Abstract
Pulmonary fibrosis is a lethal inflammatory disease. In this study, we aimed to explore the potential‐related circular RNAs (circRNAs) and genes that are associated with pulmonary fibrosis. Pulmonary fibrosis rat models were constructed and the fibrosis deposition was detected using hematoxylin and eosin and Masson staining. The differentially expressed circRNAs were obtained through RNA sequencing. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were further performed to uncover the key function and pathways in pulmonary fibrosis. The interaction networks between circRNAs and their downstream micro RNAs (miRNAs) and genes were constructed by Cytoscape Software. The quantitative polymerase chain reaction was performed to validate the expression of 10 candidate circRNAs and five of them were performed ringwise sequencing in pulmonary fibrosis rats. We further selected five candidate circRNAs target miRNAs and messenger RNAs and validated by real‐time polymerase chain reaction. The pulmonary fibrosis models were successfully constructed according to the pathological examination. circRNAs were differentially expressed between the pulmonary fibrosis and normal pulmonary tissues. GO analysis verified that the differentially expressed circRNAs were significantly clustered in the cellular component, molecular function, and biological process. In the KEGG analysis, circRNAs were enriched in the following pathways: antigen processing and presentation, phagosome, PI3K‐AKt signaling pathway, HTLV‐I infection, and Herpes simplex infection. After validation in pulmonary fibrosis rat models, it was found that five of those circRNAs (chr9:113534327|113546234 [down], chr1:200648164|200672411 [down], chr5:150850432|150865550 [up], chr20:14319170|14326640 [down], and chr10:57634023|57634588 [down]) showed a relatively consistent trend with predictions. Validation of these circRNAs target miRNAs and genes showed that chr9:113534327|113546234, chr20:14319170|14326640, and chr10:57634023|57634588 were implicated in Notch1 activated transforming growth factor‐β (TGF‐β) signaling pathway. The study demonstrated that a series of circRNAs are differentially expressed in pulmonary fibrosis rats. These circRNAs, especially TGF‐β‐ and Notch1‐related circRNAs might play an important role in regulating pulmonary fibrogenesis.
Keywords: circular RNAs, Notch1, pulmonary fibrosis, transforming growth factor‐β
1. INTRODUCTION
Pulmonary fibrosis is a potentially lethal inflammatory disease, which is characterized by the extracellular matrix components deposition excessively in the interstitium. The excessive deposition leads to the fibrotic aberrant changes in the alveoli and patients respiratory failure.1, 2 It has been reported that the incidence of pulmonary fibrosis is 13 to 14 cases per 1 00 000 persons and the current incidence in the United States is higher than that reported 10 years ago.3, 4 Usually, the risk factors of pulmonary fibrosis are aging, smoking, aberrant wound‐healing response, and certain viral or bacterial infection.5 It is considered that fibroblast growth factor (FGF) and their receptors are involved in idiopathic pulmonary fibrosis. Overexpression of FGF7/10 diminishes the extent of epithelial injury and apoptosis in pulmonary fibrosis rodents.6, 7, 8 Decreased mesenchymal expressed FGFR2 receptor relieves in bleomycin‐induced fibrosis.9 Recently, a novel type of RNA was emerging as a pathological factor in fibrosis.
Circular RNAs (circRNAs) are a type of covalently closed continuous RNA loops with covalently joined 3′‐ and 5′‐ends formed by back‐splice events.10 The circRNAs are widespread and diverse in eukaryotic cells.11, 12 circRNAs serve as epigenetic sponges of micro RNA (miRNA) which is believed to negatively regulate miRNAs and contribute to the competing endogenous RNA (ceRNA) network.12, 13 It has found that circRNAs are involved in many human diseases such as cancer.14 The relationship between circRNA and pulmonary fibrosis is rare. It has been reported that circRNA_010567 silencing increased miR‐141 and decreased transforming growth factor‐β (TGF‐β), and suppressed fibrosis‐associated proteins in diabetic mice myocardial fibrosis model.15 In one study, the circHECTD1/HECTD1 pathway causes macrophage activation and death and subsequently induces pulmonary fibroblast activation.16 However, the knowledge of circRNAs in this disease is still unclear. We speculate that more circRNAs might be involved in multiple steps of the fibrosis process.
Hence, we conducted this study to assess the expression of circRNAs and their network based on the RNA sequencing (RNA‐seq) data of pulmonary fibrosis rats, we aimed to ascertain the possible key circRNAs and target genes through comprehensive interaction network construction and function enrichment analysis. This work is promising to expound part of the underlying molecular mechanism in pulmonary fibrosis, which might contribute to the clinical therapy of pulmonary fibrosis patients.
