Many animals and protists depend on symbiotic sulfur-oxidizing bacteria as their main food source. These bacteria use energy from oxidizing inorganic sulfur compounds to make biomass autotrophically from CO2, serving as primary producers for their hosts. Here we describe a clade of nonautotrophic sulfur-oxidizing symbionts, “Candidatus Kentron,” associated with marine ciliates. They lack genes for known autotrophic pathways and have a carbon stable isotope fingerprint heavier than other symbionts from similar habitats. Instead, they have the potential to oxidize sulfur to fuel the uptake of organic compounds for heterotrophic growth, a metabolic mode called chemolithoheterotrophy that is not found in other symbioses. Although several symbionts have heterotrophic features to supplement primary production, in Kentron they appear to supplant it entirely.
KEYWORDS: gammaproteobacteria, chemosynthesis, ectosymbiont, lithoheterotrophy, meiofauna, protist
ABSTRACT
Since the discovery of symbioses between sulfur-oxidizing (thiotrophic) bacteria and invertebrates at hydrothermal vents over 40 years ago, it has been assumed that autotrophic fixation of CO2 by the symbionts drives these nutritional associations. In this study, we investigated “Candidatus Kentron,” the clade of symbionts hosted by Kentrophoros, a diverse genus of ciliates which are found in marine coastal sediments around the world. Despite being the main food source for their hosts, Kentron bacteria lack the key canonical genes for any of the known pathways for autotrophic carbon fixation and have a carbon stable isotope fingerprint that is unlike other thiotrophic symbionts from similar habitats. Our genomic and transcriptomic analyses instead found metabolic features consistent with growth on organic carbon, especially organic and amino acids, for which they have abundant uptake transporters. All known thiotrophic symbionts have converged on using reduced sulfur to gain energy lithotrophically, but they are diverse in their carbon sources. Some clades are obligate autotrophs, while many are mixotrophs that can supplement autotrophic carbon fixation with heterotrophic capabilities similar to those in Kentron. Here we show that Kentron bacteria are the only thiotrophic symbionts that appear to be entirely heterotrophic, unlike all other thiotrophic symbionts studied to date, which possess either the Calvin-Benson-Bassham or the reverse tricarboxylic acid cycle for autotrophy.
INTRODUCTION
Chemosynthetic symbioses between heterotrophic, eukaryotic hosts and bacteria that use the oxidation of inorganic chemicals or methane to fuel growth are common in marine environments. They occur in habitats ranging from deep-sea vents and seeps, where they are responsible for much of the primary production, to the interstitial pore water habitat in shallow-water sediments, where the hosts are often small and inconspicuous meiofauna. Among the energy sources for chemosynthesis are reduced sulfur species like sulfide and thiosulfate, and such sulfur-oxidizing (thiotrophic) symbioses have convergently evolved multiple times (1). They are commonly interpreted as nutritional symbioses where the symbionts fix CO2 autotrophically into biomass with the energy from sulfur oxidation and eventually serve as food for their hosts (1, 2). Indeed, several host groups have become so completely dependent on their symbionts for nutrition that they have reduced digestive systems. All sulfur-oxidizing symbioses investigated thus far possess a primary thiotrophic symbiont with genes of either the Calvin-Benson-Bassham (CBB) (3–10) or reverse tricarboxylic acid (rTCA) (11, 12) cycle for CO2 fixation, and the different pathways may relate to different ecological niches occupied by the symbioses (13). The symbionts of the vestimentiferan tubeworms are additionally able to encode both the CBB and rTCA cycles, which may be active under different environmental conditions (14–16). Beyond sulfur oxidation and carbon fixation, several thiotrophic symbionts have additional metabolic capabilities such as the uptake of organic carbon (17), the use of carbon monoxide (18) and hydrogen (19) as energy sources, and the ability to fix inorganic nitrogen (4, 5).
The thiotrophic ectosymbionts of the ciliate genus Kentrophoros constitute a distinct clade of Gammaproteobacteria named “Candidatus Kentron” (here Kentron) (20). Kentron has previously been shown with radioisotope labeling experiments to oxidize sulfide and fix CO2 (21) and to be consumed and digested by its hosts (21, 22). Unlike most ciliates, which consume their food at a specific location on the cell that bears feeding structures composed of specialized cilia, Kentrophoros has only vestiges of such cilia and instead directly engulfs its symbionts along the entire cell body (23), suggesting that Kentron bacteria are its main food source.
Given that all previous studies of thiotrophic symbionts, including Kentron, have characterized them as autotrophic, we expected that the pathways of energy and carbon metabolism used by Kentron would resemble those in other thiotrophic bacteria involved in nutritional symbioses. In this study, we used metagenomic and transcriptomic analyses of single host individuals to show that the Kentron clade lacks the canonical pathways of autotrophic CO2 fixation. Based on a metabolic reconstruction of the core genome from 11 Kentron phylotypes collected from three different sites, and results from direct protein-stable isotope fingerprinting, we propose that it is a lithoheterotrophic nutritional symbiont, relying on assimilation of organic substrates rather than fixation of inorganic carbon to feed its hosts.
RESULTS
Symbiont genome assemblies have high coverage and completeness and represent 11 phylotypes.
Genomes of Kentron symbionts were binned from 34 metagenome assemblies, each corresponding to a single Kentrophoros host ciliate individual. The genomes represented 11 symbiont phylotypes from 12 host morphospecies (Fig. 1), collected from the Mediterranean, Caribbean, and Baltic Seas (see data posted at https://doi.org/10.5281/zenodo.2575755). Most symbiont phylotypes were specific to the host morphospecies, except for one symbiont associated with two hosts (Kentrophoros sp. UNK and Kentrophoros sp. LPFa). Symbiont genome assemblies were between 3.31 and 5.02 Mbp (median, 3.91 Mbp) long but were relatively fragmented (N50: 3.52 to 37.5 kbp; median, 21.4 kbp). Genome sizes and assembly fragmentation appeared to be species/phylotype dependent (see Fig. S1A and B in the supplemental material). Nonetheless, they were relatively complete (91.4 to 94.9%; median, 93.8%) and had low contamination (0.75 to 3.56%; median, 1.87%) (see data posted at https://doi.org/10.5281/zenodo.2575767). The core genomic diversity in the clade was well sampled: 1,019 protein-encoding gene orthologs were found in all 34 genomes, and the core genome accumulation curve reached a plateau (Text S1; Fig. S1C). Kentron genome sizes were relatively large for thiotrophic symbionts and were comparable to values for “Candidatus Thiodiazotropha” spp. (4.5 Mbp) and the Gamma3 symbiont of Olavius algarvensis (4.6 Mbp).
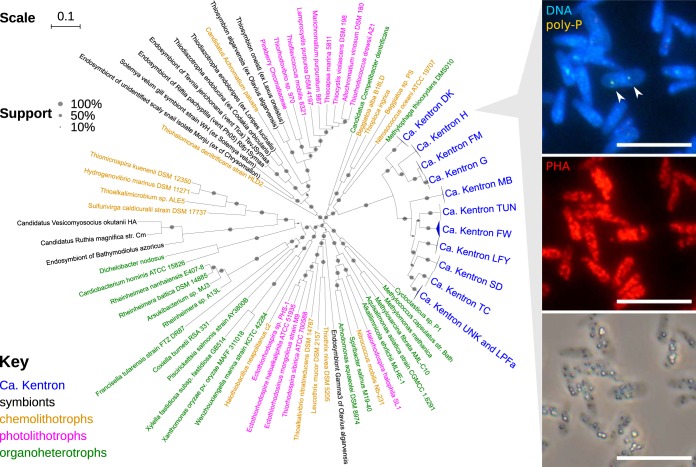
FIG 1.
Maximum-likelihood phylogeny of Kentron and basal Gammaproteobacteria from concatenated alignment of 30 conserved protein-encoding marker genes. Support values, SH-like approximate likelihood ratio test (aLRT). Branch lengths, substitutions per site. (Inset) Kentron sp. H fluorescence with DAPI stain for polyphosphate (top, arrowheads) and Nile Red stain for PHA (middle) and in bright field showing refractile sulfur globules (bottom). Bars, 10 μm.
Phylogenetics. Download Text S1, PDF file, 0.1 MB (83.3KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary statistics for Kentron genome assemblies. (A) Genome completeness per species, as assessed with gammaproteobacterial conserved marker genes by CheckM. (B) Number of contigs versus total length (Mbp) of Kentron genome bins; plot symbol areas scaled by N50 (kbp). Larger genomes are usually more fragmented (more contigs, lower N50), with the exception of Kentron sp. DK. (C) Accumulation curves for pan- and core genomes (protein-encoding genes only) of the Kentron clade, with uncertainty estimated by resampling (200 times, with replacement). “Singleton” genes were not included in an ortholog cluster. Download FIG S1, EPS file, 0.1 MB (104.6KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes for key enzymes in known autotrophic pathways are absent.
Unlike other investigated thiotrophic symbionts, genes for ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and other key enzymes in the six canonical autotrophic CO2 fixation pathways (see data posted at https://doi.org/10.5281/zenodo.2575773) were not predicted in Kentron genomes by standard annotation pipelines. Kentron sp. H has a gene for group IV RuBisCO (Fig. S2), also known as RuBisCO-like protein (RLP), which is not known to play a role in carbon fixation but participates in a variety of other pathways such as thiosulfate metabolism (24).
Maximum-likelihood phylogeny of RuBisCO proteins, including RuBisCO-like protein from Kentron phylotype H (red arrow). Download FIG S2, EPS file, 0.5 MB (493KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
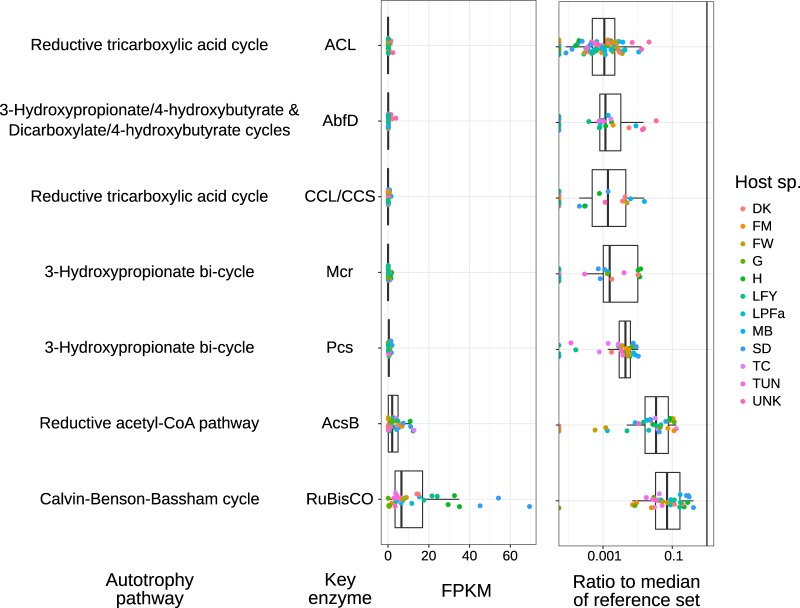
To rule out the possibility of misannotation, incomplete genome binning, or problems with genome assembly, we aligned raw, unassembled reads from Kentrophoros metagenome libraries to the curated Swiss-Prot database of protein sequences. Key autotrophy proteins had coverage values (median of 0.00, maximum of 69.3 fragments per kilobase per million [FPKM]) that were always lower than the median coverage of reference proteins from the TCA and partial 3-hydroxypropionate (3HPB) pathways (Fig. 2). In 89% of cases, the coverage was at least 50-fold lower than the reference median, and if not, the majority of reads could be attributed either to other microbial genome bins in the metagenome (mostly RuBisCO or AcsB) or to the RuBisCO-like protein in Kentron H (Fig. S3). Metatranscriptomes of two phylotypes (H and SD) were also screened with the same pipeline, and key autotrophy proteins again had coverages that were always below the median of the reference set (median, 0.00; maximum, 1.62 FPKM) (Fig. S4). We conclude that canonical autotrophy genes were indeed absent from Kentron genomes and not merely misassembled or mispredicted.
