Summary
Background
5‐aminosalicylic acid (5‐ASA) is the first‐line therapy for ulcerative colitis (UC). 5‐ASA acts locally in the colonic mucosa by numerous proposed mechanisms, and is metabolised by N‐acetyltransferase (NAT). Large variations in mucosal 5‐ASA concentrations have been reported, but the underlying mechanisms are not understood.
Aim
To study the relationship between 5‐ASA concentration, 5‐ASA formulation, NAT genotype and bacterial microbiome in patients with UC.
Methods
Patients with quiescent UC, using monotherapy of Mezavant (n = 18), Asacol (n = 14) or Pentasa (n = 10), 4.0‐4.8 g/day were included. 5‐ASA was measured in colonic mucosal biopsies and serum by ultra‐high performance liquid chromatography. NAT genotypes were determined by Sanger sequencing. Bacterial microbiome was sequenced from faeces and mucosa by 16S rRNA sequencing using Illumina Miseq.
Results
Mezavant provided the highest mucosal 5‐ASA levels (geometric mean 2.39 ng/mg), followed by Asacol (1.60 ng/mg, 33% lower, P = 0.50) and Pentasa (0.57 ng/mg, 76% lower, P = 0.033). Mucosal 5‐ASA concentration was not associated with NAT genotype, but serum 5‐ASA concentration and NAT1 genotype was associated (P = 0.044). Mucosal 5‐ASA concentration was positively associated with mucosal bacterial diversity (P = 0.0005) and bacterial composition. High mucosal 5‐ASA concentration was related to reduced abundance of pathogenic bacteria such as Proteobacteria, and increased abundance of several favourable bacteria such as Faecalibacterium.
Conclusions
Mucosal 5‐ASA concentration is positively associated with bacterial diversity and a mucosal bacterial composition that are perceived favourable in UC. Mezavant yielded higher mucosal 5‐ASA concentrations than Pentasa. 5‐ASA may have beneficial effects on the mucosal microbiome, and high concentrations possibly amend dysbiosis in UC.
1. INTRODUCTION
Mesalazine or 5‐aminosalicylic acid (5‐ASA) is the first line therapy for patients with ulcerative colitis (UC), and is effective both for inducing and maintaining remission.1, 2 5‐ASA is proposed to act through numerous mechanisms, including inhibition of pro‐inflammatory mediators such as leukotriens, prostaglandin, interleukin‐1, Nuclear factor‐κB (NF‐κB) and tumour necrosis factor alpha (TNFα), as well as a peroxisome proliferator‐activated receptor gamma (PPAR‐γ) receptor agonist (reviewed by Lichtenstein and Kamm3).
5‐ASA exerts its effect in the intestinal mucosa, and mucosal 5‐ASA concentrations are inversely correlated to disease activity.4, 5, 6, 7 It is therefore important to identify and understand the determinants of mucosal 5‐ASA concentration. In the intestinal mucosa, 5‐ASA is acetylated to its inactive metabolite N‐acetyl‐5‐ASA (Ac‐5‐ASA), mainly by the enzyme N‐acetyl‐transferase 1 (NAT1), and to a small degree by N‐acetyl‐transferase 2 (NAT2).8, 9 Orally administered unbound 5‐ASA is absorbed and inactivated in the small intestinal mucosa and in the liver, thus only small amounts 5‐ASA will reach the colonic mucosa.3, 10 Therefore, several pharmaceutical delivery systems have been developed to transport orally administered 5‐ASA to the colon. In a time‐dependent formulation (Pentasa, Ferring Pharmaceuticals, Saint‐Prex, Switzerland), 5‐ASA is coated with a semipermeable membrane of ethyl cellulose, providing a pH‐independent release of approximately 35% of its content in the small bowel, 25% in the colon, while the last 40% eliminates in the faeces.11 In a pH‐dependent delivery system (Asacol, Tillots Pharma AG, Rheinfelden, Switzerland), 5‐ASA is coated with Eudragit S which dissolves at a pH >7,12 normally occurring in the terminal ileum and caecum. The Multi Matrix System (Mezavant, Shire Pharmaceutical Contracts Ltd, in partnership with Cosmo SpA, Milan, Italy) consists of hydrophilic and lipophilic excipients, covered by a pH‐dependent coating dissolving at pH 7, causing slow diffusion of the drug into the gut lumen.3 For both Asacol and Mezavant absorption in the ileum is estimated to be around 20%.13, 14 Faecal elimination for Asacol and Pentasa has been found to be similar.12
After oral administration of 5‐ASA, mucosal concentrations are highest in the proximal colon segments and lowest in the rectum for the previously examined preparations.7, 15, 16 Higher concentrations in the rectum and left hemicolon have been achieved by combining oral and rectal 5‐ASA formulations.16, 17 However, rectal administration may be inconvenient and adherence to treatment over time is low. As 65%‐85% of patients with UC have rectosigmoid and left‐sided involvement at the time of diagnosis,18, 19 it seems essential to assure optimal concentrations of 5‐ASA in these bowel segments. Although previous studies suggest that Asacol yields higher mucosal 5‐ASA concentrations than Pentasa,4, 15 the different 5‐ASA formulations are by many considered clinically equally efficient.2 Studies comparing mucosal 5‐ASA concentrations of the more recently marketed Mezavant with other 5‐ASA formulations, have to the best of our knowledge, not been published.
Patients with UC have an altered gut microbiome, commonly called dysbiosis. Microbiome alterations in UC are characterised by reduced alpha‐diversity20, 21, 22, 23 combined with increases in the Proteobacteria phylum22, 24, 25 and decreases in the Firmicutes phylum.21, 25, 26 At genus level, Roseburia is reported to be decreased and Haemophilus increased.27 The effect of selected antibiotics, probiotics and faecal microbiota transplantation28 supports that dysbiosis is a part of the pathogenesis in UC. 5‐ASA has previously also been reported to affect intestinal bacteria, by inhibiting growth of anaerobic strains, reducing bacterial invasiveness and total faecal bacterial abundance,29, 30, 31 as well as reducing bacterial adherent biofilm thickness and Escherichia and Shigella abundances in patients with inflammatory bowel disease (IBD).21, 32 Results from a recent study also suggest that 5‐ASA acts as a polyP‐kinase inhibitor, thereby decreasing some bacteria's ability to colonise and increase their susceptibility to oxidative stress.33
In the current study, we aimed to measure and compare mucosal 5‐ASA concentrations in the left hemicolon and rectum in patients with quiescent UC using different oral 5‐ASA formulations (Mezavant, Asacol or Pentasa). We also aimed to explore underlying mechanisms which could explain variations in mucosal 5‐ASA concentration as well as exploring the interrelation between 5‐ASA mucosal concentration and bacterial composition.