2. MATERIALS AND METHODS
2.1. Rat pulmonary fibrosis models
A total of 10 female adult Sprague‐Dawley rats (about 200 g) were provided by Southern Model Animal Center and fed under controlled conditions. The animal experiments were performed in accordance with the Animal Ethics Committee of Shenzhen Luohu People's Hospital. The rats were divided into two groups: saline group and bleomycin (BLM) group. At day 0, the rats of the saline group and the BLM group were lightly anesthetized with isoflurane and injected with saline and BLM saline (5 mg/kg) intratracheally, respectively. After a week, the rats of BLM group were administered with BLM (2 mg/kg) orally.
At day 28, all of the rats were executed and dissected. The trachea was dissociated and the instilled lobes were ligated and a single lung was fixed with 4% paraformaldehyde. After fixed for 4 hours, the tissues were incubated in PBS with 15% sugar. Finally, the tissues were kept in 70% ethanol for staining.
2.2. Hematoxylin and eosin staining
The lung tissues were dehydrated in gradient alcohol and treated with xylene. The tissues were embedded with paraffin and sliced (4 μm). After treated on 40°C platform to remove paraffin, the slices were stained with hematoxylin for few minutes. The 70% and 90% ethanol were used to dehydrate. In the end, the slices were treated with eosin for several minutes and examined under a light microscope.
2.3. Masson staining
The paraffin slices were dewaxed and washed. The slices were stained using hematoxylin for 5 to 10 minutes. After being washed by H2O, the sliced were restained by Masson ponceaux for 5 to 10 minutes. The slices were embathed in 2% glacial acetic acid and treated with 1% phosphomolybdic acid for 3 to 5 minutes. Then, aniline blue was used to stain for 5 minutes and dehydrated. In the end, the slices were stained with resin.
2.4. RNA sequencing
The circRNAs data of pulmonary fibrosis was obtained from the RNA‐seq data. We performed RNA‐seq of five pulmonary fibrosis rats and five controls. Total RNA from the samples was extracted by RNeasy Mini Kit (QIAGEN, Hilden, Germany). The RNA integrity was evaluated by Agilent Bioanalyzer 2100 (Agilent, CA). RNA Clean XP Kit (Beckman Coulter, CA) and RNase‐free DNase Set (QIAGEN) were used for RNA cleanup. NanoDrop 2000 (Thermo Fisher Scientific) was used to determine the quality and concentration of the RNA. For circRNA sequencing, total RNA was digested with RNase R (Epicenter) to remove the linear RNAs. One microgram of the RNA was used for library preparation by VAHTSTM mRNA‐seq v2 Library Prep Kit for illumina (Vazyme, Nanjing, China). For messenger RNA (mRNA) sequencing, the polyA mRNA was purified via the hybridization to Dynal oligo beads. The RNA was fragmented and then the double‐strand complementary DNA (cDNA) was synthesized. End repair and A‐addition were thereby performed in order to ligate the cDNA fragments to adapters. The ligated cDNA was then subjected to polymerase chain reaction (PCR) amplification. The quality of the library was determined using Agilent Bioanalyzer 2100. Hiseq 2000 (Illumina, CA) was used for the RNA‐seq. R Software (http://www.R‐project.org) was applied for quantile normalization and subsequent data processing. The differentially expressed circRNAs or mRNAs were screened according to a fold change and q value (q < 0.05 and fold change > 2.0).
2.5. GO and pathway analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) is a comprehensive bioinformatics analysis platform.17 The circRNAs were uploaded into the function annotation portal of DAVID database. After the enrichment annotation, a list of Gene Ontology (GO) items were obtained. The related pathways of the parental genes of differential circRNAs were analyzed by the Kyoto Encyclopedia of Genes and Genomes (KEGG). P < 0.05 and false discovery rate (FDR) < 0.05 were considered as statistically significant.
2.6. Interaction analysis
Cytoscape (http://apps.cytoscape.org/apps/keggscape) is an online bioinformatics resource platform for constructing biomolecular interaction networks.17 To search the potential circRNAs that are associated with TGF‐β1 and Notch1 signaling pathways, the circRNAs and genes of TGF‐β1 and Notch1 signaling pathways were uploaded and analyzed by Cytoscape platform to identify the densely connected region in the PPI network from the STRING database.18
2.7. Real‐time polymerase chain reaction
Total RNA was isolated using the RNeasy Kit (QIAGEN) according to the manufacturer's instructions. After reversed into cDNA with MLV Reverse Transcriptase (Promega, WI), the relative expression of RNA was analyzed by ΔΔC t method. The amplification procedure is as follows: 95°C for 120 seconds; 95°C for 15 seconds; 60°C for 30 seconds; 40 cycles; melting curve: 60°C to 95°C.