FIG 2.
Read coverage (individual values and box plots) in Kentrophoros metagenomes for key enzymes of autotrophic CO2 fixation pathways, expressed as FPKM values (left) and as a fraction of the median coverage of a reference set of proteins that are expected to be present in all Kentron species (right) (see data posted at https://doi.org/10.5281/zenodo.2575773). Unassembled reads were used for alignment, to rule out the possibility of not detecting hits because of misassembly. Each point represents a separate metagenome library, colored by Kentrophoros host morphospecies. The box midline represents the median, hinges represent the interquartile range (IQR), and whiskers are data within 1.5× IQR of hinges. Abbreviations: ACL, ATP citrate lyase; AbfD, 4-hydroxybutanoyl-CoA dehydratase; CCL/CCS, citryl-CoA lyase/citryl-CoA synthase; Mcr, malonyl-CoA reductase; Pcs, propionyl-CoA synthase; AcsB, CO-methylating acetyl-CoA synthase; RuBisCO, ribulose-1,6-bisphosphate carboxylase/oxygenase.
Selected results of remapping reads with hits to canonical autotrophy enzymes to the metagenomic assemblies, for AcsB versus Kentrophoros sp. TC (left) and RuBisCO versus Kentrophoros sp. H (right). Metagenome assemblies are depicted as coverage-GC% plots, where each gray circle represents a contig, with its size scaled to the contig length. Contigs with remapping hits have solid outlines, with numbers of remapping reads indicated when >10. The majority of reads with alignments to canonical autotrophy enzymes map to scaffolds outside the Kentron symbiont genome bin (circled in blue) and could be attributed to other bacteria in the metagenomes, except for RuBisCO in Kentron sp. H, due to the RuBisCO-like protein encoded in that phylotype. Download FIG S3, TIF file, 0.3 MB (348.3KB, tif) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Read coverage in Kentrophoros metatranscriptomes for key enzymes of autotrophic CO2 fixation pathways, expressed as FPKM values (left) and as a fraction of the median coverage of a reference set of proteins (right). Each point represents a separate metatranscriptome library, colored by Kentrophoros host morphospecies. Abbreviations as in Fig. 3. Download FIG S4, EPS file, 0.1 MB (149.1KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence for lithoheterotrophic metabolism in Kentron.
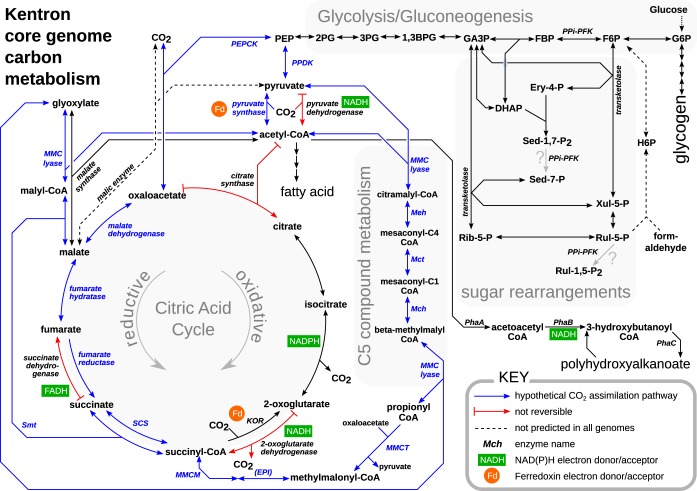
Kentron genome annotations suggested a lithoheterotrophic metabolism, in which energy is produced by oxidation of reduced sulfur and carbon is assimilated in the form of organic compounds (Fig. 3).
FIG 3.
Schematic reconstruction of carbon and central metabolism of Kentron clade, focusing on pathways discussed in the text. Compound name abbreviations: 1,3BPG, 1,3-bisphosphoglycerate; 2PG, 2-phosphoglycerate; 3PG, 3-phosphoglycerate; DHAP, dihydroxyacetone phosphate; Ery-4-P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; G6P, glucose-6-phosphate; GA3P, glyceraldehyde-3-phosphate; H6P, hexose-6-phosphate; PEP, phosphoenolpyruvate; Rib-5-P, ribose-5-phosphate; Rul-1,5-P2, ribulose-1,5-bisphosphate; Rul-5-P, ribulose-5-phosphate; Sed-1,7-P2, sedoheptulose-1,7-bisphosphate; Sed-7-P, sedoheptulose-7-phosphate; Xul-5-P, xylulose-5-phosphate. Enzyme name abbreviations: EPI, methylmalonyl-CoA epimerase; KOR, alpha-ketoglutarate oxidoreductase; Mch, mesaconyl-C1-CoA hydratase; Mct, mesaconyl-CoA C1-C4 CoA transferase; Meh, mesaconyl-C4-CoA hydratase; MMC lyase, (S)-malyl-CoA/beta-methylmalyl-CoA/(S)-citramalyl-CoA lyase; MMCM, methylmalonyl-CoA mutase; MMCT, methylmalonyl-CoA carboxytransferase; PEPCK, phosphoenolpyruvate carboxykinase; PPDK, pyruvate phosphate dikinase; PPi-PFK, pyrophosphate-dependent phosphofructokinase; Smt, succinyl-CoA:(S)-malate-CoA transferase.
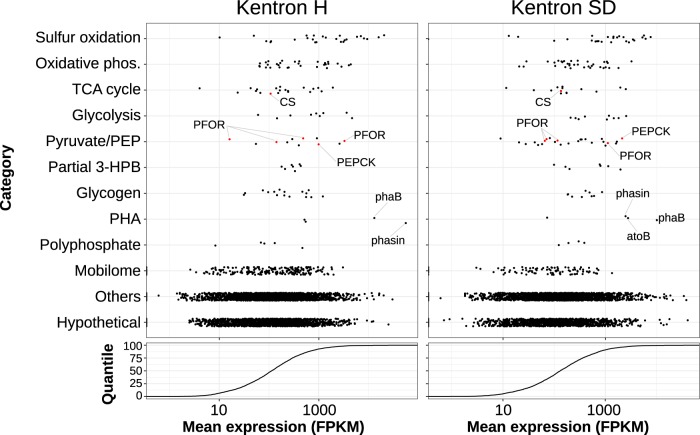
(i) Electron donors and energetics. Kentron genomes encoded a hybrid Sox-reverse Dsr pathway, similar to other symbiotic and free-living thiotrophs (e.g., Allochromatium vinosum), which would allow the oxidation of thiosulfate, elemental sulfur, and sulfide as energy sources (25, 26). They had a complete electron transport chain for oxidative phosphorylation and an FoF1-type ATP synthase. The only terminal oxygen reductase predicted was cbb3-type cytochrome c oxidase (complex IV), which has a high oxygen affinity and is typically expressed under microoxic conditions (27, 28). In the two Kentron phylotypes for which expression profiles were available, this set of functions was among the most highly expressed genes (Fig. 4).
FIG 4.
Expression of genes in selected functional categories, for two Kentron phylotypes (mean for 3 samples per phylotype, in FPKM). Cumulative distribution curves for expression per gene are shown below each dot plot. Selected genes discussed in the text are labeled; carboxylation reactions involving pyruvate or phosphoenolpyruvate (PEP) are marked in red. Abbreviations: atoB, acetyl-CoA acetyltransferase; CS, citrate synthase; PFOR, pyruvate-ferredoxin oxidoreductase (=pyruvate synthase); PEPCK, phosphoenolpyruvate carboxykinase; phaB, acetoacetyl-CoA reductase.
Four Kentron phylotypes (H, SD, FW, and G) encoded anaerobic-type Ni-dependent CO dehydrogenase precursors, adjacent to CO dehydrogenase Fe-S subunits (in FW and SD) or a CO dehydrogenase maturation factor (in G). In addition, H2 may serve as an electron donor for Kentron TC, TUN, G, and FW (one genome), which contained genes related to the oxidative-type [Ni-Fe] hydrogenase Mvh (A and G subunits), as well as auxiliary proteins for hydrogenase maturation and Ni incorporation, although they did not all occur in a single gene cluster. Both CO and H2 are known to be potential electron donors for symbiotic thiotrophs and have been measured in their habitat in Sant’ Andrea, Elba (18), where one of these Kentrophoros phylotypes (H) was collected.
Oxidoreductases for anaerobic respiration were not predicted, except for subunits NapA and -B of periplasmic nitrate reductase (in 28 and 25 genomes, respectively). Other oxidoreductases for complete denitrification or the dissimilatory nitrate reduction to ammonia pathway were absent. Some organisms can respire nitrate with only Nap, but because Nap has other potential functions, it is not possible to predict if Kentron can respire nitrate from the presence of Nap alone (29). Na+-translocating ferredoxin:NAD+ (Rnf) and NADH:ubiquinone (Nqr) oxidoreductases, which can couple reducing equivalents to the Na+ membrane potential, were also predicted.
(ii) Uptake transporters for organic substrates. Genes encoding uptake transporters for organic substrates were abundant in Kentron genomes and were also expressed in the transcriptomes (Fig. S5; see also File 2 posted at https://doi.org/10.5281/zenodo.2555833). An average of 54.1 such genes were predicted per genome (representing 18.1% of all genes with Transporter Classification Database [TCDB] hits), of which more than half had transmembrane (TM) domains (mean, 30.5 per genome). The families with the highest mean counts per genome were the ATP-binding cassette (ABC) superfamily (33.9 total, 16.4 transmembrane, counting only uptake-related subfamilies), tripartite ATP-independent periplasmic transporter (TRAP-T) family (7.2 total, 5.1 TM), and the solute:sodium symporter (SSS) family (1.6 total, 1.3 TM). Three other families—concentrative nucleoside transporter (CNT), dicarboxylate/amino acid cation symporter (DAACS), and neurotransmitter/sodium symporter (NSS)—were represented by a single gene in all Kentron genomes. Most of these families are known to target organic acids, amino acids, or small peptides. In comparison, sugar uptake transporter families were less numerous and present in only a subset of genomes (e.g., ABC subfamilies CUT1 and CUT2) or not predicted in Kentron at all (e.g., phosphotransferase system family).
