An adequate in vivo analysis of HCMV’s viral chemokine vCXCL-1 has been lacking. Here we generate recombinant MCMVs expressing vCXCL-1 to study vCXCL-1 function in vivo using MCMV as a surrogate. We demonstrate that vCXCL-1 increases MCMV dissemination kinetics for both primary and secondary dissemination. Additionally, we provide evidence, that the murine neutrophil is largely a bystander in the mouse’s response to vCXCL-1. We confirm the hypothesis that vCXCL-1 is a HCMV virulence factor. Infection of severely immunocompromised mice with MCMVs expressing vCXCL-1 was lethal in more than 50% of infected animals, while all animals infected with parental virus survived during a 12-day period. This work provides needed insights into vCXCL-1 function in vivo.
KEYWORDS: betaherpesvirus, neutrophils, vCXCL-1, viral chemokines, cytomegalovirus, MCMV
ABSTRACT
Human cytomegalovirus (HCMV) is a betaherpesvirus that is a significant pathogen within newborn and immunocompromised populations. Morbidity associated with HCMV infection is the consequence of viral dissemination. HCMV has evolved to manipulate the host immune system to enhance viral dissemination and ensure long-term survival within the host. The immunomodulatory protein vCXCL-1, a viral chemokine functioning primarily through the CXCR2 chemokine receptor, is hypothesized to attract CXCR2+ neutrophils to infection sites, aiding viral dissemination. Neutrophils harbor HCMV in vivo; however, the interaction between vCXCL-1 and the neutrophil has not been evaluated in vivo. Using the mouse model and mouse cytomegalovirus (MCMV) infection, we show that murine neutrophils harbor and transfer infectious MCMV and that virus replication initiates within this cell type. Utilizing recombinant MCMVs expressing vCXCL-1 from the HCMV strain (Toledo), we demonstrated that vCXCL-1 significantly enhances MCMV dissemination kinetics. Through cellular depletion experiments, we observe that neutrophils impact dissemination but that overall dissemination is largely neutrophil independent. This work adds neutrophils to the list of innate cells (i.e., dendritic and macrophages/monocytes) that contribute to MCMV dissemination but refutes the hypothesis that neutrophils are the primary cell responding to vCXCL-1.
INTRODUCTION
Human cytomegalovirus (HCMV) is a serious pathogen in immunocompromised populations (1, 2) and the leading cause of infectious congenital disease (3, 4). Following in utero infection, fetal abnormalities such as microcephaly or other sequelae (e.g., progressive deafness and learning disabilities) can occur (5, 6). Primary infection or latent viral reactivation in immunocompromised adults (7, 8), such as cancer therapy patients, organ transplant recipients, or HIV/AIDS patients, can cause gastroenteritis, retinitis, or organ transplant rejection (2, 9). Regardless of the host, disease due to viral infection results from viral dissemination (10). Interestingly, HCMV has evolved numerous immunomodulatory proteins that blunt normal protective immune responses, restructure inflammatory environments, and ensure long-term survival in the host (11–13). The viral chemokine, vCXCL-1, is an HCMV protein that preferentially recruits CXCR2+ neutrophils over other innate immune cells in vitro (14–16).
In addition to engaging CXCR2 (14, 15), vCXCL-1 may also signal through human CXCR1 and CX3CR1 (15, 16). CX3CR1+/CXCR1+ natural killer (NK) cells functionally respond to vCXCL-1 albeit at a significantly reduced level compared to CXCR2+/CXCR1+ neutrophils (16). Unfortunately, any in vivo evaluation of the interaction of vCXCL-1 with the immune system and its contribution to CMV pathogenesis has been complicated by the species specificity of CMV (17–19).
Mouse cytomegalovirus (MCMV) infection of mice is frequently used to study CMV dissemination. MCMV has similar pathogenesis to HCMV, contains many homologues and orthologues to HCMV genes, and disseminates via innate immune cells (20, 21). MCMV does not encode vCXCL-1 but rather encodes a C-C chemokine, MCK2, that enhances dissemination (22–24). We have previously expressed vCXCL-1 from chimpanzee CMV in MCMV and observed no salivary gland dissemination (25, 26), potentially due to inappropriate timing and expression levels of the vCXCL-1 insert. Here, we engineered human vCXCL-1 from the HCMV Toledo strain (vCXCL-1Tol) with a 2A peptide linked to the MCMV chemokine MCK2 to ensure appropriate timing and expression levels. We employed the MCMV bacmid of the Smith strain (pSM3fr-MCK2-2fl) in which MCK2 aids viral entry in addition to its role in dissemination (24). Because of MCK2's dual role, we chose to fuse vCXCL-1Tol to MCK2 instead of deleting it from the recombinant virus. We demonstrate that murine neutrophils are capable of harboring, transferring, and initiating MCMV replication and that CXCR2 stimulation is sufficient to alter MCMV dissemination kinetics. Additionally, we show that infections with recombinant MCMVs expressing vCXCL-1Tol exhibit increased viral dissemination and virulence.
RESULTS
Murine neutrophils harbor, transfer, and initiate MCMV replication.
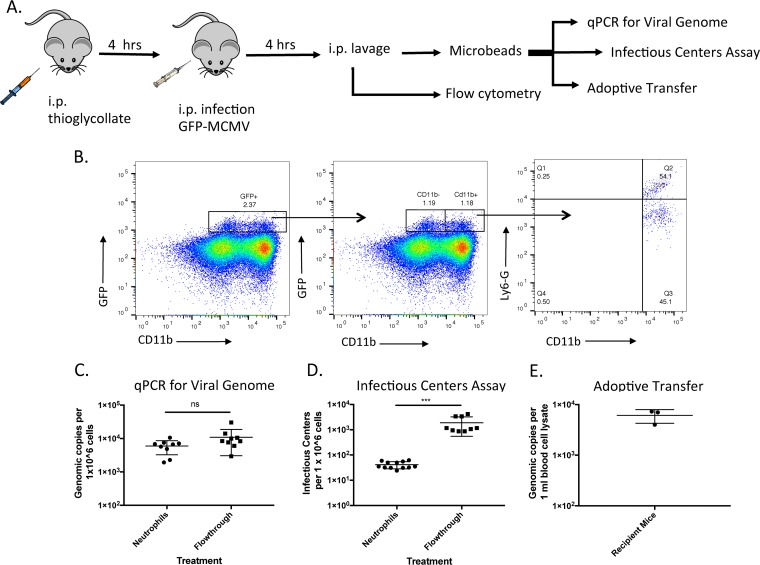
The capacity of neutrophils to impact cytomegalovirus dissemination is an area of contention. In blood of immunosuppressed patients, neutrophils harbor the largest viral burden (27, 28) and transfer infectious HCMV ex vivo and in vitro, but they cannot support productive viral replication (29, 30). The interaction between murine neutrophils and MCMV has not been evaluated. Here we use a thioglycolate inflammation model (31) to study the relationship between MCMV and murine neutrophils (Fig. 1A). Mice were infected with an MCMV encoding green fluorescent protein (GFP) under the CMV immediate early (IE) promoter (i.e., 4503) (22), and 4 h postinfection (Fig. 1B), total peritoneal exudate was harvested and analyzed by flow cytometry. Peritoneal exudate cells (PECs) in which virus has entered, uncoated, and expressed IE genes were expected to become GFP positive (GFP+). All live GFP+ cells were initially gated and further analyzed for the presence of neutrophil (Ly6G) and myeloid/granulocyte (CD11b) markers (Fig. 1B). Approximately 2% of all PECs were GFP+, with roughly half this population being CD11b+. Further analysis of the CD11b+ subpopulation indicated that ∼55% of these cells were Ly6G+ CD11b+ neutrophils and ∼45% were Ly6G− CD11b+, likely dendritic cells or patrolling (resident) monocytes (32). The GFP+ CD11b− PECs could be susceptible cell types such as epithelial and fibroblastic cells but not T or B cells (33). The gating strategy used to obtain these results is outlined (see Fig. S1B in the supplemental material). To ensure that the GFP signal is from cells that were infected and expressing GFP and not due to passive uptake or endocytosis of GFP+ virions, we infected cultured fibroblasts and used the translational inhibitor cycloheximide (34) to demonstrate that GFP expression is de novo and not due to passive uptake of GFP contamination in the viral preparation (Fig. S2).
