Abstract
Get3 in yeast or TRC40 in mammals is an ATPase that, in eukaryotes, is a central element of the GET or TRC pathway involved in the targeting of tail‐anchored proteins. Get3 has also been shown to possess chaperone holdase activity. A bioinformatic assessment was performed across all domains of life on functionally important regions of Get3 including the TRC40‐insert and the hydrophobic groove essential for tail‐anchored protein binding. We find that such a hydrophobic groove is much more common in bacterial Get3 homologs than previously appreciated based on a directed comparison of bacterial ArsA and yeast Get3. Furthermore, our analysis shows that the region containing the TRC40‐insert varies in length and methionine content to an unexpected extent within eukaryotes and also between different phylogenetic groups. In fact, since the TRC40‐insert is present in all domains of life, we suggest that its presence does not automatically predict a tail‐anchored protein targeting function. This opens up a new perspective on the function of organellar Get3 homologs in plants which feature the TRC40‐insert but have not been demonstrated to function in tail‐anchored protein targeting. Our analysis also highlights a large diversity of the ways Get3 homologs dimerize. Thus, based on the structural features of Get3 homologs, these proteins may have an unexplored functional diversity in all domains of life.
Keywords: bacteria, Chlorophyta, Embryophyta, endoplasmic reticulum, Get3p, molecular chaperone, Rhodophyta, tail‐anchored protein
1. INTRODUCTION
Tail‐anchored (TA) proteins are a class of membrane proteins that contain a C‐terminal hydrophobic transmembrane segment (TMS) and a functional N‐terminal cytosolic domain.1, 2 TA proteins are a diverse group of eukaryotic membrane proteins found among others in the secretory pathway,3 nuclear envelope,4 peroxisomes,5 mitochondria6 and in chloroplasts.7 They have a wide range of functions, such as assistance in vesicular trafficking,3 protein translocation8 and degradation9 of membrane proteins. The function of TA proteins has been shown to be essential in all domains of life and their transport to the correct biological membrane, or protein targeting, needs to be efficient and accurate as targeting errors can have detrimental cellular effects. Additionally, TMSs are prone to aggregation and their spontaneous insertion into lipid bilayers may be slow in vivo. Therefore, in order to ensure efficient and organelle‐specific insertion of TA proteins and to prevent the aggregation of TMSs in the cytoplasm, most studies to date suggest that the targeting and insertion of TA proteins involves one or more cytosolic factors.
The mechanism through which TA proteins are targeted and inserted is distinct from the co‐translational signal recognition particle (SRP)‐facilitated process by which most membrane proteins with N‐terminal or internal signals are targeted. Indeed, because the C‐terminal TMS of a TA protein emerges from the ribosome at the end of translation, TA proteins are targeted and inserted through post‐translational mechanisms. One such pathway, the guided entry of TA proteins (GET), identified a little over 10 years ago, has been shown to mediate the proper delivery of several TA proteins in mammals,10, 11 budding yeast12 and more recently in plants.13, 14
Extensive biochemical and structural studies performed over the last decade have characterized the targeting of TA proteins utilizing the yeast GET pathway (as reviewed in15). Initially, a pre‐targeting complex, consisting of a small glutamine‐rich tetratricopeptide repeat containing protein Sgt2, and Get4 and Get5 in yeast, or Bag6, SGTA, TRC35 and UBL4A in mammals, captures the TA protein following its release from the ribosome, then transfers it to the ATPase Get3 in budding yeast, or TRC40 in mammals.15, 16, 17 The TA‐bound Get3/TRC40 protects and delivers the TA protein to the ER membrane, where its receptor complex comprised of Get1 and Get2 in yeast or WRB and CAML in mammals stimulates its subsequent release into the membrane.18, 19, 20, 21
Despite the apparent complexity and necessity of the GET pathway to prevent aggregation of hydrophobic proteins, depletion of GET pathway components in budding yeast (Saccharomyces cerevisiae) and Arabidopsis thaliana is not lethal.12, 14 Yet the functional importance of the GET pathway is highlighted by the fact that GET pathway deletion S. cerevisiae strains show increased heat and oxidative stress sensitivity,12, 22 and the depletion of TRC40 is embryonically lethal in mice.23 Since Get3 was shown to possess chaperone holdase activity upon oxidation24 these phenotypes may to some extent reflect a chaperone activity not involved in targeting TA proteins during biogenesis.
2. GET3 HOMOLOGS IN THE DIFFERENT DOMAINS OF LIFE
Some phylogenetic aspects of other GET pathway components have been recently discussed, in particular the evolutionary relationships between components of the pretargeting complex comprising Get4, Get5 and Bag617 and the legacy of membrane protein biogenesis factors similar to bacterial Oxa1 that also include Get1.18 Here, we combine a review of the literature on Get3 structure and function with a comprehensive bioinformatic analysis of the structural elements of the protein involved in TA protein binding. This integration focusses on properties of Get3‐like proteins in all domains of life that render the hydrophobic cage versatile and should be considered for both functions of these proteins.
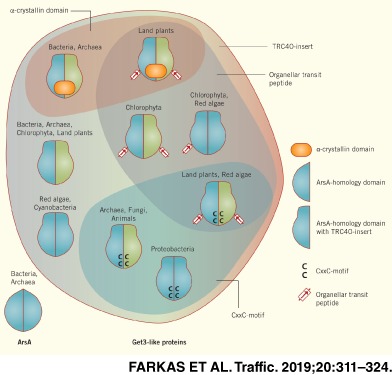
A systematic search for Get3‐ and ArsA‐homologous proteins in the KEGG and OrthoDB databases combined with further BLAST analysis yielded 2208 sequences (Supporting Information Table S1), from which 51 representative sequences were chosen to construct a phylogenetic tree (Figure 1). This analysis reveals a functionally unexplored diversity of Get3‐like proteins (Table 1). Focusing on structural aspects of different homologs such as domain organization or the presence of sequence motifs and comparing them with known structures and functions of Get3 homologs, we would like to highlight that Get3‐like chaperones from different kingdoms are more similar to each other than previously recognized based on a comparison of eukaryotic Get3 or TRC40 with prokaryotic ArsA.25 At the same time, they are remarkably diverse with respect to their modes of (pseudo)dimerization and structural features outside the well conserved ATPase domain.
Figure 1.

