Abstract
Objective: The aim of this study was to construct and validate a microRNA (miR)-based signature as a prognostic tool for lung squamous cell carcinoma (LUSC).
Materials and methods: With the use of mature miR expression profiles downloaded from The Cancer Genome Atlas database, we identified differentially expressed miRs between LUSC and matched healthy lung tissue. Thereafter, we carried out an evaluation of the association of differentially expressed miRs with overall survival (OS) with the use of univariate and multivariate Cox regression analysis. This analysis was eventually employed for the construction of a miR-based signature, which effectively predicted the prognosis. The functional enrichment analysis of the miRs included in the signature was used to explore their potential molecular mechanism in LUSC.
Results: A total of 316 miRs were differentially expressed between LUSC and matched healthy lung tissues in the training set. Following the univariate and multivariate Cox regression analysis, we found that seven miRs were independent prognostic factors. Each patient received a signature index ranging from 0 to 7. Patients with LUSC were divided into high-risk, intermediate-risk, and low-risk groups in accordance with their signature index and the OS in the three groups was significantly different. This finding remains consistent in the validation set. Besides that, this seven-miR signature remained an independent prognostic factor in comparison with routine clinicopathologic features. The seven-miR signature is a promising biomarker for predicting the 5-year survival rate of LUSC with an area under the receiver operating characteristic curveof 0.712 in the training set and 0.688 in the validation set, respectively. The target genes of seven miRs may be involved in various pathways associated with lung cancer, for instance the mitogen-activated protein kinase signaling pathway and the Wnt signaling pathway.
Conclusion: Using this signature, patients with LUSC can be divided into high-risk, intermediate-risk, and low-risk groups for more personalized management.
Keywords: lung squamous cell carcinoma, microRNA-based signature, prognosis
Introduction
Lung cancer remains the leading cause of cancer incidence and mortality across the globe,1 and approximately 80% of lung cancers are classified histopathologically as nonsmall cell lung cancer (NSCLC). NSCLC can be segregated into two major classes, which include lung nonsquamous cell carcinoma and lung squamous cell carcinoma (LUSC). In spite of the curative surgery for patients with early stage disease, approximately 40% of patients will relapse within a period of 5 years,2 with the 5-year overall survival (OS) rate amounting to 50–60%.3,4 This suggests that there are still some high-risk individuals among patients who have early stage disease. A reliable prognostic prediction model for the identification of these high-risk individuals is obviously valuable. While advanced lung nonsquamous cell cancer has greatly benefited from the detection and targeting of oncogenic alterations, for instance ALK rearrangement and EGFR mutations, LUSC has been challenging in the identification and targeting of driving mutations.5 Thus, such a prognostic prediction model is urgently needed for LUSC.
MicroRNAs (miRs) are short noncoding RNA molecules, playing crucial roles in transcriptional regulation of gene expression through several mechanisms.6 The deregulation of miRs has been shown to be associated with various cancers, including NSCLC.7–10 Although a few miR signatures have been proposed for predicting the outcome of NSCLC, including LUSC,11,12 the results of these studies are significantly inconsistent and lack validation. This may have resulted from the small sample sizes as well as the discovery of new miRs.
In this work, we propose a new miR-based signature to predict the prognosis of LUSC. With the use of this signature, LUSC can be effectively divided into high-risk, intermediate-risk, and low-risk groups. Furthermore, these results will be validated in an independent data set.
Materials and methods
Data processing
The preprocessed LUSC mature miR expression profiles in The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database, displayed as log2 converted reads per million (log2(RPM+1)), were downloaded from University of California Santa Cruz Xena (https://xenabrowser.net/datapages/, version 09-08-2017). The corresponding clinical information was downloaded from TCGA database (download date 09-26-2018). These contain two mature miR expression data, which are based on two different platforms, including 380 samples (336 LUSC tissues and 44 matched healthy lung tissues) based on the IlluminaHiSeq_miRNASeq platform (Illumina Inc., San Diego, CA, USA) and 131 LUSC tissues based on the IlluminaGA_miRNASeq platform. The samples based on the IlluminaHiSeq_miRNASeq platform were used as the training set to identify differentially expressed miRs and construct a miR-based signature for predicting prognosis, and the samples based on the IlluminaGA_miRNASeq platform were used as the validation set to verify the signature.
