44-Gb PacBio long reads. The software Quiver (based on pbsmrtpipe.pipelines.sa3_ds_resequencing in smrtlink_5.0.1; [26]) was used to polish the PacBio consensus sequence clusters. The assembly was corrected with Pilon version 1.22 (Pilon, RRID:SCR_014731) [27] using the Illumina short reads. Finally, heterozygous sequences were identified and removed using the Purge Haplotigs pipeline, with the parameters -a 75 [28, 29]. Contigs from organelle DNA sources can also be identified and filtered out during the processing with Purge Haplotigs. After the heterozygous sequences were removed, a final assembly from the PacBio reads (504.20 Mb) was generated (Table 1).
Table 1:
Overview of assembly and annotation for the yellowhorn genome
| Total length | 504,196,643 bp |
| Length of unclosed gaps | 73,800 bp |
| N50 length | |
| Initial contigs | 1,044,891 bp |
| Scaffolds | 32,173,403 bp |
| N90 length (scaffolds) | 25,069,408 bp |
| No. of scaffolds (>N90 length) | 21 |
| Largest scaffold | 40,097,451 bp |
| Guanine-cytosine content | 36.95% |
| No. predicted | |
| Protein-coding genes | 24,672 |
| Noncoding RNA genes | 1,066 |
| Content of repetitive sequences | 68.67% |
| Length of genome anchored on linkage groups | 489,286,946 bp (97.04%) |
Pseudomolecule construction and 3D chromatin conformation analysis
The Hi-C technology is an efficient strategy for pseudomolecule construction and enables the generation of genome-wide 3D chromosome architectures. We constructed Hi-C fragment libraries of 350 bp and sequenced them using the Illumina Hi-Seq platform (Illumina, San Diego, CA, USA) for chromosome pseudomolecule construction. Mapping of the Hi-C reads and assignment to restriction fragments were performed as described by Burton et al. [30]. A total of 53.39 Gb of trimmed reads, representing ∼98.70-fold coverage of the yellowhorn genome, were mapped to the assembly with the aligner BWA version 0.7.10 (BWA, RRID:SCR_010910; parameters: bwa index -a bwtsw fasta bwa aln -M 3 -O 11 -E 4 -t 2 fq1 bwa aln -M 3 -O 11 -E 4 -t 2 fq2) [31]. Only uniquely aligned reads with high alignment quality (>20) were selected for pseudomolecule construction. Duplicate removal and quality assessment were performed using HiC-Pro (version 2.8.1) with the following parameters: mapped_2hic_fragments.py -v -S -s 100 -l 1000 -a -f -r -o [32]. In total, 50.56% of the Hi-C data were grouped into valid interaction pairs. A total of 2,836 contigs (N50 length at 1.04 Mb) were assembled after error correction. LACHESIS (parameters: cluster_min_re_sites = 48; cluster_max_link_density = 2; cluster_noninformative_ratio = 2; order_min_n_res_in_trun = 14; order_min_n_res_in_shreds = 15) [30] was used to assign the order and orientation of each group, with a scaffold N50 of 32.17 Mb.
Using the 98.70-fold coverage of Hi-C reads, 489.28 Mb (97.04%) of the assembly were anchored onto the 15 pseudomolecules, which were in agreement with the yellowhorn karyotype (2n = 30) identified by Li [33]. The assembly (477.59 Mb, 94.76%) was ordered by the frequency distribution of valid interaction pairs (Table 2, Fig. S2). The coverage of the assembly reached 93.96% and the ratio of unclosed gaps was 0.15‰ (Table 1). The assembly was of sufficient quality to be used as a reference for studying yellowhorn biology and plant genomics.
Table 2:
Quantity of the contigs anchored with Hi-C
| Linkage Group | No. of anchored contigs | Sequence length (bp) |
|---|---|---|
| 1 | 68 | 40,738,791 |
| 2 | 92 | 40,039,835 |
| 3 | 38 | 37,159,809 |
| 4 | 112 | 35,552,403 |
| 5 | 84 | 35,291,867 |
| 6 | 62 | 35,706,508 |
| 7 | 66 | 33,002,525 |
| 8 | 46 | 32,947,898 |
| 9 | 66 | 30,804,552 |
| 10 | 62 | 30,699,318 |
| 11 | 68 | 29,306,026 |
| 12 | 56 | 29,390,540 |
| 13 | 47 | 29,816,145 |
| 14 | 71 | 25,601,946 |
| 15 | 72 | 23,228,783 |
| Total (ratio %) | 1,010 (35.61) | 489,286,946 (97.04) |
Transcriptome sequencing
RNA was extracted from 3 tissues (flowers, leaves, and roots) of the same individual used for DNA sequencing using the Easy Spin RNA extraction kit (Sangon Biotech, Shanghai, China; No. SK8631). The concentration of each RNA sample was checked using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., USA) and a QUBIT® Fluorometer (Life Technologies). The RNA integrity was checked using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Iso-Seq libraries were prepared according to the Isoform Sequencing protocol (Iso-Seq) using the Clontech SMARTer PCR cDNA Synthesis Kit and the BluePippin Size Selection System protocol as described by PacBio (PN 100–092-800–03). A mixed sample was sequenced on the PacBio RS II platform using P6-C4 chemistry.