Maximum likelihood rooted phylogenetic tree of three representative sequences of each group of Get3 homologs as defined in Table 1. Percentage of trees in which the sequences clustered together after applying 1000 bootstraps are indicated at nodes if the value is higher than 70%. Scale bar indicates number of substitutions per site
Table 1.
Summary of structural features present in the different groups of the phylogenetic tree (Figure 1). Based on the sequences listed in Table S1
| Species | Protein | Identified sequences in group | Localization | Hydrophobic groove/TRC40‐insert | CxC motif | CxxC motif | α‐crystallin domain | Get3 domains | Get1/2/4 binding residues | Other features/Comments | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eukaryotes | Animals, Fungi | Get3/TRC40 | 629 | Cytoplasmic | Yes | Frequent | Yes | No | Single | Yes | In 597 out of 629 sequences |
| Land plants, Chlorophytes | Get3/TRC40 | 87 | Cytoplasmic | Yes | Yes | No | No | Single | Yes | C‐terminal charged patch | |
| Red algae | Group 1 | 4 | Cytoplasmic | Yes | Frequent | No | No | Double | Yes | In three out of four sequences | |
| Red algae, Chlorophytes | Group 2 | 10 | Organellar | Yes | No | No | No | Double | No | ||
| Land plants, Red algae | Group 3 | 90 | Organellar | Yes | No | Yes | No | Single | No | ||
| Chlorophytes | Group 4 | 8 | Organellar | Yes | No | No | No | Single | No | ||
| Land plants | Group 5 | 48 | Plastidial | Yes | No | No | Yes | Single | No | ||
| Bacteria | Mainly Actinobacteria, Chloroflexi, Chlorobi Cyanobacteria, Firmicutes, some Acidobacteria, Aquificae, Bacteroidetes, Fusobacteria, Spirochaetes, Proteobacteria | Group 6 | 301 | Cytoplasmic | Yes | No | No | Yes | Single | No | Rarely: nucleotide binding site missing (such homologs excluded from current analysis) |
| Cyanobacteria | Group 7 | 29 | Cytoplasmic | Yes | No | No | No | Double | No | ||
| Myxococcales, some Proteobacteria | Group 8 | 17 | Cytoplasmic | Yes | No | Yes | No | Double | No | ||
| Mainly Actinobacteria, Firmicutes, Proteobacteria, some Aquificae, Spirochaetes, Synergistetes | Group 9 | 131 | Cytoplasmic | Yes | No | No | No | Single | No | ||
| Some Firmicutes | ArsA‐like | 18 | Cytoplasmic | No | No | No | No | Single | No | Part of arsenite resistance operon | |
| Mainly Actinobacteria, Bacteroidetes, Proteobacteria, Firmicutes, Planctomycetes | ArsA | 432 | Cytoplasmic | No | No | No | No | Double | No | Part of arsenite resistance operon | |
| Archaea | Euryarcheota | ArsA | 27 | Cytoplasmic | No | No | No | No | Double | No | |
| Euryarcheota, Crenarcheota | Group 10 | 155 | Cytoplasmic | Yes | No | No | No | Single | No | Some of the Get1/2/4 binding residues are present in some homologs | |
| Euryarcheota, Thaumarcheota, | Group 11 | 21 | Cytoplasmic | Yes | No | No | Yes | Single | No | ||
| Euryarcheota, Crenarcheota | Group 12 | 200 | Cytoplasmic | Yes | No | Yes | No | Single | No |
3. STRUCTURAL ORGANIZATION OF ARSA AND GET3 PROTEINS
A bacterial homolog of Get3, ArsA confers resistance to arsenite in Escherichia coli 26 and shows high structural similarity to Get3 (Figure 2A,B). ArsA folds such that two highly similar domains in tandem form a metal binding site and two nucleotide binding sites (NBS) at their interface.27 The NBS is similar to those found in other members of the Signal recognition particle, MinD, BioD (SIMIBI) class of P‐loop NTPases and contains conserved structural elements necessary for ATP hydrolysis including the P‐loop, Switch I and II and the A‐loop.28, 29, 30 The metal binding site involves three functionally essential cysteine residues, however, these residues are not conserved in eukaryotic Get3 homologs27 (Figure 2A, ball‐and‐stick model residues).
Figure 2.
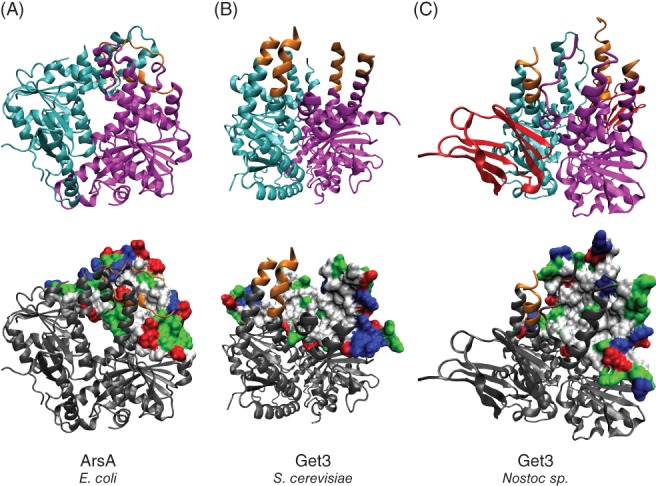
Structure of selected Get3 homologs. Top: individual domains (A) or subunits (B and C) are marked in cyan and magenta. Bottom: hydrophobic groove or homologous region shown in surface view. Hydrophobic and nonpolar residues are shown in white, polar residues shown in green, acidic residues shown in red and basic residues are shown in blue. To allow a better view of the interior of the groove, only half of the groove is shown in surface view (B and C). A, Structure of E. coli ArsA (PDB ID: 1F48). The region unique to ArsA is highlighted in orange. Heavy metal ion coordinating cysteines are shown as ball‐and‐stick models. B, Structure of S. cerevisiae Get3 (PDB ID: 4XTR). The region homologous to the one marked in orange in A is also marked in orange here. C, Structure of Get3 from a Nostoc species (PDB ID: 3IGF). The α‐crystallin domain is depicted in red
Unlike bacterial ArsA, Get3 in budding yeast (ScGet3) and other fungi and animals has a single Get3‐homology domain. Two ScGet3 monomers assemble into rotationally symmetrical homodimers to form a structure analogous to the arrangement of the two domains found in ArsA (Figure 2B). In ArsA, a short helix involved in coordinating the metal ion (orange in Figure 2A) folds into a groove, whereas the same region forms extended helices in Get3 (helix 7, 9; orange in Figure 2B), and also contains an additional stretch of amino acids dubbed the TRC40‐insert.25 Thus, a large hydrophobic surface is created (Figure 2B, bottom row) allowing Get3 to accommodate the TMSs of TA proteins. At the same time, the helix contained within the TRC40‐insert (helix 8, not visible in the structure) is thought to act as a lid that closes on captured TMSs, thus shielding them from the solvent.31