2. MATERIALS AND METHODS
2.1. Study population
Patients were recruited from the Department of Gastroenterology and Hepatology, St. Olav's Hospital, Trondheim, Norway from 2015 to 2017. Inclusion criteria were confirmed diagnosis of UC in clinical remission prior to invitation, 18‐70 years of age and use of oral 5‐ASA (Asacol, Pentasa or Mezavant) 4.0‐4.8 g once daily. Exclusion criteria were use of rectal 5‐ASA formulations, prednisolone, azathioprine, methotrexate and TNF‐α medication within the last 3 months. None of the patients had been using antibiotics or probiotics during the last 3 months prior to inclusion. The study was approved by the Regional Committee for Medical and Health Research Ethics, Central Norway (approval reference 2014/2043).
2.2. Blood samples, endoscopic procedure and collection of mucosal biopsies
Eight hours after daily 5‐ASA dose ingestion, patients underwent blood sample collection, enema bowel preparation and sigmoidoscopy. Blood haemoglobin (Hb), leucocyte concentration, plasma erythrocyte sedimentation rate (ESR) and C‐reactive protein (CRP) were analysed successively. Serum was stored at −80°C until analysis of 5‐ASA and Ac‐5‐ASA concentrations. EDTA blood was used for NAT1 and NAT2 genotyping using Sanger sequencing, as described in Supporting Information. A 240 mL sorbitol enema (Klyx, Ferring AS, Copenhagen, Denmark) was administered 30‐45 minutes before sigmoidoscopy (Olympus Exera II GIF H180, Olympus Europa GmbH, Hamburg, Germany) up to 40 cm from the anal verge. Sets of three mucosal biopsies were collected at 10, 25 and 40 cm from the anal verge (total of nine biopsies) using endoscopy forceps (EndoJaw, Olympus Medical Systems, Tokyo, Japan) after cleaning of the mucosa with water to remove visible faecal remnants. Three biopsies were put on formalin for subsequent histological scoring of inflammation. The remaining six biopsies were briefly rinsed in NaCl 9 mg/mL solution, dried, weighed, snap frozen on liquid N2, and stored at −80°C for subsequent measurement of 5‐ASA and Ac‐5‐ASA concentration and sequencing of the mucosal microbiome.
2.3. Analysis of 5‐ASA and Ac‐5‐ASA concentrations
5‐ASA and Ac‐5‐ASA concentrations were analysed in five mucosal colonic biopsies (two biopsies from 10 cm, one biopsy from 25 cm and two biopsies from 40 cm from the anal verge) and in serum from each patient, using an ultra‐high performance liquid chromatography tandem mass spectrometry (UPLC‐MS/MS) method, as described in Supporting Information.
2.4. Evaluation of disease characteristics and inflammation
Demographic and clinical characteristics were recorded. Disease activity was assessed by clinical and endoscopic Mayo score,34 Geboes histological score35 and faecal calprotectin. Endoscopic remission was defined as an endoscopic Mayo score ≤1, histological remission as a Geboes score <2.1 and remission according to total Mayo score as a total Mayo score ≤2. Patients that fulfilled both endoscopic Mayo score ≤1 and Geboes score <2.1 were classified as being in deep remission.36 Patients were instructed to bring a fresh (<24 hours) faecal sample to the appointment. An aliquot was used for calprotectin measurement using a commercial ELISA method (Calpro AS, Lysaker, Norway), normal range <50 mg/kg. Three biopsies from each patient, one from each location (10, 25 and 40 cm from the anal verge) were histologically evaluated by an experienced pathologist. The pathology assessment of inflammation was blinded and scored according to Geboes histological score.35
2.5. Microbiome analysis
The bacterial microbiome was analysed in one mucosal biopsy (sampled 25 cm from the anal verge) and in one faecal sample from each patient (stored at −80°C until analysis). Bacterial DNA was isolated using QIAamp Powerfecal (faecal samples) (Qiagen, Hilden, Germany) and DNeasy Powersoil kit (biopsy samples) (Qiagen), further described in Supporting Information. Libraries were constructed according to Illumina's 16S Metagenomic Sequencing Library Preparation. 16S rRNA amplicon sequencing was performed on an Illumina MiSeq sequencer (Illumina Inc, San Diego, CA). The short read sequencing data from both faecal and mucosal origin were analysed using the operational taxonomic unit (OTU) approach. The QIIME software pipeline (version 1.9.1)37 was used to cluster the gene sequences into OTUs based on sequence similarity. The SortMeRNA_SUMACLUST method of open‐reference OTU picking was applied to a total of 6207583 and 296047 sequencing reads and clustered into 1043 and 770 OTUs for faecal and mucosal samples respectively, at 0.8 SortMeRNA coverage threshold. The OTU taxonomy was identified using the Ribosomal Database Project classifier trained on the Greengenes database (version 13.8).
2.6. Statistical analysis
IBM SPSS Statistics version 24.0 (IBM Corp., Armonk, NY) was used to conduct the statistical analysis. Demographic and clinical characteristics are presented as median (IQR, interquartile range) for skewly distributed variables, mean value (SD) for normal distributed variables, and as % (n) for categorical variables. Accordingly, Kruskal‐Wallis test, F‐test (ANOVA) and chi‐squared test were used for comparing 5‐ASA formulations groups with respect to these measures. A multilevel linear mixed model was applied to test for difference in mean mucosal concentrations of 5‐ASA and Ac‐5‐ASA (log‐transformed data) by type of 5‐ASA formulation, adjusted for (no‐interaction model), or specific to (interaction model) sample locations in the left hemicolon and rectum (10, 25 and 40 cm from the anal verge), further described in Supporting Information. Kruskal‐Wallis test was used for comparing serum concentrations between the three 5‐ASA formulations. Regardless of 5‐ASA formulation (n = 42), mean mucosal 5‐ASA concentration (individual level defined by mean of five repeated measures, log‐transformed) and mean serum 5‐ASA concentration was compared according to NAT genotype (two phenotypes for NAT1 and three phenotypes for NAT2) and disease activity (deep remission or remission according to endoscopic or total Mayo score) using one‐way ANOVA. P‐values <0.05 were considered statistically significant. A power analysis was performed in which mean mucosal 5‐ASA concentration estimates were based on previous studies comparing Pentasa and Asacol4, 7, 15 by one‐way ANOVA and two pair‐wise comparisons. With mean 5‐ASA concentrations of 60 and 20 ng/mg, a SD of 30 ng/mg, α = 0.05 and β = 0.8, 11 patients in each group were needed.