Uptake-related transporter families found in all Kentron genomes. (a) Number of open reading frames per genome with matches to Transporter Classification families (Blastp E value <10−5, >30% amino acid identity, >70% query coverage). (b) Mean expression (FPKM) in three transcriptomes of Kentron H. (c) Mean expression (FPKM) in three transcriptomes of Kentron SD. Download FIG S5, TIF file, 0.3 MB (271KB, tif) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
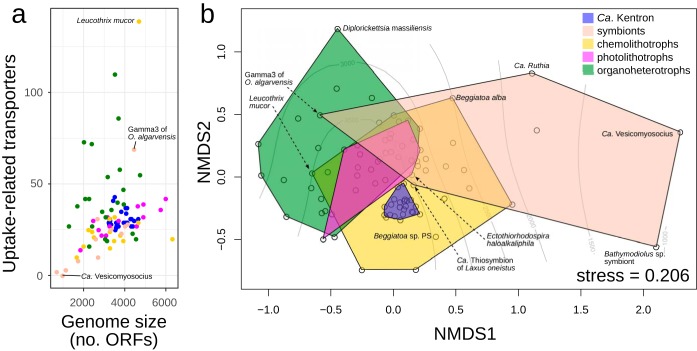
The number of organic uptake transporters in Kentron was high compared to other symbiotic thiotrophs, which had counts ranging from 2 (0 TM) in “Candidatus Vesicomyosocius okutanii” to 134 (69 TM) in the Gamma3 symbiont of Olavius algarvensis. However, larger genomes tend to have more transporters, and Kentron genomes were also relatively large (Fig. 5a). We therefore compared the content of organic-uptake-related TCDB family members per genome between Kentron and other basal gammaproteobacteria by nonmetric multidimensional scaling. Kentron overlapped the known range of variation for both phototrophs and chemolithotrophs (both free-living and symbiotic). Among heterotrophs, it was most distant from pathogens like Diplorickettsia. Among thiotrophic symbionts, it was most similar to “Candidatus Thiosymbion” but most distant from the symbionts of deep-sea bivalves (which have few uptake transporters) (Fig. 5b).
FIG 5.
Comparison of organic substrate transporters in genomes of Kentron and other basal Gammaproteobacteria, named in Fig. 1. (a) Counts of uptake-related transporters (transmembrane only) versus genome size (expressed in numbers of open reading frames). (b) Two-dimensional ordination plot (nonmetric multidimensional scaling) of genomes based on counts of uptake-related Transporter Classification families and subfamilies per genome. Bray-Curtis distance metric; stress = 0.206. Contour lines indicate approximate genome size. Colors in both plots have the same legend and represent types of metabolism.
(iii) Heterotrophic carbon metabolism. Kentron genomes encoded both glycolysis (Embden-Meyerhoff-Parnas pathway) and the oxidative tricarboxylic acid (TCA) cycle. The canonically irreversible reactions of glycolysis, pyruvate kinase and phosphofructokinase, were replaced in Kentron by reversible, pyrophosphate-dependent alternatives pyruvate phosphate dikinase and PPi-dependent phosphofructokinase (PPi-PFK), respectively. These alternatives have been found in other thiotrophic symbioses, where they may make the CBB cycle more energy efficient (30).
Genes for pyruvate dehydrogenase and the complete oxidative TCA cycle were present, including 2-oxoglutarate dehydrogenase, which is often missing in obligate autotrophs (31). The reductive equivalents for the key steps of the oxidative TCA cycle were also present, namely, ferredoxin-dependent pyruvate synthase (PFOR), ferredoxin-dependent 2-oxoglutarate synthase, and fumarate reductase. However, because neither ATP citrate lyase (ACL) nor citryl coenzyme A (CoA) lyase/citryl-CoA synthase (CCL/CCS) was predicted, a canonical autotrophic reductive TCA cycle was not predicted.
Heterotrophic carboxylases also had relatively high expression levels. PFOR was present in multiple copies per genome, of which the highest expressed were at the 98.4 and 93.0 percentiles in Kentron H and SD, respectively (Fig. 4). GDP-dependent phosphoenolpyruvate (PEP) carboxykinase (PEPCK), which can replenish oxaloacetate anaplerotically, was also highly expressed (93.0 and 96.7 percentiles) (Fig. 4). Unlike PEP carboxylase, which was not predicted, PEPCK catalyzes a reversible reaction.
Pathways that enable growth solely on acetate as both energy and carbon source were either incomplete (glyoxylate shunt) or not predicted (ethylmalonyl-CoA pathway and methylaspartate cycle).
(iv) Partial 3-hydroxypropionate bi-cycle. Genes encoding most enzymes of the 3-hydroxypropionate bi-cycle (3HPB), which is the autotrophic pathway used by members of the distant bacterial phylum Chloroflexi, were predicted in Kentron. These genes had expression levels in the 64.1 to 83.0 and 48.2 to 96.4 percentile ranges for Kentron H and SD, respectively (Fig. 4). The key enzymes malonyl-CoA reductase and propionyl-CoA synthase were absent; hence, the bi-cycle was not closed and would not function autotrophically. However, the remainder of the pathway could function in the assimilation of organic substrates (e.g., acetate and succinate) or to connect metabolite pools (acetyl-CoA, propionyl-CoA, pyruvate, and glyoxylate) (32), as previously proposed for Chloroflexus (33) and the “Ca. Thiosymbion” symbionts of gutless oligochaetes (3).
These enzymes are unusual because their genes are uncommon and have a disjunct phylogenetic distribution: Chloroflexi, at least four clades in Gammaproteobacteria (Kentron, “Ca. Thiosymbion,” “Candidatus Competibacter,” “Pink Berry” Chromatiaceae), a symbiotic Alphaproteobacteria member (“Candidatus Riegeria”) (34), and Betaproteobacteria (“Candidatus Accumulibacter”). While they were previously thought to have been horizontally transferred from Chloroflexi to the other groups (33), gene phylogenies show that the Chloroflexi probably also gained the 3HPB by horizontal transfer (35), which was supported by our analysis with Kentron homologs (Fig. S6).
Phylogenetic trees of genes of 3-hydroxypropionate bi-cycle found in Kentron, with homologs from other bacteria. Abbreviations: Mch, mesaconyl-C1-CoA hydratase; Mcl, malyl-CoA/beta-methylmalyl-CoA/citramalyl-CoA (MMC) lyase; Mct, mesaconyl-CoA C1-C4 CoA transferase; Meh, mesaconyl-C4-CoA hydratase. Download FIG S6, EPS file, 0.5 MB (538.7KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(v) Storage compounds. In addition to elemental sulfur, Kentron also has the potential to store and mobilize carbon (as polyhydroxyalkanoates [PHAs] and starch/glycogen) and phosphorus (as polyphosphate). PHA was detected with the lipophilic fluorescent dye Nile Red, and polyphosphate was detected by metachromatic yellow fluorescence emission of DAPI (Fig. 1, inset). Genes related to PHA synthesis were among the most highly expressed, namely, those encoding phasin, a protein associated with the surface of PHA granules, and putative acetoacetyl-CoA reductase (phaB) (Fig. 4). Trehalose was detected in Kentrophoros sp. H but is probably produced and accumulated by the host ciliate rather than the symbionts (Text S2).
Metabolomics. Download Text S2, PDF file, 0.1 MB (74.6KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteomics-based carbon SIF of Kentron.
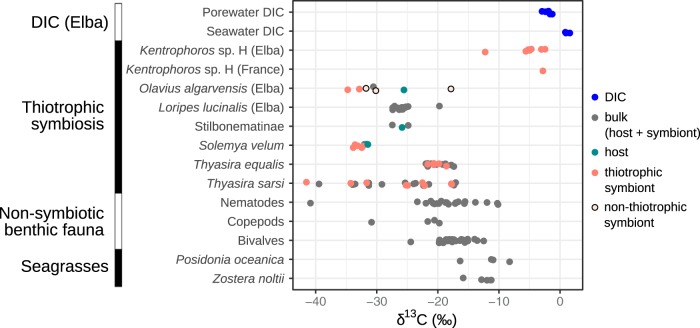
Measuring the natural abundance ratio of carbon stable isotopes 13C/12C, also known as the stable isotope fingerprint (SIF), is a challenge in Kentrophoros because of its small biomass (∼106 symbionts and ∼10 μg [wet weight] per ciliate in the largest species). Sensitive applications of isotope ratio mass spectrometry (IRMS) require at least ∼107 bacterial cells (36), and compound-specific IRMS for signatures of specific pathways like 13C enrichment in fatty acids in the rTCA cycle (11) requires considerably more. We therefore used a newly developed metaproteomics method that could distinguish the SIF of the symbiont from other biomass in the sample (37). The protein-based carbon SIF for Kentron sp. H from Elba and France ranged from −12.3 to −2.5‰ (n = 8), expressed as δ-13C values (Fig. 6; see also data posted at https://doi.org/10.5281/zenodo.2575800). In comparison, other shallow-water thiotrophic symbioses, from the same locality and elsewhere, possess the CBB cycle and have δ-13C values of <−17‰ and generally in the range of −30 to −20‰ (5, 37–40). The δ-13C of dissolved inorganic carbon (DIC) in pore water from Elba was between −2.99 and −1.32‰ (Fig. 6; see also data posted at https://doi.org/10.5281/zenodo.2575803).
FIG 6.
Carbon stable isotope δ-13C composition values in Kentrophoros sp. H (this study) and published values for other shallow-water thiotrophic symbioses (5, 37–40), nonsymbiotic benthic animals (5, 60, 61), and two Mediterranean seagrass species (60–63), compared with dissolved inorganic carbon (DIC) from pore water and seawater at Elba (this study). Values for Kentrophoros and Olavius algarvensis (except the “bulk” value) are from direct protein-SIF, and others are from isotope ratio mass spectrometry (IRMS). Values for symbiont-bearing tissue (e.g., gills) are also included under “symbiont.”
DISCUSSION
In this study, we have shown that the Kentron symbionts of Kentrophoros ciliates are unique among thiotrophic symbionts because they do not encode canonical pathways for autotrophic carbon fixation but have a variety of heterotrophic features. Their carbon stable isotope fingerprints are also substantially heavier than other thiotrophic symbioses from similar habitats. We therefore propose that Kentron bacteria are chemolithoheterotrophs (41), oxidizing inorganic compounds (in this case, reduced sulfur species) to provide energy for assimilating organic carbon as the main carbon source for growth.
Role of heterotrophic CO2 fixation.