Get3 homologs with the ability to bind TA proteins have also been found in archaea, and one out of the four archaeal homologs studied so far could deliver captured substrates to the membrane.32, 33 In bacteria, the only currently known Get3 homologs with a hydrophobic groove belong to photosynthetic bacteria and they also have an α‐crystallin domain at the C‐terminus (red in Figure 2C).34 α‐crystallin domains are key components of heat shock proteins and are essential for their chaperone function.35 Although such Get3 homologs are also found in land plants,34 their function remains unknown. Moreover, land plants, Chlorophytes and red algae have been proposed or shown to have several Get3 homologs without an α‐crystallin domain as well, some of them in chloroplasts and mitochondria.14
4. CONSERVATION OF HELIX 8, THE “LID” CLOSING THE HYDROPHOBIC GROOVE
Get3 has several hydrophobic residues necessary for its interaction with TA proteins, and they mostly converge on the C‐terminal portion of helix 7 and the short helix 8 following it.25 Recently, it has emerged that helix 8 is needed to ensure an efficient transfer of substrates from upstream components to Get3, but it has no major effect on the dissociation of substrates already captured by Get3.36 In the structures of eukaryotic Get3 homologs, the region around helix 8 is poorly defined because of its high flexibility and is heavily influenced by the overall conformation of the protein.25, 31 Although helix 8 forms a helix separate from helix 7 in fungal Get3 structures (Figure 3A), these two helices appear to line up or even merge completely in structures of archaeal homologs of Get3.32, 33 The Get3 homolog of the archaeeon Methanocaldococcus jannaschii (MjGet3) exists either as a dimer, similar to S. cerevisiae Get3 (ScGet3) or in a tetrameric form, a dimer of dimers, the assembly of which is mediated by the region corresponding to helix 8 in ScGet3 (Figure 3B).33 Although ScGet3 and its human homolog TRC40 both form tetramers under specific conditions,37, 38 these tetrameric structures remain structurally unsolved and the role of helix 8 in their assembly also remains unknown. Nevertheless, it is intriguing that based on secondary structure predictions, the regions corresponding to helix 7 and 8 of ScGet3 would be expected to form a single helix as seen in MjGet3 (Figure 3A), yet whether this region can indeed assume two distinct conformations remains to be seen.
Figure 3.
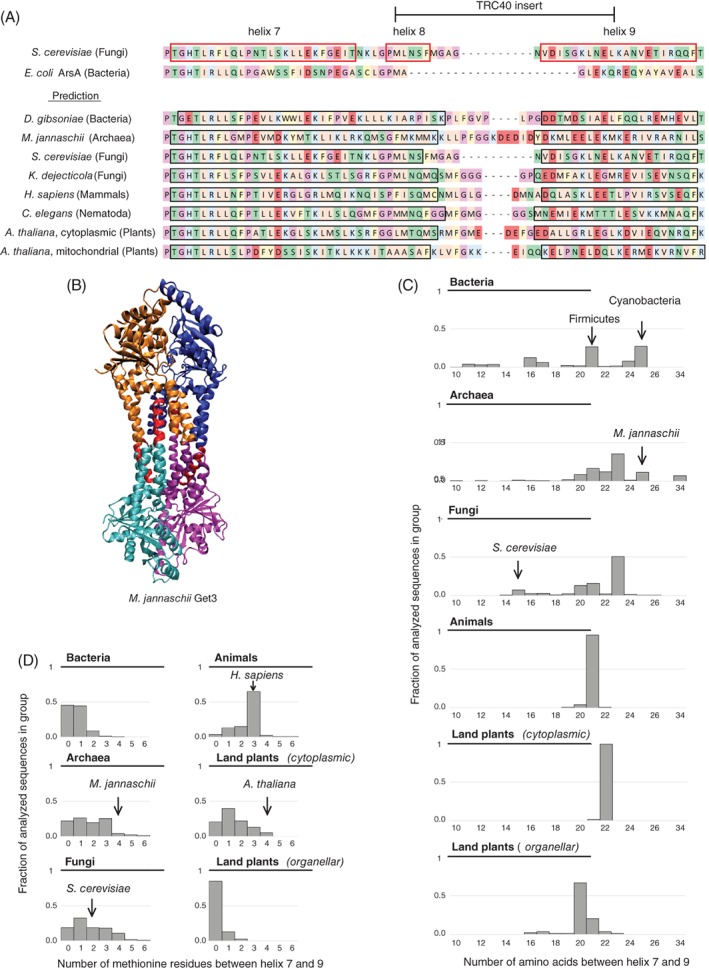
Comparison of the TRC40‐insert between species. A, Known secondary structure of ScGet3 (top) compared with the predicted structure of the same region in different species (bottom, predicted helices marked with black frame). Hydrophobic residues shown in peach, aromatic residues in ochre, basic residues in blue, acidic residues in red, hydrophilic residues in green, proline and glycine in mauve, cysteine in yellow. B, Structure of M. jannaschii Get3 (PDB ID: 3UG6). Subunits are marked with cyan, magenta, orange and blue. The region homologous to the region between helix 7 and 9 in ScGet3 is shown in red. C, Distribution of the length of the region homologous to the sequence between helix 7 and 9 in ScGet3 among the sequences used for the current analysis. All bins containing at least 1% of the sequences are shown in the chart. Number of analyzed sequences: Bacteria—299; Archaea—376; Fungi—489; Animals—140; Land plants (cytoplasmic) —78; Land plants (organellar, excluding α‐crystallin domain Get3 homologs)—87. D, Distribution of the number of methionine residues in the region homologous to the sequence between helix 7 and 9 in ScGet3 among the sequences used for the current analysis. All bins containing at least 1% of the sequences are shown in the chart. The number of sequences analyzed are as in C
Although the region linking helix 7 and 9 appears to be moderately conserved, especially at helix 8 in eukaryotes, its length varies considerably within and between phylogenetic groups (Figure 3C). Indeed, although the average length of the stretch homologous to the linker between helix 7 and 9 in ScGet3 is approximately 21 to 22 amino acids in eukaryotes, there are notable exceptions as well. For instance, ScGet3 only has 15 amino acids in this region while the archaeal MjGet3 has 25, showing that from a functional perspective, substantial variation is allowed in this region. Interestingly, unlike in bacterial ArsA (Figure 3A), the length of this region in bacterial Get3 homologs with an α‐crystallin domain is comparable to that observed in eukaryotes. For example, while the region in Firmicutes is often 21 amino acids long, just like in animals, in Cyanobacteria it is as long as in MjGet3 and in many Chlorobi bacteria almost as short as in ScGet3. However, whether this indicates any functional similarity is not known.