Screening of differentially expressed miRs
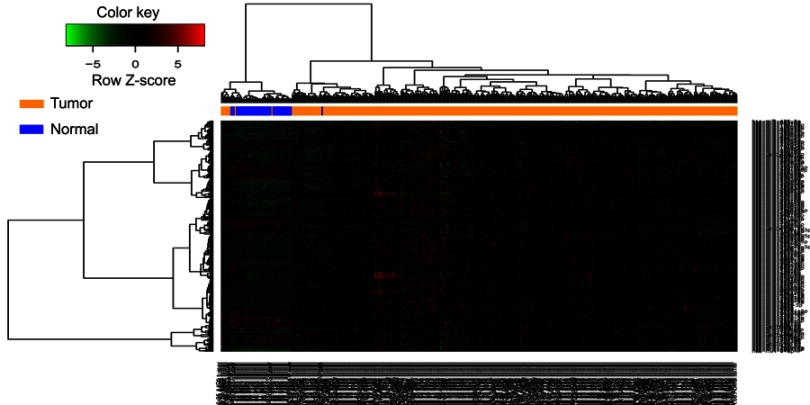
In the training set, miRs that did not express over 10% of samples were removed. The differentially expressed miRs between LUSCs and matched healthy lung tissues were analyzed using the “limma”13 package in R language. The fold changes (FCs) in the expression of individual miRs were calculated, and differentially expressed miRs with |log2FC|>0.585 and p<0.05 (adjusted by the false discovery rate) were considered significant. We applied bidirectional hierarchical clustering to differentially express miRs based on the Euclidean distance and displayed the results as a heatmap.
Construction and validation of the miR-based prognostic signature for LUSC
In the training set, patients with a survival time of less than 30 days were removed for survival analysis. The remaining patients (N=318) were separated into high and low-level groups based on the median value of the differentially expressed miRs, followed by univariate and multivariate Cox proportional hazards analyses. We found that seven miRs were independent factors of survival. We assigned each patient 1 point for the high-risk expression level of these seven miRs. Accordingly, each patient received a score ranging from 0 to 7, which we called the signature index. We considered patients having a signature index of 6 or 7 to be high risk, those with an index of 3–5 to be intermediate risk, and those with an index <3 to be low risk. The survival time was compared in the three groups using Kaplan–Meier analysis with the log-rank test. In the validation set, two patients without a survival time were removed for survival analysis. The validation set was used to confirm the robustness of the miR-based prognostic signature. Furthermore, to compare the relative prognostic value of this miR-based signature with that of routine clinicopathologic features, we carried out the univariate and multivariate Cox proportional hazards analyses in the training and validation sets. The time-dependent receiver operating characteristic (ROC) curve was used to assess the miR-based signature’s predictive value for the 5-year survival rate of LUSC and was performed using the “survivalROC” package14 in R.
Target gene prediction and functional enrichment analysis
Target gene prediction of the seven miRs was performed using the miRDB online tool (http://mirdb.org/).15 Target genes of a miR provided by miRDB are ranked by the target score. The top 50 target genes with the highest target score for each miR were extracted to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.abcc.ncifcrf.gov/home.jsp).16 p<0.05 was set as the cutoff criterion. These seven miRs and their target genes would be displayed as the miR-target network using cytoscape software.17
Statistical analysis
The χ2-test was used for the categorical data and an unpaired t-test was used to screen differentially expressed miRs. Univariate/multivariate Cox proportional hazards analyses and Kaplan–Meier survival analysis were used to compare survival between the two groups of patients. The χ2-test and survival analysis were performed using IBM SPSS statistics software program version 22.0 (IBM Corporation, Armonk, NY, USA). All tests were two-sided and p<0.05 was considered statistically significant.