The sequence data were processed using the SMRTlink 4.0 software. Circular consensus sequences were derived from the subread BAM files with the following parameters: min_length 200, max_drop_fraction 0.8, no_polish TRUE, min_zscore -999, min_passes 1, min_predicted_accuracy 0.8, max_length 18 000. Separation of the full-length and non−full-length reads was conducted using pbclassify.py (ignorepolyA false, minSeqLength 200). The non−full-length and full-length fasta files produced were then fed into the cluster step to cluster the isoforms, and subjected to final Arrow polishing with the parameters hq_quiver_min_accuracy 0.99, bin_by_primer false, bin_size_kb 1, qv_trim_5p 100, qv_trim_3p 30. The LoRDEC software (version 0.3) was used to correct sequencing errors in the consensus transcripts using the Illumina reads as a reference (parameters: -k 19 -s 3) [34]. The corrected consensus transcripts were clustered using CD-HIT (version 4.6.8) (-c 0.99 -T 6 -G 0 -aL 0.90 -AL 100 -aS 0.99 -AS 30) [35] to reduce sequence redundancy and improve the performance of other sequence analyses.
A total of 110,584 non-redundant unigenes were generated from 142,396 transcripts in the final RNA assemblies, which were used as evidence to assist with gene prediction. Among the 110,584 non-redundant transcripts, 8,466 (7.66%) were non-coding messenger RNAs (mRNAs). Each gene had an average of 2–7 transcripts, among which the longest transcript representing that gene was kept in the final gene model set.
Evaluation of assembly quality
The completeness of the final assembly was evaluated using CEGMA version 2.5 (CEGMA, RRID:SCR_015055) [36, 37] and BUSCO version 3.0.2 (BUSCO, RRID:SCR_015008) [38–40]. The CEGMA outputs showed that 94.76% of the core eukaryotic genes (235 of 248 core eukaryotic genes) were present in our assembly. The BUSCO test, referencing the embryophyta protein set (run_BUSCO.py -i plant_species.fa -o plant_species-l embryophyta_odb9/-m proteins), identified 94.7% of plant gene sets as complete (1,364 of 1,440 BUSCOs), including 89.0% single-copy and 5.7% duplicated genes (Table S2). All of these results suggested a high assembly quality for the yellowhorn genome.
Annotation of repetitive sequences
A de novo repeat database was constructed using RepeatScout version 1.0.5 (RepeatScout, RRID:SCR_014653) [41], LTR-FINDER (version 1.0.7) [42], MITE-Hunter (version 1.0) [43], and PILER (version 1.0) with default parameters [44]. The predicted repeats were classified using PASTEClassifier (version 1.0) with default parameters [45, 46]. Then, RepeatMasker version 4.0.7 (RepeatMasker, RRID:SCR_012954) [47] was used with the following parameters: “-nolow -no_is -norna -engine wublast -qq -frag 20 000” to identify repeat sequences by aligning them against known gene and genome sequences, based on Repbase (version 19.06) [48] and the de novo repeat database.
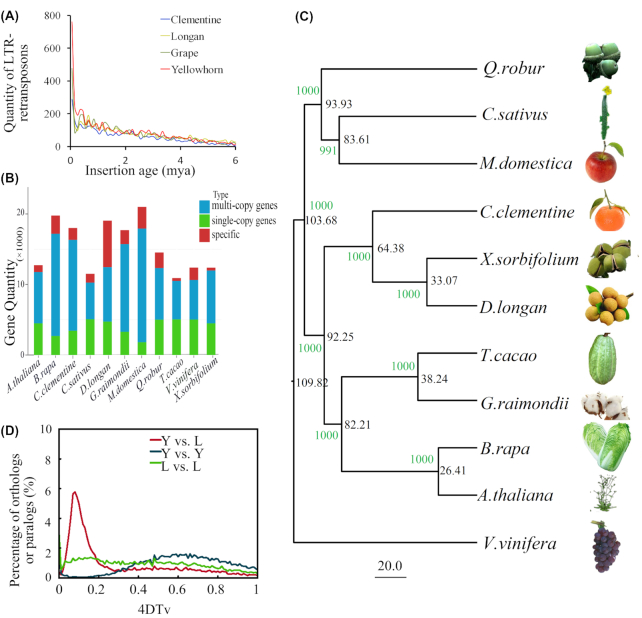
The predicted repeats represented 346.39 Mb (68.67%) of the yellowhorn genome assembly. Among these repeats, 2 types of long terminal repeat (LTR)-retrotransposons were the most abundant, including 98.68 Mb of Copia-type (19.57%) and 88.24 Mb of Gypsy-type (17.50%) repeats (Table S3). Accumulation of LTR-retrotransposons is an important contributor to genome expansion and diversity [49]. The insertion time of LTR-retrotransposons in the genome was estimated by calculating the sequence variance between the LTR arms of each LTR-retrotransposon, using a substitution rate of 1.3 × 10−8 substitutions per site per year [50]. To calculate the insertion age of each LTR retrotransposon, the 5′ and 3′ LTRs of each element were aligned with MUSCLE version 3.8.31 (MUSCLE, RRID:SCR_011812) using default setting parameters [51, 52]. Distmat (with default parameters) was used to estimate the DNA divergence between the LTR sequences with the Kimura-2-parameter base substitution model [53] and DNA divergence was converted to divergence time. A comparison of the insertion ages for LTR-retrotransposons showed similar insertion profiles among the genomes of clementine [54] (annotation version 1.0), longan [55] (annotation version 1.0), grape [56] (Vitis vinifera, annotation version GenomeScope.12X), and yellowhorn (Fig. 3A). We observed that the yellowhorn genome carried more young LTR-retrotransposons, with the highest proportion of LTR-retrotransposons with insertion ages <0.2 million years ago (mya). This might have resulted from rapid changes of its growing environment, such as the effects of pathogens and interference from human activities in recent years. The genomes sequenced by pure next-generation sequencing technology might show fewer LTR-retrotransposons because the sequence similarity between LTR arms and among different LTR-retrotransposons probably caused assembly errors in these regions, which may have led to underestimation of the number of LTR-retrotransposons in clementine and longan. Comparison of the insertion ages suggested a similar insertion age between Copia-type and Gypsy-type LTR-retrotransposons (Fig. S3).
