Abstract
Introduction
Previous studies have shown that miR-373 functions as either a tumor suppressor or an oncogene depending on which type of cancer it’s operating in. However, the functional role of miR-373 in neuroblastoma (NB) remains largely unclear.
Methods
Expression of miR-373 and SRC kinase signaling inhibitor 1 (SRCIN1) in 20 metastatic and 20 primary NB tissues was detected by quantitative real-time PCR (qRT-PCR) and Western blotting. MTT assay, flow cytometry analysis and transwell migration and invasion assays were performed to evaluate the influence of miR-373 inhibition on the growth, migration and invasion of NB cells, respectively. In vivo experiment was applied to determine the effect of miR-373 inhibition on tumor growth. Dual-luciferase reporter assay was used to confirm the interaction between miR-373 and SRCIN1.
Results
We observed a significant increase in the expression of miR-373 in metastatic NB samples compared with primary NB samples, and this was inversely correlated with SRCIN1 expression. Functional studies revealed that depletion of miR-373 inhibited in vitro NB cell growth, migration and invasion, and also suppressed tumor growth in an in vivo mouse model. Moreover, we identified that SRCIN1 was a direct and functional target gene of miR-373. Silencing of SRCIN1 partially rescued the antimiR-373-mediated inhibition of cell growth, migration and invasion.
Conclusion
The data from our study verified a potential oncogenic role of miR-373 in NB cells that occurs through direct targeting SRCIN1. The newly identified miR-373/SRCIN1 axis represents a new potential candidate for therapeutic intervention of malignant NB.
Keywords: miR-373, NB, growth, migration, invasion, SRCIN1
Introduction
Neuroblastoma (NB) is a malignant embryonic tumor derived from fetal precursors of the sympathetic nervous system, and is the most common solid extracranial malignant tumor of infants and children.1 NB accounts for approximately one-third of all childhood advanced stage incurable cancers and makes up 15% of all cancer deaths among children.2 Of NB patients, 73% have already developed malignant lesions outside the primary cancer by the time of diagnosis.3 Despite the improvements that surgery, chemoradiotherapy, stem cell transplantation and immunotherapy have provided, in cases of high-risk disease the over-all 5-year survival rate remains between 25% and 40%.4 Hence, the need for progress in the therapeutic management of this disease is an important and immediate goal that involves deciphering and clarifying the molecular mechanisms underlying NB pathogenesis and identifying novel biomarkers or therapeutic targets.
Recently, new insights into the molecular mechanisms driving NB have been made available with the use of microRNAs (miRNAs), which are a class of small endogenous non-coding RNA molecules 19–24 nucleotides in length that play a key role in the negative regulation of gene expression though targeting the 3′untranslated region (3′UTR) of specific messenger RNAs (mRNAs). This leads to inhibition or degradation of target mRNAs,5,6 and this effect of miRNAs is estimated to be responsible for the modulation of 30–60% of human gene expression. Emerging evidence has revealed that miRNAs are dysregulated in different types of human cancers including NB, and are implicated in tumorigenic processes such as cell proliferation, apoptosis, migration, cell cycle, angiogenesis, and invasion.5,7,8 Recent studies have demonstrated that miR-373 is downregulated in glioma, prostate cancer, non-small cell lung cancer (NSCLC) and bladder cancer (BCa),9–12 while it is upregulated in cervical cancer, esophageal squamous cell carcinoma, hepatocellular carcinoma (HCC) and gastric cancer.13–16 A published microarray analysis revealed that miR-373 was significantly higher in metastatic NB tissues than in primary NB tissues.2 However, the biological function and underlying molecular mechanisms of miR-373 in the pathogenesis of NB remain largely unknown.
In this study, we investigated the role of miR-373 in the development of NB and demonstrated that it was markedly upregulated in metastatic NB samples compared to primary NB samples. We also found that downregulation of miR-373 inhibited NB cell growth both in vitro and in vivo, and also suppressed cell migration and invasion. Mechanistic investigations defined SRC kinase signaling inhibitor 1 (SRCIN1) as a direct target of miR-373. Moreover, knockdown of SRCIN1 partially abolished the antimiR-373-induced inhibition of cell proliferation, migration and migration. These results indicated that miR-373 plays an important role in stimulating the proliferation, migration, and invasion of NB cells, thus highlighting it as a potential therapeutic target for NB.
Materials and methods
Patients and samples
We obtained 20 metastatic NB samples and 20 primary NB samples from Shenzhen Children’s Hospital. All tissue samples were collected, snap-frozen in liquid nitrogen, and stored at −80 °C after surgical resection. The research was approved by the Ethics Committee of Shenzhen Children’s Hospital (Permit Number: 2019 (005)), and written informed consent was obtained from a parent or legal guardian.
Cell culture and transfection
The human NB cell lines IMR-32, KELLY, LAN-5, SK-N-BE(2) and SH-SY5Y were purchased from Jennio Biotech (Guangzhou, China). IMR-32, KELLY, LAN-5 and SK-N-BE(2) cells were maintained in RPMI1640 medium (Gibco, Grand Island, NY) containing 10% FBS. SH-SY5Y cells were maintained in DMEM/F12 (Gibco) supplemented with 10% FBS. All cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. The miR-373 mimics (sequence: 5′–GAAGUGCUUCGAUUUUGGGGUGU-3′), SRCIN1 siRNAs (sequence: 5′–CACTCATCGCGCACATGTT-3′) and their corresponding controls were chemically synthesized by RiboBio (Guangzhou, China) and transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocol. The transfected cells were collected 48 h after transfection.