Results
Differentially expressed miRs between LUSC and matched healthy lung tissues
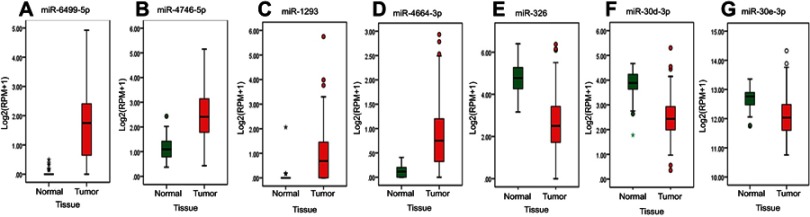
The LUSC patients’ detailed clinical characteristics including gender, age at diagnosis, and TNM stage are listed in Table 1. The training set contained more patients with early stage (I/II) disease (83.33% vs 74.42%, χ2-test p=0.033) and patients with Mx (22.96% vs 3.10%, χ2-test p=0.000) than the validation set. In accordance with the cutoff criteria (p<0.05 and |log2FC|>0.585), 316 miRs were differentially expressed between LUSC and matched healthy lung tissues in the training set. These included 223 miRs that were upregulated and 93 miRs that were downregulated in LUSC tissues. The result of the expression analysis was presented as a heatmap (Figure 1), and the result of hierarchical clustering showed that these differentially expressed miR expression patterns could basically distinguish LUSC tissues and healthy lung tissues. miR-6499-5p, miR-4746-5p, miR-1293, and miR-4664-3p were upregulated whereas miR-326, miR-30d-3p, and miR-30e-3p were downregulated in LUSC tissues (Figure 2).
Table 1.
Summary of patient cohort information
| Characteristic | Training set (N=318) | Validation set (N=129) | p-value | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Gender | 0.485 | ||||
| Male | 237 | 74.53 | 92 | 71.32 | |
| Female | 81 | 25.47 | 37 | 28.68 | |
| Age (years) | 0.201 | ||||
| ≤65 | 118 | 37.11 | 53 | 41.09 | |
| >65 | 196 | 61.64 | 76 | 58.91 | |
| Not available | 4 | 1.26 | 0 | 0.00 | |
| T stage | 0.308 | ||||
| T1–2 | 252 | 79.25 | 108 | 83.72 | |
| T3–4 | 66 | 20.75 | 21 | 16.28 | |
| Lymph node stage | 0.161 | ||||
| N0 | 205 | 64.47 | 78 | 60.47 | |
| N1–3 | 109 | 34.28 | 51 | 39.53 | |
| Nx | 4 | 1.26 | 0 | 0.00 | |
| Metastasis | 0.000 | ||||
| M0 | 243 | 76.42 | 122 | 94.57 | |
| M1 | 2 | 0.63 | 3 | 2.33 | |
| Mx | 73 | 22.96 | 4 | 3.10 | |
| Pathological stage | 0.033 | ||||
| I–II | 265 | 83.33 | 96 | 74.42 | |
| III–IV | 50 | 15.72 | 32 | 24.81 | |
| Not available | 3 | 0.94 | 0 | 0.00 | |
Note: Bold values indicate P<0.05.
Figure 1.
Hierarchical clustering dendrograms of expression patterns of differentially expressed microRNAs that can basically distinguish between lung squamous cell carcinoma and normal lung tissue. Blue, normal lung tissue; orange, tumor tissue.
Figure 2.
The expression level of the seven miRs between LUSC and normal lung tissue. (A) miR-6499-5p, (B) miR-4746-5p, (C) miR-1293, (D) miR-4664-3p, (E) miR-326, (F) miR-30d-3p, (G) miR-30e-3p.
Abbreviations: log2(RPM+1), log2 converted reads per million; LUSC, lung squamous cell carcinoma; miR, microRNA.
Construction of miR-based signature with differentially expressed miRs in the training set
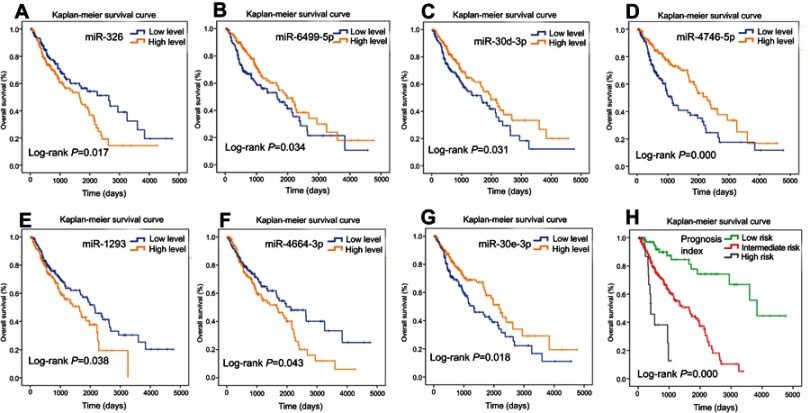