Figure 3:
Genome evolution. (A) Distribution of insertion ages of LTR-retrotransposons. The x-axis represents the estimated insertion age (mya) of the LTR-retrotransposons. The y-axis represents the number of intact LTR-retrotransposons. (B) Comparison of copy numbers in gene clusters of analysed eudicot genomes. According to the identified gene clusters, the genes were grouped into single-copy, multiple-copy, and species-specific (specific) genes. (C) Constructed phylogenetic tree and divergence time estimation. The black numbers represent estimated divergence times (mya), which are measured with a scale bar of 20 million years, and green numbers represent bootstrap values. Grape (V. vinifera) was used as an outgroup. (D) Genome duplication in eudicot genomes as revealed through 4DTv analyses. The percentages of the orthologous pairs (Y vs L) between yellowhorn (Y) and longan (L) and paralogous gene pairs within the yellowhorn (Y vs Y) and longan (L vs L) genomes are plotted against their calculated 4DTv values.
Prediction of protein-coding genes
Annotation of protein-coding genes in the yellowhorn genome was conducted by combining de novo prediction, homology information, and RNA-seq data. For the de novo prediction, Genscan (version 3.1) [57], Augustus (Augustus: Gene Prediction, RRID:SCR_008417) (version 3.1) [58], GlimmerHMM version 3.0.4 (GlimmerHMM, RRID:SCR_002654) [59], GeneID (version 1.4) [60], and SNAP (version 2006–07-28) [61] were used to analyse the repeat-masked genome with default parameters. For the similarity-based prediction, the Uniprot protein sequences from 3 sequenced plants, Arabidopsis (The Arabidopsis Information Resource [TAIR] 10, [62]), longan (V1.0, [63]), and grape (Genomescope 12×, [64]), were aligned against the ab initio gene models in the yellowhorn genome using GeMoMa (version 1.3.1) [65]. When multiple transcripts were predicted at the same location, the highest GeMoMa scoring transcript was chosen as the optimal model [66]. The RNA-seq data were aligned to the reference genome with PASA (version 2.0.2) [67] under default parameters. All predictions from the 3 methods were combined with EVidenceModeler (v1.1.1) (Mode: STANDARD S-ratio: 1.13 score>1000) [68] to produce a consensus gene set. During the EVM integration, higher weights were assigned to the predicted PASA and GeMoMa models than the ab initio models. PASA was used to modify the final gene models.
The RNA-seq reads were then aligned to the yellowhorn genome assembly with TopHat (TopHat, RRID:SCR_013035) (v2.0.10, implemented with bowtie2) [69] to identify candidate exon regions and splicing donor and acceptor sites to evaluate the gene prediction results. Infernal version 1.1 (Infernal, RRID:SCR_011809) (default parameters) [70] was used to identify non-coding ribosomal RNA (rRNA) and microRNA genes based on Rfam (version 12.1) [71] and miRbase (version 21) [72]. TRNAscan-SE (version 1.3.1) (default parameters) [73] was used to identify transfer RNA (tRNA) genes.
GenBlastA v1.0.4(-e 1e-5) was used to perform pseudogene prediction by scanning the yellowhorn genome for sequences homologous to the known protein-coding genes it contained, and premature stop codons or frame shift mutations in those sequences were identified by GeneWise version 2.4.1 (GeneWise, RRID:SCR_015054) with the parameters -both -pseudo [74, 75].
Functional annotation of the protein-coding genes was carried out by searching against the NCBI non-redundant (nr), EuKaryotic Orthologous Groups (KOG), GO, KEGG, and TrEMBL databases. Additionally, the gene models were aligned to the Pfam database using Hmmer version 3.0 (Hmmer, RRID:SCR_005305) (parameters, -E 0.00001 –domE 0.00001 –cpu 2 –noali –acc) [75–81]. Gene Ontology (GO) terms were allocated to the genes using the Blast2GO version 2.2.31 (Blast2GO, RRID:SCR_005828) pipeline [81].
In total, we predicted 24,672 protein-coding genes (Table S4) and 1,913 pseudogenes, with a mean gene length of 4,199 bp, mean intron length of 2,560 bp, and mean coding sequence length of 1,580 bp. Of these genes, 99.02% (24,429) carried ≥1 conserved functional domain (Table S5). Their functions were classified using GO terms (Fig. S4) and the KOG database (Fig. S5). For the non-coding mRNA genes, 642 tRNA, 108 microRNA, and 316 rRNA genes were predicted in the yellowhorn genome.