2.8. PCR validation
Total RNA was extracted using TRIzol Reagent (Invitrogen, WI) and reversed transcribed using GoScript RT System (Promega, Madison, WI). Ten circRNAs were obtained for validation using divergent and convergent primers. The primers were: Ppp4r1‐CF, TGGGGCCTTTCATATCCACA and Ppp4r1‐CR, GAAGTCCAGGGCTGAAGGTA; Cdc42bpa‐CF, GAAAAGGGTGTAGAGCACCGA and Cdc42bpa‐CR, TGCTGCATATTCCTGGTGAGT; Rprd1a‐CF, CTCACTCGGATGTTAGCGGAC and Rprd1a‐CR, AACTCTGGCCCCTTCCTTTTG; Fgfr2‐CF, TACGGGTCCATCAACCACAC and Fgfr2‐CR, TAGTCCAACTGATCACGGCG; Eya3‐CF, ACCACTTATCCTGGGCAGAC and Eya3‐CR, TGCGGTCTTCACAAACTGTC; Specc1l‐CF, TGGCGAGAGAATACGGAGGA, and Specc1l‐CR, GGTCTTAGCACTTTCCATAGCAG; Ick‐CF, GTTATCAGGGAGAACGACCATC, and Ick‐CR, CACCAGCACAAAGTATTCAACAG; Phldb2‐CF, GCAAGCCAGTCACATCGTTC, and Phldb2‐CR, CTTTTCCCTTTCAGCATCAAGC; Dhx33‐CF, ACAGCCTCAGCAGGATTACC, and Dhx33‐CR, ATGCCATCTGTCAGGAACTTG; Ciz1‐CF, CCCTTCTCAGCATAACCACTCAG, and Ciz1‐CR, GAGGCTGGCACTGTCATACG. The productions were sent for Sanger sequence to Holly Company for validation.
2.9. Statistical analysis
The SPSS 19.0 software (IBM Corp., NY) was used to analyze the data. All the measurements were expressed as mean ± standard deviation. The paired t test was used to compare the change before and after treatment. P < 0.05 is considered as significantly different.
3. RESULTS
3.1. BLM‐induced pulmonary fibrosis models
Pulmonary fibrosis was induced by BLM. In contrast to the control group, pulmonary fibrosis rats showed that obvious alveolar wall thickening, leukocytes infiltration, and excessive deposition of mature collagen fibers as visualized by hematoxylin and eosin (H&E) and Masson staining (Figure 1). The results indicated that pulmonary fibrosis rat models were constructed successfully.
Figure 1.
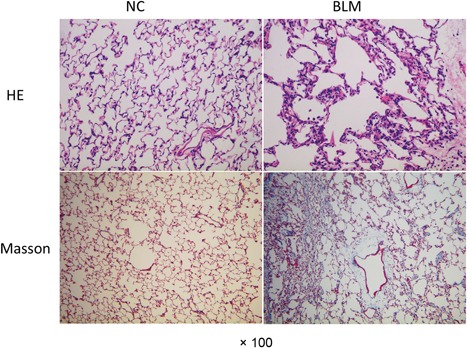
Haemotoxylin and eosin (H&E) and Masson staining of lung tissues in rat models. Pulmonary fibrosis model rats were constructed and the fibrosis deposition was detected using H&E and Masson staining (×100). BLM, bleomycin group; NC, normal group
3.2. Prediction performance of the total RNA‐Seq data
Based on the RNA‐seq data, we investigate the general information of circRNA. The RNA‐seq reads of circRNAs were shown in Figure 2A.The back‐spliced junction is a representative character and the copy number of back‐spliced junction read pairs were shown in Figure 2B. Five thousand five hundred eighty‐one and 7018 circRNAs originated from exons of BLM rats and saline rats, respectively. One thousand two hundred eighty‐four and nine circRNAs were transcribed from introns and intergenic of BLM rats. Meanwhile, 1722 and 13 circRNAs were transcribed from introns and intergenic of saline rats (Figure 2C). The chromosome distribution of differentially expressed circRNAs were displayed in Figure 2D, the upregulated circRNAs were mainly clustered in chromosomes 1, 2, and 10, and the downregulated circRNAs were mostly clustered in chromosomes 1 and 4.
Figure 2.