Associations between 5‐ASA mucosal concentrations and bacterial composition in the mucosa and faeces were examined using a linear regression model. The OTU tables generated by the QIIME pipeline were imported into the R software environment version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria) using the phyloseq package and subsequently filtered as to include only OTUs from the Bacteria kingdom and excluding OTUs classified as Mitochondria, Chloroplast or Cyanobacteria/Chlorplast. Alpha diversity values were estimated from the filtered datasets using Shannon entropy. The core microbiome was estimated by requiring 10% prevalence of detected (>1 sequence) OTUs. The core microbiome had a total of 822 and 156 OTUs in faecal and mucosal samples respectively. The count tables at a given taxonomic rank of the core microbiome were imported into the DESeq2 R package to estimate which OTUs that significantly correlated with log‐transformed mucosal 5‐ASA concentrations.38 Significant OTUs were identified using a regression model in DESeq2 using default options and P‐values were estimated using a Wald test and adjusted for multiple testing by Benjamini Hochberg false discovery rate correction.
3. RESULTS
3.1. Patient characteristics
Demographic and clinical characteristics are presented in Table 1. Overall the three 5‐ASA formulation groups were similar. There were no significant differences in total Mayo score (P = 0.055) or proportion of patients in deep remission (P = 0.750). However, the Asacol group had higher leukocyte concentrations (P = 0.036), and the Mezavant group had higher endoscopic Mayo scores (P = 0.01) and fewer patients in endoscopic remission (P = 0.021). All Pentasa patients were classified to be in endoscopic remission, while 12 (67%) Mezavant patients and 13 (93%) Asacol patients were in endoscopic remission (P = 0.021). The proportion of patients in histological remission was, however, lowest in the Pentasa group, n = 7 (70%).
Table 1.
Demographic and clinical characteristics of patients with ulcerative colitis using three different oral 5‐aminosalicylic acid (5‐ASA) formulations
| Mezavant | Asacol | Pentasa | P‐value | |
|---|---|---|---|---|
| Number of patients, n | 18 | 14 | 10 | — |
| Dose (g/d), median (IQR) | 4.8 (0.0) | 4.8 (0.2) | 4.0 (0.0) | — |
| Gender (M‐F) (n‐n) | 11‐7 | 5‐9 | 7‐3 | 0.19 |
| Age at diagnosis (y), mean (SD) | 35.8 (9.7) | 31.4 (12) | 37.2 (13.7) | 0.325 |
| Duration of disease (y), median (IQR) | 8.5 (5) | 11.0 (6) | 7.0 (15) | 0.076 |
| Disease extension, n (%) | ||||
| Rectosigmoid involvement | 6 (33) | 3 (22) | 3 (30) | 0.796 |
| Left sided | 3 (17) | 1 (7) | 1 (10) | |
| Pancolitis | 9 (50) | 10 (71) | 6 (60) | |
| Previous medical therapy (≥3 mo ago), n (%) | ||||
| Prednisolone | 17 (94) | 14 (100) | 8 (80) | — |
| Methotrexate | 0 | 1 (7) | 0 | — |
| Azathioprine | 1 (5.5) | 2 (14) | 0 | — |
| Anti‐TNFα | 0 | 2 (14) | 0 | — |
| Current co‐medication, n (%)a | ||||
| No co‐medication | 10 (56) | 10 (72) | 7 (70) | 0.802 |
| Laboratory values | ||||
| Hbb (g/dL), mean (SD) | 14.4 (1.3) | 13.9 (1.4) | 14.9 (1.1) | 0.168 |
| CRPc (mg/L), median (IQR) | <5 (0) | <5 (4) | <5 (0) | 0.074 |
| ESRd (mm/h), median (IQR) | 3,0 (4) | 7.0 (12)e | 4.5 (6) | 0.210 |
| Leukocytes (×109/L), mean (SD) | 6.7 (1.4) | 8.1 (2) | 6.6 (1.1) | 0.036f |
| Faecal calprotectin (mg/kg), median (IQR) | 33 (243) | 93 (175) | 55 (410) | 0.253 |
| Disease activity | ||||
| Total Mayo score, median (IQR) | 1.5 (3.0) | 1.0 (2.25) | 0.5 (1.25) | 0.055 |
| Clinical Mayo score, median (IQR) | 1.0 (2.0) | 0.5 (2.0) | 0.0 (1.0) | 0.311 |
| Endoscopic Mayo score, median (IQR) | 1.0 (2.0) | 0.0 (1.0) | 0.0 (1.0) | 0.010g |
| Endoscopic remissionh, n (%) | 12 (67%) | 13 (93%) | 10 (100%) | 0.021i |
| Histologic Geboes score, median (IQR) | 1.1 (1.1) | 1.1 (0.1) | 0.1 (1.3) | 0.328 |
| Histologic remissionj , n (%) | 14 (78) | 11 (79) | 7 (70) | 0.873 |
| Deep remissionk, n (%) | 12 (67) | 11 (79) | 7 (70) | 0.750 |
Percentage of patients on monotherapy with oral 5‐ASA and no co‐medication. For patients using co‐medication, the following drugs were used: amiloride/hydrochlorothiazide (n = 1), lisinopril/hydrochlorothiazide (n = 1), losartan/ hydrochlorothiazide(n = 1), calcium and cholecalciferol (n = 3), zoledronate (n = 1), alendronate (n = 2), pantoprazole (n = 1), esomeprazole (n = 1), drospirenone/ethinylestradiol (n = 1), estradiol/norethindrone acetate (n = 1), ethinylestradiol/levonorgestrel (n = 1), venlafaxine (n = 2), levothyroxine (n = 2), tiotropium bromide (n = 1), simvastatin (n = 1), cetirizine (n = 1), paracetamol (n = 1), zolmitriptan (n = 1), sumatriptan (n = 1), desoximetasone crème 2.5 mg/g (n = 1), terbinafine (n = 1).
Blood haemoglobin.
C‐reactive protein.
Plasma erythrocyte sedimentation rate.
For one Asacol patient SR‐value is missing.
Asacol group had higher leukocyte concentration.
Mezavant group had higher endoscopic Mayo scores.
Defined as a endoscopic Mayo score ≤1.
Mezavant patients had lower rates of endoscopic remission.
Defined as a Geboes score <2.1.
Defined as a endoscopic Mayo score ≤1 and a Geboes score <2.
3.2. 5‐ASA drug formulation was associated with mucosal 5‐ASA concentration
A graphical display of the individual 5‐ASA concentrations at different locations (Figure 1) illustrates large inter‐individual variations, but small intra‐individual variation between segments. Boxplots of mucosal 5‐ASA concentration at all locations are shown in Figure S4.
Figure 1.
