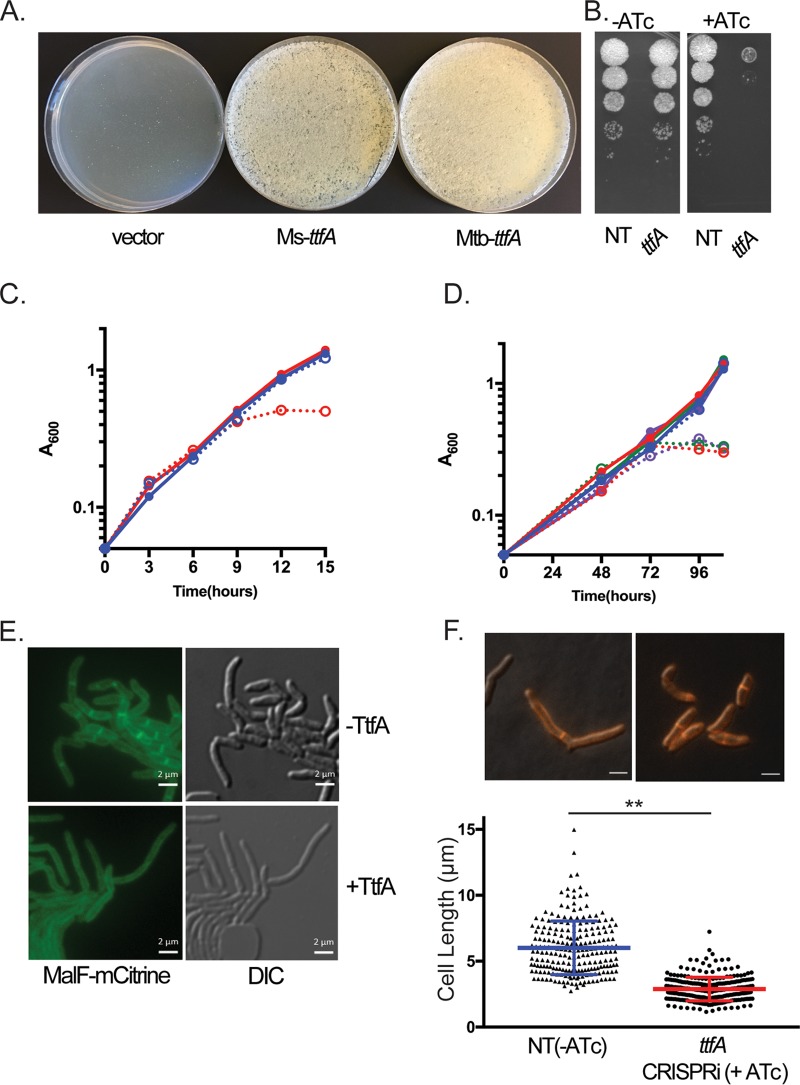
FIG 2.
MsTtfA/MtbTtfA is required for mycobacterial growth and cell elongation. (A) M. smegmatis strains with deletion of chromosomal ttfA and carrying a copy of ttfA at the attB phage integration site were subjected to marker exchange with attB integrating vectors. Δttfa attB::ttfA strep (MGM6414) strains transformed with pMV306kan (vector), pAJF792 (encoding MsTtfA), or pAJF793 (MtbTtfA) are shown on kanamycin agar. (B) Ten-fold dilutions of M. smegmatis carrying ATc-inducible CRISPRi nontargeting control (NT [MGM6418]) or ttfA (MGM6419) on agar media with and without ATc. (C) Growth curves of nontargeting (MGM6418, blue) and ttfA-targeting (MGM6419, red) CRISPRi M. smegmatis strains grown under uninduced (solid, closed circles) and ATc-induced (dashed, empty circles) conditions. (D) Growth curves of nontargeting (MGM6715, blue) or three distinct M. tuberculosis ttfA-targeting CRISPRi M. tuberculosis strains (MGM6675, red; MGM6677, green; MGM6679, purple) grown under uninduced (solid, closed circles) and ATc-induced (dashed, empty circles) conditions. (E) Fluorescence microscopy of an M. smegmatis ttfA-targeting CRISPRi strain marked with MalF(1,2)-mCitrine (MGM6433) 15 h post-CRISPRi induction with ATc (top, −TtfA) or, an uninduced control at 15 h (+TtfA, bottom). YFP (left) and DIC (right) images shown. Exposure times for YFP, 250 ms, 40% LED. (F) Loss of TtfA leads to short cells. Cell lengths of nontargeting (MGM6418, triangles) and TtfA-targeting (MGM6419, circles) CRISPRi strains induced for 12 h. Representative DIC/FM 4-64 images used for quantitation shown above the graph. Error bars are standard deviation and ** indicates P < 0.001.