For each of the 316 differentially expressed miRs, we used the median expression level as a cutoff point to stratify the 318 patients into a high-level group and a low-level group. The univariate Cox proportional hazards regression analysis revealed that a total of 10 miRs had prognostic value. We then applied a multivariate Cox proportional hazards regression analysis to identify seven miRs (Table 2) – miR-326, miR-6499-5p, miR-30d-3p, miR-4746-5p, miR-1293, miR-4664-3p, and miR-30e-3p – as independent prognostic factors (Figure 3A–G). We scored the seven-miR signature by the value assigned for each miR. Each patient was assigned one score for low expression of miR-6499-5p, miR-30d-3p, miR-4746-5p, and miR-30e-3p, respectively, and one score for high expression of miR-326, miR-1293, and miR-4664-3p, respectively. Then, we summed the scores for each patient, resulting in a signature index ranging from 0 to 7. Moreover, we considered patients with a signature index of 6 or 7 to be high risk, those with an index of 3–5 to be intermediate risk, and those with an index <3 to be low risk. The comparison of the survival time was carried out in the three groups using Kaplan–Meier analysis with the log-rank test. The OS in the low-risk (N=70), intermediate-risk (N=229), and high-risk (N=19) patients was 3838 days (95% CI 3236–4439 days), 1655 days (95% CI 1223–2086 days), and 408 days (95% CI 233–582 days), respectively. The OS in the three groups was significant (log-rank p=0.000, Figure 3H). This indicated that the seven-miR signature created was associated with survival in LUSC.
Table 2.
Univariate and multivariate analyses of 10 microRNAs in lung squamous cell carcinoma patients
| microRNA | Univariate Cox analysis | Multivariate Cox analysis | |||
|---|---|---|---|---|---|
| p-value | HR (95% CI) | β | p-value | HR (95% CI) | |
| miR-326 | 0.018 | 1.077~2.204 | 0.454 | 0.033* | 1.038~2.389 |
| miR-6499-5p | 0.035 | 0.480~0.974 | −0.568 | 0.004* | 0.386~0.832 |
| miR-193b-5p | 0.012 | 1.103~2.247 | 0.321 | 0.107 | 0.933~2.034 |
| miR-30d-3p | 0.032 | 0.477~0.968 | −0.377 | 0.047* | 0.472~0.995 |
| miR-4746-5p | 0.000 | 0.361~0.738 | −0.632 | 0.002* | 0.358~0.788 |
| miR-1293 | 0.039 | 1.019~2.102 | 0.489 | 0.015* | 1.102~2.413 |
| miR-3607-3p | 0.018 | 1.077~2.202 | 0.076 | 0.708 | 0.725~1.606 |
| miR-4664-3p | 0.044 | 1.010~2.056 | 0.504 | 0.011* | 1.122~2.445 |
| let-7e-3p | 0.037 | 1.022~2.079 | 0.085 | 0.684 | 0.722~1.643 |
| miR-30e-3p | 0.019 | 0.460~0.934 | −0.467 | 0.021* | 0.421~0.932 |
Note: *P<0.05.
Figure 3.
Survival analysis using Kaplan–Meier curves with the log-rank test in the training set. (A) miR-326, (B) miR-6499-5p, (C) miR-30d-3p, (D) miR-4746-5p, (E) miR-1293, (F) miR-4664-3p, (G) miR-30e-3p. (H) Seven-miR signature prognosis index.
Abbreviation: miR, microRNA.
Validation of this seven-miR signature in the validation set
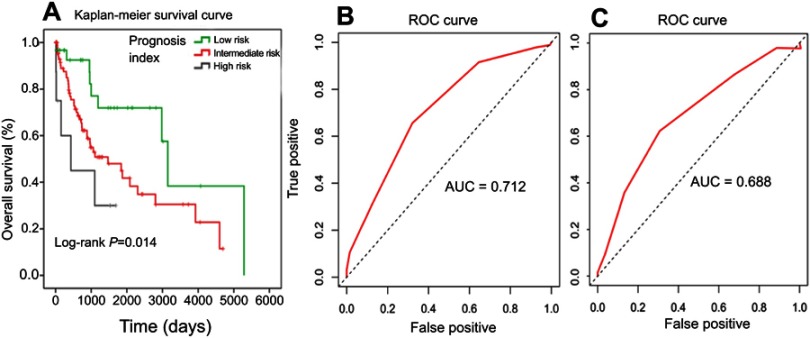
Just as in the training set, patients in the validation set were divided into the low-risk, intermediate-risk, and high-risk groups according to this seven-miR signature index and Kaplan–Meier analysis was used to compare OS. The OS in the low-risk (N=30), intermediate-risk (N=91), and high-risk (N=8) patients was 3149 days (95% CI 2825–3472 days), 1470 days (95% CI 521–2418 days), and 345 days (95% CI 0–1104 days), respectively. The OS in the three groups was significantly different (log-rank p=0.014, Figure 4A).