Besides the length of the linker between helix 7 and 9, its amino acid composition also shows variation within and between phyla (Figure 3D). It has been suggested that the methionine‐rich nature of the hydrophobic groove is important for the accommodation of the TMS.25 This hypothesis is further strengthened by the analogy with SRP, where the methionine‐rich M domain of Srp54 is essential for binding the signal peptide.39 In Get3 helix 8, there are two and three methionine residues in ScGet3 and human TRC40, respectively, and their combined loss in ScGet3 leads to decreased substrate binding.25 Consistent with the idea that the presence of the methionine residues is related to the TA targeting function, in homologs not expected to be involved in TA protein targeting (bacterial and plastidial‐mitochondrial Get3 homologs without an α‐crystallin domain in land plants), there is mostly no or just a single methionine in the corresponding region. However, looking at Get3 homologs known to bind or target TA proteins, it becomes clear that although eukaryotic and archaeal homologs tend to have at least one or more methionine residues in this stretch, there are several species without any as well (Figure 3D). Taken together, although helix 8 may have become enriched in methionine in certain species to support TA protein targeting, the presence of methionine residues does not seem to be a requirement for helix 8 to fulfill its function.
5. HELICES LINING THE HYDROPHOBIC GROOVE
One of the defining features of Get3 with respect to bacterial ArsA is the presence of the TRC40‐insert, which corresponds to helix 8 in ScGet3 and the amino acids linking it to helix 9 (Figure 4A).25 The TRC40 insert with an extended helix 7 and 9, together with helices 4, 5 and 6 creates a hydrophobic area so that TMSs can be accommodated and shielded from solvents in the resulting groove. Mutational studies have revealed that some of the hydrophobic residues of helix 7 and 8 are important for substrate binding by Get3.25 Interestingly, while the residues that show the strongest effect in mutational studies of ScGet3 are not conserved in bacterial ArsA, other hydrophobic residues in helix 7 are universally conserved in eukaryotic Get3 homologs and bacterial ArsA as well (Figure 4A).40 Therefore, the presence of these crucial hydrophobic residues and the TRC40‐insert may be indicative of functional similarity between eukaryotic Get3 and any given bacterial homolog. Indeed, several bacterial phyla have Get3 homologs with an α‐crystallin domain that also have the TRC40‐insert, and the surrounding helices often contain periodic hydrophobic amino acids that are even positionally conserved (Figure 4A). Although previously only described in photosynthetic bacteria and land plants,34 such Get3 homologs can be found in diverse groups of non‐photosynthetic bacteria as well, including Actinobacteria, Aquificae, Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria.
Figure 4.
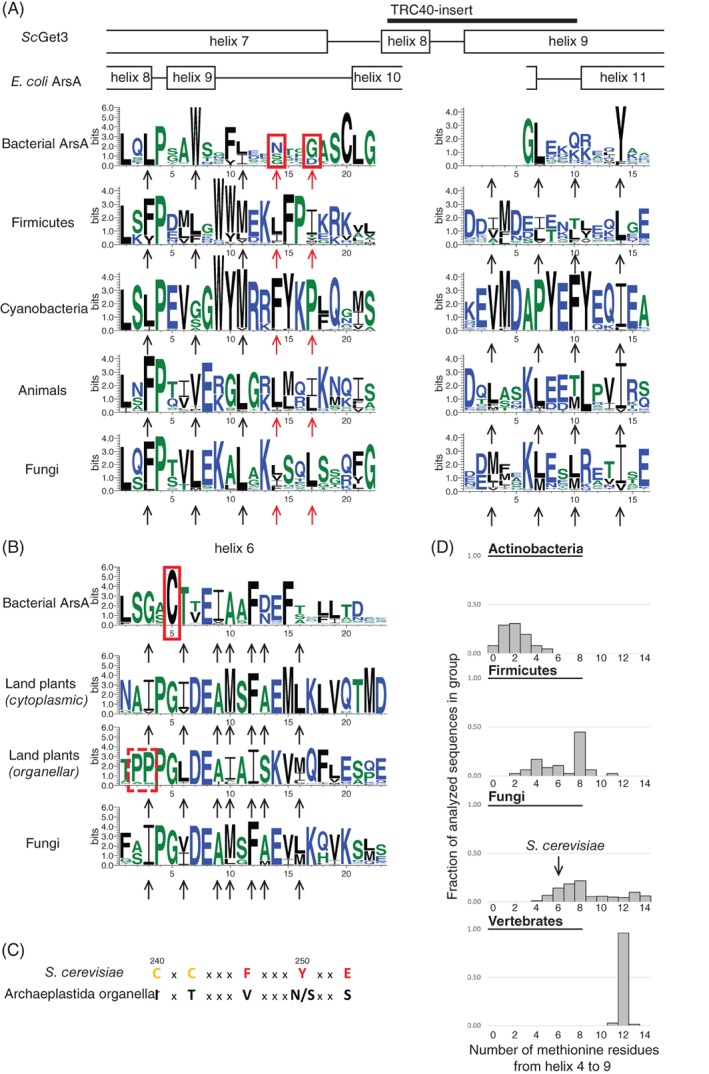
Consensus sequence and important features of the helices flanking the hydrophobic groove and of the region C‐terminally adjacent to it. A, Consensus sequence of the region homologous to ScGet3 helix 7 and 9 in different groups of Get3 homologs. Residues flanking the hydrophobic groove in ScGet3 are marked with an arrow. Residues shown to be important for TA protein binding are marked with a red arrow. Corresponding residues are highlighted with a red rectangle in the ArsA consensus sequence. B, Consensus sequence of the bottom of the hydrophobic groove (ScGet3 helix 6) in different groups of Get3 homologs. Residues facing the hydrophobic groove in ScGet3 are marked with an arrow. Heavy metal ion coordinating cysteine in ArsA and additional proline residues in organellar Get3 homologs highlighted by red boxes. C, Comparison of the CxC motif and key Get1/Get2/Get4 residues between ScGet3 and organellar homologs of Get3 in land plants. D, Distribution of the number of methionine residues in the region homologous to the sequence from helix 4 to 9 in ScGet3 among the sequences used for the current analysis. All bins containing at least 1% of the sequences are shown in the chart. Number of analyzed sequences: Actinobacteria—62; Firmicutes—47; Fungi—489; Vertebrates—70
The helix lying at the bottom of the hydrophobic groove, helix 6, shows a high overall similarity between bacterial ArsA and Get3, and mutations of hydrophobic residues in this helix mostly affect the ATPase activity of Get3 but not substrate binding.25 Since one of the cysteines involved in coordinating the metal ion is close to the N‐terminal part of this helix, its presence, coupled with the lack of the Get3‐/TRC40‐insert, is expected to be a strong indicative feature of ArsA homologs (Figure 4B). Indeed, most such ArsA homologs in our analysis are highly similar to E. coli ArsA and have two domains in tandem. However, some Firmicutes bacteria seem to be unique in that they possess two copies of such ArsA homologs, but each with only a single domain instead of two (Table 1). In this case, one of them is similar to the first domain of E. coli ArsA, and the other is similar to the second. However, because there is no obvious feature that would mediate dimerization, it is unknown whether they actually do form dimers and function as a bona fide ArsA in vivo.