FIG 1.
Neutrophils harbor and transfer MCMV ex vivo and in vivo. (A) Experimental design. (B) PECs were evaluated for the presence of GFP-expressing virus. Live GFP-expressing cells were gated for CD11b+ cells followed by Ly6-G+ gating. (C) Total DNA was extracted from 1 × 106 purified neutrophils or Flowthrough cells. qPCR was performed to evaluate the number of MCMV genomes present. (D) Infectious center assay for determining the number of infected neutrophils. A total of 1 × 106 purified neutrophils were incubated with an uninfected fibroblast monolayer for 5 days, and the number of plaques was evaluated. (E) MCMV genomes were quantified from whole blood isolated from mice that were adoptively transferred 1 × 106 neutrophils from infected mice. Values are average titers ± standard deviations (SD) (error bars). Statistical significance was determined by Student’s t test and indicated as follows: ns, not significant; ***, P ≤ 0.001.
Gating strategy used to identify GFP-positive cells. (A) Neutrophil (Ly6G+, CD11b+) purity following microbead isolation was evaluated with flow cytometry. Data are representative of two experiments with six or seven mice per experiment. Live cells were gated. (B) Following identification of live cells, single cells were analyzed for GFP expression. Gates were set on unstained infected peritoneal exudate cells. Download FIG S1, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
De novo GFP expression. MEF 10.1 cells were infected with RM4503 at an MOI of 3 at 4 hours postinfection. GFP expression was determined via flow cytometry. (A to C) Uninfected cells (A), infected cells treated with 50 μg/ml of cycloheximide (CHX) for 1 hour prior to infection and 4 hours postinfection (B), or infected but untreated cells (vehicle control) (C). Download FIG S2, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We further evaluated the contribution of neutrophils to MCMV infection. Using anti-Ly6G microbeads, a 97% pure neutrophil population was prepared from peritoneal exudate from thioglycolate-treated, MCMV-infected mice (Fig. S1A). This population included approximately 25% of the total GFP+ cells as described above. The remaining population of peritoneal exudate cells (designated Flowthrough) represented ∼75% of all the GFP+ cells. When both populations were assayed for viral genome via quantitative PCR (qPCR) (Fig. 1C), we did not observe a significant difference in viral genome content between the two populations, indicating that neutrophils and other cell populations harbor MCMV genome 4 h postinfection (p.i.). As neutrophils are phagocytes and may be in the process of destroying virions, we carried out an infectious center assay. Consistent with this notion, the Ly6G+ population had dramatically less infectious MCMV compared with Flowthrough (Fig. 1D). To determine whether this neutrophil population was capable of infecting a new host, isolated neutrophils were adoptively transferred into uninfected immunocompetent mice. Three days after neutrophil transfer, blood was harvested and assayed for MCMV genome via qPCR. Transfer of infected neutrophils was sufficient to infect a new host (Fig. 1E), although it is possible that a contaminating cell population could contribute this infectivity as well.
CXCR2 stimulation alters normal MCMV dissemination kinetics.
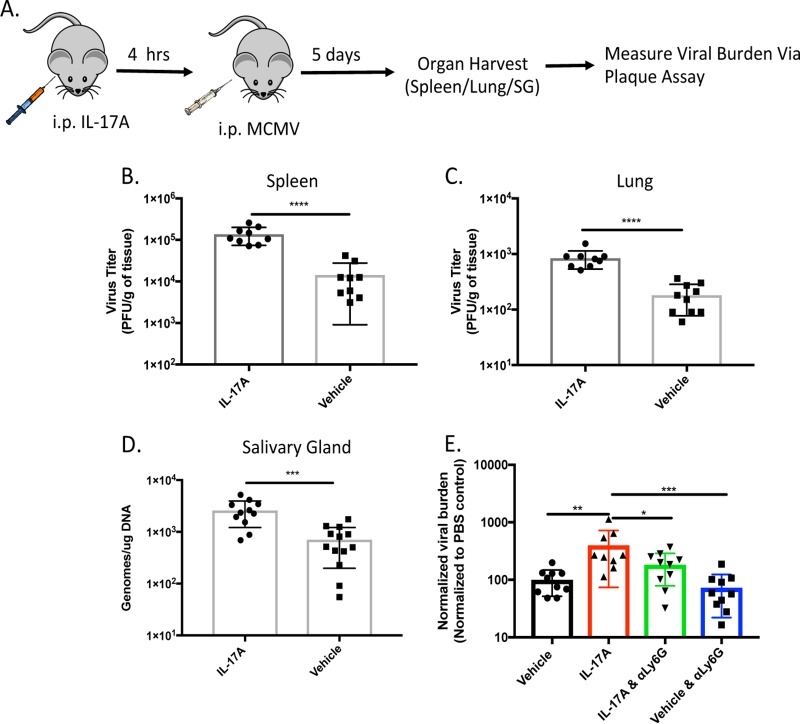
Because vCXCL-1Tol functions primarily through CXCR2 (14, 15, 35), we sought to understand whether a targeted CXCR2 response could alter normal MCMV dissemination kinetics. When mice are injected intraperitoneally (i.p.) with interleukin 17A (IL-17A), mesothelial cells release host CXCL-1 (i.e., Groα) eliciting a neutrophilic, CXCR2-mediated response (36). We confirmed reports that IL-17A treatment alters only the neutrophil population in the peritoneum 4 h following treatment (data not shown). We exploited this experimental setup to mimic the viral chemokine’s function in vivo and evaluate the contribution of the vCXCL-1/CXCR2 signaling axis on viral dissemination (Fig. 2A). Mice treated with IL-17A prior to i.p. infection with MCMV exhibited significantly greater viral burden in spleen and lungs 5 days postinfection (dpi) compared to control animals treated with vehicle control (Fig. 2B and C). IL-17A-treated animals also had greater levels of salivary gland (SG)-associated virus seeding 5 dpi (Fig. 2D). Although there was faster seeding, IL-17A-treated animals did not exhibit a significant difference in viral titers within this organ 14 dpi compared to control animals (data not shown).
FIG 2.
CXCR2 stimulation increases MCMV dissemination. (A) MCMV was administered i.p. during peak neutrophil response induced with IL-17A or vehicle control (not treated). (B and C) Viral titers of whole-organ homogenates were determined in the spleen (B) and lungs (C) of mice treated with IL-17A 5 days postinfection. n = 3 with 4 or 5 mice per group. (D) The salivary glands (SGs) of mice were harvested 5 days postinfection (dpi). Total DNA was harvested and subjected to qPCR for total MCMV burden in the SG. (E) Depletion of neutrophils reduces IL-17A-enhanced MCMV dissemination to the spleen. Plaque assays of spleens of mice 5 dpi. that received phosphate-buffered saline (PBS; i.e., vehicle), IL-17A alone, neutrophil-depleting antibody (anti-Ly6G [αLy6G]) coupled with IL-17A treatment (IL-17A & αLy6G), or neutrophil-depleting antibody alone prior to infection with MCMV. Data are from two pooled experiments. Data are normalized to the mean PFU/gram of tissue of the PBS control for each experiment. Values are average titers ± SD. Statistical significance was determined by Student’s t test or one-way ANOVA with Tukey’s multiple comparison of means and indicated as follows: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Neutrophils were depleted 2 days prior to IL-17A treatment and MCMV infection, then treated every other day after infection with anti-Ly6G or vehicle control (Fig. S3A). The titers of the virus in the spleens were determined 5 dpi (Fig. 2E). Neutrophil depletion resulted in a decreased viral burden in the spleens of IL17A-treated mice, suggesting that in this inflammatory milieu, neutrophils play a role in dissemination perhaps in addition to normal lymphatic drainage from the peritoneum (37).
Confirmation of neutrophil depletion. Neutrophil depletion was evaluated by flow cytometry. (A) Four hours after IL-17A treatment, mice were euthanized, and PECs were harvested. PECs were assayed for the presence of neutrophils (Ly6G+, CD11b+). (B) Whole blood was assayed for the presence of neutrophils (Ly6G+, CD11b+) from neutrophil-depleted or vehicle control-treated mice prior to infection with recombinant MCMVs. Data are representative of two experiments with three animals per group. Download FIG S3, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of recombinant MCMV expressing vCXCL-1Tol.