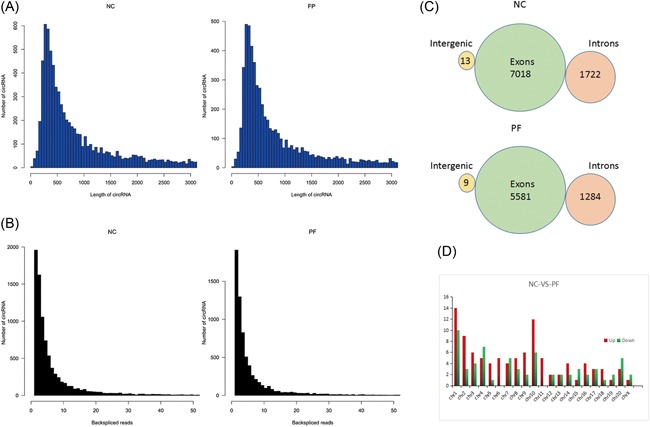
Prediction performance of the total RNA‐Seq data. A, The RNA‐seq reads of circRNAs. B, The back‐spliced junction read pairs of circRNAs. C, The distribution of circRNAs among intergenic exons and introns. D, The chromosome distribution of differentially expressed circRNAs. circRNAs, circular RNAs; NC, normal group; RNA‐seq, RNA sequencing
3.3. The enrichment GO terms and KEGG
GO analysis of 16 circRNAs were analyzed on DAVID. The significantly enriched terms (P < 0.05 and FDR < 0.05) were displayed (Figure 3B). The results from GO verified that circRNAs were significantly clustered in cellular component (CC), molecular function (MF), and biological process (BP) (Figure 3B). The three most significant GO terms in CC were cell, cell part, and organelle. In MF, the four most significant terms were binding, catalytic, enzyme regulator, and molecular transducer; the enzyme regulator and molecular transducer have equal rate. In BP, target genes were enriched in the following GO terms: cellular process, biological regulation, metabolic process, and multicellular organismal process; the metabolic process and multicellular organismal process have equal rates. In the KEGG analysis, the targets were enriched in the following pathways: antigen processing and presentation, phagosome, PI3K‐AKt signaling pathway, HTLV‐I infection, and Herpes simplex infection (Figure 3A).
Figure 3.
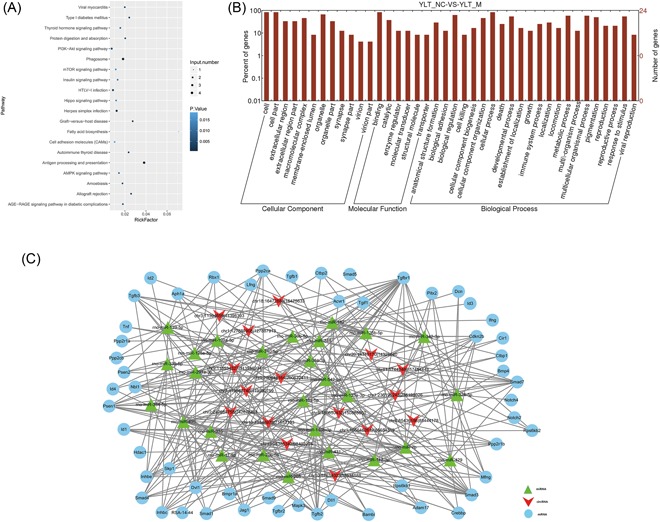
GO and KEGG analyses showed the interaction among target genes. A, GO analysis showed that circRNAs were significantly clustered in the cellular component, molecular function, and biological process. B, KEGG analysis of differentially expressed circRNAs. C, The interaction network predicted the interaction among the target genes. circRNAs, circular RNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes
3.4. The interaction network
The interaction network incorporating 97 nodes was generated by inputting the 16 circRNAs into the Cytospace (Figure 3C). Subsequently, 10 circRNAs (chr9:113534327|113546234, chr13:98386342|98403224, chr18:16472884|16475633, chr1:200648164|200672411, chr5:150850432|150865550, chr20:14319170|14326640, chr8:85436186|85444178, chr11:57442466|57454443, chr10:57634023|57634588, and chr3:11394659|11396393) were verified according to the most densely connected region in the network module analysis based on topology (Table 1). These 16 circRNAs connected 27 miRNA and one single circRNA connected several miRNAs, such as circRNA (chr3:11394659|11396393) interacted with miR‐340‐5p, miR‐125b‐5p, and miR‐125a‐5p. Through the interaction network construction, 54 target genes were considered to be related with pulmonary fibrosis (Figure 3C). In these target genes, TGF‐β family and Notch family genes were found to be highly frequent (Figure 3C).
Table 1.
The information of 10 candidate circRNAs
| Sl no. | circRNAs | PF/NC | Genes | Length, bp |
|---|---|---|---|---|
| 1 | chr9:113534327|113546234 | Down | Ppp4r1 | 4371 |
| 2 | chr13:98386342|98403224 | Up | Cdc42bpa | 1529 |
| 3 | chr18:16472884|16475633 | Up | Rprd1a | 1415 |
| 4 | chr1:200648164|200672411 | Down | Fgfr2 | 1383 |
| 5 | chr5:150850432|150865550 | Up | Eya3 | 1149 |
| 6 | chr20:14319170|14326640 | Down | Specc1l | 804 |
| 7 | chr8:85436186|85444178 | Up | Ick | 530 |
| 8 | chr11:57442466|57454443 | Up | Phldb2 | 486 |
| 9 | chr10:57634023|57634588 | Down | Dhx33 | 399 |
| 10 | chr3:11394659|11396393 | Down | Ciz1 | 331 |
Abbreviations: circRNA, circular RNA; NC, normal group; PF, pulmonary fibrosis models group.