RNA extraction and qRT-PCR
Total RNA was isolated from tissues and cultured cells using TRIzol reagent (Invitrogen) according to the instructions supplied by the manufacturer. Total RNA was reverse transcribed and cDNA was amplified using a TaKaRa Reverse Transcription Kit (TaKaRa, Dalian, China) with stem-loop primers for miR-373 or random primers for SRCIN1, U6 and β-actin. qRT-PCR was carried out using SYBR Green master mix (TaKaRa) on an ABI PRISM 7500 sequence detection System (Applied Biosystems, Foster City, CA). The relative expression value of miR-373 or SRCIN1 was normalized to U6 or β-actin expression and calculated using the 2−ΔΔCT method.15 The primers used in this study were as follows: miR-373 RT primers: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACACCC-3′; PCR primers: forward 5′-GCCAGAAGTGCTTCGATTTTG-3′, reverse 5′-GTGCAGGGTCCGAGGT −3′; U6 primers: forward 5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-AACGCTTCACGAATTTGCGT-3′; SRCIN1 primers: forward 5′-GTCCGCACCTGGGGAGAGC-3′, reverse 5′-AGGATGAACCAACAAAGGCAAA-3′; β-actin primers: forward 5′-CTCCATCCTGGCCTCGCTGT-3′, reverse 5′-ACTAAGTCATAGTCCGCCTAGA-3′.
Plasmid construction
The SRCIN1 3′UTR containing the putative miR-373 binding sites was amplified from the genome of SK-N-BE(2) cells using PCR. Site-specific mutations were introduced using a site-directed mutagenesis PCR method. The wild-type and mutated SRCIN1 3′UTRs were subcloned into the psiCHECK2 luciferase vector (Promega, Madison, WI). All constructs were verified by DNA sequencing.
Lentiviruses and cell infection
The antimiR-373 and negative control lentiviruses carrying green fluorescent protein were packaged by Genechem (Shanghai, China). SK-N-BE(2) and SH-SY5Y cells were cultured in 6-well plates and infected with antimiR-373 or negative control lentiviruses for 24 h without FBS. The supernatant was removed and fresh culture medium containing 10% FBS was added to each well. After 48 h, transfected cells were collected and under expression efficiencies were confirmed by qRT-PCR.
MTT (3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide) assay
The transfected cells were plated at a density of 5×103 cells/well in 96-well plates. After 1, 2, 3, 4 and 5 days, 20 μl of 5 mg/ml MTT (Sigma, St.Louis, MO) was added to each well. Then, the 96-well plates were incubated for 4 h at 37 ºC. After removal of the supernatant, 150 μl of DMSO was added to dissolve the formazan crystals. Finally, the absorbance was measured using a microplate reader (Thermo Fisher Scientific, Waltham, MA) at the wave length of 490 nm. Experiments were performed in triplicate.
Cell cycle and apoptosis analysis
For cell cycle analysis, the transfected cells were collected and fixed with 75% ethanol at 4 °C overnight, then incubated with RNase (1 mg/ml) at 37 °C for 30 min, and finally stained with PI (20 µg/ml) for 30 min at room temperature. The stained cells were filtered to remove cell clumps and analyzed by flow cytometry (Becton Dickinson, Franklin Lakes, NJ). For apoptosis analysis, the transfected cells were processed by using Annexin V-FITC/PI Apoptosis Detection Kit (Beyotime Biotechnology, Shanghai, China) according to the instruction of the manufacturer and subjected to flow cytometric analysis (Becton Dickinson). Experiments were carried out in triplicate.
Transwell migration and invasion assays
The migration and invasion assays were carried out in 24-well transwell insert chambers with 8 μm pore size polycarbonate membranes (Corning, New York, NY). Briefly, 1×105 transfected cells in serum-free medium were plated into the upper chamber coated without or with Matrigel. Medium containing 20% FBS in the lower chamber acts as a nutritional attractant. After 24 h, nonmigrating or noninvading cells were removed from the top surface of the insert with a cotton swab. Cells that migrated or invaded to the lower surface of filter were fixed in pre-chilled 70% ethanol for 30 min and stained with 0.1% crystal violet for 10 min. Five visual fields per filter were randomly chosen and counted under a light microscope.17 Experiments were performed in triplicate.
Tumor xenograft model
Healthy 4-weeks-old NOG mice were purchased from the Medical Experimental Animal Center of Guangdong Province. All animal experiments were approved by the Animal Care Committee of Shenzhen Children’s Hospital (Permit Number: 2019 (005)). For the xenograft experiment, mice were divided into 2 groups (n=4), and injected with 5×106 antimiR-373-transfected or antimiR-NC-transfected SK-N-BE(2) cells subcutaneously in the right flank. The tumor size was measured once a week using a vernier caliper. After 10 weeks, mice were sacrificed, and tumors were weighed and photographed. The tumor volume (V) was calculated using the following standard formula: V (mm3) = (D × d 2)/2, where D is the longest diameter and d is the shortest diameter.