Another special feature in helix 6 is found in plastidial and mitochondrial Get3 homologs without an α‐crystallin domain in land plants, Chlorophyta and red algae, that is, the Archaeplastida clade. Besides the Get3 homologs already shown to localize to the chloroplast and mitochondria,14 similar organellar Get3 homologs are predicted to exist in other groups within the Archaeplastida clade as well (Table S2). In spite of overall sequence similarity to ScGet3, many of these homologs have several proline residues at the N‐terminus of helix 6 (Figure 4B), the relevance of which is currently unknown. Furthermore, such homologs uniformly lack the CxC motif found on the beta strand following helix 9, a feature strongly, although not universally conserved among eukaryotic cytoplasmic Get3 homologs (Figure 4C). Considering that these homologs also lack key residues required for binding Get1, Get2 and Get4 (Figure 4C, Table 1), it is clear that such organellar Get3 homologs should fulfill a related yet distinct function compared to cytoplasmic Get3 homologs.
Looking at the hydrophobic groove as a whole, its methionine‐rich nature has been thought to be a feature related to the TA protein targeting function of Get3.25 As stated above, the presence of methionine residues in helix 8 is probably not a prerequisite for TA protein targeting. However, counting all the methionine residues that could potentially flank the hydrophobic groove (from helix 4 to helix 9), it becomes clear that despite considerable variety, all fungi have at least four methionine residues in this region (ScGet3 has six), and most vertebrates have three times as many (Figure 4D). On the other hand, many bacterial Get3 homologs with an α‐crystallin domain also have multiple methionine residues (Table S3), with several Firmicutes homologs having as many as eight (Figure 4D). As an exception, homologs in Actinobacteria tend to have fewer or no methionine residues at all (Figure 4D). Taken together, the fact that many bacterial homologs have as many methionine residues as some fungi do, and that there has been no indication so far that these homologs target TA proteins in bacteria, it is likely that the methionine‐rich nature of the hydrophobic groove had already been present before the TA protein targeting function of Get3 was acquired. Then, as eukaryotic Get3 became more and more specialized to target TA proteins, it may have acquired further methionine residues in the groove to facilitate the binding of TMSs.
6. GET3 HOMOLOGS IN THE EUKARYOTIC GROUP ARCHAEPLASTIDA
In yeast and most other eukaryotes, Get3 functions as a rotationally symmetrical homodimer because dimerization is necessary for both its ATPase activity and the formation of the TMS binding hydrophobic groove.22, 25 In fungi and mammals, a conserved CxxC motif in each subunit aligns to coordinate a zinc ion (Figure 5A), which is necessary for dimer formation.22, 25 Bacterial ArsA homologs are very similar structurally, but the two halves of the dimer are produced as two domains in tandem in a single polypeptide chain. Each domain corresponds to a subunit in a eukaryotic dimeric Get3.27 It is likely that because of the two domains being part of a single protein, the interaction between them is stable enough so that no CxxC is required in ArsA homologs.
Figure 5.
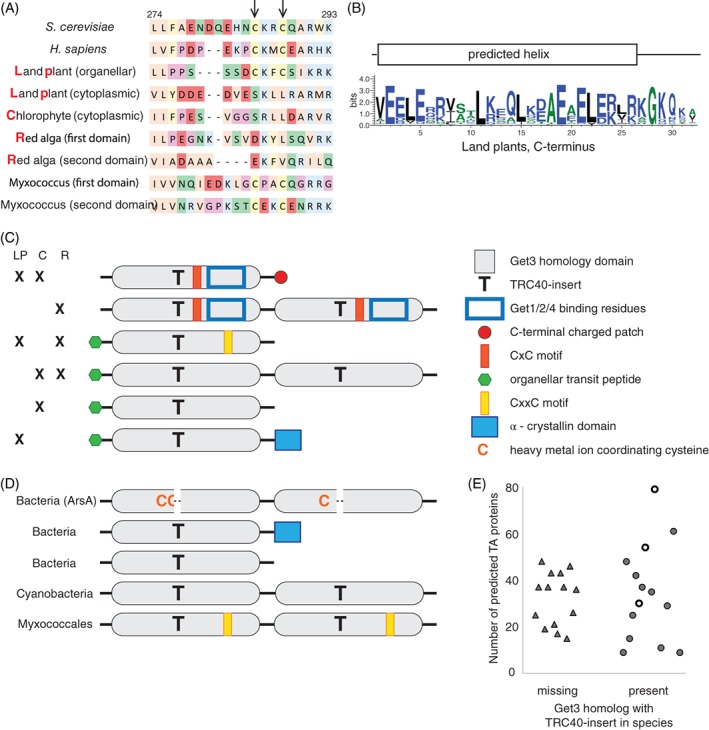
Get3 homologs use various strategies to form dimers. A, Comparison of the sequence adjacent to the CxxC motif in ScGet3 and homologs from other organisms. B, Consensus sequence and secondary structure prediction of the charged C‐terminal helix found in cytoplasmic Get3 homologs in land plants. C, Graphical representation of main structural features of land plant (LP), chlorophyte (C) and red algal (R) Get3 homologs. D, Graphical representation of main structural features of bacterial Get3 homologs. E, Comparison of the presence or absence of a TRC40‐insert containing Get3 homolog in bacterial species with the number of predicted TA proteins in the given species. Empty circles represent Proteobacteria and Cyanobacteria with TRC40‐insert containing Get3 homologs arranged as two domains in a single polypeptide
Surprisingly, cytoplasmic homologs in land plants and Chlorophytes lack the CxxC motif, yet they are functional in targeting TA proteins13 and they form dimers.14 An analysis of the sequence of these homologs provides clues as to how dimerization could happen in the absence of the CxxC motif. On the one hand, a pronounced acidic patch composed of three to five acidic residues is located in land plant homologs adjacent to the site where the CxxC motif would be (Figure 5A). Barring a few exceptions, an ExxE motif is found in this sequence, and such motifs are known to be able to coordinate iron ions.41, 42 Therefore, although land plant homologs are lacking a CxxC motif, they could still utilize metal ions to stabilize the dimer. On the other hand, most land plant and Chlorophyte homologs have a short, ca. 30 amino acid long, strongly charged extension missing in all other phyla, which may be involved in dimerization (Figure 5B and C). Finally, cytoplasmic Get3 homologs in red algae are distinct from land plants or Chlorophytes, in that they form a single polypeptide chain containing two domains in tandem, just like bacterial ArsA, and they similarly lack the CxxC motif as well (Figure 5A and C).