Figure 4.
Kaplan–Meier curves with the log-rank test of the seven-miR signature prognosis index in the validation set (A). ROC curves of seven-miR signature prognosis index in the training set (B) and the validation set (C).
Abbreviations: AUC, area under the ROC curve; miR, microRNA; ROC, receiver operating characteristic.
Prognostic value of this seven-miR signature
The time-dependent ROC curve was used to assess this seven-miR signature’s predictive value of the 5-year survival rate for LUSC in both the training set and the validation set. The seven-miR signature index is a promising biomarker for predicting the 5-year-survival rate of LUSC with an area under the ROC curve (AUC) of 0.712 (Figure 4B) in the training set and 0.688 in the validation set (Figure 4C), respectively. Furthermore, this seven-miR signature index remained an independent prognostic factor in comparison with routine clinicopathologic features in both the training set (Table 3) and the validation set (Table 4).
Table 3.
Univariate and multivariate analysies of clinicopathological features of the seven-microRNA signature in the training set
| Factor | Univariate Cox analysis | Multivariate Cox analysis | ||
|---|---|---|---|---|
| p-value | HR (95% CI) | p-value | HR (95% CI) | |
| Gender (male/female) | 0.791 | 0.622~1.436 | ||
| Age (>65 years/≤65 years) | 0.692 | 0.744~1.562 | ||
| T stage (T3–4/T1–2) | 0.032* | 1.040~2.351 | 0.611 | 0.661~2.021 |
| Lymph node stage (N1–3/N0) | 0.462 | 0.798~1.644 | ||
| Metastasis (M1/M0) | 0.009* | 1.600~27.030 | 0.166 | 0.641~13.300 |
| Pathological stage (III–IV/I–II) | 0.032* | 1.040~2.427 | 0.214 | 0.807~2.601 |
| PI (≥4/≤3 scores) | 0.000* | 2.146~4.550 | 0.000* | 1.985~4.699 |
Note: *P<0.05.
Table 4.
Univariate and multivariate analyses of clinicopathological features of the seven-microRNA signature in the validation set
| Factor | Univariate Cox analysis | Multivariate Cox analysis | ||
|---|---|---|---|---|
| p-value | HR (95% CI) | p-value | HR (95% CI) | |
| Gender (male/female) | 0.221 | 0.370~1.258 | ||
| Age (>65 years/≤65 years) | 0.191 | 0.823~2.647 | ||
| T stage (T3–4/T1–2) | 0.004* | 1.340~4.724 | 0.135 | 0.823~4.286 |
| Lymph node stage (N1–3/N0) | 0.247 | 0.798~2.396 | ||
| Metastasis (M1/M0) | 0.449 | 0.418~7.203 | ||
| Pathological stage (III–IV/I–II) | 0.025* | 1.087~3.429 | 0.486 | 0.613~2.794 |
| PI (≥4/≤3 scores) | 0.001* | 1.492~4.439 | 0.001* | 1.411~4.227 |
Note: *P<0.05.