We have previously generated recombinant MCMVs that overexpress vCXCL-1 or host CXCL-1. These viruses disseminated normally to the spleen, liver, and lung but were defective in SG dissemination (25), leading to a hypothesis that levels and/or timing of chemokine expression hindered SG dissemination. Therefore, we engineered a recombinant MCMV in which vCXCL-1 from the HCMV Toledo strain (vCXCL-1Tol) was expressed as a cleavable fusion protein with the MCMV chemokine, MCK2. A picornavirus 2A self-cleaving peptide enabled the coexpression of the two chemokines (38) (Fig. 3A). The recombinant MCMV was generated by coupling classical bacterial recombineering with bacterial artificial chromosome from the Smith strain (pSM3fr-MCK2-2fl) (39) and CRISPR/Cas9 technology. HindIII RFLP analysis confirmed galK insertion into pSM3fr-MCK2-2fl, resulting in a 1.5-kb size increase of the 7.1-kb band into the top of the triplet band (Fig. 3B, boxed in red). The 500-bp addition of vCXCL-1Tol into the 7.1-kb band resulted in the creation of a doublet in the middle band of the HindIII triplet (Fig. 3B). The supernatants of pSM3fr-MCK2-2fl-infected MEF 10.1 cells were subjected to Western blot analysis using anti-FLAG antibodies to identify MCK2 and anti-HIS antibodies to identify vCXCL-1Tol (Fig. 3C). Glycosylated MCK2 was detected at ∼40 kDa (40, 41) and vCXCL1 was detected at ∼17 kDa (26). These data indicate the self-cleaving 2A-peptide efficiently generated two independent proteins. In order to evaluate whether insertion of vCXCL-1Tol alters replication in vitro, both single-step and multistep growth curves were conducted (Fig. 3D and E). These analyses demonstrate that insertion of the vCXCL-1Tol gene does not alter viral replication in vitro.
FIG 3.
Generation of recombinant MCMV expressing vCXCL-1Tol. (A) Bacterial artificial chromosome (BAC) schematic of vCXCL-1Tol insertion into the mck2 locus. (B) HindIII RFLP analysis of the BACs. The triplet banding patterns are boxed in red. (C) Western blot analysis of the viral supernatant of MCK2-2A-vCXCL-1Tol MCMV. (D) Single-step growth curve (MOI of 5.0). DPI, day postinfection. (E) Multistep growth curves (MOI of 0.05). Values are average titers ± SD (error bars).
vCXCL-1Tol expression alters normal MCMV dissemination kinetics.
To test the hypothesis that vCXCL-1Tol alters normal dissemination, we infected BALB/c mice with pSM3fr-MCK2-2fl expressing vCXCL-1Tol (called RMvCXCL-1Tol) compared to parental pSM3fr-MCK2-2fl (Smith) and a rescued virus (called RMvCXCL-1Tol RQ). RQ has had the MCK2-vCXCL1Tol fusion replaced with the pSM3fr-MCK2-2fl mck2 locus and serves as a control for adventitious mutations that could have arisen during recombineering. In addition, whole-genome sequencing was performed on recombinant MCMVs without revealing any unexpected mutations (data not shown). Even though mice inoculated in the footpad (FP) with recombinant MCMVs did not exhibit any difference in viral burden at the site of inoculation 3 dpi (Fig. 4A), RMvCXCL-1Tol levels were ∼100-fold higher in the popliteal lymph node at this time compared to either pSM3fr-MCK2-2fl or RMvCXCL-1Tol RQ (Fig. 4B). Therefore, the addition of vCXCL-1Tol to MCMV does not alter replication at the infection site, but it does increase the levels of virus in the draining lymph node (LN). Importantly, vCXCL-1Tol expression dramatically increased secondary dissemination to SG. Plaque assays of whole-organ homogenates revealed ∼100-fold increased viral levels in SG at 14 dpi compared to either wild-type (WT) or RQ viruses (Fig. 4C). These data indicate that vCXCL-1Tol significantly increases both primary and secondary dissemination kinetics of MCK2-repaired pSM3fr bacmid-derived virus.
FIG 4.
Infection of mice with vCXCL-1Tol-expressing MCMVs increases primary and secondary dissemination. Mice were infected by injecting 1 × 106 PFU of either virus Smith, RMvCXCL-1Tol, or RMvCXCL-1Tol RQ into their footpad (FP). (A to C) Plaque assays were performed on FP (A), lymph nodes (LN) (B), and salivary glands (SG) (C) that were harvested 3 (FP and LN) and 14 (SG) dpi. n = 3 to 5 experiments with at least three mice per group. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison of the means. (D and E) Mice were infected either intranasally (i.n.) (D) or intraperitoneally (i.p.) (E), and plaque assays were performed on SG harvested 14 dpi. Results from two independent experiments with three or four mice per group are presented. Values are average titers ± SD (error bars). Statistical significance was determined by Student’s t test and indicated as follows: ns, not significant; **, P ≤ 0.01; ****, P ≤ 0.0001.
It has been reported that different routes of inoculation produce different dissemination patterns (32). To evaluate this possibility with our recombinant viruses, mice were inoculated intranasally (i.n.) (Fig. 4D) or i.p. (Fig. 4E) with RMvCXCL-1Tol or Smith, and SGs were harvested for plaque assays at 14 dpi. Regardless of the inoculation route, expression of vCXCL-1Tol enhances MCMV SG dissemination and/or replication.
Analysis of cellular subsets responding to vCXCL-1Tol in vivo early during infection.
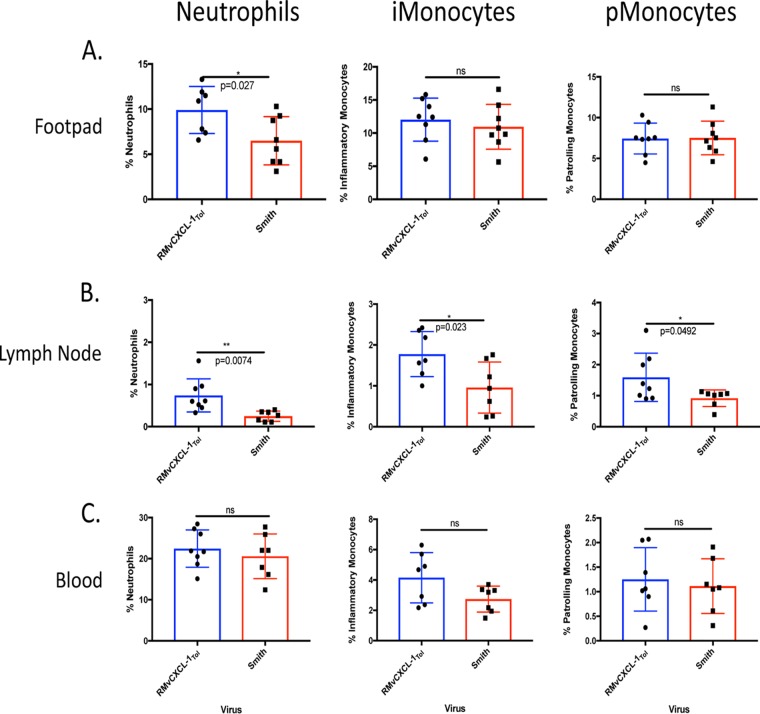
Because vCXCL-1Tol significantly increased dissemination independent of the inoculation site, we evaluated leukocytes recruited to the inoculation site, draining LN, and bloodstream following a FP infection early during infection to identify inflammatory cells potentially contributing to increased dissemination. Mice were infected as in Fig. 4, and FP, LN, and blood samples from infected animals were analyzed by flow cytometry at 3 dpi. In the FP (Fig. 5A), expression of vCXCL-1Tol induced a significantly higher neutrophil influx compared to control virus, without altering the levels of either inflammatory monocytes (Ly6G− Ly6Chi CD11b+ CD11c−) or patrolling monocytes (Ly6G− Ly6Clo CD11b+ CD11c+) (32). Within the draining LN, all three leukocyte populations were significantly increased by vCXCL-1Tol compared to control (Fig. 5B), while blood showed no difference (Fig. 5C). There was no difference in total leukocyte influx into the LN and blood (Fig. S4). These data suggest that vCXCL-1 recruitment of neutrophils to the infection site and draining LN or monocytes in LN could be responsible for increased dissemination to SG.