Our results conflict with a previous study that concluded that Kentron is an autotroph, based on uptake of 14C-labeled bicarbonate, at a maximum rate of 0.117 bacterial cell carbon h−1 under oxic conditions (21). The CO2 fixation rates were lower than expected, but this was attributed to the artificial experimental conditions. To rule out the possibility that only some species are autotrophs, we collected Kentrophoros matching the described morphology from the same site (Nivå Bay, Denmark), but their symbionts (phylotype DK) lacked canonical autotrophic pathways like other Kentron phylotypes.
However, the ability to fix CO2 alone is insufficient evidence for autotrophy, which is defined as the ability to grow with inorganic carbon as the sole or major carbon source (42). Heterotrophs can also fix CO2 to some extent, e.g., via anaplerotic reactions in the oxidative TCA cycle (43). Such heterotrophic fixation can account theoretically for up to a third of total cell carbon (44) and has been measured at 10% or more in some bacteria (45, 46). The previous study (21) measured assimilation only of CO2, but not of total carbon, nor the growth rate of the cells. Instead, the authors assumed that all CO2 fixed was autotrophic and argued that the resulting calculated growth rate was low but plausible.
We reinterpret their results as heterotrophic CO2 fixation during the mobilization of the storage polymer polyhydroxyalkanoate (PHA) to biomass, which involves the heterotrophic carboxylation of acetyl-CoA to pyruvate by ferredoxin-dependent pyruvate synthase (PFOR). This is plausible because their experiments were performed with freshly collected cells that had visible cellular inclusions, and Kentron uses PHA as a carbon store (Fig. 1 and 3). In our data, genes for PFOR and another heterotrophic carboxylase, PEP carboxykinase (PEPCK), were highly expressed, suggesting that these enzymes are indeed present and functionally important (Fig. 4).
Furthermore, the previous study (21) also observed that adding sulfide or thiosulfate decreased the CO2 fixation rates, although sufficient electron acceptor was available. A lithoautotroph should instead fix more CO2 under such conditions, as observed in other thioautotrophic symbioses (47). This decrease is, however, consistent with our interpretation of heterotrophic CO2 fixation during PHA mobilization, because PHA conversion to biomass is a net oxidation reaction (see Text S3 in the supplemental material), which would not be favored under reducing conditions.
Metabolic reconstruction details. Download Text S3, PDF file, 0.1 MB (110.4KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Could Kentron use a novel autotrophic CO2 fixation pathway?
Different autotrophic carbon fixation pathways each have characteristic degrees of isotope fractionation discriminating against the heavier isotope 13C, resulting in biomass that is relatively depleted in 13C (i.e., more negative δ-13C values) (48). Kentron bacteria were more enriched in 13C than other shallow-water thiotrophic symbioses and showed only a modest 13C depletion relative to DIC from the same site (Fig. 6), which ruled out the possibility that they use a pathway with strong isotope fractionation (ε), such as the CBB cycle (ε = 10 to 22‰) or the reductive acetyl-CoA pathway (ε = 15 to 36‰) (49). Other pathways such as the reverse TCA cycle (ε = 4 to 13‰) or 3-hydroxypropionate bicycle (ε ≈ 0‰) may still fall in this range, but given that the key genes for these pathways were not detected, this possibility would require the postulation of hitherto-unknown enzymes.
In two different thermophilic bacteria, the oxidative TCA cycle has recently been found to function in the autotrophic direction without using ACL or CCL/CCS, but instead by reversing the citrate synthase reaction. Such a “reversed oxidative TCA” (roTCA) cycle would not be distinguishable from the oxidative TCA by genome sequences alone (50, 51). However, both roTCA bacteria require anoxic conditions with hydrogen as the energy source for autotrophic growth. They are also facultative autotrophs and switch to heterotrophic growth when suitable substrates like acetate are available. Citrate synthase is also highly expressed in the roTCA, whereas in Kentron the gene has only moderate expression (Fig. 4, 51.2 and 56.3 percentiles in Kentron H and SD, respectively). For these reasons, it is unlikely that a microaerophilic sulfur oxidizer like Kentron uses the roTCA for autotrophic growth.
Alternatively, it is possible to combine several reactions predicted in Kentron, including elements of the partial 3HPB, into a hypothetical autotrophic CO2 fixation pathway, without proposing any novel enzymes or biochemical reactions, with PFOR and PEPCK as carboxylases (Fig. 3; Text S3). Although it is stoichiometrically and energetically feasible, it is unlikely that this is a truly functional pathway, as the same enzymes are predicted in other bacteria, including “Ca. Thiosymbion,” a thiotrophic symbiont that has clear genetic (3) and isotopic (37) signatures of autotrophic CO2 fixation with the Calvin cycle (Text S3).
The autotrophy-heterotrophy spectrum in thiotrophic symbiosis.
Thiotrophic symbioses are most commonly found in nutrient-limited environments, and their symbionts are assumed to provide the hosts with nutrition through the autotrophic fixation of CO2. Indeed, the symbionts of deep-sea bivalves Bathymodiolus and Calyptogena show characteristic features of obligate autotrophy in their genomes, namely, an incomplete TCA cycle and the lack of organic uptake transporters (Table 1) (19, 52–54). This appears to be the exception, however, as other symbiont clades possess heterotrophic features to various degrees (Table 1). Some features, e.g., glycolysis, are involved in the mobilization of storage compounds, but abundant presence and expression of organic uptake transporters, as we observed in Kentron in this study, are a clearer marker of heterotrophic assimilation (55). Mixotrophic potential in other symbionts has been variously suggested to be a strategy to cope with carbon limitation by recycling host waste, as a nutritional supplement to autotrophy, or to be retained for a hypothetical free-living stage of the symbiont life cycle (3, 7, 30). Thus, we propose a spectrum among thiotrophic symbionts between obligate autotrophs and heterotrophic Kentron, with various degrees of mixotrophy in between.
TABLE 1.
Comparison of metabolic features predicted in thiotrophic symbiont genomesa
| Feature | Host habitat and organism |
||||||
|---|---|---|---|---|---|---|---|
| Hydrothermal vents and seeps |
Shallow water sediment interstitial |
||||||
| Bathy symbiont | Ruthia | Endoriftia | Thiodiazotropha | Solemya symbiont | Thiosymbion | Kentron | |
| Autotrophy | |||||||
| CBB cycle (RuBisCO, phosphoribulokinase) | + | + | + | + | + | + | |
| PPi-phosphofructokinase | + | + | + | + | + | + | + |
| rTCA (citrate cleavage) | + | ||||||
| Diazotrophy, nitrogenase | + | (+) | |||||
| TCA cycle | |||||||
| Oxidative TCA | + | + | + | + | + | ||
| Kor and Frd (reductive TCA) | + | + | + | + | |||
| Glyoxylate shunt | + | + | + | ||||
| Central metabolism | |||||||
| Pyruvate phosphate dikinase | + | + | + | + | + | ||
| PEP synthase | + | + | + | + | |||
| Pyruvate synthase (PFOR) | + | + | + | + | + | ||
| Pyruvate carboxylase | + | + | |||||
| PEP carboxylase | + | + | |||||
| PEP carboxykinase (GTP) | + | + | + | ||||
| Malic enzyme | + | + | + | + | + | + | (+) |
| C5 reactions of p3HPB | + | + | |||||
| Energy | |||||||
| Rnf transporter | + | + | + | + | + | + | + |
| V-type ATPase | + | + | + | ||||
| Cyt c oxidase cbb3 type | + | + | + | + | + | + | + |
| Cyt c oxidase aa3 type | + | + | + | + | + | ||
| Storage compounds | |||||||
| Glycogen | + | + | + | + | + | ||
| Polyhydroxyalkanoates | + | + | + | + | |||
| Polyphosphate synthesis | + | + | + | + | |||
Symbols: +, present; (+), not in all genomes. Abbreviations: Bathy, Bathymodiolus; CBB, Calvin-Benson-Bassham cycle; Cyt c, cytochrome c; Frd, fumarate reductase; Kor, 2-oxoglutarate:ferredoxin oxidoreductase; PEP, phosphoenolpyruvate; PPi, pyrophosphate; rTCA, reverse TCA cycle.
Lithoheterotrophy is not uncommon for free-living thiotrophs and appears to be more prevalent among those that have the Sox pathway (i.e., thiosulfate oxidizers) than those with the rDsr/Sox pathway (i.e., thiotrophs that can store and oxidize elemental sulfur) (Text S3). Moreover, some free-living thiotrophs that possess the CBB cycle may nonetheless grow only when supplied with organic substrates, e.g., freshwater Beggiatoa (56). Functional heterotrophy may therefore be underestimated as it is not necessarily apparent from genomic predictions.
Host biology constrains the feasibility of autotrophy for a thiotrophic symbiont. To meet nutritional requirements by chemoautotrophy alone, the host must provide high O2 flux to its symbionts, beyond what it requires itself (57). This is metabolically demanding, and it is telling that the bathymodioline and vesicomyid bivalves, whose symbionts have the most autotrophic features, are relatively large animals with intracellular symbionts that are located in their gill tissues, which can better maintain ventilation and homeostasis than smaller hosts that have extracellular symbionts. Specialization for high autotrophic production rates is also seen in the preconcentration of CO2 by the bivalve Bathymodiolus azoricus for its symbionts and in its thiotrophic symbiont’s metabolic dependence on the animal to replenish TCA cycle intermediates (58).
Meiofaunal hosts like Kentrophoros and stilbonematine nematodes, in contrast, are much smaller, cannot span substrate gradients, and must be able to tolerate fluctuating anoxia. Their smaller sizes (100-μm to 1-mm range) also mean that they are less constrained by diffusion limitation in nutrient uptake. Given that shallow-water coastal environments also receive more organic input, for example, from land or from seagrass beds, than deep-sea hydrothermal environments, it is not surprising that the shallow-water meiofaunal symbioses have more heterotrophic features than the deep-sea ones (Table 1).
Ecophysiological model of the Kentrophoros symbiosis.
Based on our results and previous descriptions of morphology and behavior in Kentrophoros and other symbioses, we propose the following model for the ecophysiology of this symbiosis.
Kentrophoros fuels its growth by the phagocytosis and digestion of its symbionts (22). There has to be a net input of energy and organic carbon from environmental sources for the overall growth of the host-symbiont system, and heterotrophic carboxylation may also be a substantial carbon source. To give its symbionts access to these substrates, Kentrophoros likely shuttles between oxic and anoxic zones in marine sediment, like other motile, sediment-dwelling hosts with thiotrophic symbionts (38). In anoxic sediment, both the predicted energy and carbon sources, namely, sulfide and organic acids, are produced by microbial activity (59). Many organic acids, such as acetate and succinate, are more oxidized than average biomass (see data posted at https://doi.org/10.5281/zenodo.2575777), and the electron donor to assimilate them would be sulfide. However, if this were to occur under anoxic conditions, sulfide would not be completely oxidized to sulfate. Complete oxidation of sulfide to sulfate requires oxygen, so elemental sulfur would be the intermediate, partly oxidized form accumulated when Kentron assimilates organic acids. The synthesis of PHA from small organic acids like acetate can also function as both an additional electron sink for sulfide oxidation and a carbon store. Hydrolysis of polyphosphate and mobilization of glycogen are also potential sources of energy in the absence of oxygen.