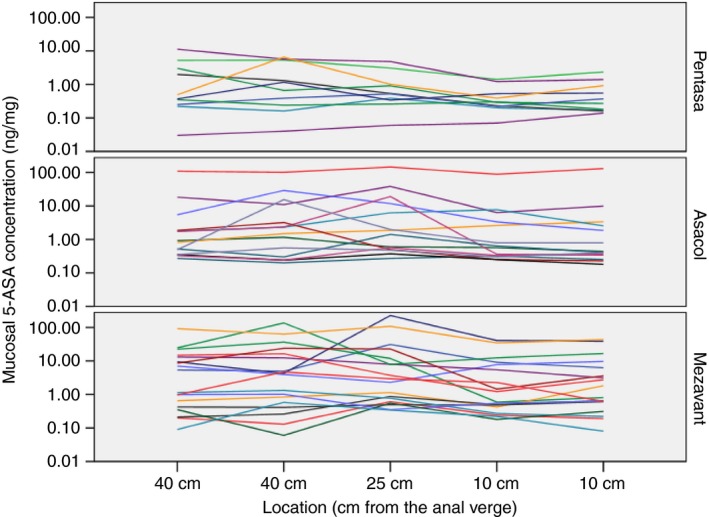
Mucosal 5‐aminosalicylic acid (5‐ASA) concentration in five colonic biopsies sampled 40, 25 and 10 cm from the anal verge in patients with ulcerative colitis using three different oral 5‐ASA formulations. Each line represents one patient
The overall test for difference in adjusted mean 5‐ASA concentration (averaged over location) between the three 5‐ASA formulation groups (Table 2) did not reach statistical significance (P = 0.099), but pairwise comparisons revealed a significant higher mean concentration in patients using Mezavant compared with Pentasa (geometric mean 2.39 ng/mg vs 0.57 ng/mg, P = 0.033) and a nonsignificant higher mean value compared to the Asacol group (2.39 ng/mg vs 1.60 ng/mg, P = 0.50). Recalculated into % difference, the Pentasa patients had 76% lower 5‐ASA concentrations compared with Mezavant patients, and Asacol patients had in average 33% lower concentrations. The mean 5‐ASA concentration level differed significantly by location (P ≤ 0.001), with the lowest concentrations in the rectum regardless of formulation. Despite a more clear decreasing trend according to location in the Pentasa group, the test for interaction between location and formulation did not reach statistical significance (P = 0.68). Consequently, the difference between the three 5‐ASA formulations was consistently observed at each location, although power of test within subgroups was low. Adjustment for deep remission did not change the results notably, but provided more precise estimates (slightly lower P‐values and smaller CI, results not shown).
Table 2.
Mucosal concentrations (ng/mg) of 5‐aminosalicylic acid (5‐ASA) and acetylated 5‐ASA (Ac‐5‐ASA) in the left hemicolon and rectum in patients with ulcerative colitis using three different oral 5‐ASA formulations
| Geometric mean (95% confidence interval)a | P‐value | ||||
|---|---|---|---|---|---|
| Mezavant (n = 18) | Asacol (n = 14) | Pentasa (n = 10) | By locationb | By formulationc | |
| 5‐ASA concentrations | |||||
| Adjusted meand | 2.39 (1.09‐5.28) | 1.60 (0.65‐3.91) | 0.57 (0.20‐1.64) | 0.099 | |
| Difference (%) | 1.0 (reference) | −33.2 (−79.8, 119.7) | −76.3 (−93.7, −11.2) | ||
| P‐valuee | 0.50 | 0.033 | |||
| By location (cm from the anal verge) | <0.001 | ||||
| 40 | 2.71 (1.20‐6.10) | 1.71 (0.68‐4.31) | 0.76 (0.26‐2.26) | 0.24 | |
| 25 | 3.22 (1.36‐7.59) | 2.31 (0.88‐6.10) | 0.58 (0.18‐1.81) | 0.064 | |
| 10 | 1.61 (0.71‐3.63) | 1.09 (0.43‐2.74) | 0.37 (0.13‐1.10) | 0.081 | |
| P‐value, locationf | 0.013 | 0.011 | 0.006 | ||
| P ‐value, interactiong | 0.68 | ||||
| Ac‐5‐ASA concentrations | |||||
| Adjusted meand | 1.56 (0.98‐2.49) | 1.03 (0.61‐1.74) | 1.19 (0.64‐2.22) | 0.47 | |
| Difference (%) | 1.0 (reference) | −34.3 (−67.4, 32.4) | −23.9(−64.9, 65.2) | ||
| P‐valuee | 0.23 | 0.48 | |||
| By location (cm from the anal verge): | <0.001 | ||||
| 40 | 2.03 (1.24‐3.30) | 1.68 (0.97‐2.93) | 2.02 (1.05‐3.89) | 0.85 | |
| 25 | 1.50 (0.88‐2.57) | 0.88 (0.48‐1.61) | 1.05 (0.52‐2.15) | 0.47 | |
| 10 | 1.29 (0.79‐2.10) | 0.71 (0.41‐1.24) | 0.78 (0.40‐1.50) | 0.27 | |
| P‐value, locationf | 0.009 | <0.001 | <0.001 | ||
| P‐value, interactiong | 0.23 | ||||
Results based on analyses of log‐transformed data in multilevel linear mixed model, with and without interaction between 5‐ASA formulation type and location, and with additional adjustments for pinch biopsy site and replicates (replicate samples at 40 and 10 cm).
F‐tests for difference in mean values between locations; 10, 25 and 40 cm from the anal verge (no‐interaction model).
F‐tests for difference in mean values between 5‐ASA formulation groups, overall (no‐interaction model) and at each specific location (interaction model).
Overall mean, averaged over location and order of sampling (estimated marginal means from no‐interaction model).
Pairwise tests (each group compared with Mezavant).
F‐tests for difference in mean values between locations within each 5‐ASA formulation (interaction model).
F‐test for interaction between location and 5‐ASA formulation type.
The mucosal concentration of Ac‐5‐ASA decreased significantly from oral to anal direction, and did not differ between the formulations overall (P = 0.47) or in pairwise comparisons (Mezavant vs Pentasa P = 0.23 and Mezavant vs Asacol P = 0.48). There were no significant differences in serum 5‐ASA (P = 0.20) or serum Ac‐5‐ASA (P = 0.20) between the different 5‐ASA formulations (Table 3).
Table 3.
Serum concentrations (ng/mL) of 5‐aminosalicyclic acid (5‐ASA) and acetylated 5‐ASA (Ac‐5‐ASA) in patients with ulcerative colitis using three different oral 5‐ASA formulations
| Mezavant | Asacol | Pentasa | P‐valuea | |
|---|---|---|---|---|
| Serum concentrations (ng/mL), median (interquartile range) | ||||
| 5‐ASA | 3420 (5085) | 1658 (3347) | 3028 (6317) | 0.202 |
| Ac‐5‐ASA | 2319 (2769) | 1770 (2439) | 4634 (5555) | 0.200 |
Compared using Kruskal‐Wallis test.