Target gene prediction and functional enrichment analysis of these seven miRs
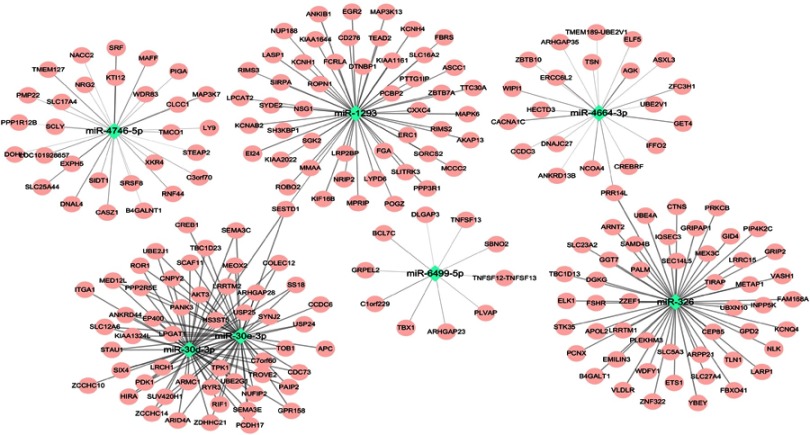
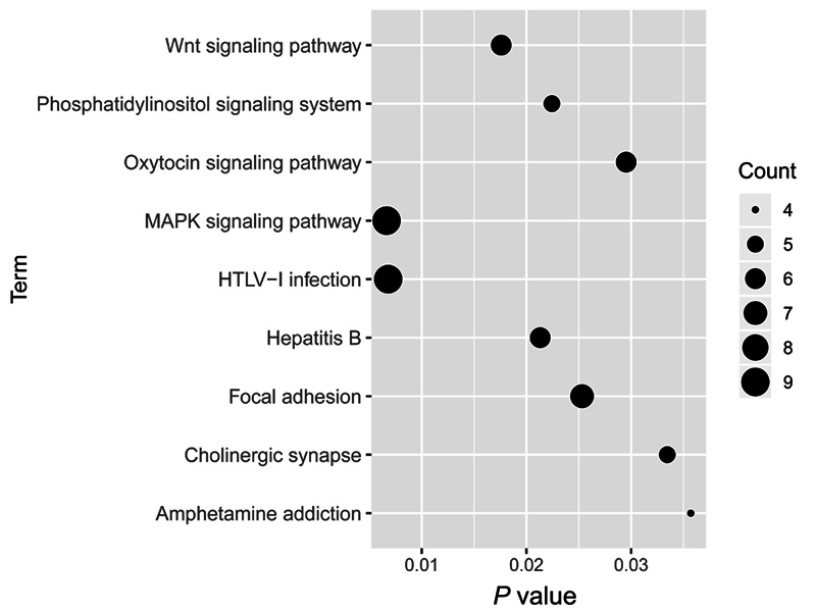
Target gene prediction of these seven miRs was performed using the miRDB online tool. These seven miRs and their respective top 50 target genes (if targets ≥50) were displayed as the miR-target network (Figure 5). Moreover, biological functions of these seven miRs were explored by KEGG pathway enrichment analyses for their target genes using the DAVID. The target genes were involved in various pathways associated with lung cancer, for instance the mitogen-activated protein kinase signaling pathway and the Wnt signaling pathway (Figure 6).
Figure 5.
The network of the seven microRNAs and their target genes.
Figure 6.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses of target genes of the seven microRNAs.
Abbreviation: MAPK, mitogen-activated protein kinase.
Discussion
LUSC accounts for 20–30% of lung cancer cases,18 and the ratio is larger in the countries and regions where tobacco is not controlled, such as the People's Republic of China. The management of LUSC remains dependent on the stage of disease of individual patients and without suggested personalized biomarkers.19 miR expression profiles are highly specific to individual types of cells, tissues, and organs20 and may serve as potential biomarkers of clinical relevance.9,21,22 In our present study, we proposed a new seven-miR signature to predict the prognosis of LUSC. In the same manner as that of several other studies, patients with a survival time of less than 30 days were removed for survival analysis in order to rule out acute death from nontumor causes.23 Patients with LUSC could be divided into high-risk, intermediate-risk, and low-risk groups using this seven-miR signature. The OS in the three groups was significantly different in both the training set and the validation set. Furthermore, this seven-miR signature remained an independent prognostic factor in comparison with routine clinicopathologic features. The seven-miR signature is a promising biomarker for predicting the 5-year-survival rate of LUSC with an AUC of 0.712 in the training set and 0.688 in the validation set, respectively. In addition, it is worth noting that the training set contained more patients with early stage (I/II) disease and patients with Mx than the validation set, and the miR expression profiles of the training set and validation set were based on different platforms. Therefore, this might indicate that this seven-miR signature is still robust in different populations and suitable for different platforms. Based on the predictive seven-miR signature, high-risk patients could be followed up more frequently and accept more active management than low-risk patients.