Compared to cytoplasmic homologs, predicted organellar Get3 variants without an α‐crystallin domain in Archaeplastida display an even greater diversity. Namely, these proteins lack the CxC motif and key Get1, Get2, Get4 binding residues and they often contain extra prolines in helix 6. Even so, they are still hypothesized to dimerize and use several different ways to achieve this (Figure 5C). In organellar homologs in land plants and red algae, a CxxC motif is present, and is likely used to form a dimer. However, red algae have other homologs as well, as do Chlorophytes, that contain two domains in a single protein. Intriguingly, additional homologs of Get3 can be found in Chlorophytes that have no apparent dimerization motif, which does not exclude the possibility that they still form dimers in unexpected ways.
It has to be noted that an organellar homolog from Chlamydomonas reinhardtii predicted here to be organellar has been previously proposed to be cytoplasmic.43 However, the protein is highly similar to other homologs in land plants that have been shown to be organellar,14 and homologous proteins from other Chlorophytes are consistently predicted to be organellar as well (Table S2). Therefore, the localization of these homologs in Chlorophytes remains uncertain for the moment. The picture is further complicated by the fact that some of the Get3 homologs in Archaeplastida are highly similar in sequence to other organellar homologs, yet are predicted to be cytoplasmic because of a lack of a transit peptide (Table S2). This could either indicate a further cytoplasmic group of such homologs or simply reflect an inaccurate bioinformatic prediction of the N‐terminus of the proteins based on genomic sequences.
Besides the above‐mentioned homologs, land plants also have a plastidial Get3 homolog that is closer in similarity to cyanobacterial homologs than it is to eukaryotic ones (Figures 5C and 1 and Table 1).34 Accordingly, this is the only eukaryotic Get3 homolog currently known to have an α‐crystallin domain at its C‐terminus like the one seen in the structure of NostocGet3 (Figure 2C).
The fact that land plants have Get3 homologs in the chloroplast with hypothetically two different ways to dimerize (one with an α‐crystallin domain, one with a CxxC‐motif) raises the question of what advantage having two such close homologs may bring. A possibility would be that the different modes of dimerization allow the organism to regulate the activity or various functions of the protein. Nonetheless, as no study has been carried out on these proteins to date, their function remains elusive as of now.
7. PREVIOUSLY UNNOTICED BACTERIAL GET3 HOMOLOGS
As mentioned above, several major groups of bacteria have a Get3 homolog with the TRC40‐insert similar to NostocGet3 (Figure 2C), with an α‐crystallin domain attached to the C‐terminus and a nucleotide binding site present, which is missing in NostocGet3 (Figure 5D, Table 1). Although it is known that α‐crystallin domains can mediate dimerization and act as a chaperone,35 it is not clear from the available structure of NostocGet3 whether or how it contributes to the stabilization of the Get3 dimer. The possibility that it may have a different function is supported by the fact that several groups of bacteria have Get3 homologs with the TRC40‐insert but no α‐crystallin domain (Figure 5D and Table 1). These are highly similar to archaeal Get3 homologs lacking a CxxC motif, at least one of which has been demonstrated to be able to form dimers and bind TA proteins.33 Therefore, it is highly likely that they can also dimerize and bind hydrophobic sequences.
Furthermore, uniquely among bacteria, Cyanobacteria, Myxococcal species and some further Proteobacteria contain Get3 homologs with two domains in tandem where both domains contain a TRC40‐insert (Figure 5D). Except for Cyanobacteria, they also have the CxxC motif, which makes these homologs unique not only among bacteria but in all domains of life.
It is currently unclear what the functions of these Get3 homologs are. Taking into consideration that all of the above‐mentioned bacterial homologs have the TRC40‐insert, current theory would predict their involvement in TA protein biogenesis. Since they have not been characterized yet, we can only rely on predictions. If they indeed insert TA proteins into the membrane, one would expect that bacterial species with more TA proteins would be more likely to have a TRC40‐insert containing Get3 homolog than species that have fewer TA proteins. Indeed, Proteobacteria and Cyanobacteria with Get3 homologs arranged as two domains in tandem tend to have more predicted TA proteins than other bacteria (empty circles in Figure 5E and Table S4). However, comparing the abundance of predicted TA proteins between different bacterial species and the presence or absence of other Get3 homologs with a TRC40‐insert reveals no correlation (Figure 5E and Table S4). Furthermore, it has been shown that other chaperones are responsible for TA protein targeting in bacteria, at least in E. coli. 44
Considering that ScGet3 can act as a more general chaperone under specific conditions,38 that a large group of bacterial Get3 homologs have an α‐crystallin domain with expected chaperone activity, and that all the TRC40‐insert‐containing homologs mentioned above are predicted to have a hydrophobic groove, it is possible that these homologs act more as general chaperones than TA protein targeting factors. From this perspective, the TA protein targeting activity of cytoplasmic eukaryotic Get3 homologs could represent an adaptation of an ancient, more general chaperoning function. On the same note, it would be interesting to know whether the above‐mentioned Get3 homologs in chloroplasts and mitochondria have a similar function to those found in bacteria or represent a third group of Get3‐like chaperones with unexpected functions. As summarized in Table 1, it is clear that Get3‐like chaperones are widespread and structurally diverse and much remains to be discovered about the dynamic structure and function of these proteins.