3.3. Mucosal 5‐ASA concentration was positively associated to abundance of numerous beneficial bacteria
Mucosal 5‐ASA concentration was significantly associated to the mucosal bacterial abundance on all taxonomic levels (Table 4), whereas the association to bacterial abundance in faecal samples was weaker (Table 5). Mucosal 5‐ASA concentration and alpha‐diversity was positively associated in mucosal biopsies (P = 4.7 × 10−4), whereas no association was found in faecal samples (Figure 2). Regression analysis of bacterial abundances on phylum level and mucosal 5‐ASA concentration (Figure 3) revealed that mucosal 5‐ASA mucosal concentrations were associated with lower abundances of the Proteobacteria phylum (P = 1.2 × 10−15) and higher abundances of Firmicutes (P = 2.6 × 10−6) and Bacteroidetes (P = 3.1 × 10−4) in mucosal biopsies. There were no associations between faecal bacterial abundance on phylum level and 5‐ASA mucosal concentration.
Table 4.
Mucosal bacteria abundances significantly associated with 5‐aminosalicylic acid (5‐ASA) concentration on different taxonomic levels (regression analysis)
| Phylum | Class | Order | Family | Genus | P | B |
|---|---|---|---|---|---|---|
| Proteobacteria | — | — | — | — | 1.2 × 10−15 | −1.29 |
| Firmicutes | — | — | — | — | 2.6 × 10−6 | 0.48 |
| Bacteroidetes | — | — | — | — | 3.1 × 10−4 | 0.99 |
| Firmicutes | Clostridia | — | — | — | 6.1 × 10−21 | 2.33 |
| Bacteroidetes | Bacteroidia | — | — | — | 6.9 × 10−9 | 2.10 |
| Proteobacteria | Alphaproteobacteria | — | — | — | 1.2 × 10−6 | −0.87 |
| Firmicutes | Bacilli | — | — | — | 1.2 × 10−5 | −0.70 |
| Proteobacteria | Betaproteobacteria | — | — | — | 5.5 × 10−5 | −0.65 |
| Bacteroidetes | Flavobacteriia | — | — | — | 4.8 0.10−3 | −2.34 |
| Firmicutes | Clostridia | Clostridiales | — | — | 2.4 × 10−20 | 2.62 |
| Bacteroidetes | Bacteroidia | Bacteroidales | — | — | 8.2 × 10−10 | 2.10 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | — | — | 1.3 × 10−4 | 1.94 |
| Firmicutes | Bacilli | Lactobacillales | — | — | 1.4 × 10−4 | 3.37 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | — | — | 9.8 × 10−4 | −0.57 |
| Firmicutes | Bacilli | Bacillales | — | — | 0.002 | −0.49 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | — | 7.8 × 10−13 | 1.73 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | — | 1.6 × 10−12 | 2.01 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | — | 9.8 × 10−12 | −1.27 |
| Firmicutes | Bacilli | Bacilliales | Bacillaceae | — | 6.2 × 10−12 | −1.32 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Brucellaceae | — | 2.8 × 10−8 | −1.51 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | — | 3.9 × 10−5 | 1.46 |
| Bacteroidetes | Flavobacteriia | Flavobacteriales | Weeksellaceae | — | 5.7 × 10−4 | −2.95 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | — | 8.2 × 10−4 | 2.04 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | — | 0.011 | 2.38 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | — | 0.015 | −1.11 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | — | 0.015 | 2.10 |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | — | 0.015 | 1.76 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | — | 0.029 | −0.74 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | — | 0.031 | 3.60 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | — | 0.031 | 0.99 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | — | 0.035 | 1.87 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Faecalibacterium | 4.8 × 10−16 | 3.16 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia | 2.1 × 10−14 | 2.60 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 1.8 × 10−10 | 2.72 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Coprococcus | 3.6 × 10−10 | 2.41 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Dorea | 1.8 × 10−8 | 3.05 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Ruminococcus | 6.1 × 10−8 | 3.97 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillospira | 6.2 × 10−8 | 3.94 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Roseburia | 9.6 × 10−8 | 3.39 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Porphyromonadaceae | Parabacteroides | 1.0 × 10−7 | 3.40 |
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | 1.9 × 10−5 | 2.10 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 5.5 × 10−5 | 3.55 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 1.2 × 10−4 | 3.36 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospira | 1.2 × 10−4 | 2.02 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus | 2.1 × 10−4 | 2.76 |
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | Collinsella | 1.2 × 10−3 | 2.17 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Odoribacteraceae | Odoribacter | 9.3 × 10−3 | 2.48 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 0.021 | 4.51 |
| Firmicutes | Clostridia | Clostridiales | Veillonellaceae | Dialister | 0.028 | 1.40 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Weeksellaceae | Cloacibacterium | 0.048 | −1.62 |
Bacteria within each taxonomic level are listed by increasing P‐values. P = adjusted P‐value, B = regression coefficient. Red text = increased abundance (positive association), blue = decreased abundance (negative association).
Table 5.
Faecal bacteria abundances significantly associated with 5‐aminosalicylic acid (5‐ASA) concentration on different taxonomic levels (regression analysis)
| Phylum | Class | Order | Family | Genus | P | B |
|---|---|---|---|---|---|---|
| Proteobacteria | Betaproteobacteria | — | — | — | 0.004 | −2.02 |
| Bacteroidetes | Betaproteobacteria | Burkholderiales | — | — | 0.007 | −2.02 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | — | 7.7 × 10−5 | −5.17 |
| Firmicutes | Betaproteobacteria | Burkholderiales | Alcaligenaceae | — | 0.02 | −1.92 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Prevotellaceae | Prevotella | 7.2 × 10−5 | −5.27 |
| Firmicutes | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Sutterella | 0.025 | −1.91 |
Bacteria within each taxonomic level are listed by increasing P‐values. P = adjusted P‐value, B = regression coefficient, Blue text = decreased abundance (negative association).
Figure 2.
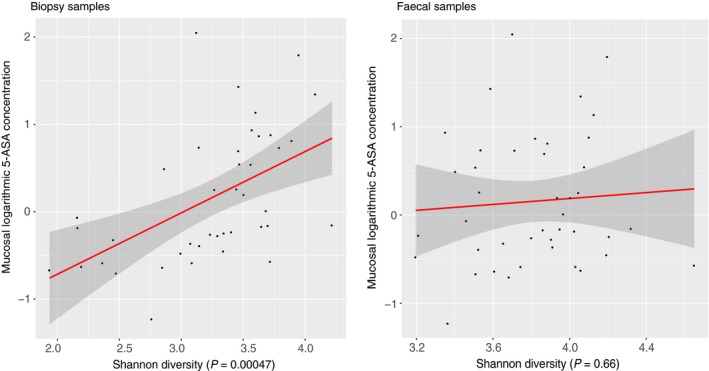
Bacterial diversity analysis in biopsies and faecal samples in patients with ulcerative colitis using 5‐aminosalicylic (5‐ASA). Red line = regression line, dark grey area = 95% confidence interval
Figure 3.