Immunohistochemistry
Tumors were fixed in a 4% formaldehyde solution, embedded in paraffin, and cut into 4 μm-thick sections. After deparaffinization, sections were rehydrated, subjected to antigen retrieval by heating in 10 mM citrate buffer for 10 min, treated with 3% H2O2/methanol for 10 min, and blocked with 2% BSA for 30 min. Sections were then incubated at 4 °C overnight with Ki67 antibody (1:400; Cell Signaling Technology, Danvers, MA). After washing with PBS, sections were incubated with a secondary antibody for 1 h at room temperature, followed by reaction with DAB and counterstained with 10% hematoxylin. Finally, sections were digitally imaged with a light microscope (Nikon, Tokyo, Japan). Image Pro Plus 6.0 (Media Cybernetics, Inc., Rockville, MD) was used to count Ki67 positive cells in five random fields per section.
Dual-luciferase reporter assay
SK-N-BE(2) and SH-SY5Y cells were seeded into 24-well plates and transfected with the wild-type or mutant luciferase reporters along with miR-373 or negative control mimics using Lipofectamine 2,000. After transfection for 48 h, cells were lysed, and luciferase activities were measured with a Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions. The relative luciferase activity in the cells was expressed as the ratio of renilla luciferase activity to firefly luciferase activity.
Western blotting
Protein lysates were prepared, subjected to 10% SDS-PAGE, and transferred onto PVDF membranes.18 After blocking in 5% non-fat milk at room temperature for 1 h, the membranes were incubated at 4 °C overnight with SRCIN1 antibody (1:1,000; Cell Signaling Technology) and GAPDH antibody (1:10,000; Abcam, Cambridge, MA). Membranes were then incubated with peroxidase-conjugated secondary antibodies at room temperature for 2 h. Proteins were visualized with ECL detection reagents.
Statistical analysis
All data are expressed as the means ± standard deviation (SD). The statistical significance of the differences between two groups was analyzed using the Student’s t-test. The correlation between miR-373 and SRCIN1 expression was analyzed by pearman’s correlation. p-values less than 0.05 were considered statistically significant. Statistical analysis was performed using SPSS 19.0 software.
Results
Expression of miR-373 and SRCIN1 in human NB tissues and cell lines
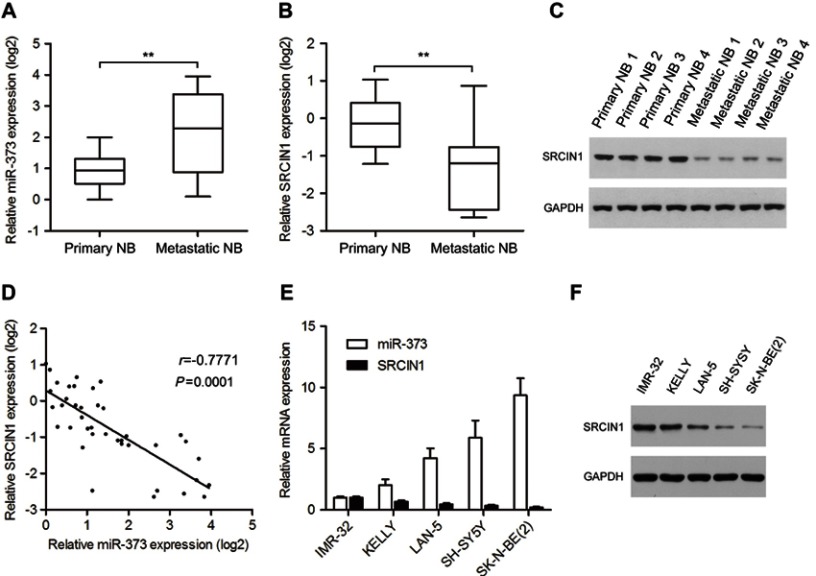
In an attempt to explore the potential involvement of the miR-373 in the process of NB metastasis, we used qRT-PCR analysis to measure the miR-373 expression levels in 20 primary and 20 metastatic NB tissues. The results showed that the level of miR-373 expression was significantly higher in metastatic NB tissues than in primary NB tissues (Figure 1A), consistent with previously published microarray data.2 Because SRCIN1 had been identified as a candidate target for miR-373 using the TargetScan and Pictar databases,19,20 we examined the same clinical samples for SRCIN1 mRNA and protein expression using qRT-PCR and Western blotting. Compared with primary NB samples, metastatic NB samples exhibited decreased mRNA and protein expression of SRCIN1 (Figure 1B and C). Pearson’s correlation analysis revealed that there was an inverse correlation between miR-373 and SRCIN1 mRNA levels (Figure 1D). Furthermore, a significant anticorrelation was also observed between miR-373 and SRCIN1 mRNA levels in a series of NB cell lines. Specifically, the cells with the relatively high levels of miR-373 exhibited low levels of SRCIN1 as determined by qRT-PCR and Western blotting (Figure 1E and F). Together, these data indicated the presence of a significant anti-correlation between miR-373 and SRCIN1 expression in NB tissues and cell lines.
Figure 1.