Under oxic conditions, elemental sulfur can be further oxidized to sulfate for energy, and PHA can be mobilized for biosynthesis. Glycogen and polyphosphate reserves can also be regenerated. The various storage inclusions in Kentron, namely, elemental sulfur, PHA, glycogen, and polyphosphate, hence represent pools of energy, reducing equivalents, and carbon that function as metabolic buffers for the symbiont living in a fluctuating environment.
The symbionts may also bring a syntrophic benefit to their hosts under anoxic conditions, when the ciliates can yield energy only by fermentation. By assimilating fermentation waste products and keeping their concentrations low in their host, the symbionts can improve the energy yields for their hosts and allow them to better tolerate periods of anoxia. This could also be a form of resource recycling under carbon-limited conditions, which has been proposed for other thiotrophic symbionts with the potential to assimilate organic acids (3, 7).
Kentron is relatively enriched in 13C compared to nonsymbiotic shallow-water benthic fauna, such as nematodes and bivalves (δ-13C ∼ −20 to −10‰) (5, 60, 61), and to the seagrasses (δ-13C ∼ −15 to −10‰) (60–63) that are the main primary producers in the habitat of Kentrophoros (Fig. 6). The higher values in Kentron could be partly caused by preferring specific substrates with higher 13C content, such as acetate, which has a wide range of δ-13C (−2.8 to −20.7‰) in marine pore waters depending on the dominant microbial processes at the site (64). Given how close the δ-13C of Kentron is to DIC, it is possible that heterotrophic CO2 fixation contributes to this 13C signature, but the isotope fractionation values of the heterotrophic carboxylases have not been characterized, to our knowledge. Repeated internal recycling of host waste products, as we postulate, could also cause accumulation of 13C in the host-symbiont system.
Our metabolic model has parallels to free-living thiotrophs and to heterotrophic bacteria involved in enhanced biological phosphorus removal (EBPR) from wastewater, which also use storage inclusions like polyphosphate, glycogen, and PHA as metabolic buffers for fluctuating oxygen and nutrient conditions (65–68).
Conclusion.
We have shown that a diverse and widespread clade of symbiotic sulfur bacteria lacks genes encoding canonical enzymes for autotrophic CO2 fixation, despite being a food source for their hosts. This is unlike all other thiotrophic symbionts sequenced to date, which possess the CBB or rTCA cycles for autotrophy. We propose a lithoheterotrophic model for the Kentrophoros nutritional symbiosis, which challenges the chemoautotrophic paradigm usually applied to thiotrophic symbiosis. Uptake of organic substrates from the environment, heterotrophic carboxylation, and recycling of host waste may play a bigger part in thiotrophic symbioses than previously thought. Our results suggest that nutritional symbioses can also be supported by chemolithoheterotrophy and that thiotrophic symbioses fall on a spectrum between autotrophy and heterotrophy. We speculate that heterotrophy is feasible for Kentron because its hosts are relatively small and flat and have the highest known symbiont-host biomass ratio among thiotrophs; larger hosts will experience more diffusion limitation and can fulfill only a smaller fraction of their carbon needs heterotrophically.
MATERIALS AND METHODS
Sample collection.
Specimens of Kentrophoros were collected in 2013 and 2014 from Elba, Italy (Mediterranean Sea); in 2015 from Twin Cayes, Belize (Caribbean Sea); and in 2016 from Nivå Bay, Denmark (Øresund Strait between Baltic and North Seas) (see data at https://doi.org/10.5281/zenodo.2575755), as previously described (20).
Microscopy.
Live Kentrophoros sp. H was macerated to release bacteria and fixed with 4% (wt/vol) formaldehyde in seawater on glass slides. Cells were rinsed and stained with DAPI (1 μg/ml) and Nile Red (2.5 μg/ml) for 20 min, rinsed, and imaged under epifluorescence with a long-pass DAPI and fluorescein isothiocyanate (FITC)/enhanced green fluorescent protein (EGFP) long-pass filter cube, respectively, and a Canon EOS 700D camera.
DNA/RNA extraction and sequencing.
Samples for DNA and RNA extraction, comprising single ciliate cells and their symbionts, were fixed in RNAlater (Ambion) and stored at 4°C. Before DNA extraction, samples were centrifuged (8,000 × g, 5 min), and excess RNAlater was removed by pipetting. DNA was extracted with the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's instructions and eluted in 50 μl of buffer AE. DNA concentration was measured fluorometrically with the Qubit DNA high-sensitivity kit (Life Technologies). Each DNA sample was screened by PCR with eukaryotic 18S rRNA primers EukA/EukB (69) followed by capillary sequencing to identify the Kentrophoros phylotype, as previously described (20). Libraries for metagenomic sequencing were prepared with the Ovation Ultralow Library System V2 kit (NuGEN) according to the manufacturer's protocol. Libraries were sequenced as either 100- or 150-bp paired-end reads on the Illumina HiSeq 2500 platform.
RNA was extracted with the RNeasy Plus Micro kit (Qiagen) according to the manufacturer's protocol and eluted in 15 μl RNase-free water. cDNA was synthesized with the Ovation RNASeq System v2 (NuGEN) according to the manufacturer's protocol, sheared to 350-bp target size with the Covaris microTUBE system, and cleaned up with the Zymo Genomic DNA Clean & Concentrator kit. The sequencing library was prepared from cDNA with the NEBNext Ultra DNA library preparation kit for Illumina and sequenced on the Illumina HiSeq 2500 platform as 100-bp single-end reads.
Assembly, binning, and annotation of symbiont genomes.
Reads were trimmed from both ends to remove fragments matching TruSeq adapters and to remove bases with Phred quality scores of <2, using either Nesoni v0.111 (https://github.com/Victorian-Bioinformatics-Consortium/nesoni) or BBmap v34+ (https://sourceforge.net/projects/bbmap/). Trimmed reads were error corrected with BayesHammer (70). Error-corrected reads were initially assembled with IDBA-UD v1.1.1 (71) or SPAdes v3.5.0+ (70). The reference coverage per contig was obtained by mapping the error-corrected read set against the assembly with BBmap (“fast” mode). Conserved marker genes in the assembly were identified and taxonomically classified with Amphora2 (72) or Phyla-Amphora (73). 16S rRNA genes were identified with Barrnap v0.5 (https://github.com/tseemann/barrnap) and classified by searching against the Silva SSU-Ref NR 119 database (74) with Usearch v8.1.1831 (75). Differential coverage information (76) was obtained by mapping reads from other samples of the same host morphospecies onto the assembly with BBmap. Contigs belonging to the primary Kentrophoros symbiont (the “primary symbiont bin”) were heuristically identified by a combination of differential coverage, assembly graph connectivity, GC%, affiliation of conserved marker genes, and affiliation of 16S rRNA sequence using gbtools v2.5.2 (77). Reads mapping to the primary symbiont bin were reassembled with SPAdes. Binning and reassembly of the primary symbiont genome were iteratively repeated for each metagenome sample until the primary symbiont bin appeared to contain only a single genome, based on the number and taxonomic affiliation of conserved marker genes and 16S rRNA. For final genome bins, summary statistics were computed with Quast v4.4 (78), and completeness and contamination were estimated with CheckM v1.0.11 (79) using the Gammaproteobacteria taxonomy workflow. Average amino acid identity (AAI) and average nucleotide identity (ANI) values between genomes were calculated with CompareM v0.0.21 (https://github.com/dparks1134/CompareM) and jSpecies v1.2.1, respectively (80).
Genome bins were annotated with the IMG/M pipeline for downstream analyses (81). Metabolic pathways were predicted from the annotated proteins with the PathoLogic module (82) of Pathway Tools v20.5 (83), followed by manual curation. Metabolic modules from KEGG (84) were also predicted with the KEGG Mapper tool (http://www.kegg.jp/kegg/mapper.html, accessed January 2017) from KEGG Orthology terms in the IMG annotation.
Transcriptome analysis.
Metatranscriptome reads for Kentrophoros sp. H and SD were mapped onto symbiont genome assemblies from the respective species (IMG genome IDs 2609459750 and 2615840505) using BBmap (minimum identity, 0.97). Read counts per genomic feature were calculated with featureCounts v1.5.2 (85) and transformed into FPKM values (fragments per kilobase pair reference per million reads mapped).
Verifying absence of key genes for autotrophic pathways.
Key enzymes that are diagnostic for known autotrophic pathways were identified from the literature (48, 86–88) (see data posted at https://doi.org/10.5281/zenodo.2575773). These were absent from Kentron genome annotations, with the exception of a RuBisCO-like protein (RLP) in Kentron sp. H (see below). To verify that the absence of autotrophy-related sequences was not caused by incomplete genome bins, misprediction of open reading frames, or misassembly of the reads, we aligned raw reads from host-symbiont metagenomes and metatranscriptomes against the UniProt Swiss-Prot database (release 2017_01) (89) using diamond blastx (v0.8.34.96, “sensitive” mode) (90). Sequences for certain key enzymes were absent from Swiss-Prot, so representative sequences from UniProtKB were manually added to the database (see data posted at https://doi.org/10.5281/zenodo.2575773 and File 4 at https://doi.org/10.5281/zenodo.2555833). Reads with hits to target enzymes (identified by EC number or from the list of additional sequences) were counted, extracted, and mapped against the initial metagenomic assembly for the corresponding library. Raw counts of reads were transformed to FPKM values using three times the mean amino acid length of the target proteins as the reference length. As a comparison, FPKM values were also calculated for a reference set of enzymes of the TCA cycle and partial 3HPB pathway (see data posted at https://doi.org/10.5281/zenodo.2575773), which were expected to be present in all Kentron genomes.
Identification of transporter genes for substrate uptake.
Families and subfamilies of transporter proteins from the Transporter Classification Database (TCDB, accessed 2 February 2017) (91) that were described as energy-dependent uptake transporters for organic substrates were shortlisted (see data posted at https://doi.org/10.5281/zenodo.2575779). Translated open reading frames (ORFs) for Kentron and selected genomes of other symbiotic and free-living basal Gammaproteobacteria (see data posted at https://doi.org/10.5281/zenodo.2575781) were aligned with BLASTP (92) (best-scoring hit with E value of <10−5, >30% amino acid sequence identity, and >70% coverage of reference sequence, parameters from reference 55) against TCDB. As TCDB also includes nonmembrane proteins that are involved in transport (e.g., the ATPase subunit of ABC transporters), we also counted how many hits contained transmembrane domains, predicted with tmhmm v2.0c (93). To compare the transporter content between genomes, the tabulated counts of organic substrate uptake TC family hits per genome were analyzed by nonmetric multidimensional scaling (NMDS) with the metaMDS function in the R package vegan v2.5.1 (https://CRAN.R-project.org/package=vegan) (Bray-Curtis distance, 2 dimensions, 2,000 runs).