Chromosome synteny between the yellowhorn and reference genomes
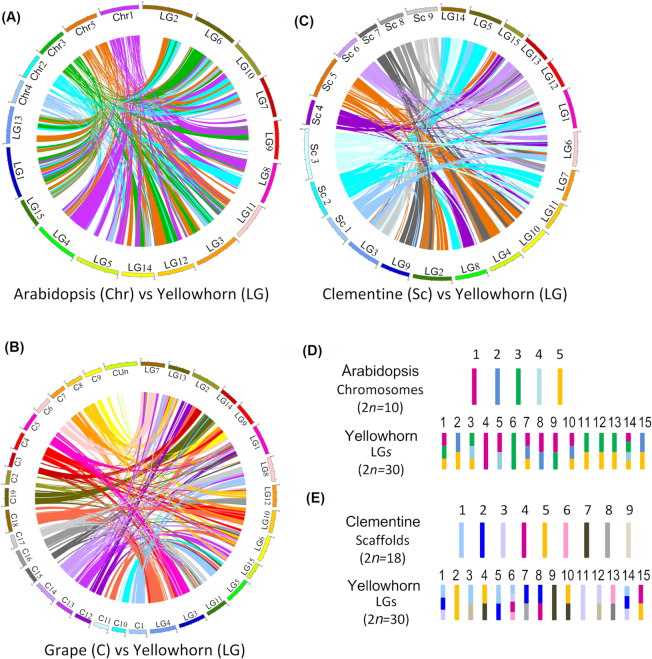
To investigate the evolution of the yellowhorn chromosomes, gene collinearity was determined by anchoring the aligned yellowhorn genes to the reference genomes of clementine, Arabidopsis, and grape using MCscan (version 0.8) [82]. The parameters of the MCscan alignment were as follows: $/MCScanX xxx.blast$-s 10 –b $2 (inter-species) blastp -query b.fa -db adb -out xyz.blast -evalue 1e-10 -num_threads 16 -outfmt 6 -num_alignments 5. A total of 367, 409, and 386 syntenic blocks were identified on the basis of the orthologous gene orders, corresponding to 28,372, 18,650, and 23,400 genes in each genome, respectively. The mean gene number per block was 77.3, 45.6, and 60.6 genes, respectively. This suggested that yellowhorn and clementine shared the highest collinearity, which was consistent with their close phylogenetic relationship as members of the Sapindales clade. The alignments of syntenic chromosomes were visualized between yellowhorn and the other genomes. The frequency of large-scale fragment rearrangements between yellowhorn and clementine, including inversions and translocations, was considerably lower than between yellowhorn and the other 2 genomes (Fig. 4). In particular, structural variation between yellowhorn and grape was so frequent that it was too difficult to speculate on the syntenic relationships among the chromosomes (Fig. 4B). The chromosome alignments between yellowhorn linkage groups (LGs) and clementine pseudomolecules revealed that most of the cross-chromosome rearrangements were different from those between yellowhorn and Arabidopsis (Fig. 4D and E). Yellowhorn LGs 2 and 11 were found to be syntenic to single clementine pseudomolecules, Scaffold 5 and 3, respectively, and LGs 3, 4, 5, 7, 8, 10, 12, 14, and 15 were each aligned to 2 reference chromosomes of clementine. Comparatively, frequency of chromosome rearrangement was a little higher between the yellowhorn LGs and Arabidopsis chromosomes. Arabidopsis Chromosome 1 was predominantly syntenic to yellowhorn LG 4, which demonstrated that the yellowhorn genome contained some conserved genome structure from its originals (Fig. 4D). Intriguingly, similar chromosomal fusion events were found among some chromosomes. Aligned fragments of Arabidopsis Chromosomes 1, 3, and 5 were fused to form yellowhorn LGs 1 and 14, similarly to clementine Scaffolds 1, 2, and 3. Yellowhorn LG 6 was aligned to clementine scaffolds 1, 3, 4, and 6 but had extensive collinearity with Arabidopsis Chromosome 3 (Fig. 4D, E). However, phylogenetic analysis suggested a distant relationship between Arabidopsis and yellowhorn. These findings suggested that Arabidopsis and yellowhorn share a chromosome of their origins, despite extensive rearrangements. Overall, these findings shed new light on the evolution of eudicot plant chromosomes.
Figure 4:
Chromosome synteny. The circularized blocks represent the chromosomes of yellowhorn and other genomes. Aligned genes identified by MCscanX are connected by lines, with their chromosome locations shown in different colours. (A) Chromosome alignment of yellowhorn and Arabidopsis. (B) Chromosome alignment of yellowhorn and grape. (C) Chromosome alignment of yellowhorn and clementine. Coloured ribbons connect the aligned genes. Yellowhorn linkage groups are labelled LG 1−15, Arabidopsis chromosomes labelled Chr 1−5, grape chromosomes are labelled C1−19 and CUn (chromosome location unknown), and clementine scaffolds are labelled Sc 1−9. Scale, 10 Mb. (D) Chromosome rearrangements between Arabidopsis and yellowhorn. (E) Chromosome rearrangements between clementine and yellowhorn. Arabidopsis and clementine chromosomes are represented as bars of different colours. Synteny and rearrangement of the yellowhorn chromosomes are indicated by different blocks, corresponding to the reference Arabidopsis and clementine chromosomes.