FIG 5.
vCXCL-1Tol alters cellular infiltrate in primary dissemination organs. Mice were infected via the footpads as in Fig. 4. (A to C) Footpads (A), lymph nodes (B), and peripheral blood (C) were harvested 3 dpi and evaluated by flow cytometry. Cells were characterized as follows: neutrophils, Ly6G+ CD11b+; inflammatory monocytes (iMonocytes), Ly6G− Ly6Chi CD11b+ CD11c−; patrolling monocytes (pMonocytes), Ly6G− Ly6Clo CD11b+ CD11c+. Values are average titers ± SD (error bars). Statistical significance was determined by Student’s t test and indicated as follows: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01.
Total cell counts in LN and blood. At 3 days postinfection, LN (A) and blood (B) were harvested, and total leukocytes were counted. Blood was harvested via cardiac puncture and placed in lithium heparin-treated tubes. ACK lysis was performed to remove red blood cells, and leukocytes were counted. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Removal of specific cellular subsets reveals insights into vCXCL-1Tol function.
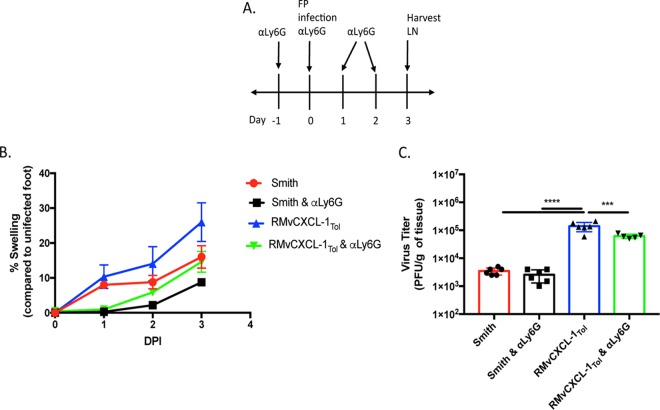
Using depleting antibodies, the importance of different cell types was tested. Neutrophil depletion with anti-Ly6G antibody prior to and during MCMV infection (Fig. 6A and Fig. S3B) reduced the level of vCXCL-1-induced swelling to levels observed with control virus in the absence of depletion (Fig. 6B). Neutrophil depletion also decreased to the level of swelling in controls. Thus, the increase in swelling due to vCXCL-1Tol was dependent on neutrophils. To examine whether neutrophil depletion alters viral burden in different tissues, LNs were harvested from MCMV-infected mice that had either been neutrophil depleted or left untreated, and virus titers in the draining LN were determined 3 dpi (Fig. 6C). Neutrophil-depleted mice infected with RMvCXCL-1Tol had less virus in the LN than non-neutrophil-depleted mice, but viral titers remained higher than the viral titers in the controls. These data indicate that neutrophils are partially responsible for vCXCL-1Tol-mediated dissemination, but they are not the major cell type responding to vCXCL-1Tol.
FIG 6.
Neutrophils are partially responsible for vCXCL-1Tol-induced inflammation and dissemination. (A) Neutrophil depletion experimental design. αLy6G, anti-Ly6G antibody; FP, footpad; LN, lymph node. (B) Footpad swelling of infected footpads up to 3 dpi normalized to uninfected footpads. (C) Plaque assays were performed on LN harvested from depleted or control mice at 3 dpi. Values are averages of two experiments with three mice per group. Bars represent the average titers ± SD (error bars). Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison of means and indicated as follows: ***, P ≤ 0.001; ****, P ≤ 0.0001.
vCXCL-1Tol increases virulence.
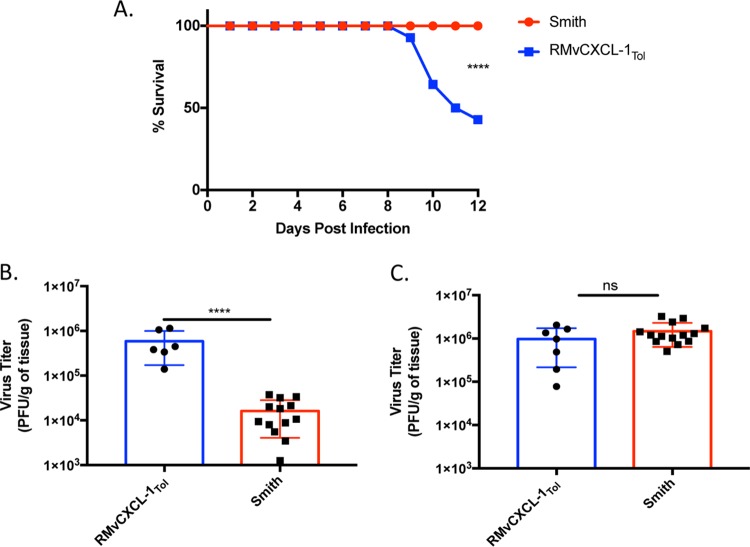
Because there is no effective method for depleting monocytes without depleting other cellular subsets (42), we infected NOD−SCID−IL-2 gamma chain-deficient (NSG) mice. These mice lack T, B, and NK cells and have defective macrophages and dendritic cells (43). Although these mice are highly immunodeficient, monocytes and neutrophils remain functional. The number of mice surviving daily was recorded and plotted as a Kaplan-Meier survival curve (Fig. 7A). Interestingly, many mice infected with RMvCXCL-1Tol died on or before 10 dpi, while all mice infected with the control virus survived to 12 dpi at which point SGs and spleens were harvested. These data indicate that vCXCL-1Tol is a virulence factor, as 58% of mice infected with recombinant MCMVs expressing vCXCL-1Tol died compared to mice infected with the Smith strain, which all survived. We also observed significantly higher viral titers in the spleens of mice during infection with RMvCXCL-1Tol compared to the control group (Fig. 7B), even though there was no difference in SG viral titer for mice in both these groups at 12 dpi (Fig. 7C). These data point to either viral load in the spleen, inflammatory infiltrates into the liver (Fig. S5), or another unknown effect of vCXCL-1Tol as a determinant for mortality in NSG mice.
FIG 7.
vCXCL-1Tol increases virulence of MCMV in NSG mice which correlates with increased viral loads in the spleen. (A) Kaplan-Meier survival curve comparing NSG mice infected with either RMvCXCL-1Tol or Smith MCMV. (B and C) At the termination of the experiment (12 days postinfection), MCMV titers in the spleen (B) or SG (C) were determined. Bars represent the average titers ± SD (error bars). Each symbol shows the viral titer for an individual mouse. Data are from three separate experiments with four to six mice per group for each virus. Statistical significance was ascertained using the Student’s t test and indicated as follows: ns, not significant; ****, P ≤ 0.0001.
vCXCL-1Tol increases MCMV liver inflammation in NSG mice. (A) Liver samples from NSG animals harvested at day 12 postinfection were sectioned and stained with H&E using standard procedures. Representative sections from animals infected with strain Smith and RMvCXCL-1Tol are shown. (B) Two independent sections of liver from each infected animal were counted using an Evos XL Core Microscope. The graph depicts the number of infiltrates quantified from each animal as well as an overall average from five or six animals in each group. Horizontal bars (± SD) represent the average infiltrate counts, and the numerical average is shown to the left of the bar. Each symbol represents the infiltrate count from an individual mouse. Download FIG S5, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
As most HCMV infections are asymptomatic, studying its dissemination from primary infection to the establishment of latency in humans has been limited to in vitro and ex vivo analysis (1, 2). These studies revealed that innate immune cells are major reservoirs of HCMV in blood and contribute to viral dissemination (28). HCMV’s reliance on innate immune cells has resulted in the evolution of immunomodulatory proteins such as chemokines that are capable of activating and recruiting innate immune cells (13). The discovery of the HCMV chemokine vCXCL-1 led to the hypothesis that vCXCL-1 recruits innate immune cells to the infection site and that these recruited cells contribute to HCMV dissemination (29, 44, 45). An adequate in vivo analysis of vCXCL-1 function is lacking. Here we employed a mouse model using recombinant MCMV expressing HCMV vCXCL-1 in vivo. A major limitation of the MCMV model and the reason recombinant MCMVs were generated is that some of the immunomodulatory proteins of MCMV and HCMV differ. MCMV encodes a C-C chemokine which also aids viral entry in MCMV Smith strain (24). However, HCMV encodes C-C and C-X-C chemokines (13), enhancing recruitment of multiple leukocyte subtypes (i.e., monocytes, neutrophils, and NK cells) in vitro (15, 16, 46). vCXCL-1 attracts neutrophils and potentially NK cells (15, 16). These observations suggest that neutrophils could be major players in HCMV dissemination.