Protein extraction and peptide preparation.
Samples of Kentrophoros sp. H for proteomics were collected by decantation from sediment adjacent to seagrass meadows at Sant’ Andrea, Isola d’Elba, Italy on 3 June 2014 and from Pampelonne Beach, Provence-Alpes-Côte d’Azur, France, in July 2018. Ciliates were individually fixed in RNAlater and subsequently stored at 4°C and then at −80°C. One individual Kentrophoros sp. H specimen and nine pooled samples of four or five individuals each (see data posted at https://doi.org/10.5281/zenodo.2575800) were used to prepare tryptic digests following the filter-aided sample preparation (FASP) protocol (94) with minor modifications (95). Samples were lysed in 30 μl of SDT-lysis buffer (4% [wt/vol] SDS, 100 mM Tris-HCl pH 7.6, 0.1 M DTT) by heating to 95°C for 10 min. To avoid sample losses, we did not clear the lysate by centrifugation after lysis. Instead, we loaded the whole lysate on to the 10-kDa filter units used for the FASP procedure. The Qubit protein assay kit (Thermo Fisher Scientific, Life Technologies) was used to determine peptide concentrations, according to the manufacturer’s instructions. Peptide concentrations were below the detection limit in all samples.
1D-LC-MS/MS.
All peptide samples were analyzed by 1D-LC-MS/MS as previously described (96), with the modification that a 75-cm analytical column was used. Briefly, an UltiMate 3000 RSLCnano liquid chromatograph (Thermo Fisher Scientific) was used to load peptides with loading solvent A (2% acetonitrile, 0.05% trifluoroacetic acid) onto a 5-mm, 300-μm-internal-diameter (i.d.) C18 Acclaim PepMap100 precolumn (Thermo Fisher Scientific). Since peptide concentrations were very low, complete peptide samples (80 μl) were loaded onto the precolumn. Peptides were eluted from the precolumn onto a 75-cm by 75-μm analytical EASY-Spray column packed with PepMap RSLC C18, 2-μm material (Thermo Fisher Scientific) heated to 60°C. Separation of peptides on the analytical column was achieved at a flow rate of 225 nl min−1 using a 460-min gradient going from 98% buffer A (0.1% formic acid) to 31% buffer B (0.08% formic acid, 80% acetonitrile) in 363 min and then to 50% B in 70 min and to 99% B in 1 min and ending with 26 min 99% B. Eluting peptides were analyzed in a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Carryover was reduced by running two wash runs (injection of 20 μl acetonitrile) between samples. Data acquisition in the Q Exactive Plus was done as previously described (5).
Protein identification and quantification.
Protein sequences predicted from Kentron genomes described above were clustered at 98% identity with CD-HIT v4.7 (97). Representative sequences, proteins predicted from a preliminary host transcriptome, and the cRAP protein sequence database (http://www.thegpm.org/crap/) of common lab contaminants were used as a database for protein identification, which contained 5,715 protein sequences. For protein identification, MS/MS spectra were searched against this database using the Sequest HT node in Proteome Discoverer version 2.2 (Thermo Fisher Scientific) as previously described (5).
Direct protein-SIF.
Stable carbon isotope fingerprints (SIFs = δ-13C values) for Kentron symbiosis were determined from proteomic data (37). Briefly, human hair with a known δ-13C value was used as reference material to correct for instrument fractionation. A tryptic digest of the reference material was prepared as described above and analyzed with the same 1D-LC-MS/MS method as the samples. The peptide-spectrum match (PSM) files generated by Proteome Discoverer were exported in tab-delimited text format. 1D-LC-MS/MS raw files were converted to mzML format using the MSConvertGUI available in the ProteoWizard tool suite (98). Only the MS1 spectra were retained in the mzML files, and the spectra were converted to centroided data by vendor algorithm peak picking. The PSM and mzML files were used as input for the Calis-p software (https://sourceforge.net/projects/calis-p/) to extract peptide isotope distributions and to compute the direct protein-SIF δ-13C value for Kentron and the human hair reference material (37). Direct protein-SIF δ-13C values were corrected for instrument fragmentation by applying the offset between the direct protein-SIF δ-13C value and known δ-13C value of the reference material. We obtained between 50 and 499 peptides with sufficient intensity for direct protein-SIF from seven of the nine pooled samples (see data posted at https://doi.org/10.5281/zenodo.2575800). These samples were thus well above the necessary number of peptides needed to obtain an accurate estimate. Due to the low biomass of the individual Kentrophoros specimen (∼10 μg), only 14 peptides with sufficient intensity were obtained for this sample.
Dissolved inorganic carbon δ-13C.
Seawater and pore water samples were collected from the vicinity of seagrass meadows at Sant’ Andrea, Elba, Italy, in July 2017 to determine the δ-13C of dissolved inorganic carbon (DIC). Seawater was sampled at the surface from a boat, whereas pore water was sampled at 15-cm sediment depth with a steel lance. Samples were drawn into 20-ml plastic syringes; 6 ml of each was fixed with 100 μl of 300 mM ZnCl2 and stored at 4°C until processing. δ-13C was measured with a Finnigan MAT 252 gas isotope ratio mass spectrometer with Gasbench II (Thermo Scientific), using Solnhofen limestone as a standard and 8 technical replicates per sample.
Code availability.
Scripts used to screen for autotrophy-related genes in metagenome libraries, to classify transporter families, and to calculate phylogenetic trees are available at https://github.com/kbseah/mapfunc, https://github.com/kbseah/tcdbparse_sqlite, and https://github.com/kbseah/phylogenomics-tools, respectively.
Data availability.
Annotated genomes are in the Joint Genome Institute GOLD database (https://gold.jgi.doe.gov/) study Gs0114545. Metagenomes and metatranscriptomes are in European Nucleotide Archive studies PRJEB25374 and PRJEB25540. The mass spectrometry metaproteomics are in ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) data set PXD011616.
ACKNOWLEDGMENTS
We thank our fieldwork hosts: the HYDRA Institute on Elba, especially Miriam Weber, Hannah Kuhfuß, and Matthias Schneider; the Smithsonian CCRE program and staff at the Carrie Bow Caye field station; and Lasse Riemann and the Marine Biological Section, University of Copenhagen. Mario Schimak, Oliver Jäckle, Juliane Wippler, Judith Zimmermann, Miriam Sadowski, Silke Wetzel, Nikolaus Leisch, and Anne-Christin Kreutzmann assisted in sample collection. Library preparation and sequencing were performed at the Max Planck Genome Centre Cologne. We thank Marc Strous for access to proteomics equipment. We thank Dolma Michellod and Alexander Gruhl for collecting DIC samples and Henning Kuhnert and the MARUM Stable Isotope Laboratory team for DIC IRMS measurements. We also thank Roland Dieterich for computational support; Caitlin Petro, Erik Puskas, Tora Gulstad, and Frantisek Fojt for mass spectrometry support; and Lizbeth Sayavedra, Oliver Müller-Cajar, Monika Bright, Jörg Ott, Verena Carvalho, Cameron Callbeck, Marc Mußmann, and members of the Symbiosis Department for useful discussions and comments.
The proteomics and direct protein-SIF work was supported by the Campus Alberta Innovation Chair Program, the Canadian Foundation for Innovation, and a discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (above to Marc Strous) and the NC State Chancellor’s Faculty Excellence Program Cluster on Microbiomes and Complex Microbial Communities (M.K.). Financial support was provided by the Max Planck Society; the Humboldt Foundation to C.P.A.; a Marie Curie Fellowship to H.R.G.-V.; the Gordon and Betty Moore Foundation Marine Microbial Initiative Investigator Award to N.D. (grant GBMF3811); SFB 987 “Microbial Diversity in Environmental Signal Response” to L.S.V.B. and T.J.E.; and FET-Open grant 686330 (Future Agriculture) to J.Z.
B.K.B.S., N.D., and H.R.G.-V. designed the study. B.K.B.S. and H.R.G.-V. performed fieldwork. B.H. prepared sequencing libraries with B.K.B.S. and coordinated sequencing. M.L. performed metabolomics mass spectrometry analyses. A.K. prepared samples and generated data for proteomics. M.K. and A.K. processed and analyzed proteomics data. M.K. performed protein-SIF analysis. B.K.B.S., C.P.A., J.Z., L.S.V.B., T.J.E., M.L., and H.R.G.-V. analyzed genomics and transcriptomics data. B.K.B.S. wrote the manuscript draft. All authors participated in revising the manuscript.
Footnotes
Citation Seah BKB, Antony CP, Huettel B, Zarzycki J, Schada von Borzyskowski L, Erb TJ, Kouris A, Kleiner M, Liebeke M, Dubilier N, Gruber-Vodicka HR. 2019. Sulfur-oxidizing symbionts without canonical genes for autotrophic CO2 fixation. mBio 10:e01112-19. https://doi.org/10.1128/mBio.01112-19.