3.5. Significantly differentially expressed circRNAs validation
The expression of 10 circRNAs were validated in six pairs of rat tissues (normal pulmonary tissues and pulmonary fibrosis tissues), and one pair of these was discarded because of extreme variation. Since chr5:150850432|150865550 is upregulated in pulmonary fibrosis tissues and eight circRNAs are downregulated in pulmonary. The expression of five of those circRNAs (chr9:113534327|113546234 [down], chr1:200648164|200672411 [down], chr5:150850432|150865550 [up], chr20:14319170|14326640 [down], and chr10:57634023|57634588 [down]) showed a relatively consistent trend with predictions (Figure 4). We sequenced PCR product and determined circular sites in five differentially expressed circRNA in BLM model (Figure 5). Next, we validated whether the above‐mentioned five differentially expressed circRNA had the property of circularity. Results in Figure 6 indicated that all five circRNA had circularity property.
Figure 4.
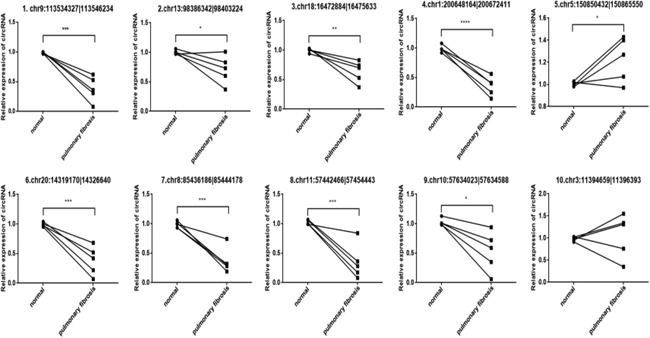
Validation of significantly differentially expressed circRNAs by real‐time polymerase chain reaction. circRNA, circular RNA
Figure 5.
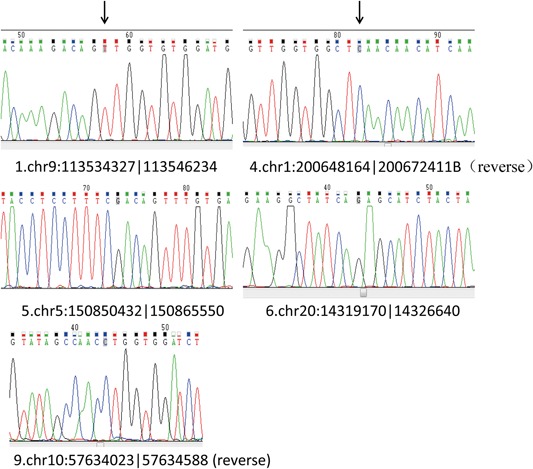
The circular sites of the circRNAs. The circular sites of five differentially expressed circRNAs in the bleomycin model were analyzed with the next‐generation sequencing. circRNA, circular RNA
Figure 6.
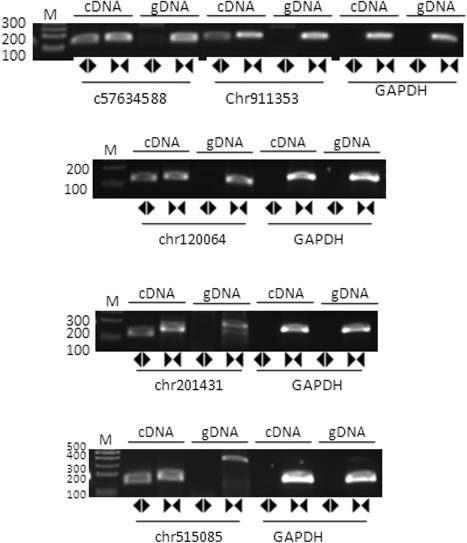
The circularity of the circRNAs. The circularity of five circRNAs in bleomycin models were detected by real‐time polymerase chain reaction, using the GAPDH as the negative control. cDNA, complementary DNA; circRNA, circular RNA; GADPH, glyceraldehyde‐3‐phosphate dehydrogenase; gDNA, genomic DNA
3.6. Validation of circRNA target genes
We further selected the five circRNAs target genes (miRNAs and mRNAs) based on the interaction network and validated in five pairs of normal and pulmonary fibrosis tissues via real‐time PCR (RT‐PCR). As shown in Figure7, chr9:113534327|113546234, chr20:14319170|14326640, and chr10:57634023|57634588 were significantly downregulated in pulmonary fibrosis tissues as predicted, their target miRNAs rno‐miR‐30b‐5p, rno‐miR‐7b, and rno‐miR‐429 were significantly upregulated, and the expression level of target mRNAs Cdkn2b, Samd3, and Tgfbr1 were significantly decreased in pulmonary fibrosis tissues, which showed a consistent trend with predictions. The validation of chr1:200648164|200672411 and chr5:150850432|150865550 target miRNA and mRNAs were not consistent with our prediction, which may be the reason that the two circRNAs interaction with other miRNA and mRNAs.