Bacterial phyla correlated to 5‐aminosalicylic acid (5‐ASA) mucosal concentration in biopsies and faecal samples in patients with ulcerative colitis (UC). P: phylum, P = adjusted P‐value, B = regression coefficient, Rlog = regularised logarithm transformation. Blue line = regression line. Dark grey area = 95% confidence interval
In the mucosal biopsies, 10 bacterial families were positively and six families were negatively associated to mucosal 5‐ASA concentration (Figures S5 and S6), the most significant association was seen for Lachnospiraceae and Ruminococcaceae families (Figure 4A,B). Totally 19 mucosal bacterial genera were associated with 5‐ASA concentration. The bacterial genus in the mucosa that was most significantly associated to 5‐ASA concentration was Faecalibacterium (4.8 × 10−16) (Figure 4C), followed by the genera Blautia (Figure 4D), Bacteroides, Coprococcus and Dorea. Of the genera associated with 5‐ASA concentration, 18 of 19 were positively associated (Figure S7). Cloacibacterium was the only genus negatively associated (P = 0.048) with mucosal 5‐ASA concentration in the mucosal biopsies (Figure 4F). On species level, Faecalibacterium prausnitzii was positively correlated with mucosal 5‐ASA concentration (P = 1.3 × 10−6) (Figure 4E). In faeces, two bacterial genera were associated with 5‐ASA mucosal concentration; Prevotella (P = 7.2 × 10−5) and Sutterella (P = 0.03), both genera were negatively associated with 5‐ASA concentrations (Table 5).
Figure 4.
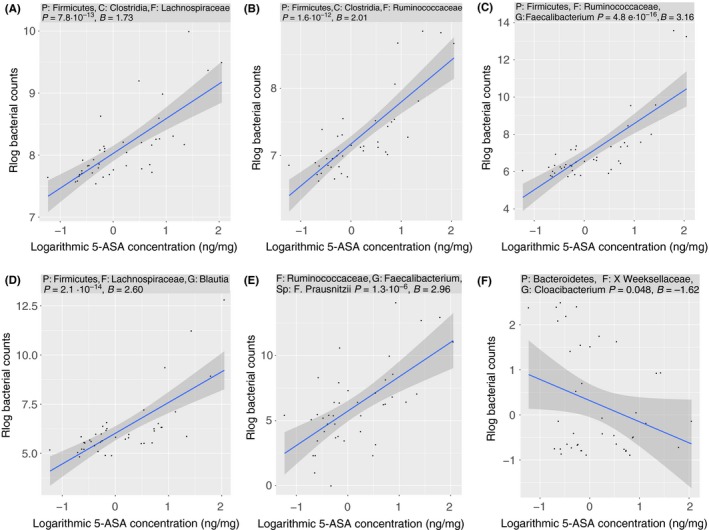
Selected bacterial families, genera and species significantly correlated to 5‐aminosalicylic acid (5‐ASA) mucosal concentration in mucosal biopsies in patients with ulcerative colitis; (A) Lachnospiraceae, (B) Ruminococcaceae, (C) Faecalibacterium, (D) Blautia, (E) Faecalibacterium prausnitzii, (F) Cloacibacterium (for complete list see Figures S5‐S7). P, phylum; C, class; F, family; G, genus; Sp, species. P = adjusted P‐value; B = regression coefficient, Rlog = regularised logarithm transformation. Blue line = regression line. Dark grey area = 95% confidence interval
3.4. Mucosal 5‐ASA concentration was not associated with mucosal inflammation
Mucosal 5‐ASA concentrations and disease activity were not significantly associated in our patients. Mucosal 5‐ASA concentration was neither associated with deep remission (P = 0.106), total Mayo score (P = 0.114) nor endoscopic Mayo score (P = 0.055). The inflammation variables tended to be higher in patients with high mucosal 5‐ASA concentration.
3.5. NAT1 genotype influences serum, but not mucosal 5‐ASA concentration
All patients were successfully genotyped for NAT1 and NAT2. Eight patients (19.1%) had a slow acetylator NAT1 phenotype (Table 6). Thirty‐one patients (73.8%) had a slow acetylator NAT2 phenotype (Table S3). Patients with NAT1 slow acetylators status had significantly higher 5‐ASA serum concentrations (P = 0.044) than patients with NAT1 rapid acetylator status. There was no significant association between serum 5‐ASA concentration and NAT2 genotype (P = 0.708). Mucosal 5‐ASA concentration was not significantly associated with neither NAT1 (P = 0.276) nor NAT2 (P = 0.488) genotype.
Table 6.
Frequencies of N‐acetyl‐transferase 1 (NAT1) genotypes and phenotype/acetylator status in patients with ulcerative colitis using oral 5‐aminosalicylic acid
| NAT1 Genotype | Number of patients (n) | Phenotype/Acetylator status | Observed frequency (%) |
|---|---|---|---|
| NAT1*3/*4 | 1 | Rapid | 2.4 |
| NAT1*4/*4 | 24 | Rapid | 57.1 |
| NAT1*4/*10 | 7 | Rapid | 16.7 |
| NAT1*4/*14B | 2 | Slow | 4.8 |
| NAT1*4/*15 | 1 | Slow | 2.4 |
| NAT1*4/*16 | 1 | Rapid | 2.4 |
| NAT1*4/*17 | 4 | Slow | 9.5 |
| NAT1*10/*10 | 1 | Rapid | 2.4 |
| NAT1*10/*14A | 1 | Slow | 2.4 |
4. DISCUSSION
This is the first study to measure mucosal 5‐ASA concentration in UC patients using different oral 5‐ASA formulations, and correlating it to NAT genotype and bacterial microbiota. Concurring with previous findings, our patients had declining concentrations of 5‐ASA towards the rectum.16 A novel finding was that patients using Mezavant had higher overall mucosal 5‐ASA concentrations, compared to patients using Pentasa, while there was no significant difference between patients using Mezavant and Asacol. All patients used high doses of 5‐ASA, however the manufacturers' maximal recommended dose of Pentasa (4.0 g) is 17% lower than of Mezavant (4.8 g) and Asacol (4.8 g), but this could only explain a minor proportion of the observed differences. The large inter‐individual variation in concentration was consistent with previous reports,4, 5, 6, 7 whereas the intra‐individual variation between bowel segments was small. Factors such as colonic transit time, intraluminal pH and disease pattern may affect dissolution and uptake of 5‐ASA in the colon39 and could explain some of the inter‐individual variation in 5‐ASA concentration reported in the present and previous studies.4, 15, 16 The mucosal Ac‐5‐ASA concentration reflects the amount of 5‐ASA metabolised by NAT1 in the colonic mucosa. There were no significant differences in Ac‐5‐ASA mucosal concentrations between the three 5‐ASA formulations. Similarly, the serum concentrations of 5‐ASA or Ac‐5‐ASA did not differ between the formulations.