Expression of miR-373 and SRCIN1 in human NB tissues and cell lines. The expression of miR-373 (A) and SRCIN1 (B) was measured byqRT-PCR in 20 primary and 20 metastatic NB tissues. (C) SRCIN1 protein levels were detected by Western blotting. (D) Pearson’s correlation analysis of miR-373 and SRCIN1 expression in NB tissues. (E) Relative miR-373 and SRCIN1 mRNA expression levels of five NB cell lines were examined with qRT-PCR. (F) Western blotting analysis of SRCIN1 protein levels. The results are expressed as mean ± SD (n=3). **p<0.01.
Abbreviations: SRCIN1, SRC kinase signaling inhibitor 1; qRT-PCR, quantitative real-time PCR; SD, standard deviation.
Downregulation of miR-373 inhibited the proliferation, migration and invasion of NB cells
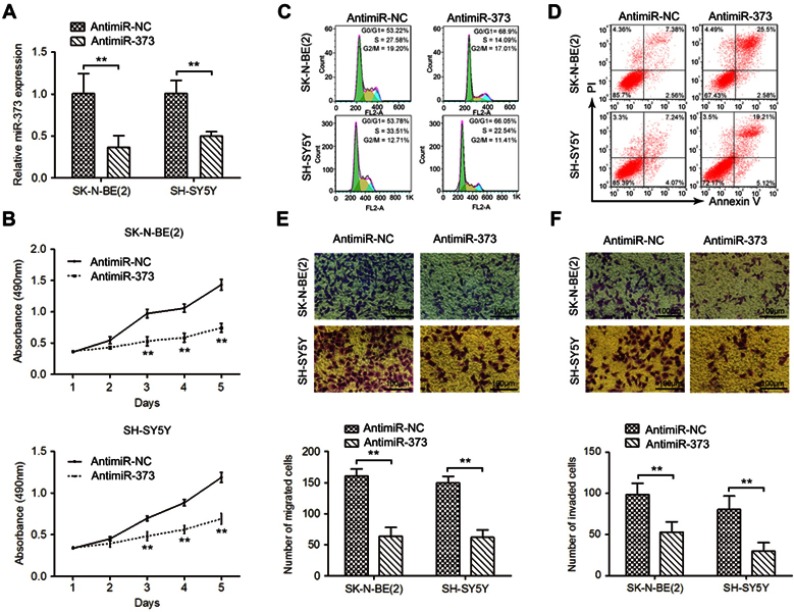
To explore the potential role of miR-373 in NB tumorigenesis, SK-N-BE(2) and SH-SY5Y cells, which expressed highest miR-373, were chosen to perform loss-of-function experiments. qRT-PCR confirmed that antimiR-373 lentivirus could decrease miR-373 expression in both SK-N-BE(2) and SH-SY5Y cells, compared with the corresponding controls (Figure 2A). Results from MTT assay revealed a significant reduction in the cell viability over time in the antimiR-373-transfected cells when compared to the control cells (Figure 2B). To confirm this result, we analyzed cell cycle progression using flow cytometry and found that inhibition of miR-373 led to a significant increase in the percentage of cells in G0/G1 phase and a dramatic decrease in the percentage of cells in S phase (Figure 2C). In addition, the proportion of apoptotic cells was significantly higher in the antimiR-373-transfected cells (Figure 2D). Moreover, transwell migration and invasion assays showed that downregulation of miR-373 markedly retarded the migrative and invasive capability of NB cells (Figure 2E and F). Collectively, our data demonstrated that miR-373 serves as an oncomiR and enhances cell growth, invasion and migration in vitro.
Figure 2.
Downregulation of miR-373 inhibited the proliferation, migration, and invasion of NB cells. (A) qRT-PCR analysis of miR-373 expression in SK-N-BE(2) and SH-SY5Y cells infected with antimiR-373 or negative control lentivirus. (B) Cell viability was measured with MTT assay. Flow cytometry was used to evaluate the effects of miR-373 on cell cycle distribution (C) and apoptosis (D). (E) The migration ability of antimiR-373-treated cells as determined by transwell migration assay. Bars represent the number of migrated cells. (F) Representative pictures and quantification of transwell invasion assay. The results are expressed as mean ± SD (n=3). **p<0.01.
Abbreviations: qRT-PCR, quantitative real-time PCR; MTT, 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide; SD, standard deviation.