Figure 7.
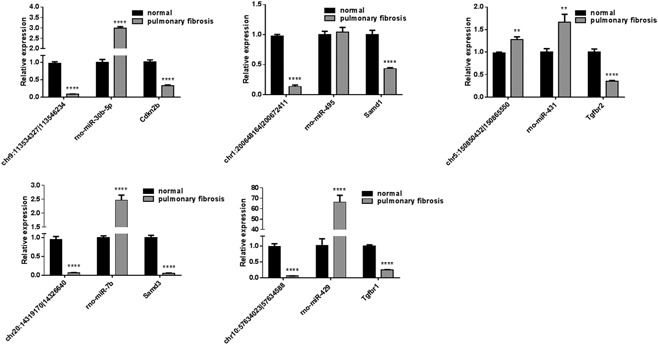
Validation of circRNA target genes. The candidate of five circRNAs target genes (miRNAs and mRNAs) in five pair of normal and pulmonary fibrosis tissues were detected by real‐time polymerase chain reaction. circRNA, circular RNA; miRNA, micro RNA; mRNA, messenger RNA. **P < 0.01, ****P < 0.0001
4. DISCUSSION
In this study, we constructed pulmonary fibrosis rat models and compared the circRNA profiles of models and controls. Comparison analyses were performed to systematically evaluate the differences of circRNAs from total RNA‐Seq data. The result showed that several biological function modules and specific circRNAs were involved in the pathogenesis of pulmonary fibrosis.
Pulmonary fibrosis is an irreversible, progressive, and lethal lung disorder. Pulmonary fibrosis involves fibroblast destruction, extracellular matrix remodeling, and deposition, and collagen accumulation.19 Accumulating evidence indicates that TGF‐β and Notch1 are involved in epithelial‐to‐mesenchymal transition and fibroblast activation, resulting in various types of tissue fibrosis including kidney fibrosis, pulmonary fibrosis and cardiac fibrosis.20, 21, 22, 23 Inhibition of Notch1 can relieve fibrosis through suppressing Notch‐mediated TGF‐β signaling activation.24 There are five Notch ligands and four Notch receptors in mammals.25 The Notch receptors catalyzed by the secretase combination to release Notch intracellular domain (NICD), which can induce the target genes transcription. The process can lead to the fibrosis via activation of the TGF‐β signaling pathway and be inhibited by secretase inhibitor.26, 27 It was considered that myofibroblasts are the key effector cells and α‐smooth muscle actin (α‐SMA) has an aberrant expression in the fibrosis development.28, 29 Myofibroblasts are the main source of fibrogenic cytokines and type I collagen in fibrotic lesions and contribute to the pulmonary altered mechanical properties.30, 31 The growth factors believed to be important for fibrosis include TGF‐β, vascular endothelial growth factor (VEGF), FGF‐2, connective tissue growth factor (CTGF), epidermal growth factor (EGF), insulin‐like growth factor (IGF), interleukin‐18 (IL‐18), and endothelin (ET).32 TGF‐β could induce expression of α‐SMA via a mechanism that involves ET‐1.33, 34 In addition, TGF‐β could induce myofibroblast differentiation, including CTGF, which is a common target of TGF‐β and ET‐1.35
In pulmonary fibrosis, the interaction of circRNAs and Notch signaling pathway was unknown. In our analysis, to search the potential circRNAs that are associated with TGF‐β1 and Notch1 signaling pathways, the circRNAs and genes of TGF‐β1 and Notch1 signaling pathways were uploaded and analyzed by the Cytoscape platform. Results showed that these 16 circRNAs connected 27 miRNA and one single circRNA connected several miRNAs. Through the interaction network construction, 54 target genes were considered to be related with pulmonary fibrosis (Figure 3C). In these target genes, TGF‐β family and Notch family genes were found to be high frequent. Validation of the five candidate circRNAs target genes by RT‐PCR revealed that chr9:113534327|113546234, chr20:14319170|14326640, and chr10:57634023|57634588 exhibited a significantly lower expression in pulmonary fibrosis tissues. Since circRNAs are the sponge of miRNA, lower level of chr9:113534327|113546234, chr20:14319170|14326640, and chr10:57634023|57634588 could upregulate the miRNA rno‐miR‐30b‐5p, rno‐miR‐7b, and rno‐miR‐429, thus to inhibit the expression of target genes Cdkn2b, Samd3, and Tgfbr1, the results in Figure 7 showed a consistent trend with predictions. It is reported that silencing Cdkn2b expression by siRNA resulted in the increased fibroblasts cell proliferation, which contribute to the pulmonary fibrosis.36 Samd3 acts as a signaling molecule of TGF‐β signaling pathway, inhibition of TGF‐β1‐mediated nuclear translocation of pSMAD3 by miR‐26a could suppress TGF‐β1‐induced proliferation and differentiation of lung fibroblasts.37 Tgfbr1 is a member of the TGF‐β superfamily, which promotes pulmonary fibrosis via activation of the TGF‐β signaling pathway.38 These evidence suggested that chr9:113534327|113546234, chr20:14319170|14326640, and chr10:57634023|57634588 may be involved in pulmonary fibrosis via regulating Notch‐activated TGF‐β signaling pathway, while it needs to be further investigated.