The intestinal bacterial composition and metabolic products may affect disease activity in UC. 5‐ASA has been found to inhibit in vitro growth of Mycobacterium avium paratuberculosis as well as anaerobic bacteria.31, 40 5‐ASA also influence bacterial gene expression causing reduced bacterial invasiveness.30 Furthermore, in patients with irritable bowel syndrome, 5‐ASA reduces overall faecal bacterial abundance, increases Firmicutes and decreases Bacteroidetes abundances.29 In the present study, mucosal 5‐ASA concentration was remarkably associated with alterations in the mucosal bacterial composition, but to a lesser degree with alterations in faecal microbiota. High mucosal 5‐ASA concentration was associated with high bacterial diversity, decreased mucosal abundances of Proteobacteria phylum and increased mucosal abundances of Firmicutes and Bacteroidetes phyla. Reduced bacterial diversity is a hallmark of dysbiosis in UC, whereas increased bacterial diversity is perceived beneficial.20, 21, 25, 41 Proteobacteria are increased in patients with UC,25, 42, 43 and linked to increased disease activity and relapse frequency.24, 42 Overall, the bacterial genera positively associated with high 5‐ASA concentrations have previously been associated with beneficial effects in IBD: (a) the abundances of the butyrate producing Faecalibacterium and Roseburia have been inversely correlated to disease activity in UC.44 (b) Blautia, Bacteroides, Parabacteroides and Sutterella have all been reported to be inversely correlated with inflammation in patients with ileal‐pouch anal anastomosis.45 (c) Ruminococcaceae and Ruminococcus spp in donor faeces transplanted to UC patients is associated with induction of remission46 and (d) the Lachnospiraceae and Ruminococcaceae family, to which most genera in Table 4 belong, may protect against UC as they are increased in healthy twins discordant for UC.22 One genus, Cloacibacterium, was found to be negatively associated to mucosal 5‐ASA concentration, recently this genus has been reported to be more abundant in inflamed vs. non‐inflamed tissue of UC patients.23
Although 16S rRNA sequencing may not be the preferred method for analysing bacteria on species level, the mucosal 5‐ASA concentration was positively associated with the mucosal abundance of F. prausnitzii. Faecalibacterium prausnitzii is found to be depleted in UC patients and inversely correlated with disease activity, whereas increased abundance is associated with long‐term remission.44, 47
The mucosal 5‐ASA concentration was not associated with alterations in the faecal bacteriome to the same extent as the mucosal bacteriome. The faecal bacterial diversity, and bacterial abundance on phylum level, were not associated with mucosal 5‐ASA concentrations. However, abundances of the Betaproteobacteria class and Burkholderiales order in faeces were negatively associated to mucosal 5‐ASA concentration, the former association was also found in the mucosa and is presumed favourable in UC.24, 42
5‐ASA concentration have previously been found to be inversely correlated to disease activity index scores,5, 7 endoscopic and histologic remission4, 6 in UC patients with mild to moderate disease activity. In the present study, patients had low disease activity, and in contrast to previous findings, the 5‐ASA concentration was not significantly associated with the combined parameters of disease activity. Such associations may not be evident within a relatively homogenous patient group with low disease activity. In fact, there was a trend towards higher mucosal 5‐ASA concentrations in patients with higher endoscopic scores and it is unlikely that the microbiome features associated with mucosal 5‐ASA concentration were confounded by the degree of inflammation. NAT1 and NAT2 genotypes have previously not been found to influence treatment efficacy,48 but studies correlating NAT genotypes to 5‐ASA mucosal concentrations have previously not been published. We found no correlation between NAT genotypes and mucosal 5‐ASA concentration, thus NAT1 genotype cannot explain the large inter‐individual variations in mucosal 5‐ASA concentrations in our study. Interestingly we found that patients with the NAT1 slow acetylator genotype had significantly higher serum 5‐ASA concentrations compared to patients with NAT1 rapid acetylator genotype, which could theoretically influence the risk of concentration dependent systemic adverse effects.
5. CONCLUSIONS
Patients using Mezavant had significantly higher mucosal 5‐ASA concentrations than patients using Pentasa. Our results suggest that 5‐ASA increases bacterial diversity, favours numerous beneficial bacteria and inhibits disadvantageous bacteria in UC patients. In conclusion, our novel findings indicate that 5‐ASA may have positive effects on the mucosal microbiome and could amend dysbiosis in UC patients.
Supporting information
ACKNOWLEDGMENTS
The authors would like to thank Bjørn Munkvold for preparation of histological sections. The bacterial sequencing was provided by the Genomics Core Facility (GCF), NTNU. GCF is funded by the Faculty of Medicine and Health Sciences at NTNU and RHA. Financial support was given from the Liaison Committee between Central Norway Regional Health Authority (RHA) and Norwegian University of Science and Technology (NTNU) and from St. Olav's Hospital, Trondheim University Hospital.
Declaration of personal interests: None.
AUTHORSHIP
Guarantor of the article: Reidar Fossmark.
Author contributions: TCM and RF designed the study. MO, TCM and RF have collected the data. OS, AF, WRB, GA, ESR, and BG have contributed to analyses of biological material and data. MO, OS, AF, AvBG, GA, AKS, TCM and RF have interpreted the results and drafted the manuscript. All authors have approved the final version of the manuscript including the author list.
Olaisen M, Spigset O, Flatberg A, et al. Mucosal 5‐aminosalicylic acid concentration, drug formulation and mucosal microbiome in patients with quiescent ulcerative colitis. Aliment Pharmacol Ther. 2019;49:1301–1313. 10.1111/apt.15227
The Handling Editor for this article was Dr Colin Howden, and it was accepted for publication after full peer‐review.
Funding information
This study was funded in full by the Liaison Committee between Central Norway Regional Health Authority (RHA) and Norwegian University of Science and Technology (NTNU) and by St. Olav's Hospital, Trondheim University Hospital.