REFERENCES
- 1.Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 2.Stewart FJ, Newton ILG, Cavanaugh CM. 2005. Chemosynthetic endosymbioses: adaptations to oxic–anoxic interfaces. Trends Microbiol 13:439–448. doi: 10.1016/j.tim.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J, Chang Y-J, Shah M, VerBerkmoes NC, Zarzycki J, Fuchs G, Markert S, Hempel K, Voigt B, Becher D, Liebeke M, Lalk M, Albrecht D, Hecker M, Schweder T, Dubilier N. 2012. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci U S A 109:E1173–E1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.König S, Gros O, Heiden SE, Hinzke T, Thürmer A, Poehlein A, Meyer S, Vatin M, Mbéguié-A-Mbéguié D, Tocny J, Ponnudurai R, Daniel R, Becher D, Schweder T, Markert S. 2016. Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat Microbiol 2:16193. doi: 10.1038/nmicrobiol.2016.193. [DOI] [PubMed] [Google Scholar]
- 5.Petersen JM, Kemper A, Gruber-Vodicka H, Cardini U, van der Geest M, Kleiner M, Bulgheresi S, Mußmann M, Herbold C, Seah BKB, Antony CP, Liu D, Belitz A, Weber M. 2016. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat Microbiol 2:16195. doi: 10.1038/nmicrobiol.2016.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Distel DL, Altamia MA, Lin Z, Shipway JR, Han A, Forteza I, Antemano R, Limbaco M, Tebo AG, Dechavez R, Albano J, Rosenberg G, Concepcion GP, Schmidt EW, Haygood MG. 2017. Discovery of chemoautotrophic symbiosis in the giant shipworm Kuphus polythalamia (Bivalvia: Teredinidae) extends wooden-steps theory. Proc Natl Acad Sci U S A 114:E3652–E3658. doi: 10.1073/pnas.1620470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dmytrenko O, Russell SL, Loo WT, Fontanez KM, Liao L, Roeselers G, Sharma R, Stewart FJ, Newton IL, Woyke T, Wu D, Lang J, Eisen JA, Cavanaugh CM. 2014. The genome of the intracellular bacterium of the coastal bivalve, Solemya velum: a blueprint for thriving in and out of symbiosis. BMC Genomics 15:924. doi: 10.1186/1471-2164-15-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber-Vodicka HR, Dirks U, Leisch N, Baranyi C, Stoecker K, Bulgheresi S, Heindl NR, Horn M, Lott C, Loy A, Wagner M, Ott J. 2011. Paracatenula, an ancient symbiosis between thiotrophic Alphaproteobacteria and catenulid flatworms. Proc Natl Acad Sci U S A 108:12078–12083. doi: 10.1073/pnas.1105347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinke C, Schmitz-Esser S, Loy A, Horn M, Wagner M, Bright M. 2009. High genetic similarity between two geographically distinct strains of the sulfur-oxidizing symbiont ‘Candidatus Thiobios zoothamnicoli.’ FEMS Microbiol Ecol 67:229–241. doi: 10.1111/j.1574-6941.2008.00628.x. [DOI] [PubMed] [Google Scholar]
- 10.Assié A, Leisch N, Meier DV, Gruber-Vodicka H, Tegetmeyer HE, Meyerdirks A, Kleiner M, Hinzke T, Joye S, Saxton M, Dubilier N, Petersen JM. 2018. Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria). bioRxiv doi: 10.1101/437616. [DOI] [PMC free article] [PubMed]
- 11.Suzuki Y, Sasaki T, Suzuki M, Nogi Y, Miwa T, Takai K, Nealson KH, Horikoshi K. 2005. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl Environ Microbiol 71:5440–5450. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell BJ, Stein JL, Cary SC. 2003. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana, a hydrothermal vent polychaete. Appl Environ Microbiol 69:5070–5078. doi: 10.1128/aem.69.9.5070-5078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, Gartman A, Luther GW, Seewald JS, Fisher CR, Girguis PR. 2012. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc Natl Acad Sci U S A 109:E3241–E3250. doi: 10.1073/pnas.1202690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hügler M, Albrecht D, Robidart J, Bench S, Feldman RA, Hecker M, Schweder T. 2007. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315:247–250. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 15.Thiel V, Hügler M, Blümel M, Baumann HI, Gärtner A, Schmaljohann R, Strauss H, Garbe-Schönberg D, Petersen S, Cowart DA, Fisher CR, Imhoff JF. 2012. Widespread occurrence of two carbon fixation pathways in tubeworm endosymbionts: lessons from hydrothermal vent associated tubeworms from the Mediterranean Sea. Front Microbiol 3:423. doi: 10.3389/fmicb.2012.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin-Blum M, Dubilier N, Kleiner M. 2019. Genetic evidence for two carbon fixation pathways (the Calvin-Benson-Bassham cycle and the reverse tricarboxylic acid cycle) in symbiotic and free-living bacteria. mSphere 4:e00394-18. doi: 10.1128/mSphere.00394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponsard J, Cambon-Bonavita M-A, Zbinden M, Lepoint G, Joassin A, Corbari L, Shillito B, Durand L, Cueff-Gauchard V, Compère P. 2013. Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host-shrimp Rimicaris exoculata. ISME J 7:96–109. doi: 10.1038/ismej.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleiner M, Wentrup C, Holler T, Lavik G, Harder J, Lott C, Littmann S, Kuypers MMM, Dubilier N. 2015. Use of carbon monoxide and hydrogen by a bacteria-animal symbiosis from seagrass sediments. Environ Microbiol 17:5023–5035. doi: 10.1111/1462-2920.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, Hourdez S, Girguis PR, Wankel SD, Barbe V, Pelletier E, Fink D, Borowski C, Bach W, Dubilier N. 2011. Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 20.Seah BKB, Schwaha T, Volland J-M, Huettel B, Dubilier N, Gruber-Vodicka HR. 2017. Specificity in diversity: single origin of a widespread ciliate-bacteria symbiosis. Proc Biol Sci 284:20170764. doi: 10.1098/rspb.2017.0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenchel T, Finlay BJ. 1989. Kentrophoros: a mouthless ciliate with a symbiotic kitchen garden. Ophelia 30:75–93. [Google Scholar]
- 22.Raikov IB. 1971. Bactéries épizoiques et mode de nutrition du cilié psammophile Kentrophoros fistulosum Fauré-Fremiet (étude au microscope électronique). Protistologica 7:365–378. [Google Scholar]
- 23.Foissner W. 1995. Kentrophoros (Ciliophora, Karyorelictea) has oral vestiges: a reinvestigation of K. fistulosus (Fauré-Fremiet, 1950) using protargol impregnation. Arch Für Protistenkd 146:165–179. doi: 10.1016/S0003-9365(11)80107-7. [DOI] [Google Scholar]
- 24.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahl C, Friedrich CG. 2008. Microbial sulfur metabolism. Springer, Berlin, Germany. [Google Scholar]
- 26.Ghosh W, Dam B. 2009. Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev 33:999–1043. doi: 10.1111/j.1574-6976.2009.00187.x. [DOI] [PubMed] [Google Scholar]
- 27.Pitcher RS, Watmough NJ. 2004. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta 1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Ducluzeau A-L, Ouchane S, Nitschke W. 2008. The cbb3 oxidases are an ancient innovation of the domain Bacteria. Mol Biol Evol 25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 29.Sparacino-Watkins C, Stolz JF, Basu P. 2014. Nitrate and periplasmic nitrate reductases. Chem Soc Rev 43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleiner M, Petersen JM, Dubilier N. 2012. Convergent and divergent evolution of metabolism in sulfur-oxidizing symbionts and the role of horizontal gene transfer. Curr Opin Microbiol 15:621–631. doi: 10.1016/j.mib.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Wood AP, Aurikko JP, Kelly DP. 2004. A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol Rev 28:335–352. doi: 10.1016/j.femsre.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs G, Berg IA. 2014. Unfamiliar metabolic links in the central carbon metabolism. J Biotechnol 192:314–322. doi: 10.1016/j.jbiotec.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Zarzycki J, Fuchs G. 2011. Coassimilation of organic substrates via the autotrophic 3-hydroxypropionate bi-cycle in Chloroflexus aurantiacus. Appl Environ Microbiol 77:6181–6188. doi: 10.1128/AEM.00705-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jäckle O, Seah BKB, Tietjen M, Leisch N, Liebeke M, Kleiner M, Berg JS, Gruber-Vodicka HR. 2019. Chemosynthetic symbiont with a drastically reduced genome serves as primary energy storage in the marine flatworm Paracatenula. Proc Natl Acad Sci U S A 116:8505–8514. doi: 10.1073/pnas.1818995116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shih PM, Ward LM, Fischer WW. 2017. Evolution of the 3-hydroxypropionate bicycle and recent transfer of anoxygenic photosynthesis into the Chloroflexi. Proc Natl Acad Sci U S A 114:10749–10754. doi: 10.1073/pnas.1710798114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eek KM, Sessions AL, Lies DP. 2007. Carbon-isotopic analysis of microbial cells sorted by flow cytometry. Geobiology 5:85–95. doi: 10.1111/j.1472-4669.2006.00094.x. [DOI] [PubMed] [Google Scholar]
- 37.Kleiner M, Dong X, Hinzke T, Wippler J, Thorson E, Mayer B, Strous M. 2018. Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities. Proc Natl Acad Sci U S A 115:E5576–E5584. doi: 10.1073/pnas.1722325115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott JA, Novak R, Schiemer F, Hentschel U, Nebelsick M, Polz M. 1991. Tackling the sulfide gradient: a novel strategy involving marine nematodes and chemoautotrophic ectosymbionts. Mar Ecol 12:261–279. doi: 10.1111/j.1439-0485.1991.tb00258.x. [DOI] [Google Scholar]
- 39.Conway N, Capuzzo JM, Fry B. 1989. The role of endosymbiotic bacteria in the nutrition of Solemya velum: evidence from a stable isotope analysis of endosymbionts and host. Limnol Oceanogr 34:249–255. doi: 10.4319/lo.1989.34.1.0249. [DOI] [Google Scholar]
- 40.Dando PR, Spiro B. 1993. Varying nutritional dependence of the thyasirid bivalves Thyasira sarsi and T. equalis on chemoautotrophic symbiotic bacteria, demonstrated by isotope ratios of tissue carbon and shell carbonate. Mar Ecol Prog Ser 92:151. doi: 10.3354/meps092151. [DOI] [Google Scholar]
- 41.Boden R, Hutt LP. 2018. Chemolithoheterotrophy: means to higher growth yields from this widespread metabolic trait, p 1–25. In Rojo F. (ed), Aerobic utilization of hydrocarbons, oils and lipids. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 42.Kelly DP, Wood AP. 2013. The chemolithotrophic prokaryotes, p 275–287. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 43.Owen OE, Kalhan SC, Hanson RW. 2002. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 44.Schönheit P, Buckel W, Martin WF. 2016. On the origin of heterotrophy. Trends Microbiol 24:12–25. doi: 10.1016/j.tim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Perez RC, Matin A. 1982. Carbon dioxide assimilation by Thiobacillus novellus under nutrient-limited mixotrophic conditions. J Bacteriol 150:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roslev P, Larsen MB, Jørgensen D, Hesselsoe M. 2004. Use of heterotrophic CO2 assimilation as a measure of metabolic activity in planktonic and sessile bacteria. J Microbiol Methods 59:381–393. doi: 10.1016/j.mimet.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Volland J-M, Schintlmeister A, Zambalos H, Reipert S, Mozetič P, Espada-Hinojosa S, Turk V, Wagner M, Bright M. 2018. NanoSIMS and tissue autoradiography reveal symbiont carbon fixation and organic carbon transfer to giant ciliate host. ISME J 12:714–727. doi: 10.1038/s41396-018-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hügler M, Sievert SM. 2011. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu Rev Mar Sci 3:261–289. doi: 10.1146/annurev-marine-120709-142712. [DOI] [PubMed] [Google Scholar]
- 49.Hayes JM. 2001. Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev Mineral Geochem 43:225–277. doi: 10.2138/gsrmg.43.1.225. [DOI] [Google Scholar]