In summary, we compared the circRNA profiles in rat pulmonary tissues and pulmonary fibrosis tissues and found a series of circRNAs related to pulmonary fibrosis. We demonstrated that circRNAs exerts pulmonary fibrogenesis mediated by the TGF‐β and Notch signaling pathway.
AUTHOR CONTRIBUTIONS
LY designed the study and wrote the draft. LY, XL, and NZ conducted the experiments. LC, JX, and WT collected and analyzed the data. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
ACKNOWLEDGMENT
This work was supported by the Key specialty funds from Luohu District in Shenzhen City (100710).
Yang L, Liu X, Zhang N, Chen L, Xu J, Tang W. Investigation of circular RNAs and related genes in pulmonary fibrosis based on bioinformatics analysis. J Cell Biochem. 2019;120:11022‐11032. 10.1002/jcb.28380
References
REFERENCES
- 1. Kikuchi N, Ishii Y, Morishima Y, et al. Nrf2 protects against pulmonary fibrosis by regulating the lung oxidant level and Th1/Th2 balance. Respir Res. 2010;11:31 10.1186/1465-9921-11-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Turgut NH, Kara H, Elagoz S, Deveci K, Gungor H, Arslanbas E. The protective effect of naringin against bleomycin‐induced pulmonary fibrosis in Wistar rats. Pulm Med. 2016;2016:7601393 10.1155/2016/7601393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch J, 3rd , Huynh R, Fishbein M, Saggar R, Belperio J, Weigt S. Idiopathic pulmonary fibrosis: epidemiology, clinical features, prognosis, and management. Semin Respir Crit Care Med. 2016;37(3):331‐357. 10.1055/s-0036-1582011 [DOI] [PubMed] [Google Scholar]
- 4. Raghu G, Chen SY, Yeh WS, et al. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001‐11. Lancet Respir Med. 2014;2(7):566‐572. 10.1016/S2213-2600(14)70101-8 [DOI] [PubMed] [Google Scholar]
- 5. Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N Engl J Med. 2001;345(7):517‐525. 10.1056/NEJMra003200 [DOI] [PubMed] [Google Scholar]
- 6. Lindahl G, Stock C, Leoni P, et al. Transforming growth factor‐beta‐induced expression of inhibin beta a (activin A), a potential therapeutic target in lung fibrosis, is blocked by bromodomain inhibitor Jq1 in primary adult pulmonary fibroblasts. Am J Respir Crit Care Med. 2014;189:A2015. [Google Scholar]
- 7. Marchand‐Adam S, Plantier L, Bernuau D, et al. Keratinocyte growth factor expression by fibroblasts in pulmonary fibrosis– poor response to interleukin‐1 beta. Am J Respir Cell Mol Biol. 2005;32(5):470‐477. 10.1165/rcmb.2004-0205OC [DOI] [PubMed] [Google Scholar]
- 8. Gupte VV, Ramasamy SK, Reddy R, et al. Overexpression of fibroblast growth factor‐10 during both inflammatory and fibrotic phases attenuates bleomycin‐induced pulmonary fibrosis in mice. Am J Respir Crit Care Med. 2009;180(5):424‐436. 10.1164/rccm.200811-1794OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J, Yu ZH, Zhou ZY, et al. Inhibition of alpha‐SMA by the ectodomain of FGFR2c attenuates lung fibrosis. Mol Med. 2012;18(6):992‐1002. 10.2119/molmed.2011.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607‐613. [DOI] [PubMed] [Google Scholar]
- 11. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141‐157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333‐338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 13. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384‐388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 14. Bachmayr‐Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057 10.1038/srep08057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR‐141 by targeting TGF‐beta1. Biochem Biophys Res Commun. 2017;487(4):769‐775. 10.1016/j.bbrc.2017.04.044 [DOI] [PubMed] [Google Scholar]
- 16. Zhou Z, Jiang R, Yang X, et al. circRNA mediates silica‐induced macrophage activation via HECTD1/ZC3H12A‐dependent ubiquitination. Theranostics. 2018;8(2):575‐592. 10.7150/thno.21648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Papageorgiou I, Court MH. Identification and validation of microRNAs directly regulating the UDP‐glucuronosyltransferase 1A subfamily enzymes by a functional genomics approach. Biochem Pharmacol. 2017;137:93‐106. 10.1016/j.bcp.2017.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498‐2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. American Thoracic Society, European Respiratory Society . American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165(2):277‐304. 10.1164/ajrccm.165.2.ats01 [DOI] [PubMed] [Google Scholar]