Inhibition of miR-373 suppressed tumor growth in a mouse xenograft model
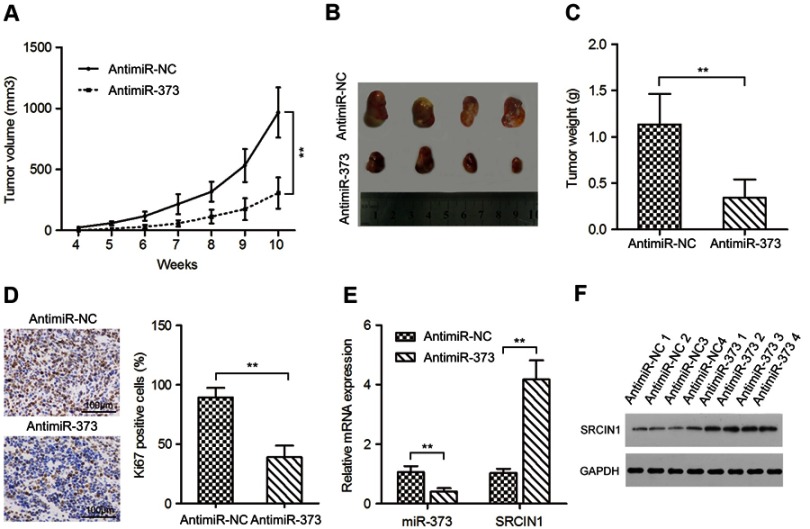
Our in vitro results indicated that downregulation of miR-373 impaired NB cell growth. We next want to explore whether this also occur in vivo. SK-N-BE(2) cells transfected with antimiR-373 or negative control lentivirus were subcutaneously injected into 4-week-old NOG mice. Tumors formed from the antimiR-373-transfected SK-N-BE(2) cells grew more slowly than tumors induced by the control cells (Figure 3A). Additionally, after 10 weeks, the weight and size of the transplanted tumors in the antimiR-373 group were significantly lighter and smaller than those in the control group (Figure 3B and C). Furthermore, we examined NB proliferation and miR-373 and SRCIN1 expression in the transplanted tumors and found that antimiR-373 lentivirus decreased tumor miR-373 expression, increased SRCIN1 mRNA and protein levels and decreased cell proliferation marker Ki67 protein expression in xenograft tumors when compared with the control group (Figure 3D-F). Taken together, these findings indicated that miR-373 plays an important role in promoting the tumorigenicity of NB cells in vivo.
Figure 3.
Inhibition of miR-373 suppressed tumor growth in a mouse xenograft model. (A) SK-N-BE(2) cells infected with antimiR-373 or negative control lentivirus were implanted subcutaneously into NOG mice. The volumes of xenograft tumors were calculated every week. (B) Representative images of xenograft tumors. (C) The average weight of xenograft tumors. (D) Immunohistochemistry were performed to measure the protein levels of Ki67 in xenograft tumors. Tumor levels of miR-373 and SRCIN1 were measured by qRT-PCR (E) and Western blotting (F), respectively. The results are expressed as mean ± SD (n=3).**p<0.01.
Abbreviations: qRT-PCR, quantitative real-time PCR; SRCIN1, SRC kinase signaling inhibitor 1; SD, standard deviation.
MiR-373 targeted SRCIN1 expression directly via its 3′UTR
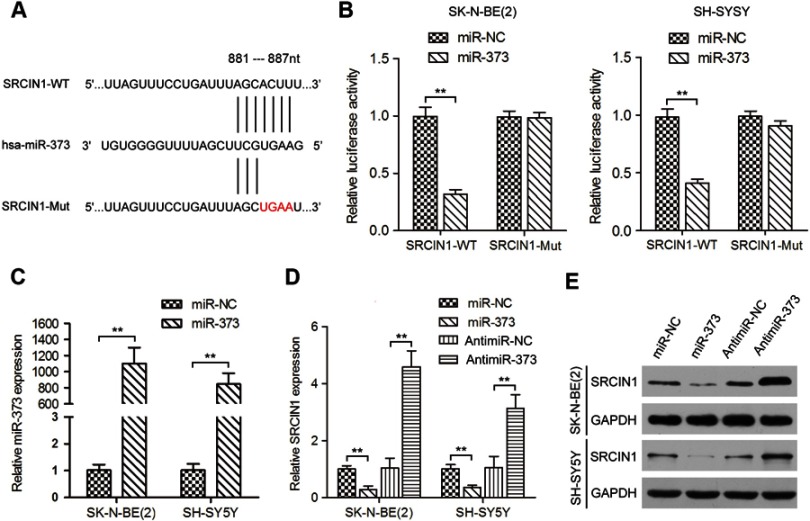
To investigate the molecular mechanism by which miR-373 facilitates NB cell proliferation and motility, we predicted the potential targets of miR-373 using three algorithms including TargetScan and Pictar.19,20 The SRCIN1 3′UTR was found to contain sequences complementary to the miR-373 seed sequence. To determine whether SRCIN1 is a direct target of miR-373, we first constructed a wild-type luciferase reporter plasmid carrying the predicted binding site of the SRCIN1 3′UTR and a mutant luciferase reporter plasmid in which the predicted binding site was mutated (Figure 4A). The wild-type or mutant luciferase reporters were co-transfected with miR-373 or negative control mimics into SK-N-BE(2) and SH-SY5Y cells for 48 h, and followed by examination of luciferase activity in the transfected cells. As illustrated in Figure 4B, miR-373 yielded an obvious decrease in the luciferase activity for the SRCIN1 wild-type 3′UTR, whereas the mutant luciferase reporter activity was unchanged. Next, we determined whether the changes in the expression of miR-373 would regulate SRCIN1 expression. To this end, SK-N-BE(2) and SH-SY5Y cells were transfected with miR-373 or negative control mimics, and the overexpression of miR-373 was confirmed by qRT-PCR (Figure 4C). The expression levels of SRCIN1 mRNA (Figure 4D) and protein (Figure 4E) were diminished by miR-373 mimics in both cell lines. However, reducing endogenous miR-373 levels using the antimiR-373 lentivirus had the opposite effect. Together, the above results suggest that miR-373 repressed the expression of SRCIN1 through directly binding to the 3′UTR of SRCIN1.