Identification of gene clusters and duplication
Gene clustering was conducted using OrthoMCL version 5 (OrthoMCL DB: Ortholog Groups of Protein Sequences, RRID:SCR_007839, parameters: Pep_length 10 Stop_coden 20 PercentMatchCutoff 50 EvalueExponentCutoff -5 Mcl 1.5 #1.2∼4.0) [83] among the protein sequences of 10 high-quality typical eudicot genomes representative of important families, including D. longan (Sapindaceae, Sapindales) [54], Citrus clementina (Rutaceae, Sapindales) [55], Brassica rapa (Brassicaceae, Brassicales), Arabidopsis thaliana (Brassicaceae, Brassicales) [84, 85], Theobroma cacao (Sterculiaceae, Malvales) [86], Gossypium raimondii (Malvaceae, Malvales) [87], Quercus robur (Fagaceae, Fagales) [88], V. vinifera (Vitaceae, Vitales) [56], Cucumis sativus (Cucurbitaceae, Cucurbitales) [89], and Malus × domestica (Rosaceae, Rosales) [90], as well as yellowhorn (Additional file: Table S6). The yellowhorn genes were clustered into a total of 14,828 families, including 169 yellowhorn-specific gene families (Additional file: Table S7). Comparison of gene copy numbers among the 11 eudicot genomes indicated that the yellowhorn genome had a similar proportion of single- and multiple-copy genes to the other analysed genomes (Fig. 3B). Intriguingly, the species-specific genes of yellowhorn were similar to those of T. cacao, which implied that the yellowhorn genes might have conserved the similar gene structure with their origins.
More than 300 one-to-one single-copy genes shared by all 11 genomes were identified and used to construct a phylogenetic tree using PhyML (version 3.0) (Fig. 3C) [91]. The TIM2+I+G model was used to construct the evolutionary tree as determined by jmodeltest. The software Muscle (version 3.8.31) [51, 52] was used to align the orthologs. The alignment outputs were treated with Gblocks (version 14.1) with the parameters -t = p -b5 = h -b4 = 5 -b3 = 15 -d = y -n = y [92]. Divergence times were estimated using MCMCtree (version 4.7a) [93, 94] with the following parameters: burn-in = 10 000, sample-number = 100 000, sample-frequency = 2. The TimeTree database [95], r8s (parameter: r8s -b -f r8s_in.txt > r8s_out.txt), and divergence time (Whelan and Goldman [96] and Yang et al. [97]) were used to calibrate the time. The fossil calibration times used in the evolutionary trees were as follows: (((Qrob, (Csat, Mdom)), ((Ccle, (Xsor, Dlon)), ((Tcac, Grai), (Brapa, Atha)“<30.9>20.4”))), Vvin)“<115>105”. The credibility intervals for the divergence time estimates were as follows: UTREE 1 = (((Qrob: 93.929608, (Csat: 83.608799, Mdom: 83.608799) [&95% = {67.268, 96.218}]: 10.320809) [&95% = {78.104, 105.034}]: 9.748170, ((Ccle: 64.380901, (Xsor: 33.069679, Dlon: 33.069679) [&95% = {18.376, 48.565}]: 31.311222) [&95% = {46.354, 81.164}]: 27.870851, ((Tcac: 38.243394, Grai: 38.243394) [&95% = {21.870, 56.407}]: 43.965024, (Brapa: 26.409279, Atha: 26.409279) [&95% = {20.721, 30.886}]: 55.799139) [&95% = {67.279, 94.364}]: 10.043334) [&95% = {77.382, 103.299}]: 11.426026) [&95% = {89.679, 113.000}]: 6.145826, Vvin: 109.823604) [&95% = {104.966, 114.982}]. Yellowhorn and longan in the Sapindaceae family showed the closest relationship, with the divergence time estimated at ∼33.07 mya. Using the orthologous gene pairs of yellowhorn and longan identified by gene collinearity and paralogous pairs identified by gene clustering, 4DTv (4-fold degenerate synonymous sites of the third codons) values were calculated for all of the duplicated pairs. A species divergence peak (4DTv ∼ 0.1) was observed in the yellowhorn vs longan ortholog 4DTv distribution, but no obvious peak could be seen in the yellowhorn and longan paralog curves (Fig. 3D). In a self-alignment of the chromosomes based on gene synteny, no large-scale gene duplications were found in the yellowhorn genome (Fig. S2), suggesting that the yellowhorn genome has not undergone whole-genome or large-fragment duplication.
Availability of supporting data and materials
The raw sequence data have been deposited in NCBI under project accession No. PRJNA483857. The Biosample Number of transcriptome sequencing, Pacbio SMRT sequencing, Illumina short-read sequencing and Illumina sequencing for Hi-C were SAMN09748200, SAMN11653335, SAMN11653337 and SAMN11653336. The SRA accession No. of transcriptome sequencing, PacBio SMRT sequencing, Illumina short-read sequencing, and Illumina sequencing for Hi-C was SRR7768197, SRR7768198, SRR7768199, and SRR7768201, respectively (in SRP159119). The accession No. of Xanthoceras sorbifolium genome sequencing and assembly was QUWJ 00000000. All supplementary figures and tables are provided in Additional Files. Additional supporting data, including the genome assembly, annotations and phylogenetic tree files, are available via the GigaScience database GigaDB [98].