- 50.Nunoura T, Chikaraishi Y, Izaki R, Suwa T, Sato T, Harada T, Mori K, Kato Y, Miyazaki M, Shimamura S, Yanagawa K, Shuto A, Ohkouchi N, Fujita N, Takaki Y, Atomi H, Takai K. 2018. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science 359:559–563. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 51.Mall A, Sobotta J, Huber C, Tschirner C, Kowarschik S, Bačnik K, Mergelsberg M, Boll M, Hügler M, Eisenreich W, Berg IA. 2018. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359:563–567. doi: 10.1126/science.aao2410. [DOI] [PubMed] [Google Scholar]
- 52.Kuwahara H, Yoshida T, Takaki Y, Shimamura S, Nishi S, Harada M, Matsuyama K, Takishita K, Kawato M, Uematsu K, Fujiwara Y, Sato T, Kato C, Kitagawa M, Kato I, Maruyama T. 2007. Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr Biol 17:881–886. doi: 10.1016/j.cub.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 53.Newton ILG, Woyke T, Auchtung TA, Dilly GF, Dutton RJ, Fisher MC, Fontanez KM, Lau E, Stewart FJ, Richardson PM, Barry KW, Saunders E, Detter JC, Wu D, Eisen JA, Cavanaugh CM. 2007. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315:998–1000. doi: 10.1126/science.1138438. [DOI] [PubMed] [Google Scholar]
- 54.Sayavedra L, Kleiner M, Ponnudurai R, Wetzel S, Pelletier E, Barbe V, Satoh N, Shoguchi E, Fink D, Breusing C, Reusch TB, Rosenstiel P, Schilhabel MB, Becher D, Schweder T, Markert S, Dubilier N, Petersen JM. 2015. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. Elife 4:e07966. doi: 10.7554/eLife.07966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yelton AP, Acinas SG, Sunagawa S, Bork P, Pedrós-Alió C, Chisholm SW. 2016. Global genetic capacity for mixotrophy in marine picocyanobacteria. ISME J 10:2946–2957. doi: 10.1038/ismej.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nelson DC, Williams CA, Farah BA, Shively JM. 1988. Occurrence and regulation of Calvin cycle enzymes in non-autotrophic Beggiatoa strains. Arch Microbiol 151:15–19. doi: 10.1007/BF00444662. [DOI] [Google Scholar]
- 57.Childress JJ, Girguis PR. 2011. The metabolic demands of endosymbiotic chemoautotrophic metabolism on host physiological capacities. J Exp Biol 214:312–325. doi: 10.1242/jeb.049023. [DOI] [PubMed] [Google Scholar]
- 58.Ponnudurai R, Kleiner M, Sayavedra L, Petersen JM, Moche M, Otto A, Becher D, Takeuchi T, Satoh N, Dubilier N, Schweder T, Markert S. 2017. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J 11:463–477. doi: 10.1038/ismej.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jørgensen BB. 2000. Bacteria and marine biogeochemistry, p 173–207. In Schulz HD, Zabel M (ed), Marine geochemistry, 2nd ed Springer, Berlin, Germany. [Google Scholar]
- 60.Vafeiadou A-M, Materatski P, Adão H, De Troch M, Moens T. 2014. Resource utilization and trophic position of nematodes and harpacticoid copepods in and adjacent to Zostera noltii beds. Biogeosciences 11:4001–4014. doi: 10.5194/bg-11-4001-2014. [DOI] [Google Scholar]
- 61.Vizzini S, Sara G, Michener RH, Mazzola A. 2002. The role and contribution of the seagrass Posidonia oceanica (L.) Delile organic matter for secondary consumers as revealed by carbon and nitrogen stable isotope analysis. Acta Oecol 23:277–285. doi: 10.1016/S1146-609X(02)01156-6. [DOI] [Google Scholar]
- 62.Cooper LW, DeNiro MJ. 1989. Stable carbon isotope variability in the seagrass Posidonia oceanica: evidence for light intensity effects. Mar Ecol Prog Ser 50:225–229. doi: 10.3354/meps050225. [DOI] [Google Scholar]
- 63.McMillan C, Parker PL, Fry B. 1980. 13C/12C ratios in seagrasses. Aquat Bot 9:237–249. doi: 10.1016/0304-3770(80)90025-X. [DOI] [Google Scholar]
- 64.Heuer V, Elvert M, Tille S, Krummen M, Mollar XP, Hmelo LR, Hinrichs K-U. 2006. Online δ13C analysis of volatile fatty acids in sediment/porewater systems by liquid chromatography-isotope ratio mass spectrometry. Limnol Oceanogr Methods 4:346–357. doi: 10.4319/lom.2006.4.346. [DOI] [Google Scholar]
- 65.Høgslund S, Revsbech NP, Kuenen JG, Jørgensen BB, Gallardo VA, van de Vossenberg J, Nielsen JL, Holmkvist L, Arning ET, Nielsen LP. 2009. Physiology and behaviour of marine Thioploca. ISME J 3:647–657. doi: 10.1038/ismej.2009.17. [DOI] [PubMed] [Google Scholar]
- 66.Winkel M, Salman-Carvalho V, Woyke T, Richter M, Schulz-Vogt HN, Flood BE, Bailey JV, Mußmann M. 2016. Single-cell sequencing of Thiomargarita reveals genomic flexibility for adaptation to dynamic redox conditions. Front Microbiol 7:964. doi: 10.3389/fmicb.2016.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seviour RJ, McIlroy S. 2008. The microbiology of phosphorus removal in activated sludge processes—the current state of play. J Microbiol 46:115–124. doi: 10.1007/s12275-008-0051-0. [DOI] [PubMed] [Google Scholar]
- 68.Schulz HN, Schulz HD. 2005. Large sulfur bacteria and the formation of phosphorite. Science 307:416–418. doi: 10.1126/science.1103096. [DOI] [PubMed] [Google Scholar]
- 69.Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 70.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng Y, Leung HCM, Yiu SM, Chin F. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 72.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 73.Wang Z, Wu M. 2013. A phylum-level bacterial phylogenetic marker database. Mol Biol Evol 30:1258–1262. doi: 10.1093/molbev/mst059. [DOI] [PubMed] [Google Scholar]
- 74.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 76.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 77.Seah BKB, Gruber-Vodicka HR. 2015. gbtools: interactive visualization of metagenome bins in R. Front Microbiol 6:1451. doi: 10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Markowitz VM, Chen I-M, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, Huntemann M, Billis K, Varghese N, Tennessen K, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC. 2014. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karp PD, Latendresse M, Caspi R. 2011. The Pathway Tools pathway prediction algorithm. Stand Genomic Sci 5:424–429. doi: 10.4056/sigs.1794338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karp PD, Latendresse M, Paley SM, Krummenacker M, Ong QD, Billington R, Kothari A, Weaver D, Lee T, Subhraveti P, Spaulding A, Fulcher C, Keseler IM, Caspi R. 2016. Pathway Tools version 19.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform 17:877–890. doi: 10.1093/bib/bbv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 86.Bar-Even A, Noor E, Milo R. 2012. A survey of carbon fixation pathways through a quantitative lens. J Exp Bot 63:2325–2342. doi: 10.1093/jxb/err417. [DOI] [PubMed] [Google Scholar]
- 87.Berg IA. 2011. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fuchs G. 2011. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life?. Annu Rev Microbiol 65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 89.Magrane M, UniProt Consortium. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 91.Saier MH, Reddy VS, Tamang DG, Vastermark A. 2014. The Transporter Classification Database. Nucleic Acids Res 42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Krogh A, Larsson B, von Heijne G, Sonnhammer E. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 94.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. 2009. Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 95.Hamann E, Gruber-Vodicka H, Kleiner M, Tegetmeyer HE, Riedel D, Littmann S, Chen J, Milucka J, Viehweger B, Becker KW, Dong X, Stairs CW, Hinrichs K-U, Brown MW, Roger AJ, Strous M. 2016. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature 534:254–258. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kleiner M, Thorson E, Sharp CE, Dong X, Liu D, Li C, Strous M. 2017. Assessing species biomass contributions in microbial communities via metaproteomics. Nat Commun 8:1558. doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetics. Download Text S1, PDF file, 0.1 MB (83.3KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary statistics for Kentron genome assemblies. (A) Genome completeness per species, as assessed with gammaproteobacterial conserved marker genes by CheckM. (B) Number of contigs versus total length (Mbp) of Kentron genome bins; plot symbol areas scaled by N50 (kbp). Larger genomes are usually more fragmented (more contigs, lower N50), with the exception of Kentron sp. DK. (C) Accumulation curves for pan- and core genomes (protein-encoding genes only) of the Kentron clade, with uncertainty estimated by resampling (200 times, with replacement). “Singleton” genes were not included in an ortholog cluster. Download FIG S1, EPS file, 0.1 MB (104.6KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maximum-likelihood phylogeny of RuBisCO proteins, including RuBisCO-like protein from Kentron phylotype H (red arrow). Download FIG S2, EPS file, 0.5 MB (493KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selected results of remapping reads with hits to canonical autotrophy enzymes to the metagenomic assemblies, for AcsB versus Kentrophoros sp. TC (left) and RuBisCO versus Kentrophoros sp. H (right). Metagenome assemblies are depicted as coverage-GC% plots, where each gray circle represents a contig, with its size scaled to the contig length. Contigs with remapping hits have solid outlines, with numbers of remapping reads indicated when >10. The majority of reads with alignments to canonical autotrophy enzymes map to scaffolds outside the Kentron symbiont genome bin (circled in blue) and could be attributed to other bacteria in the metagenomes, except for RuBisCO in Kentron sp. H, due to the RuBisCO-like protein encoded in that phylotype. Download FIG S3, TIF file, 0.3 MB (348.3KB, tif) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Read coverage in Kentrophoros metatranscriptomes for key enzymes of autotrophic CO2 fixation pathways, expressed as FPKM values (left) and as a fraction of the median coverage of a reference set of proteins (right). Each point represents a separate metatranscriptome library, colored by Kentrophoros host morphospecies. Abbreviations as in Fig. 3. Download FIG S4, EPS file, 0.1 MB (149.1KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Uptake-related transporter families found in all Kentron genomes. (a) Number of open reading frames per genome with matches to Transporter Classification families (Blastp E value <10−5, >30% amino acid identity, >70% query coverage). (b) Mean expression (FPKM) in three transcriptomes of Kentron H. (c) Mean expression (FPKM) in three transcriptomes of Kentron SD. Download FIG S5, TIF file, 0.3 MB (271KB, tif) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phylogenetic trees of genes of 3-hydroxypropionate bi-cycle found in Kentron, with homologs from other bacteria. Abbreviations: Mch, mesaconyl-C1-CoA hydratase; Mcl, malyl-CoA/beta-methylmalyl-CoA/citramalyl-CoA (MMC) lyase; Mct, mesaconyl-CoA C1-C4 CoA transferase; Meh, mesaconyl-C4-CoA hydratase. Download FIG S6, EPS file, 0.5 MB (538.7KB, eps) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolomics. Download Text S2, PDF file, 0.1 MB (74.6KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic reconstruction details. Download Text S3, PDF file, 0.1 MB (110.4KB, pdf) .
Copyright © 2019 Seah et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Annotated genomes are in the Joint Genome Institute GOLD database (https://gold.jgi.doe.gov/) study Gs0114545. Metagenomes and metatranscriptomes are in European Nucleotide Archive studies PRJEB25374 and PRJEB25540. The mass spectrometry metaproteomics are in ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) data set PXD011616.