To establish our model, we needed to understand MCMV-neutrophil interactions and how/whether neutrophils have a role in MCMV dissemination. This is important because neutrophils transfer HCMV (29), and vCXCL-1 alters human neutrophil functions (15). We show in a thioglycolate-induced model that murine neutrophils harbor and transfer MCMV to new cells although not as efficiently as other cells in the Flowthrough. This could be due to the neutrophil’s shorter half-life ex vivo or to the fact that neutrophils have phagocytosed viral genomes that are not infectious. We also show that viral transcription/translation begins in neutrophils (i.e., GFP expression) (Fig. 1), demonstrating that murine neutrophils can harbor MCMV.
Because vCXCL-1 is expressed late in HCMV’s life cycle (1), it is likely that infectious viral particles are being released simultaneously with chemokine expression. Therefore, we sought to mimic this environment. IL-17A injected i.p. into mice induces CXCL-1 (i.e., Groα) expression (36). CXCL-1 is a host chemokine that uses the CXCR2 receptor (47), inducing primarily a neutrophilic influx into the peritoneal cavity. Stimulating the CXCR2 signaling axis during infection increased viral burden in primary dissemination organs (i.e., spleen and lungs) and resulted in increased SG seeding. However, IL-17A treatment prior to infection did not lead to increased viral burden in SGs 14 dpi. This potentially could be due to a variety of factors, including localization or longevity of CXCR2 stimulation, as IL-17A treatment is transient. Therefore, it is not surprising that IL-17A treatment impacted only primary dissemination. Interestingly, when neutrophils were depleted from the IL-17A inflammation model, there was only a slight decrease in viral burden compared to nondepleted animals. This points to another CXCR2+ cell or another chemokine/cytokine-responsive cell responding to IL-17A. While neutrophils are the only statistically significant population that changes in response to IL-17A (36; data not shown), there could be a biologically relevant subset of CXCR2+ cells (48–50) responding to CXCL1 and aiding viral dissemination. It will be important to determine what cell types (e.g., inflammatory monocytes, NK cells, macrophages, etc.) are becoming infected immediately after IL-17A treatment.
Previously, we have demonstrated that overexpressing vCXCL-1 in the context of MCMV infection did not change primary dissemination but inhibited secondary dissemination (25). We speculated that the timing and quantity of viral chemokine expression induced an abnormal inflammatory environment, resulting in expedited or premature viral clearance. To alleviate this concern, we generated a recombinant MCMV expressing vCXCL-1 at relatively normal physiological times and levels by linking it to the MCMV protein MCK2. When mice were infected with recombinant vCXCL-1Tol MCMV, there was a significant difference in viral dissemination kinetics for both primary and secondary dissemination compared to controls. Because different inoculation routes have different dissemination mechanisms, it was important to determine whether the phenotype observed was inoculation route dependent (32, 51). We showed that the RMvCXCL-1Tol dissemination phenotype was still present whether mice were inoculated by the more natural inoculation route (i.e., intranasal [51]) or the more common intraperitoneal route. Because vCXCL-1Tol alters neutrophil functions (15), murine neutrophils were depleted to dissect this relationship in vivo. As with IL-17A neutrophil depletion, neutrophil depletion coupled with recombinant viral infection significantly reduced viral dissemination but did not return viral burden to WT MCMV levels. These data indicate that there is another cellular target through which vCXCL-1Tol is functioning.
As monocytes have also been shown to express CXCR2 under certain conditions (48, 52), the next logical experiment would be to deplete monocytes. Unfortunately, specific depletion of monocytes is not efficient (42). Instead, we chose to utilize the NSG mouse model. These mice are highly immunocompromised and lack mature B, T, and NK cells (43) and have defective macrophages and dendritic cells, which could participate in MCMV dissemination (32, 51, 53). However, NSG mice have functional monocytes. Because neutrophils were not the major cell type aiding dissemination (Fig. 6), we hypothesized that if viral dissemination of vCXCL-1Tol-expressing recombinant MCMV were still increased compared to WT MCMV in NSG mice, it would point toward a monocytic response to vCXCL-1Tol. Surprisingly, infection of NSG mice with RMvCXCL-1Tol and not strain Smith resulted in death (Fig. 7). This was not expected, but these results support the claim that vCXCL-1Tol is a virulence factor (12, 15, 26, 54). Additionally, Fig. 7 shows that primary dissemination (i.e., dissemination to the spleen) is significantly different between the two viruses, but there is no difference in secondary dissemination. This could indicate that primary dissemination and secondary dissemination are separate events mediated by different cell types, similar to our findings with CXCL-1-overexpressing MCMVs (25).
While neutrophils are not the major cell type aiding dissemination, our results provide evidence that murine neutrophils contribute to MCMV dissemination. We also report that vCXCL-1Tol significantly enhances MCMV dissemination and pathogenesis. However, the hypothesis that neutrophils are the sole responders to vCXCL-1Tol is incorrect. Other CXCR2-positive cells such as monocytes (48, 52) and myeloid-derived suppressor cells (55, 56) could be other targets for vCXCL-1. This hypothesis supports previous findings in which monocytes are major drivers of CMV dissemination (1, 2, 32, 57). Interestingly, the deaths of NSG mice infected with vCXCL-1Tol MCMV provide supporting evidence that this protein is a virulence factor albeit in NSG mice. We have previously shown that vCXCL-1 from different HCMV strains vary in their ability to activate and induce neutrophil recruitment (15), but now we have a system to test their virulence in vivo.
MATERIALS AND METHODS
Plasmids.
pCas9 was a gift from Luciano Marraffini (Addgene plasmid 42876) (58). The selectable marker was replaced with kanamycin using Gibson Assembly. The p2A-Tol open reading frame (ORF) was synthesized (Genscript) after adding the P2A sequence (Addgene). This plasmid contains 250 bp of MCK2 that is FLAG tagged followed by the 2A peptide and vCXCL-1 from the HCMV Toledo strain, which is 6× HIS tagged. pcDNA-MCMV IE1 was generated as a qPCR standard.
Cells and mice.
All experiments were performed with low-passage (<20) cells. Mouse embryonic fibroblast 10.1 (MEF 10.1) (59) were cultured in Dulbecco modified Eagle medium (DMEM) (Corning) supplemented with 10% Fetalclone III serum (HyClone, Logan, UT), 1% penicillin-streptomycin, and 1% l-glutamine.
BALB/c mice were purchased from Jackson Laboratory (Bar Harbor, ME) and housed under specific-pathogen-free conditions. Four- to 5-week-old nonobese diabetic severe combined immunodeficient, IL-2 common γ chain null (NSG) mice were purchased from Jackson Laboratory (Bar Harbor, ME). The Institutional Animal Care and Use Committees (IACUCs) at the University of Tennessee and University of Cincinnati approved all animal procedures.
Viruses, BAC mutagenesis, and recombinant virus generation.
RM4503 virus was a gift from Ed Mocarski, Emory University (22), and MCMV Smith strain virus was derived from the pSM3fr-MCK2-2fl bacterial artificial chromosome (BAC) (originally from B. Adler) was a gift from Chris Benedict (60). MCMV was produced in vitro using MEF 10.1 cells. All viruses were stored at −80°C until use. Viral titer was assessed by plaque assay (described below) on MEF 10.1 cells.