Figure 4.
miR-373 targeted SRCIN1 expression directly via its 3′UTR. (A) Schematic illustration of the putative miR-373 targeting sequences in the 3′UTR of SRCIN1. Mutant sequences are shown in red type. (B) miR-373 or negative control mimics and WT or Mut luciferase reporters were co-transfected into SK-N-BE(2) and SH-SY5Y cells. The luciferase activity was measured by dual-luciferase reporter assay. (C) Expression of miR-373 in SK-N-BE(2) and SH-SY5Y cells transfected with miR-373 or negative control mimics. qRT-PCR (D) and Western blotting (E) were performed to examine SRCIN1 mRNA and protein expression in cells transfected with miR-373 mimics, negative control mimics, antimiR-373 lentivirus or negative control lentivirus. **p<0.01.
Abbreviations: SRCIN1, SRC kinase signaling inhibitor 1; 3′UTR, 3′untranslated region; WT, wild-type; Mut, mutant; qRT-PCR, quantitative real-time PCR; SD, standard deviation.
SRCIN1 could partially counter act the phenotypes caused by miR-373
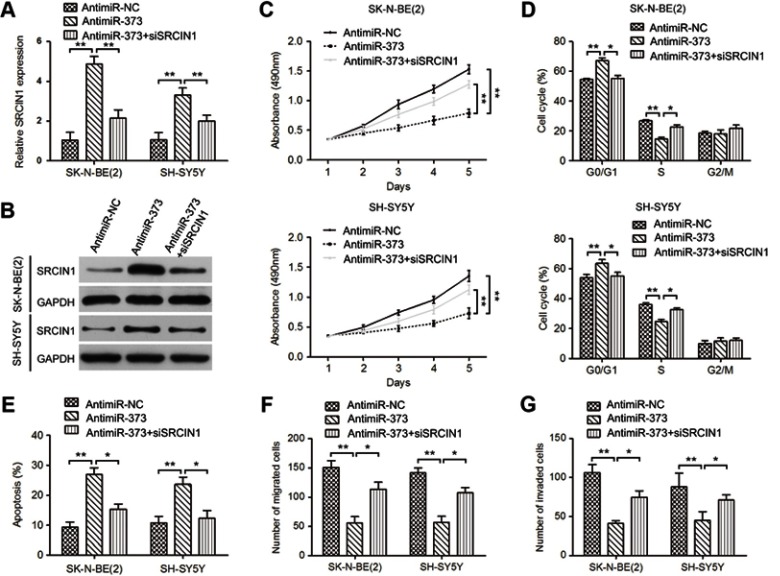
Given that miR-373 could reduce SRCIN1 expression, we wondered if SRCIN1 could be responsible for the miR-373-induced malignant phenotypes observed in NB cells. A SRCIN1 siRNA was transfected into antimiR-373-transfected SK-N-BE(2) and SH-SY5Y cells. qRT-PCR and Western blotting analysis showed that silencing of SRCIN1 by SRCIN1 siRNA in antimiR-373-treated cells could partially reverse the effect of SRCIN1 upregulation resulted from antimiR-373 (Figure 5A and B). As expected, knockdown of SRCIN1 partially restored the inhibitory effects of antimiR-373 on NB cell growth, migration and invasion (Figure 5C-G). Taken together, these results indicated that SRCIN1 is a functionally important target of miR-373 that is involved in the growth and metastasis of NB cells.
Figure 5.
SRCIN1 could counteract the phenotypes caused by miR-373. qRT-PCR (A) and Western blotting (B) analysis of SRCIN1 mRNA and protein expression in antimiR-373-treated cells transiently infected with SRCIN1 siRNA. MTT, Flow cytometry, Transwell migration and invasion assays were used to analyze cell viability (C), cell cycle distribution (D), apoptosis (E), migration (F) and invasion (G). The results are expressed as mean ± SD (n=3). *p<0.05; **p<0.01.
Abbreviations: qRT-PCR, quantitative real-time PCR; SRCIN1, SRC kinase signaling inhibitor 1; MTT, 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide; SD, standard deviation.
Discussion
Accumulating evidence has implicated the aberrant expression of miRNAs as a crucial player in the development and progression of cancer. In this study, we found significantly more miR-373 in metastatic tumor tissues compared with primary tumors, and that suppression of miR-373 restrained NB cell proliferation, cell cycle progression, migration and invasion in vitro, as well as inhibited tumorigenesis in vivo. Furthermore, miR-373 inhibited SRCIN1 expression by directly targeting its 3′UTR. We also found that SRCIN1 partially counteracted the phenotypes induced by miR-373. These findings suggested that miR-373 could facilitate the proliferation, migration and invasion of NB cells through controlling SRCIN1 expression.