Editor's Note
Please note, another Data Note presenting a genome assembly of Xanthoceras sorbifolium has been published back-to-back with this one in GigaScience [99].
Additional Files
Additional file 1: Tables S1−S7
Table S1: PacBio data statistics.
Table S2: Genome quality assessed by the BUSCO test.
Table S3: Repetitive sequence content.
Table S4: Prediction of protein-coding genes.
Table S5: Function annotation of protein-coding genes.
Table S6: Data used in orthoMCL analysis.
Table S7: Annotation and locus information of 169 yellowhorn-specific gene families.
Additional file 2: Figures S1−S5
Figure S1: Length distribution of the 3 types of PacBio reads produced.
Figure S2: Interaction frequency distribution of Hi-C links among chromosomes.
Figure S3. Distribution of insertion ages of Copia-type and Gypsy-type LTR-retrotransposons.
Figure S4: Function classification of protein-coding genes against the GO term database.
Figure S5: KOG function classification of protein-coding genes.
John Mackay, Ph.D. -- 10/28/2018 Reviewed
Laura Kelly -- 11/11/2018 Reviewed
Laura Kelly -- 4/18/2019 Reviewed
Pedro Martinez Garcia -- 11/30/2018 Reviewed
Abbreviations
4DTv: 4-fold degenerate synonymous sites of the third codons; bp: base pair; BUSCO: Benchmarking Universal Single-Copy Orthologs; BWA: Burrows-Wheeler Aligner; CD-HIT: Cluster Database at High Identity with Tolerance; CEGMA: Core Eukaryotic Genes Mapping Approach; EVM: EVidenceModeler; Gb: gigabases; GCE: Genomic Character Estimator; GeMoMa: Gene Model Mapper; GO: Gene Ontology; kb: kilobases; KEGG: Kyoto Encyclopedia of Genes and Genomes; KOG: EuKaryotic Orthologous Groups; LG: linkage group; LTR: long terminal repeat; Mb: megabases; mRNA: messenger RNA; mya: million years ago; MCscan: Multiple Collinearity Scan toolkit; MITE: miniature inverted-repeat transposable element; MUSCLE: MUltiple Sequence Comparison by Log-Expectation; NCBI: National Center for Biotechnology Information; NR: non-redundant; PacBio: Pacific Biosciences; PE: paired-end; Pfam: Protein Families; PhyML: Phylogeny Maximum Likelihood; RNA-Seq: RNA sequencing; rRNA: ribosomal RNA; SMRT: Single-Molecule Real-Time; SNAP: Semi-HMM-based Nucleic Acid Parser; SRA: Sequence Read Archive; TAIR: The Arabidopsis Information Resource; tRNA: transfer RNA.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was financially supported by the Central Public-Interest Scientific Institution Basal Research Fund (CAFYBB2019QD001), the National “12th Five-Year” Plan for Science & Technology Support of China (2015BAD07B0106), the National Natural Science Foundation of China (31800571, 31870594, 31760213), the National Key Research and Development Plan of China (2016YFC050080506) and Key Research Development Program of Liaoning Province (2017204001).
Authors' contributions
Q.B., H.Y., Y.Lu, C.R., and L.W. conceived and designed the study; T.C., X.L., Y.Li, S.F., X.H., G.F., Y.C., J.D., D.C., Z.Z., and Z.L. prepared materials and conducted the experiments; Q.B., Y.Z., W.D., Y.Lu, and L.G. wrote the manuscript.
References
- 1. Wang Q, Yang L, Ranjitkar S, et al.. Distribution and in situ conservation of a relic Chinese oil woody species yellowhorn Xanthoceras sorbifolium Bunge. Can J For Res. 2017;47:1450–6. [Google Scholar]
- 2. Board E. Flora of China. In: Xia NH, Paul AG.Sapindacea.Beijing:Science press;1989.P.7.1985;47:72. [Google Scholar]
- 3. Yu HY, Fan SQ, Bi QX, et al.. Seed morphology, oil content and fatty acid composition variability assessment in yellow horn (Xanthoceras sorbifolium Bunge) germplasm for optimum biodiesel production. Ind Crop Prod. 2017;97:425–30. [Google Scholar]
- 4. Yao ZY, Qi JH, Yin LM, et al.. Biodiesel production from Xanthoceras sorbifolia in China: Opportunities and challenges. Renew Sust Energy Rev. 2013;24:57–65. [Google Scholar]
- 5. Venegas-Calerón M, Ruíz-Méndez MV, Martínez-Force E, et al.. Characterization of Xanthoceras sorbifolium Bunge seeds: Lipids, proteins and saponins content. Ind Crop Prod. 2017;109:192–8. [Google Scholar]
- 6. Ruan CJ, Yan R, Wang BX, et al. The importance of yellow horn (Xanthoceras sorbifolium) for restoration of arid habitats and production of bioactive seed oils. Ecol Engineer2017;99:504-12. [Google Scholar]
- 7. Taylor DC, Guo Y, Katavic V, et al.. New seed oils for improved human and animal health: genetic manipulation of the brassicaceae for oils enriched in nervonic acid. In: Modification of Seed Composition to Promote Health and Nutrition, Agronomy Monographs 51. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2009:219–232., doi: 10.2134/agronmonogr51.c10. [DOI] [Google Scholar]
- 8. Qi Y, Ji XF, Chi TY, et al.. Xanthoceraside attenuates amyloid β peptide 1–42 -induced memory impairments by reducing neuroinflammatory responses in mice. Eur J Pharmacol. 2017;820:18–30. [DOI] [PubMed] [Google Scholar]
- 9. Ji XF, Chi TY, Liu P, et al.. The total triterpenoid saponins of Xanthoceras sorbifolia improve learning and memory impairments through against oxidative stress and synaptic damage. Phytomedicine. 2017;25:15–24. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Y, Xiao LU, Xiao B, et al.. Research progress and application prospect of Xanthoceras sorbifolia for treating Alzheimer's disease. Drug Eval Res. 2018;25:912–7. [Google Scholar]
- 11. Galbraith DW, Harkins KR, Maddox JM, et al.. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–51. [DOI] [PubMed] [Google Scholar]