Tan et al24 proposed a five-miR (miR-210, miR-182, miR-486-5p, miR-30a, and miR-140-3p) signature for LUSC diagnosis and miR-31 for prognosis based on LUSCs from the People's Republic of China in 2011. Interestingly, these six miRs are not included in our miR signature, suggesting that the tumor heterogeneity of LUSC in races may also be reflected in the expression patterns of miRs. Huang et al25 proposed that miR-140-3p was a positive prognostic biomarker for LUSC. Filipska et al26 proposed that miR-192 and miR-662 enhance chemoresistance and invasiveness of LUSC. Luo et al27 found that miR-223-3p functions as a tumor suppressor in LUSC by the miR-223-3p-mutant p53 regulatory feedback loop. However, these miRs have not been involved in our miR signature after univariate and multivariate analyses. Taking into account the sample size of these studies, this may suggest that these miRs may not be a reliable prognostic marker, although they have crucial biological functions. Gao et al28 proposed a seven-miR (miR-101-2, miR-139, miR-182, miR-183, miR-190, miR-326, and miR-944) signature using miR stem loop expression profiles in TCGA database to divide patients into high-risk and low-risk groups, and the AUC of their signature model for the training set (N=224) and the test set (N=223) was only 0.604 and 0.610, respectively. Beer et al found that miR-146b alone may be a biomarker for predicting prognosis in LUSC in 2009.11 Wang et al29 suggested a seven-miR (miR-148b, miR-365, miR-32, miR-375, miR-21, miR-125b, and miR-155) prognostic signature for NSCLC including LUSC. However, there is no comparability between our seven-miR signature and these miR-based signatures because our signature was constructed based on mature miR rather than miR stem loop expression profiles. More meaningfully, the present study is the first miR-based signature that could effectively divide patients with LUSC into high-risk, intermediate-risk, and low-risk groups for more personalized management.
Among our seven miRs, miR-326 was identified as a tumor suppressor miR in various cancers30–32 and was downregulated in LUSC, but high expression of miR-326 was a high risk factor of LUSC in our study. This indicates that the molecular mechanism of miR-326 is complex, and its function in different tumors may be inconsistent. miR-30d was found to be significantly downregulated in LUSC compared with normal lung tissues in a previous study,33 which is consistent with the results of our study. Another study reported that exosomal miR-30e-3p was lung adenocarcinoma specific, rather than LUSC specific.34 However, our results indicated that miR-30e-3p in tumor tissue was a prognostic factor for LUSC. This is an indication of the fact that the expression of miR-30e-3p in LUSC tissues and exosomes may be different. The functions of the other four miRs are still rarely reported. To explore the potential biological functions of the seven miRs, their target genes were predicted respectively using miRDB online analysis tools. The target genes were involved in various pathways associated with lung cancer, for instance the mitogen-activated protein kinase signaling pathway and the Wnt signaling pathway. Given their prognostic value in LUSC, further exploration of the molecular function of these seven miRs is encouraged.
There exist some limitations to this work. There were more LUSC tissues than healthy lung tissues. Despite the fact that a preliminary exploration was carried out, the molecular function of these seven miRs in LUSC was unknown and further experimental validation was lacking. Therefore, it is not clear whether these seven miRs are causal or merely markers for predicting prognosis in LUSC. It may be essential to validate or even improve this seven-miRNA signature in a larger independent cohort.
Conclusion
We propose the first seven-miR signature to predict the prognosis of LUSC. With the use of this signature, patients with LUSC can be divided into high-risk, intermediate-risk, and low-risk groups for personalized management.
Acknowledgments
This work was supported by the Youth Science Foundation of Guangxi Medical University (Grant number: GXMUYSF 201716) and the Guangxi Natural Science Foundation (Grant number: 2018GXNSFBA281091 and 2018GXNSFAA281091).