The miR-373 is located on chromosomal band 19q13.4.21 It belongs to the miR-520/-373 family, which consists of three different miRNA clusters, miR-302/-367, miR-371/-372/-373 and miR-520.22 Accumulating evidence shows that miR-373 can function as oncogenes or tumor suppressors in human cancers by regulating specific genes and therefore play key roles in tumorigenesis.11–13,23,24 As a tumor suppressor, overexpression of miR-373 results in attenuation of NSCLC cell proliferation, migration and invasion through IRAK2 and LAMP1 axes.11 Enforcing miR-373 expression also exhibits a robust capacity to suppress BCa cell proliferation, migration and invasion by activating E-cadherin expression, and obviously represses tumor growth and metastasis in nude mice.12 Additionally, ectopic expression of miR-373 distinctly reduces cell propagation, migration and invasion, and boosted apoptosis in gemcitabine resistance pancreatic carcinoma cells by targeting CCND2.23 On the other hand, Wang et al showed that upregulation of miR-373 enhances cervical cancer cell proliferation and colony formation in vitro, and tumorigenicity in vivo by directly targeting YOD1.13 Huang et al have also reported that miR-373 stimulates breast cancer cell migration and invasion in vitro and in vivo by suppression of CD44.24 In this study, our data indicated that miR-373 was overexpressed in metastatic tumor tissues compared to primary tumor tissues, and inhibition of miR-373 decreased NB cell growth in vitro and in vivo, as well as reduced cell migration and invasion. However, the potential molecular mechanism by which miR-373 promotes the malignant behaviors of NB cells is still unknown.
Using bioinformatics analysis, we found that SRCIN1 is a target of miR-373. Dual-luciferase assay further confirmed that miR-373 suppressed the expression of SRCIN1 by binding to its 3′UTR. SRCIN1, also named p140CAP (p140 Cas-associated protein), contains two coiled-coil domains, two proline-rich regions, and two regions carrying highly charged amino acids.25 SRCIN1 is downregulated in multiple types of cancers such as lung cancer, osteosarcoma, gastric cancer and breast cancer, and plays an essential role in the development and progression of cancer.25–28 The targeting and suppression of SRCIN1 by miR-150 was found to be a cause of lung cancer cell growth and metastasis.26 Xu et al demonstrated that miR-374a’s repression of SRCIN1 in gastric cancer promotes cell proliferation, migration and invasion in vitro, and tumor growth in vivo.27 Moreover, SRCIN1 has been reported to inhibit breast cancer cell proliferation, colony formation, migration and invasion, and promote apoptosis.28,29 Additionally, SRCIN1 has also been shown to repress osteosarcoma cell growth, invasion and EMT.23 In our study, silencing of SRCIN1 partially abrogated the inhibitory roles of antimiR-373 in NB cell growth and motility. What’s more, we found a negative correlation between miR-373 and SRCIN1 expression in NB samples. These data indicated that SRCIN1 is a functional mediator for miR-373 in NB.
Recently, several studies have shown that specific miRNAs and genes correlate with MYCN amplification in NB.2,30,31 Brouwer et al reported that Dickkopf-3 is regulated by the MYCN-induced miR-17–92 cluster in NB.30 Tivnan et al demonstrated that MYCN directly regulated miR-34a expression.31 In our study, IMR-32, KELLY, and SK-N-BE(2) cell lines are MYCN-amplified cell lines, whether MYCN correlated with miR-373 and/or SRCIN1 in NB cells and/or NB tissues need further investigation.
In conclusion, our findings have provided evidence that miR-373 is upregulated in metastatic tumor tissues when compared with primary tumor tissues, and ectopic expression of miR-373 can efficiently promote the growth and metastasis of NB cells through the direct targeting of the 3′UTR of SRCIN1. This newly identified miR-373/SRCIN1 axis provides compelling mechanistic insight and might provide important direction for future therapeutic approaches in the treatment of NB.
Acknowledgment
This work was funded by Shenzhen’s Sanming Project (SZSM 201512033).
Disclosure
The authors report no competing financial interests or conflicts of interest in this work.
References
- 1.Zhang H, Pu J, Qi T, et al. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene. 2014;33(3):387–397. doi: 10.1038/onc.2012.574 [DOI] [PubMed] [Google Scholar]
- 2.Bienertova-Vasku J, Mazanek P, Hezova R, et al. Extension of microRNA expression pattern associated with high-risk neuroblastoma. Tumour Biol. 2013;34(4):2315–2319. doi: 10.1007/s13277-013-0777-0 [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Pan M, Han L, Lu H, Hao X, Dong Q. miR-338-3p suppresses neuroblastoma proliferation, invasion and migration through targeting PREX2a. FEBS Lett. 2013;587(22):3729–3737. doi: 10.1016/j.febslet.2013.09.044 [DOI] [PubMed] [Google Scholar]
- 4.Wu K, Yang L, Chen J, et al. miR-362-5p inhibits proliferation and migration of neuroblastoma cells by targeting phosphatidylinositol 3-kinase-C2beta. FEBS Lett. 2015;589(15):1911–1919. doi: 10.1016/j.febslet.2015.05.056 [DOI] [PubMed] [Google Scholar]
- 5.Xu YF, Mao YP, Li YQ, et al. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett. 2015;363(2):146–155. doi: 10.1016/j.canlet.2015.04.006 [DOI] [PubMed] [Google Scholar]