BAC mutagenesis was performed on pSM3fr-MCK2-2fl BAC by coupling galK recombineering (39) with CRISPR/Cas9 technology. Briefly, Escherichia coli SW105 containing pSM3fr-MCK2-2fl was induced to express lambda red recombinase, and galK was inserted into the MCK2 locus, resulting in pSM3fr-MCK2-2fl-galK. SW105 cells containing pSM3fr-MCK2-2fl-galK were induced by heat and pCas9 with a gRNA targeting galK along with a PCR product containing the MCK2-2A-vCXCL-1Tol DNA sequence was transformed into them. Transformants were plated on chloramphenicol and kanamycin. The following day, colonies were streaked onto MacConkey agar base (Difco) containing 1% galactose. Colonies that retained the galK gene were pink, while transformants with the desired recombination were white. SW105 cells containing recombinant BACs were further assessed via HindIII restriction fragment length polymorphism (RFLP) analysis, and the 2A-Tol insert was sequenced via Sanger sequencing after PCR amplification.
vCXCL-1Tol and control BACs were transfected into MEF 10.1 cells using LT1 transfection reagent (Mirus). BAC origin excision was achieved through serial passage as previously described (61). Confirmation of loss of the BAC origin was assessed in purified viral particles instead of infected cells and then evaluated with PCR. Approximately 10 serial passages were needed to effectively excise the BAC origin.
Western blotting.
Roller bottle (850 cm2) (Coring) supernatant from infected MEF 10.1 cells was harvested at 100% cytopathic effect (CPE). Ni-NTA beads were used to purify 6× His proteins (vCXCL-1Tol) and anti-FLAG agarose beads (Sigma) were used to purify FLAG-tagged proteins (MCK2) from approximately 60 ml of supernatant. Purified proteins were combined 1:1 with 6× His and FLAG. Proteins were run on a 15% SDS-PAGE and blotted onto an AZURE Biosystems membrane. Anti-His and anti-FLAG AZURE Western kits were used. Membranes were developed using an Odyssey Clx Li-Cor. Blots were analyzed using Image Studio v4.0.
Sequencing and genome assembly.
Sequencing was conducted as previously described (62). Genome assembly was conducted with Geneious version 11.1.5. Briefly, purified viral DNA was harvested using quick -gDNA miniprep kit (Zymo Research). The sequencing was designed to ensure a 40× genome coverage. Illumina sequencing reads (150 × 150 paired-end sequencing reads) were mapped to the parental MCMV C5X reference genome (wild-type [WT] Smith strain). Geneious software’s SNP-finding function was used to find mutations and single nucleotide polymorphisms (SNPs). All resequencing data are available from authors upon request.
qPCR MCMV quantification.
SYBR green real-time quantitative PCR (qPCR) was performed to measure viral load using primers designed to detect MCMV IE1 (63): IE1 Forward (5′-AGCCACCAACATTGACCACGCAC-3′) and IE1 Reverse (5′-GCCCCAACCAGGACACACAACTC-3′). Copy number was standardized using pcDNA-MCMV IE1. PCR was performed using a Chromo4 DNA engine PCR system (Bio-Rad). Quantification of viral DNA (IE1) was carried out using MJ Opticon Monitor analysis software version 3.1.
Peritoneal inflammation models and neutrophil purification.
Thioglycolate-induced peritoneal inflammation was conducted as previously described (31). Briefly, mice were injected with 3% Brewer’s thioglycolate (BD). Four hours after the mice were injected with BD, the mice were infected with 1 × 106 PFU of MCMV. Three hours later, the mice were euthanized, and peritoneal exudate cells (PECs) were harvested. Inflammation induced by IL-17A (Shenandoah Biotechnology Inc.) was performed as previously described (36).
Neutrophil purification was conducted using an anti-Ly6G microbead purification kit (Miltenyi Biotec). Briefly, 9 or 10 mice were administered 3% thioglycolate, and PECs were harvested 4 h postinjection. Peritoneal exudate was pooled from all mice, and the resulting single-cell suspension was subjected to microbead purification. Purity was determined by flow cytometry.
Flow cytometry.
For FP infections, feet (cut at the ankles) were minced into small pieces (∼3 mm) and incubated on a rotatory shaker at 37°C for 1 h in a 0.5% (wt/vol) solution of type I collagenase (Worthington). The suspension of cells, LN, or PECs were passed through a 40-μm cell strainer (Fisher Scientific). Red blood cells were lysed with ACK (i.e., ammonium chloride-potassium) lysis buffer. Cells were stained for flow cytometry analysis with the following fluorochrome-conjugated antibodies for cellular subsets: anti-Ly6G (1A8), anti-Ly6C (HK1.4), anti-F4/80 (BM8), and anti-CD11c (N418) (all from BioLegend), anti-CD49b (DX5) from eBioscience, and anti-CD11b (M1/70) from BD Pharmingen. Data were acquired on BD LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo software, version 10.1.
Neutrophil depletion.
In vivo depletion of neutrophils was performed using 1A8 (anti-Ly6G) (BioXcell) as previously described (64). Briefly, depleting antibodies were administered every day starting at 2 days prior to MCMV infection or IL-17A treatment and then every other day until harvest. Neutrophils were depleted using 0.25 mg/inoculation with anti-Ly6G. Flow cytometry was used to confirm depletion.
Plaque assay.
Plaque formation assay on MEF 10.1 cells was used to determine viral titers in organs. Briefly, MEF 10.1 cells were plated in a six-well dish. Organs were harvested and homogenized. Homogenate was serially diluted and added to MEF 10.1 cells and incubated for 1 h. After incubation, diluted virus was removed, and cells were overlaid with carboxylmethyl cellulose (CMC) medium and incubated for 5 days. CMC was removed, and plates were stained with Coomassie blue.
Adoptive transfer of neutrophils.
Four hours after i.p. thioglycolate injection, 9 or 10 mice were infected i.p. with MCMV (RM4503) (22). Four hours postinfection, the mice were euthanized, and PECs were isolated. Neutrophils were purified using MACs beads as described above. Neutrophils (1 × 106) were injected into the footpads of naive mice, and the amount of MCMV in the blood was quantified from 250 μl of whole blood via qPCR at 3 days after transfer.
In vitro growth assay.
MEF 10.1 cells were plated in triplicate in a six-well dish and infected with Smith or RMvCXCL-1Tol recombinants for either a multistep (multiplicity of infection [MOI] of 0.05) or single-step (MOI of 5) growth analysis. Supernatants were collected at the indicated times p.i. and sonicated prior to assessing the titer. The titers of viruses were determined via plaque assay.
Infectious center assay.
The infectious center assay was performed as previously described (65). Briefly, PECs were harvested, and red blood cells were lysed with ACK lysis buffer. PECs or purified neutrophils (1 × 106) were incubated for 12 h on an uninfected monolayer and overlaid with CMC medium. Plaques were counted after 5 days.
Virulence studies.
NSG mice (four to six animals per virus strain per experiment) were infected i.p. with 5 × 105 PFU of either RMvCXCL-1Tol or Smith. Mice were monitored daily for weight loss and administered supportive care if necessary. Mice were euthanized at day 12, SG and spleens were removed, and virus titers in organs were determined by plaque assay.
Statistical analysis.
Statistical significance was determined using Student’s t test or one-way analysis of variance (ANOVA) with Tukey’s multiple comparison of means using Prism 7 (GraphPad Software, Inc.).
ACKNOWLEDGMENTS
We thank Edward Mocarski for his critical review of the manuscript and editoral comments during the crafting of this article. We thank Jeremiah Johnson for his aid in sequencing the recombinant viruses.
This work was supported through grants from the NIH: grant R15 AI144683-0 (T.E.S.) and grants R01 AI121028 and R21 DE026267 (W.E.M.).
Footnotes
Citation Jackson JW, Hancock TJ, LaPrade E, Dogra P, Gann ER, Masi TJ, Panchanathan R, Miller WE, Wilhelm SW, Sparer TE. 2019. The human cytomegalovirus chemokine vCXCL-1 modulates normal dissemination kinetics of murine cytomegalovirus in vivo. mBio 10:e01289-19. https://doi.org/10.1128/mBio.01289-19.
Contributor Information
Thomas Shenk, Princeton University.
Chris Benedict, La Jolla Institute for Immunology.
Daniel Streblow, Oregon Health & Science University.