- 12. Pellicer J, Leitch IJ. The application of flow cytometry for estimating genome size and ploidy level in plants. Methods Mol Biolo (Clifton, NJ). 2014;1115:279–307. [DOI] [PubMed] [Google Scholar]
- 13. Schmutz J, Cannon SB, Schlueter J, et al.. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:78–83. [DOI] [PubMed] [Google Scholar]
- 14. Tuskan GA, Difazio S, Jansson S, et al.. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2016;313:1596–604. [DOI] [PubMed] [Google Scholar]
- 15. Dolezel J, Greilhuber J, Suda J, et al.. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2007;2:2233–44. [DOI] [PubMed] [Google Scholar]
- 16. Toh H, Shirane K, Miura F, et al.. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics. 2017;18:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alberto CM, Sanso AM, Xifreda CC, et al.. Chromosomal studies in species of Salvia (Lamiaceae) from Argentina. Bot J Linn Soc. 2015;141:483–90. [Google Scholar]
- 18. Bolger AM, Lohse M, Usadel B, et al.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17:10–2. [Google Scholar]
- 20. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu BH, Shi YJ, Yuan JY, et al.. Estimation of genomic characteristics by analyzing kmer frequency in de novo genome projects.arXivprint 2013. arXiv:1308.2012. [Google Scholar]
- 22. Kajitani R, Toshimoto K, Noguchi H, et al.. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-Greater-Than-30-kb-SMRTbell-Libraries-Using-Needle-Shearing-and-BluePippin-Size-Selection-on-Sequel-and-RSII-Systems.pdf. [Google Scholar]
- 24. Li R, Fan W, Tian G, et al.. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chin J. Falcon Genome Assembly Tool Kit Manual. https://github.com/PacificBiosciences/FALCON/wiki/Manual. [Google Scholar]
- 26. How to use pbsmrtpipe. http://pbsmrtpipe.readthedocs.io/en/master/getting_started.html. [Google Scholar]
- 27. Pilon. https://github.com/broadinstitute/pilon/wiki. [Google Scholar]
- 28. Roach M. purge_haplotigs. https://bitbucket.org/mroachawri/purge_haplotigs. [Google Scholar]
- 29. Roach MJ, Schmidt S, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19:60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Burton J N, Adey A, Patwardhan RP, et al.. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31:1119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Servant N, Varoquaux N, Lajoie BR, et al.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li MX. Karyotype analysis of some oil plants. Acta Bot Boreali-Occident Sin. 1987;7:246–51. [Google Scholar]
- 34. Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu L, Niu B, Zhu Z, et al.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parra G, Bradnam K, Korf I, et al.. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7. [DOI] [PubMed] [Google Scholar]
- 37. CEGMA Website. http://korflab.ucdavis.edu/datasets/cegma/. Accessed 2nd April 2019. [Google Scholar]
- 38. Simao FA, Waterhouse RM, Ioannidis P, et al.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- 39. Nishimura O, Hara Y, Kuraku S. Volante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics. 2017;33:3635–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. gVolante: Completeness Assessment of Genome/Transcriptome Sequences. https://gvolante.riken.jp/analysis.html. Accessed 2nd April 2019. [Google Scholar]
- 41. Price AL, Jones NC, Pevzner PA, et al.. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:i351–8. [DOI] [PubMed] [Google Scholar]
- 42. Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Edgar RC, Myers EW. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–8. [DOI] [PubMed] [Google Scholar]
- 45. Wicker T, Sabot F, Hua-Van A, et al.. A unified classification system for eukaryotic transposbale elments. Nat Rev Genet. 2007;8:973–82. [DOI] [PubMed] [Google Scholar]
- 46. Hoede C, Arnoux S, Moisset M, et al.. PASTEC: An Automatic Transposable Element Classification Tool. PLoS One. 2014;9:e91929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2009, doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 48. Jurka J, Kapitonov VV, Pavlicek A, et al.. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. [DOI] [PubMed] [Google Scholar]
- 49. Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A. 1997;94:7704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zuccolo A, Sebastian S, Yu Y, et al.. Assessing the extent of substitution rate variation of retrotransposon long terminal repeat sequences in Oryza sativa and Oryza glaberrima. Rice. 2010;3:242–50. [Google Scholar]
- 51. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. MUSCLE. https://www.ebi.ac.uk/Tools/msa/muscle/. [Google Scholar]
- 53. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. [DOI] [PubMed] [Google Scholar]
- 54. Lin Y, Min J, Lai R, et al.. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. Gigascience. 2017;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu GA, Prochnik S, Jenkins J, et al.. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat Biotechnol. 2014;32:656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jaillon O, Aury JM, Noel B, et al.. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. [DOI] [PubMed] [Google Scholar]
- 57. Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. [DOI] [PubMed] [Google Scholar]
- 58. Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19:215–25. [DOI] [PubMed] [Google Scholar]