- 6.Guo J, Dong Q, Fang Z, et al. Identification of miRNAs that are associated with tumor metastasis in neuroblastoma. Cancer Biol Ther. 2010;9(6):446–452. doi: 10.4161/cbt.9.6.10894 [DOI] [PubMed] [Google Scholar]
- 7.Peres J, Kwesi-Maliepaard EM, Rambow F, Larue L, Prince S. The tumour suppressor, miR-137, inhibits malignant melanoma migration by targetting the TBX3 transcription factor. Cancer Letters. 2017;405:111–119. doi: 10.1016/j.canlet.2017.07.018 [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Chen S, Xiu YL, Sun KX, Zong ZH, Zhao Y. RhoC is a major target of microRNA-93-5P in epithelial ovarian carcinoma tumorigenesis and progression. Mol Cancer. 2015;14:31. doi: 10.1186/s12943-014-0278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Yu H, Liu Y, et al. Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the MiR-373/EGFR axis. Cell Physiol Biochem. 2018;45(1):131–147. doi: 10.1159/000486253 [DOI] [PubMed] [Google Scholar]
- 10.Qiu X, Zhu J, Sun Y, et al. TR4 nuclear receptor increases prostate cancer invasion via decreasing the miR-373-3p expression to alter TGFbetaR2/p-Smad3 signals. Oncotarget. 2015;6(17):15397–15409. doi: 10.18632/oncotarget.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seol HS, Akiyama Y, Shimada S, et al. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014;353(2):232–241. doi: 10.1016/j.canlet.2014.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Wang C, Miao S, Li C, Chen Z, Li F. Enhancing E-cadherin expression via promoter-targeted miR-373 suppresses bladder cancer cells growth and metastasis. Oncotarget. 2017;8(55):93969–93983. doi: 10.18632/oncotarget.21400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LQ, Zhang Y, Yan H, Liu KJ, Zhang S. MicroRNA-373 functions as an oncogene and targets YOD1 gene in cervical cancer. Biochem Biophys Res Commun. 2015;459(3):515–520. doi: 10.1016/j.bbrc.2015.02.138 [DOI] [PubMed] [Google Scholar]
- 14.Lee KH, Goan YG, Hsiao M, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315(15):2529–2538. doi: 10.1016/j.yexcr.2009.06.001 [DOI] [PubMed] [Google Scholar]
- 15.Wu N, Liu X, Xu X, et al. MicroRNA-373, a new regulator of protein phosphatase 6, functions as an oncogene in hepatocellular carcinoma. Febs J. 2011;278(12):2044–2054. doi: 10.1111/j.1742-4658.2011.08120.x [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Li X, Tan Z, et al. MicroRNA-373 is upregulated and targets TNFAIP1 in human gastric cancer, contributing to tumorigenesis. Oncol Lett. 2013;6(5):1427–1434. doi: 10.3892/ol.2013.1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Liu Y, Shu XO, Cai Q. MiR-374a suppresses lung adenocarcinoma cell proliferation and invasion by targeting TGFA gene expression. Carcinogenesis. 2016;37(6):567–575. doi: 10.1093/carcin/bgw038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao WT, Ye YP, Zhang NJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232(4):415–427. doi: 10.1002/path.4309 [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 20.Krek A, Grun D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- 21.Wei F, Cao C, Xu X, Wang J. Diverse functions of miR-373 in cancer. J Transl Med. 2015;13:162. doi: 10.1186/s12967-015-0541-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eyking A, Reis H, Frank M, Gerken G, Schmid KW, Cario E. MiR-205 and MiR-373 are associated with aggressive human mucinous colorectal cancer. PloS One. 2016;11(6):e0156871. doi: 10.1371/journal.pone.0156871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu W, Liu Q, Pan J, Sui Z. MiR-373-3p enhances the chemosensitivity of gemcitabine through cell cycle pathway by targeting CCND2 in pancreatic carcinoma cells. Biomed Pharmacother. 2018;105:887–898. doi: 10.1016/j.biopha.2018.05.091 [DOI] [PubMed] [Google Scholar]
- 24.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681 [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Wang H, Li X, Liu Y, Zhao C, Zhu D. SRCIN1 suppressed osteosarcoma cell proliferation and invasion. PloS One. 2016;11(8):e0155518. doi: 10.1371/journal.pone.0155518 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Cao M, Hou D, Liang H, et al. miR-150 promotes the proliferation and migration of lung cancer cells by targeting SRC kinase signalling inhibitor 1. Eur J Cancer. 2014;50(5):1013–1024. doi: 10.1016/j.ejca.2013.12.024 [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Wang W, Su N, et al. miR-374a promotes cell proliferation, migration and invasion by targeting SRCIN1 in gastric cancer. FEBS Lett. 2015;589(3):407–413. doi: 10.1016/j.febslet.2014.12.027 [DOI] [PubMed] [Google Scholar]
- 28.Yang F, Luo LJ, Zhang L, et al. MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene. 2017;600:21–28. doi: 10.1016/j.gene.2016.11.037 [DOI] [PubMed] [Google Scholar]
- 29.Di Stefano P, Damiano L, Cabodi S, et al. p140Cap protein suppresses tumour cell properties, regulating Csk and Src kinase activity. Embo J. 2007;26(12):2843–2855. doi: 10.1038/sj.emboj.7601724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Brouwer S, Mestdagh P, Lambertz I, et al. Dickkopf-3 is regulated by the MYCN-induced miR-17-92 cluster in neuroblastoma. Int J Cancer. 2012;130(11):2591–2598. doi: 10.1002/ijc.26295 [DOI] [PubMed] [Google Scholar]
- 31.Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011;11:33. doi: 10.1186/1471-2407-11-33 [DOI] [PMC free article] [PubMed] [Google Scholar]