REFERENCES
- 1.Liu L. 2014. Fields Virology, 6th Edition. Clin Infect Dis 59:613. doi: 10.1093/cid/ciu346. [DOI] [Google Scholar]
- 2.Britt W. 2007. Virus entry into host, establishment of infection, spread in host, mechanisms of tissue damage In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: https://www.ncbi.nlm.nih.gov/books/NBK47407/. [PubMed] [Google Scholar]
- 3.Demmler GJ. 1994. Congenital cytomegalovirus infection. Semin Pediatr Neurol 1:36–42. [PubMed] [Google Scholar]
- 4.Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. 2013. The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schleiss MR. 2013. Cytomegalovirus in the neonate: immune correlates of infection and protection. Clin Dev Immunol 2013:501801. doi: 10.1155/2013/501801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stagno S, Pass RF, Dworsky ME, Britt WJ, Alford CA. 1984. Congenital and perinatal cytomegalovirus infections: clinical characteristics and pathogenic factors. Birth Defects Orig Artic Ser 20:65–85. [PubMed] [Google Scholar]
- 7.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J Clin Virol 41:180–185. doi: 10.1016/j.jcv.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Sinclair J, Poole E. 2014. Human cytomegalovirus latency and reactivation in and beyond the myeloid lineage. Future Virol 9:557–563. doi: 10.2217/fvl.14.34. [DOI] [Google Scholar]
- 9.Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J Gen Virol 87:1763–1779. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 10.Jackson JW, Sparer T. 2018. There is always another way! Cytomegalovirus' multifaceted dissemination schemes. Viruses 10:E383. doi: 10.3390/v10070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mocarski ES., Jr. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol 10:332–339. doi: 10.1016/S0966-842X(02)02393-4. [DOI] [PubMed] [Google Scholar]
- 12.Miller-Kittrell M, Sparer TE. 2009. Feeling manipulated: cytomegalovirus immune manipulation. Virol J 6:4. doi: 10.1186/1743-422X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McSharry BP, Avdic S, Slobedman B. 2012. Human cytomegalovirus encoded homologs of cytokines, chemokines and their receptors: roles in immunomodulation. Viruses 4:2448–2470. doi: 10.3390/v4112448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penfold ME, Dairaghi DJ, Duke GM, Saederup N, Mocarski ES, Kemble GW, Schall TJ. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc Natl Acad Sci U S A 96:9839–9844. doi: 10.1073/pnas.96.17.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heo J, Dogra P, Masi TJ, Pitt EA, de Kruijf P, Smit MJ, Sparer TE. 2015. Novel human cytomegalovirus viral chemokines, vCXCL-1s, display functional selectivity for neutrophil signaling and function. J Immunol 195:227–236. doi: 10.4049/jimmunol.1400291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamin R, Lecker LSM, Weisblum Y, Vitenshtein A, Le-Trilling VTK, Wolf DG, Mandelboim O. 2016. HCMV vCXCL1 binds several chemokine receptors and preferentially attracts neutrophils over NK cells by interacting with CXCR2. Cell Rep 15:1542–1553. doi: 10.1016/j.celrep.2016.04.042. [DOI] [PubMed] [Google Scholar]
- 17.Lafemina RL, Hayward GS. 1988. Differences in cell type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J Gen Virol 69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 18.Lilja AE, Shenk T. 2008. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci U S A 105:19950–19955. doi: 10.1073/pnas.0811063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang QY, Maul GG. 2006. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J Virol 80:7510–7521. doi: 10.1128/JVI.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dogra P, Sparer TE. 2014. What we have learned from animal models of HCMV. Methods Mol Biol 1119:267–288. doi: 10.1007/978-1-62703-788-4_15. [DOI] [PubMed] [Google Scholar]
- 21.Hudson JB. 1979. The murine cytomegalovirus as a model for the study of viral pathogenesis and persistent infections. Arch Virol 62:1–29. doi: 10.1007/BF01314900. [DOI] [PubMed] [Google Scholar]
- 22.Saederup N, Aguirre SA, Sparer TE, Bouley DM, Mocarski ES. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J Virol 75:9966–9976. doi: 10.1128/JVI.75.20.9966-9976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saederup N, Lin YC, Dairaghi DJ, Schall TJ, Mocarski ES. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc Natl Acad Sci U S A 96:10881–10886. doi: 10.1073/pnas.96.19.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner FM, Brizic I, Prager A, Trsan T, Arapovic M, Lemmermann NAW, Podlech J, Reddehase MJ, Lemnitzer F, Bosse JB, Gimpfl M, Marcinowski L, MacDonald M, Adler H, Koszinowski UH, Adler B. 2013. The viral chemokine MCK-2 of murine cytomegalovirus promotes infection as part of a gH/gL/MCK-2 complex. PLoS Pathog 9:e1003493. doi: 10.1371/journal.ppat.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dogra P, Miller-Kittrell M, Pitt E, Jackson JW, Masi T, Copeland C, Wu S, Miller WE, Sparer T. 2016. A little cooperation helps murine cytomegalovirus (MCMV) go a long way: MCMV co-infection rescues a chemokine salivary gland defect. J Gen Virol 97:2957–2972. doi: 10.1099/jgv.0.000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller-Kittrell M, Sai J, Penfold M, Richmond A, Sparer TE. 2007. Functional characterization of chimpanzee cytomegalovirus chemokine, vCXCL-1(CCMV). Virology 364:454–465. doi: 10.1016/j.virol.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerna G, Zipeto D, Percivalle E, Parea M, Revello MG, Maccario R, Peri G, Milanesi G. 1992. Human cytomegalovirus infection of the major leukocyte subpopulations and evidence for initial viral replication in polymorphonuclear leukocytes from viremic patients. J Infect Dis 166:1236–1244. doi: 10.1093/infdis/166.6.1236. [DOI] [PubMed] [Google Scholar]
- 28.Hassan-Walker AF, Mattes FM, Griffiths PD, Emery VC. 2001. Quantity of cytomegalovirus DNA in different leukocyte populations during active infection in vivo and the presence of gB and UL18 transcripts. J Med Virol 64:283–289. doi: 10.1002/jmv.1048. [DOI] [PubMed] [Google Scholar]
- 29.Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. 1998. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil-endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis 177:1465–1474. doi: 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- 30.Gerna G, Percivalle E, Baldanti F, Sozzani S, Lanzarini P, Genini E, Lilleri D, Revello MG. 2000. Human cytomegalovirus replicates abortively in polymorphonuclear leukocytes after transfer from infected endothelial cells via transient microfusion events. J Virol 74:5629–5638. doi: 10.1128/jvi.74.12.5629-5638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron EJ, Proctor RA. 1982. Elicitation of peritoneal polymorphonuclear neutrophils from mice. J Immunol Methods 49:305–313. doi: 10.1016/0022-1759(82)90130-2. [DOI] [PubMed] [Google Scholar]
- 32.Daley-Bauer LP, Roback LJ, Wynn GM, Mocarski ES. 2014. Cytomegalovirus hijacks CX3CR1hi patrolling monocytes as immune-privileged vehicles for dissemination in mice. Cell Host Microbe 15:351–362. doi: 10.1016/j.chom.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaroskova L, Holub M, Hajdu I, Trebichavsky I. 1975. Adherent and non-adherent mouse peritoneal exudate cells. Folia Biol (Praha) 21:111–116. [PubMed] [Google Scholar]
- 34.Cifarelli A, Pepe G, Paradisi F, Piccolo D. 1979. The influence of some metabolic inhibitors on phagocytic activity of mouse macrophages in vitro. Res Exp Med (Berl) 174:197–204. doi: 10.1007/BF01851332. [DOI] [PubMed] [Google Scholar]
- 35.Luttichau HR. 2010. The cytomegalovirus UL146 gene product vCXCL1 targets both CXCR1 and CXCR2 as an agonist. J Biol Chem 285:9137–9146. doi: 10.1074/jbc.M109.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witowski J, Pawlaczyk K, Breborowicz A, Scheuren A, Kuzlan-Pawlaczyk M, Wisniewska J, Polubinska A, Friess H, Gahl GM, Frei U, Jorres A. 2000. IL-17 stimulates intraperitoneal neutrophil infiltration through the release of GRO alpha chemokine from mesothelial cells. J Immunol 165:5814–5821. doi: 10.4049/jimmunol.165.10.5814. [DOI] [PubMed] [Google Scholar]
- 37.Hsu KM, Pratt JR, Akers WJ, Achilefu SI, Yokoyama WM. 2009. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J Gen Virol 90:33–43. doi: 10.1099/vir.0.006668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibrahimi A, Vande Velde G, Reumers V, Toelen J, Thiry I, Vandeputte C, Vets S, Deroose C, Bormans G, Baekelandt V, Debyser Z, Gijsbers R. 2009. Highly efficient multicistronic lentiviral vectors with peptide 2A sequences. Hum Gene Ther 20:845–860. doi: 10.1089/hum.2008.188. [DOI] [PubMed] [Google Scholar]