- 59. Majoros WH, Pertea M, Salzberg SL, et al.. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–9. [DOI] [PubMed] [Google Scholar]
- 60. Blanco E, Parra G, Guigó R, et al.. Using geneid to identify genes. Cur Protoc Bioinformatics. 2007;18:3–4. [DOI] [PubMed] [Google Scholar]
- 61. Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. BrassicaDB Arabidopsis thaliana. http://brassicadb.org/brad/datasets/pub/BrassicaceaeGenome/Arabidopsis_thaliana/. [Google Scholar]
- 63. Lin Y, Min J, Lai R, et al.. Supporting data for "Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics.". GigaScience Database. 2017. 10.5524/100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vitis vinifera(wine grape). https://www.ncbi.nlm.nih.gov/genome/?term=Vitis+vinifera+genome. [Google Scholar]
- 65. Jens K, Michael W, Erickson J L, et al.. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Campbell MA, Haas BJ, Hamilton JP, et al.. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang S, Lomsadze A, Borodovsky M, et al.. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2014;43:58–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Haas BJ, Papanicolaou A, Yassour M, et al.. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Trapnell C, Pachter L, Salzberg SL, et al.. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Griffiths-Jones S, Moxon S, Marshall M, et al.. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Griffithsjones S, Grocock RJ, Van DS, et al.. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. She R, Chu JK, Pei J, et al.. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Birney E, Clamp M, Durbin R, et al.. GeneWise and Genomewise. Genome Res. 2004;14:988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Marchlerbauer A, Lu S, Anderson JB, et al.. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tatusov RL, Natale DA, Garkavtsev IV, et al.. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dimmer EC, Huntley RP, Alamfaruque Y, et al.. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40:565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Du JL, Yuan ZF, Ma ZW, et al.. KEGG-PATH: Kyoto Encyclopedia of Genes and Genomes-based pathway analysis using a path analysis model. Mol Biosystems. 2014;10:2141–7. [DOI] [PubMed] [Google Scholar]
- 80. Boeckmann B, Bairoch A, Apweiler R, et al.. The Swiss-Prot knowledgebase and its supplement TREMBL in 2003. Nucleic Acids Res. 2003;31:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Altschul S, Gish W, Miller W, et al.. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- 82. Tang HB, Bowers JE, Wang XY, et al.. Synteny and collinearity in plant genomes. Science. 2008;320:486–8. [DOI] [PubMed] [Google Scholar]
- 83. Li L, Jr SC, Roos DS, et al.. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang XW, Wang HZ, Wang J, et al.. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–40. [DOI] [PubMed] [Google Scholar]
- 85. Theologis A, Ecker JR, Palm CJ, et al.. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature. 2000;408:816–20. [DOI] [PubMed] [Google Scholar]
- 86. Argout X, Salse J, Aury JM, et al.. The genome of Theobroma cacao. Nat Genet. 2013;43:101–8. [DOI] [PubMed] [Google Scholar]
- 87. Wang K, Wang Z, Li F, et al.. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44:1098–103. [DOI] [PubMed] [Google Scholar]
- 88. Plomion C, Aury JM, Amselem J, et al.. Oak genome reveals facets of long lifespan. Nat Plants. 2017;4:440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Huang S, Li R, Zhang Z, et al.. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–81. [DOI] [PubMed] [Google Scholar]
- 90. Velasco R, Zharkikh A, Affourtit J, et al.. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 2010;42:833–9. [DOI] [PubMed] [Google Scholar]
- 91. Guindon S, Dufayard JF, Lefort V, et al.. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. [DOI] [PubMed] [Google Scholar]
- 92. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77. [DOI] [PubMed] [Google Scholar]
- 93. Yang Z. Phylogenetic Analysis by Maximum Likelihood (PAML). http://abacus.gene.ucl.ac.uk/software/paml.html. [Google Scholar]
- 94. Battistuzzi FU, Billingross P, Paliwal A, et al.. Fast and slow implementations of relaxed-clock methods show similar patterns of accuracy in estimating divergence times. Mol Biol Evol. 2011;28:2439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. TimeTree. http://www.timetree.org/. Accessed 2nd Aprl 2019. [Google Scholar]
- 96. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol Biol Evol. 2001;18:691–9. [DOI] [PubMed] [Google Scholar]
- 97. Yang Z, Nielsen R, Hasegawa M. Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol. 1998;15:1600–11. [DOI] [PubMed] [Google Scholar]
- 98. Bi Q, Zhao Y, Du W, et al.. Supporting data for “Pseudomolecule-level assembly of the Chinese oil tree yellowhorn (Xanthoceras sorbifolium) genome.”. GigaScience Database. 2019. 10.5524/100606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liang Q, Li H, Li S, et al.. The genome assembly and annotation of yellowhorn (Xanthoceras sorbifolium Bunge). GigaScience. 2019.doi:10.1093/gigascience/giz071 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
John Mackay, Ph.D. -- 10/28/2018 Reviewed
Laura Kelly -- 11/11/2018 Reviewed
Laura Kelly -- 4/18/2019 Reviewed
Pedro Martinez Garcia -- 11/30/2018 Reviewed