- 20. Plantier L, Crestani B, Wert SE, et al. Ectopic respiratory epithelial cell differentiation in bronchiolised distal airspaces in idiopathic pulmonary fibrosis. Thorax. 2011;66(8):651‐657. 10.1136/thx.2010.151555 [DOI] [PubMed] [Google Scholar]
- 21. Russell JL, Goetsch SC, Gaiano NR, Hill JA, Olson EN, Schneider JW. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ Res. 2011;108(1):51‐59. 10.1161/CIRCRESAHA.110.233262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bielesz B, Sirin Y, Si H, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120(11):4040‐4054. 10.1172/JCI43025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schnaper HW, Jandeska S, Runyan CE, et al. TGF‐beta signal transduction in chronic kidney disease. Front Biosci. 2009;14:2448‐2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao Z, Zhang J, Peng X, et al. The Notch gamma‐secretase inhibitor ameliorates kidney fibrosis via inhibition of TGF‐beta/Smad2/3 signaling pathway activation. Int J Biochem Cell Biol. 2014;55:65‐71. 10.1016/j.biocel.2014.08.009 [DOI] [PubMed] [Google Scholar]
- 25. Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19(6):781‐791. [DOI] [PubMed] [Google Scholar]
- 26. Milano J, McKay J, Dagenais C, et al. Modulation of notch processing by gamma‐secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82(1):341‐358. 10.1093/toxsci/kfh254 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y, Shen RW, Han B, et al. Notch signaling mediated by TGF‐beta/Smad pathway in concanavalin A‐induced liver fibrosis in rats. World J Gastroenterol. 2017;23(13):2330‐2336. 10.3748/wjg.v23.i13.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Phan SH. The myofibroblast in pulmonary fibrosis. Chest. 2002;122(6 suppl):286S‐289S. [DOI] [PubMed] [Google Scholar]
- 29. Desmoulière A, Darby IA, Gabbiani G. Normal and pathologic soft tissue remodeling: role of the myofibroblast, with special emphasis on liver and kidney fibrosis. Lab Invest. 2003;83(12):1689‐1707. [DOI] [PubMed] [Google Scholar]
- 30. Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127(3):526‐537. 10.1038/sj.jid.5700613 [DOI] [PubMed] [Google Scholar]
- 31. Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J Pathol. 2003;200(4):500‐503. 10.1002/path.1427 [DOI] [PubMed] [Google Scholar]
- 32. Chaudhary NI, Roth GJ, Hilberg F, et al. Inhibition of PDGF, VEGF and FGF signalling attenuates fibrosis. Eur Respir J. 2007;29(5):976‐985. 10.1183/09031936.00152106 [DOI] [PubMed] [Google Scholar]
- 33. Rodríguez‐Pascual F, Redondo‐Horcajo M, Lamas S. Functional cooperation between Smad proteins and activator protein‐1 regulates transforming growth factor‐beta‐mediated induction of endothelin‐1 expression. Circ Res. 2003;92(12):1288‐1295. 10.1161/01.RES.0000078491.79697.7F [DOI] [PubMed] [Google Scholar]
- 34. Rodriguez‐Pascual F, Reimunde FM, Redondo‐Horcajo M, Lamas S. Transforming growth factor‐beta induces endothelin‐1 expression through activation of the Smad signaling pathway. J Cardiovasc Pharmacol. 2004;44(suppl 1):S39‐S42. [DOI] [PubMed] [Google Scholar]
- 35. Clozel M, Salloukh H. Role of endothelin in fibrosis and anti‐fibrotic potential of bosentan. Ann Med. 2005;37(1):2‐12. [DOI] [PubMed] [Google Scholar]
- 36. Huang SK, Scruggs AM, McEachin RC, White ES, Peters‐Golden M. Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome‐wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung. PLOS One. 2014;9(9):e107055 10.1371/journal.pone.0107055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajasekaran S, Rajaguru P, Sudhakar Gandhi PS. MicroRNAs as potential targets for progressive pulmonary fibrosis. Front Pharmacol. 2015;6:254 10.3389/fphar.2015.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vittal R, Mickler EA, Fisher AJ, et al. Type V collagen induced tolerance suppresses collagen deposition, TGF‐beta and associated transcripts in pulmonary fibrosis. PLOS One. 2013;8(10):e76451 10.1371/journal.pone.0076451 [DOI] [PMC free article] [PubMed] [Google Scholar]