- 39.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDonald MR, Burney MW, Resnick SB, Virgin HW IV.. 1999. Spliced mRNA encoding the murine cytomegalovirus chemokine homolog predicts a beta chemokine of novel structure. J Virol 73:3682–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pontejo SM, Murphy PM. 2017. Two glycosaminoglycan-binding domains of the mouse cytomegalovirus-encoded chemokine MCK-2 are critical for oligomerization of the full-length protein. J Biol Chem 292:9613–9626. doi: 10.1074/jbc.M117.785121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swiecki M, Gilfillan S, Vermi W, Wang Y, Colonna M. 2010. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8+ T cell accrual. Immunity 33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, Greiner DL, Handgretinger R. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol 174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 44.Waldman WJ, Knight DA, Huang EH, Sedmak DD. 1995. Bidirectional transmission of infectious cytomegalovirus between monocytes and vascular endothelial cells: an in vitro model. J Infect Dis 171:263–272. doi: 10.1093/infdis/171.2.263. [DOI] [PubMed] [Google Scholar]
- 45.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng Q, Tao R, Gao HH, Xu J, Shang SQ, Zhao N. 2012. HCMV-encoded UL128 enhances TNF-alpha and IL-6 expression and promotes PBMC proliferation through the MAPK/ERK pathway in vitro. Viral Immunol 25:98–105. doi: 10.1089/vim.2011.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahuja SK, Lee JC, Murphy PM. 1996. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B. Determinants of high affinity binding and receptor activation are distinct. J Biol Chem 271:225–232. doi: 10.1074/jbc.271.1.225. [DOI] [PubMed] [Google Scholar]
- 48.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, Dewor M, Georgiev I, Schober A, Leng L, Kooistra T, Fingerle-Rowson G, Ghezzi P, Kleemann R, McColl SR, Bucala R, Hickey MJ, Weber C. 2007. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med 13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 49.Gerszten RE, Garcia-Zepeda EA, Lim YC, Yoshida M, Ding HA, Gimbrone MA Jr, Luster AD, Luscinskas FW, Rosenzweig A. 1999. MCP-1 and IL-8 trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Nature 398:718–723. doi: 10.1038/19546. [DOI] [PubMed] [Google Scholar]
- 50.Połeć A, Ráki M, Åbyholm T, Tanbo TG, Fedorcsák P. 2011. Interaction between granulosa-lutein cells and monocytes regulates secretion of angiogenic factors in vitro. Hum Reprod 26:2819–2829. doi: 10.1093/humrep/der216. [DOI] [PubMed] [Google Scholar]
- 51.Farrell HE, Lawler C, Oliveira MT, Davis-Poynter N, Stevenson PG. 2016. Alveolar macrophages are a prominent but nonessential target for murine cytomegalovirus infecting the lungs. J Virol 90:2756–2766. doi: 10.1128/JVI.02856-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonecchi R, Facchetti F, Dusi S, Luini W, Lissandrini D, Simmelink M, Locati M, Bernasconi S, Allavena P, Brandt E, Rossi F, Mantovani A, Sozzani S. 2000. Induction of functional IL-8 receptors by IL-4 and IL-13 in human monocytes. J Immunol 164:3862–3869. doi: 10.4049/jimmunol.164.7.3862. [DOI] [PubMed] [Google Scholar]
- 53.Farrell HE, Bruce K, Lawler C, Oliveira M, Cardin R, Davis-Poynter N, Stevenson PG. 2017. Murine cytomegalovirus spreads by dendritic cell recirculation. mBio 8:e01264-17. doi: 10.1128/mBio.01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pontejo SM, Murphy PM, Pease JE. 2018. Chemokine subversion by human herpesviruses. J Innate Immun 10:465–478. doi: 10.1159/000492161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu H, Gu Y, Xue Y, Yuan M, Cao X, Liu Q. 2017. CXCR2+ MDSCs promote breast cancer progression by inducing EMT and activated T cell exhaustion. Oncotarget 8:114554–114567. doi: 10.18632/oncotarget.23020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. 2013. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith MS, Bentz GL, Alexander JS, Yurochko AD. 2004. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J Virol 78:4444–4453. doi: 10.1128/jvi.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harvey DM, Levine AJ. 1991. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev 5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 60.Jordan S, Krause J, Prager A, Mitrovic M, Jonjic S, Koszinowski UH, Adler B. 2011. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary glands due to a fixed mutation of MCK-2. J Virol 85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol 73:7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelley BR, Ellis JC, Hyatt D, Jacobson D, Johnson J. 2018. Isolation and whole-genome sequencing of environmental Campylobacter. Curr Protoc Microbiol 51:e64. doi: 10.1002/cpmc.64. [DOI] [PubMed] [Google Scholar]
- 63.Kamimura Y, Lanier LL. 2014. Rapid and sequential quantitation of salivary gland-associated mouse cytomegalovirus in oral lavage. J Virol Methods 205:53–56. doi: 10.1016/j.jviromet.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. 2008. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol 83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 65.Bittencourt FM, Wu SE, Bridges JP, Miller WE. 2014. The M33 G protein-coupled receptor encoded by murine cytomegalovirus is dispensable for hematogenous dissemination but is required for growth within the salivary gland. J Virol 88:11811–11824. doi: 10.1128/JVI.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gating strategy used to identify GFP-positive cells. (A) Neutrophil (Ly6G+, CD11b+) purity following microbead isolation was evaluated with flow cytometry. Data are representative of two experiments with six or seven mice per experiment. Live cells were gated. (B) Following identification of live cells, single cells were analyzed for GFP expression. Gates were set on unstained infected peritoneal exudate cells. Download FIG S1, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
De novo GFP expression. MEF 10.1 cells were infected with RM4503 at an MOI of 3 at 4 hours postinfection. GFP expression was determined via flow cytometry. (A to C) Uninfected cells (A), infected cells treated with 50 μg/ml of cycloheximide (CHX) for 1 hour prior to infection and 4 hours postinfection (B), or infected but untreated cells (vehicle control) (C). Download FIG S2, TIF file, 1.9 MB (2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confirmation of neutrophil depletion. Neutrophil depletion was evaluated by flow cytometry. (A) Four hours after IL-17A treatment, mice were euthanized, and PECs were harvested. PECs were assayed for the presence of neutrophils (Ly6G+, CD11b+). (B) Whole blood was assayed for the presence of neutrophils (Ly6G+, CD11b+) from neutrophil-depleted or vehicle control-treated mice prior to infection with recombinant MCMVs. Data are representative of two experiments with three animals per group. Download FIG S3, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Total cell counts in LN and blood. At 3 days postinfection, LN (A) and blood (B) were harvested, and total leukocytes were counted. Blood was harvested via cardiac puncture and placed in lithium heparin-treated tubes. ACK lysis was performed to remove red blood cells, and leukocytes were counted. Download FIG S4, TIF file, 1.7 MB (1.7MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
vCXCL-1Tol increases MCMV liver inflammation in NSG mice. (A) Liver samples from NSG animals harvested at day 12 postinfection were sectioned and stained with H&E using standard procedures. Representative sections from animals infected with strain Smith and RMvCXCL-1Tol are shown. (B) Two independent sections of liver from each infected animal were counted using an Evos XL Core Microscope. The graph depicts the number of infiltrates quantified from each animal as well as an overall average from five or six animals in each group. Horizontal bars (± SD) represent the average infiltrate counts, and the numerical average is shown to the left of the bar. Each symbol represents the infiltrate count from an individual mouse. Download FIG S5, TIF file, 2.2 MB (2.2MB, tif) .
Copyright © 2019 Jackson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.