8. MATERIALS AND METHODS
8.1. Retrieval and processing of sequences and structures
Identifiers of Get3 and ArsA homologs were retrieved from KEGG Database (https://www.genome.jp/kegg/) and OrthoDB45 (https://www.orthodb.org). Identifiers of Get3 homologs with an α‐crystallin domain in land plants were retrieved using a blast search in land plants using the sequence of NostocGet3. Sequences were retrieved from Uniprot (www.uniprot.org) based on the identifiers collected from the databases and the blast search and aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Alignment was visualized and manually adjusted using Jalview.46 Incomplete sequences were filtered out based on missing major regions compared to other homologs. Secondary structure predictions were carried out using JPred4 (http://www.compbio.dundee.ac.uk/jpred4/).47 Logos of consensus sequences were visualized using WebLogo 3.48 Structures were retrieved from RCSB (https://www.rcsb.org/) and visualized using VMD (http://www.ks.uiuc.edu/Research/vmd/).49
8.2. Construction of the phylogenetic tree
All analyses related to the phylogenetic tree were carried out in Mega X.50 Sequences were aligned using the MUSCLE algorithm with default settings and manually adjusted when necessary. The evolutionary history was inferred by using the Maximum Likelihood method and Whelan and Goldman + Freq. model.51 The tree with the highest log likelihood (−46 225.64) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches if they clustered together in more than 70% of the trees. Initial trees for the heuristic search were obtained automatically by applying Neighbor‐Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 1.3658]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 51 amino acid sequences. There were a total of 1055 positions in the final dataset.
8.3. Prediction of TA proteins in bacteria
The proteome of each species was download from Uniprot (www.uniprot.org). Transmembrane domains in the whole proteome were predicted using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).52 Proteins with a single TMS and less than 30 amino acids between the TMS and the C‐terminus were considered candidates. These were tested for the presence of an N‐terminal signal sequence using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/) with a cutoff value set to the value recommended by the software for the given bacterial phylum.53
Supporting information
TABLE S1. List of Get3 homologs used in the current study. The classification of each homolog based on the groups listed in Table 1 and the species they are found in are provided
TABLE S2. List of Get3 homologs with a potential plastidial or mitochondrial localization used in the current study. The classification of each homolog based on the groups listed in Table 1 and their predicted localization based on TargetP 1.1 are provided. Abbreviations of predictions: C, chloroplast; M, mitochondria; −, no organelle predicted
TABLE S3. Distribution of the number of methionine residues in the region homologous to the sequence from helix 4 to 9 in ScGet3 among bacteria containing Get3 homologs with an α‐crystallin domain.
TABLE S4. Analysis of presence or absence of different types of Get3 or ArsA homologs, number of proteins, membrane proteins and tail‐anchored proteins in the indicated bacterial taxa.
ACKNOWLEDGMENTS
We thank Nica Borgese, Ursula Jakob and Kathrin Ulrich as well as members of the Schwappach lab for extremely useful comments and discussion. The work was funded by the Deutsche Forschungsgemeinschaft (SFB1190 P04 to B.S. and A.F.).
Farkas Á, De Laurentiis EI, Schwappach B. The natural history of Get3‐like chaperones. Traffic. 2019;20:311–324. 10.1111/tra.12643
Funding information Deutsche Forschungsgemeinschaft, Grant/Award Number: SFB1190 P04
Peer Review The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12643
REFERENCES
- 1. Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C‐terminal anchor. Trends Cell Biol. 1993;3:72‐75. [DOI] [PubMed] [Google Scholar]
- 2. Borgese N, Fasana E. Targeting pathways of C‐tail‐anchored proteins. Biochim Biophys Acta Biomembr. 2011;1808(3):937‐946. [DOI] [PubMed] [Google Scholar]
- 3. Hardwick KG, Pelham HRB. SED5 encodes a 39‐kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119(3):513‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Manilal S, Man NT, Sewry CA, Morris GE. The Emery‐Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet. 1996;5(6):801‐808. 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- 5. Elgersma Y, Kwast L, van den Berg M, et al. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S.Cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16(24):7326‐7341. 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase‐mediated mitochondrial fission is a multi‐step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367‐380. 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Loeffelholz O, Kriechbaumer V, Ewan RA, et al. OEP61 is a chaperone receptor at the plastid outer envelope. Biochem J. 2011;438(1):143‐153. 10.1042/BJ20110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finke K, Plath K, Panzner S, et al. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae . 1996;15(7):1482‐1494. [PMC free article] [PubMed] [Google Scholar]
- 9. Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365(6442):176‐179. 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 10. Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128(6):1147‐1159. 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 11. Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail‐anchored (TA) proteins. J Cell Sci. 2008;121(11):1832‐1840. 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schuldiner M, Metz J, Schmid V, et al. The GET complex mediates insertion of tail‐anchored proteins into the ER membrane. Cell. 2008;134(4):634‐645. 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srivistava R, Zalisko BE, Keenan RJ, Howell SH. The GET system inserts the tail‐anchored SYP72 protein into endoplasmic reticulum membranes. Plant Physiol. December 2016;2016:00928‐01145. 10.1104/pp.16.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xing S, Mehlhorn DG, Wallmeroth N, et al. Loss of GET pathway orthologs in Arabidopsis thaliana causes root hair growth defects and affects SNARE abundance. Proc Natl Acad Sci. 2017;114(8):E1544‐E1553. 10.1073/pnas.1619525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mateja A, Keenan RJ. A structural perspective on tail‐anchored protein biogenesis by the GET pathway. Curr Opin Struct Biol. 2018;51:195‐202. 10.1016/j.sbi.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mock J‐Y, Chartron JW, Zaslaver M, Xu Y, Ye Y, Clemons WM. Bag6 complex contains a minimal tail‐anchor–targeting module and a mock BAG domain. Proc Natl Acad Sci. 2015;112(1):106‐111. 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mock J‐Y, Xu Y, Ye Y, Clemons WM. Structural basis for regulation of the nucleo‐cytoplasmic distribution of Bag6 by TRC35. Proc Natl Acad Sci. 2017;114(44):11679‐11684. 10.1073/pnas.1702940114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anghel SA, McGilvray PT, Hegde RS, Keenan RJ. Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 2017;21(13):3708‐3716. 