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet. 2000;355(9202):479–485. doi: 10.1016/S0140-6736(00)82038-3 [DOI] [PubMed] [Google Scholar]
- 3.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P. The international association for the study of lung cancer staging project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4(7):792–801. doi: 10.1097/JTO.0b013e3181a7716e [DOI] [PubMed] [Google Scholar]
- 4.Sawabata N, Asamura H, Goya T, et al. Japanese lung cancer registry study: first prospective enrollment of a large number of surgical and nonsurgical cases in 2002. J Thorac Oncol. 2010;5(9):1369–1375. doi: 10.1097/JTO.0b013e3181e452b9 [DOI] [PubMed] [Google Scholar]
- 5.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8(7):823–859. doi: 10.1097/JTO.0b013e318290868f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43(10):1529–1544. doi: 10.1016/j.ejca.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Bica-Pop C, Cojocneanu-Petric R, Magdo L, Raduly L, Gulei D, Berindan-Neagoe I. Overview upon miR-21 in lung cancer: focus on NSCLC. Cell Mol Life Sci. 2018;75(19):3539–3551. doi: 10.1007/s00018-018-2877-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legras A, Pecuchet N, Imbeaud S, et al. Epithelial-to-Mesenchymal transition and MicroRNAs in lung cancer. Cancers (Basel). 2017;9(8):101. doi: 10.3390/cancers9080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol. 2017;12(2):179–200. doi: 10.1007/s11523-017-0478-5 [DOI] [PubMed] [Google Scholar]
- 10.Lin Y, Lv Y, Liang R, et al. Four-miRNA signature as a prognostic tool for lung adenocarcinoma. Onco Targets Ther. 2018;11:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69(14):5776–5783. doi: 10.1158/0008-5472.CAN-09-0587 [DOI] [PubMed] [Google Scholar]
- 12.Landi MT, Zhao Y, Rotunno M, et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin Cancer Res. 2010;16(2):430–441. doi: 10.1158/1078-0432.CCR-09-1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. [DOI] [PubMed] [Google Scholar]
- 15.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi: 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 19.Soldera SV, Leighl NB. Update on the treatment of metastatic squamous non-small cell lung cancer in new era of personalized medicine. Front Oncol. 2017;7:50. doi: 10.3389/fonc.2017.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 21.Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. doi: 10.1016/j.ccr.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 22.Dvinge H, Git A, Graf S, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497(7449):378–382. doi: 10.1038/nature12108 [DOI] [PubMed] [Google Scholar]
- 23.Lin P, Wen DY, Li Q, He Y, Yang H, Chen G. Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell Physiol Biochem. 2018;48(5):1953–1967. doi: 10.1159/000492519 [DOI] [PubMed] [Google Scholar]
- 24.Tan X, Qin W, Zhang L, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17(21):6802–6811. doi: 10.1158/1078-0432.CCR-11-0419 [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Wang Y, Li Q, Fei X, Ma H, Hu R. miR-140-3p functions as a tumor suppressor in squamous cell lung cancer by regulating BRD9. Cancer Lett. 2019;446:81–89. doi: 10.1016/j.canlet.2019.01.007 [DOI] [PubMed] [Google Scholar]
- 26.Filipska M, Skrzypski M, Czetyrbok K, et al. MiR-192 and miR-662 enhance chemoresistance and invasiveness of squamous cell lung carcinoma. Lung Cancer. 2018;118:111–118. doi: 10.1016/j.lungcan.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 27.Luo P, Wang Q, Ye Y, et al. MiR-223-3p functions as a tumor suppressor in lung squamous cell carcinoma by miR-223-3p-mutant p53 regulatory feedback loop. J Exp Clin Cancer Res. 2019;38(1):74. doi: 10.1186/s13046-019-1079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, Wu Y, Yu W, Li H. Identification of a seven-miRNA signature as prognostic biomarker for lung squamous cell carcinoma. Oncotarget. 2016;7(49):81670–81679. doi: 10.18632/oncotarget.13164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Guo H, Dong Z, et al. Establishment and validation of a 7-microRNA prognostic signature for non-small cell lung cancer. Cancer Manag Res. 2018;10:3463–3471. doi: 10.2147/CMAR.S170481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nawaz Z, Patil V, Paul Y, et al. PI3 kinase pathway regulated miRNome in glioblastoma: identification of miR-326 as a tumour suppressor miRNA. Mol Cancer. 2016;15(1):74. doi: 10.1186/s12943-016-0557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang X, Li Z, Men Q, Li Y, Li H, Chong T. miR-326 functions as a tumor suppressor in human prostatic carcinoma by targeting Mucin1. Biomed Pharmacother. 2018;108:574–583. doi: 10.1016/j.biopha.2018.09.053 [DOI] [PubMed] [Google Scholar]
- 32.Jadideslam G, Ansarin K, Sakhinia E, Alipour S, Pouremamali F, Khabbazi A. The MicroRNA-326: autoimmune diseases, diagnostic biomarker, and therapeutic target. J Cell Physiol. 2018;233:9209s–9222. doi: 10.1002/jcp.v233.12 [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Li X, Yang Q, et al. The role of microRNA in human lung squamous cell carcinoma. Cancer Genet Cytogenet. 2010;200(2):127–133. doi: 10.1016/j.cancergencyto.2010.03.014 [DOI] [PubMed] [Google Scholar]
- 34.Jin X, Chen Y, Chen H, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]