10.1016/j.celrep.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang F, Chan C, Weir NR, Denic V. The Get1/2 transmembrane complex is an endoplasmic‐reticulum membrane protein insertase. Nature. 2014;512(7515):441‐444. 10.1038/nature13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vilardi F, Lorenz H, Dobberstein B. WRB is the receptor for TRC40/Asna1‐mediated insertion of tail‐anchored proteins into the ER membrane. J Cell Sci. 2011;124(8):1301‐1307. 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail‐anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48(3):387‐397. 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 22. Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem. 2006;281(1):410‐417. 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 23. Mukhopadhyay R, Ho Y‐S, Swiatek PJ, Rosen BP, Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580(16):3889‐3894. 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 24. Voth W, Schick M, Gates S, et al. The protein targeting factor Get3 functions as ATP‐independent chaperone under oxidative stress conditions. Mol Cell. 2014;56(1):116‐127. 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mateja A, Szlachcic A, Downing ME, et al. The structural basis of tail‐anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361‐366. 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C‐M, Misra TK, Silver S, Rosen BP. Nucleotide sequence of the structural genes for an anion pump. J Biol Chem. 1986;261(32):15030‐15038. [PubMed] [Google Scholar]
- 27. Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19(17):4838‐4845. 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bange G, Sinning I. SIMIBI twins in protein targeting and localization. Nat Struct Mol Biol. 2013;20(7):776‐780. 10.1038/nsmb.2605. [DOI] [PubMed] [Google Scholar]
- 29. Zhou T, Radaev S, Rosen BP, Gatti DL. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J Biol Chem. 2001;276(32):30414‐30422. 10.1074/jbc.M103671200. [DOI] [PubMed] [Google Scholar]
- 30. Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P‐loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41‐72. 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 31. Mateja A, Paduch M, Chang H‐Y, et al. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347(6226):1152‐1155. 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sherrill J, Mariappan M, Dominik P, Hegde RS, Keenan RJ. A conserved archaeal pathway for tail‐anchored membrane protein insertion. Traffic. 2011;12(9):1119‐1123. 10.1111/j.1600-0854.2011.01229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suloway CJM, Rome ME, Clemons WM. Tail‐anchor targeting by a Get3 tetramer: the structure of an archaeal homologue. EMBO J. 2012;31(3):707‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chartron JW, Clemons WM, Suloway CJM. The complex process of GETting tail‐anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22(2):217‐224. 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hochberg GKA, Ecroyd H, Liu C, et al. The structured core domain of B‐crystallin can prevent amyloid fibrillation and associated toxicity. Proc Natl Acad Sci. 2014;111(16):E1562‐E1570. 10.1073/pnas.1322673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chio US, Chung S, Weiss S, Shan S. A chaperone lid ensures efficient and privileged client transfer during tail‐anchored protein targeting. Cell Rep. 2019;26(1):37‐44.e7. 10.1016/j.celrep.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurdi‐Haidar B, Heath D, Aebi S, Howell SB. Biochemical characterization of the human arsenite‐stimulated ATPase (hASNA‐I). J Biol Chem. 1998;273(35):22173‐22176. 10.1074/jbc.273.35.22173. [DOI] [PubMed] [Google Scholar]
- 38. Bozkurt G, Stjepanovic G, Vilardi F, et al. Structural insights into tail‐anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci. 2009;106(50):21131‐21136. 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lütcke H, High S, Römisch K, Ashford AJ, Dobberstein B. The methionine‐rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992;11(4):1543‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borgese N, Righi M. Remote origins of tail‐anchored proteins. Traffic. 2010;11(7):877‐885. 10.1111/j.1600-0854.2010.01068.x. [DOI] [PubMed] [Google Scholar]
- 41. Wösten MMSM, Kox LFF, Chamnongpol S, Soncini FC, Groisman EA. A signal transduction system that responds to extracellular iron. Cell. 2000;103(1):113‐125. 10.1016/S0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 42. Stearman R, Yuan DS, Yamaguchi‐Iwai Y, Klausner RD, Dancis A. A permease‐oxidase complex involved in High‐affinity iron uptake in yeast. Science. 1996;271(5255):1552‐1557. 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 43. Formighieri C, Cazzaniga S, Kuras R, Bassi R. Biogenesis of photosynthetic complexes in the chloroplast of Chlamydomonas reinhardtii requires ARSA1, a homolog of prokaryotic arsenite transporter and eukaryotic TRC40 for guided entry of tail‐anchored proteins. Plant J. 2013;73(5):850‐861. 10.1111/tpj.12077. [DOI] [PubMed] [Google Scholar]
- 44. Peschke M, Le Goff M, Koningstein GM, et al. SRP, FtsY, DnaK and YidC are required for the biogenesis of the E. coli tail‐anchored membrane proteins DjlC and Flk. J Mol Biol. 2018;430(3):389‐403. 10.1016/j.jmb.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 45. Kriventseva EV, Kuznetsov D, Tegenfeldt F, et al. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47(D1):D807‐D811. 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2‐‐a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189‐1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389‐W394. 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Crooks GE, Hon G, Chandonia J‐M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188‐1190. 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33‐38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 50. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547‐1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum‐likelihood approach. Mol Biol Evol. 2001;18(5):691‐699. 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 52. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol. 2001;305(3):567‐580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 53. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785‐786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1. List of Get3 homologs used in the current study. The classification of each homolog based on the groups listed in Table 1 and the species they are found in are provided
TABLE S2. List of Get3 homologs with a potential plastidial or mitochondrial localization used in the current study. The classification of each homolog based on the groups listed in Table 1 and their predicted localization based on TargetP 1.1 are provided. Abbreviations of predictions: C, chloroplast; M, mitochondria; −, no organelle predicted
TABLE S3. Distribution of the number of methionine residues in the region homologous to the sequence from helix 4 to 9 in ScGet3 among bacteria containing Get3 homologs with an α‐crystallin domain.
TABLE S4. Analysis of presence or absence of different types of Get3 or ArsA homologs, number of proteins, membrane proteins and tail‐anchored proteins in the indicated bacterial taxa.