REFERENCES
- 1. Wang Y, Parker CE, Bhanji T, Feagan BG, MacDonald JK. Oral 5‐aminosalicylic acid for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2016;4:CD000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Y, Parker CE, Feagan BG, MacDonald JK. Oral 5‐aminosalicylic acid for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2016;5:CD000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lichtenstein GR, Kamm MA. Review article: 5‐aminosalicylate formulations for the treatment of ulcerative colitis–methods of comparing release rates and delivery of 5‐aminosalicylate to the colonic mucosa. Aliment Pharmacol Ther. 2008;28:663‐673. [DOI] [PubMed] [Google Scholar]
- 4. D'Inca R, Paccagnella M, Cardin R, et al. 5‐ASA colonic mucosal concentrations resulting from different pharmaceutical formulations in ulcerative colitis. World J Gastroenterol. 2013;19:5665‐5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D'haens G, Hommes D, Engels L, et al. Once daily MMX mesalazine for the treatment of mild‐to‐moderate ulcerative colitis: a phase II, dose‐ranging study. Aliment Pharmacol Ther. 2006;24:1087‐1097. [DOI] [PubMed] [Google Scholar]
- 6. Frieri G, Giacomelli R, Pimpo M, et al. Mucosal 5‐aminosalicylic acid concentration inversely correlates with severity of colonic inflammation in patients with ulcerative colitis. Gut. 2000;47:410‐414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Naganuma M, Iwao Y, Ogata H, et al. Measurement of colonic mucosal concentrations of 5‐aminosalicylic acid is useful for estimating its therapeutic efficacy in distal ulcerative colitis: comparison of orally administered mesalamine and sulfasalazine. Inflamm Bowel Dis. 2001;7:221‐225. [DOI] [PubMed] [Google Scholar]
- 8. Hein DW, Doll MA, Rustan TD, et al. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633‐1638. [DOI] [PubMed] [Google Scholar]
- 9. Hickman D, Pope J, Patil Sd, et al. Expression of arylamine N‐acetyltransferase in human intestine. Gut. 1998;42:402‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harris MS, Lichtenstein GR. Review article: delivery and efficacy of topical 5‐aminosalicylic acid (mesalazine) therapy in the treatment of ulcerative colitis. Aliment Pharmacol Ther. 2011;33:996‐1009. [DOI] [PubMed] [Google Scholar]
- 11. Rasmussen SN, Bondesen S, Hvidberg EF, et al. 5‐aminosalicylic acid in a slow‐release preparation: Bioavailability, plasma level, and excretion in humans. Gastroenterology. 1982;83:1062‐1070. [PubMed] [Google Scholar]
- 12. Sandborn WJ, Hanauer SB. Systematic review: the pharmacokinetic profiles of oral mesalazine formulations and mesalazine pro‐drugs used in the management of ulcerative colitis. Aliment Pharmacol Ther. 2003;17:29‐42. [DOI] [PubMed] [Google Scholar]
- 13. Brunner M, Assandri R, Kletter K, et al. Gastrointestinal transit and 5‐ASA release from a new mesalazine extended‐release formulation. Aliment Pharmacol Ther. 2003;17:395‐402. [DOI] [PubMed] [Google Scholar]
- 14. Sandborn WJ, Hanauer SB, Buch A. Comparative pharmacokinetics of equimolar doses of 5‐aminosalicylate administered as oral mesalamine (Asacol) and balsalazide: a randomized, single‐dose, crossover study in healthy volunteers. Aliment Pharmacol Ther. 2004;19:1089‐1098. [DOI] [PubMed] [Google Scholar]
- 15. De Vos M, Verdievel H, Schoonjans R, Praet M, Bogaert M, Barbier F. Concentrations of 5‐ASA and Ac‐5‐ASA in human ileocolonic biopsy homogenates after oral 5‐ASA preparations. Gut. 1992;33:1338‐1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frieri G, Pimpo MT, Palumbo GC, et al. Rectal and colonic mesalazine concentration in ulcerative colitis: oral vs. oral plus topical treatment. Aliment Pharmacol Ther. 1999;13:1413‐1417. [DOI] [PubMed] [Google Scholar]
- 17. Frieri G, Pimpo M, Galletti B, et al. Long‐term oral plus topical mesalazine in frequently relapsing ulcerative colitis. Dig Liver Dis. 2005;37:92‐96. [DOI] [PubMed] [Google Scholar]
- 18. Henriksen M, Jahnsen J, Lygren I, et al. Ulcerative colitis and clinical course: results of a 5‐year population‐based follow‐up study (the IBSEN study). Inflamm Bowel Dis. 2006;12:543‐550. [DOI] [PubMed] [Google Scholar]
- 19. Ungaro R, Mehandru S, Allen PB, Peyrin‐Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756‐1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alipour M, Zaidi D, Valcheva R, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. 2016;10:462‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lepage P, Häsler R, Spehlmann ME, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227‐236. [DOI] [PubMed] [Google Scholar]
- 23. Hirano A, Umeno J, Okamoto Y, et al. Comparison of the microbial community structure between inflamed and non‐inflamed sites in patients with ulcerative colitis. J Gastroenterol Hepatol. 2018;33:1590-1597. [DOI] [PubMed] [Google Scholar]
- 24. Haberman Y, Tickle TL, Dexheimer PJ, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617‐3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Imhann F, Vich Vila A, Bonder MJ, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67:108‐119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Walker AW, Sanderson JD, Churcher C, et al. High‐throughput clone library analysis of the mucosa‐associated microbiota reveals dysbiosis and differences between inflamed and non‐inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shah R, Cope JL, Nagy‐Szakal D, et al. Composition and function of the pediatric colonic mucosal microbiome in untreated patients with ulcerative colitis. Gut Microb. 2016;7:384‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McIlroy J, Ianiro G, Mukhopadhya I, Hansen R, Hold GL. Review article: the gut microbiome in inflammatory bowel disease‐avenues for microbial management. Aliment Pharmacol Ther. 2018;47:26‐42. [DOI] [PubMed] [Google Scholar]
- 29. Andrews CN, Griffiths TA, Kaufman J, Vergnolle N, Surette MG, Rioux KP. Mesalazine (5‐aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea‐predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2011;34:374‐383. [DOI] [PubMed] [Google Scholar]
- 30. Kaufman J, Griffiths TA, Surette MG, Ness S, Rioux KP. Effects of mesalamine (5‐aminosalicylic acid) on bacterial gene expression. Inflamm Bowel Dis. 2009;15:985‐996. [DOI] [PubMed] [Google Scholar]
- 31. Sandberg‐Gertzen H, Kjellander J, Sundberg‐Gilla B, Jarnerot G. In vitro effects of sulphasalazine, azodisal sodium, and their metabolites on Clostridium difficile and some other faecal bacteria. Scand J Gastroenterol. 1985;20:607‐612. [DOI] [PubMed] [Google Scholar]
- 32. Swidsinski A, Loening‐Baucke V, Bengmark S, Lochs H, Dorffel Y. Azathioprine and mesalazine‐induced effects on the mucosal flora in patients with IBD colitis. Inflamm Bowel Dis. 2007;13:51‐56. [DOI] [PubMed] [Google Scholar]
- 33. Dahl J‐U, Gray MJ, Bazopoulou D, et al. The anti‐inflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol. 2017;2:16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5‐aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625‐1629. [DOI] [PubMed] [Google Scholar]
- 35. Geboes K, Riddell R, Ost A, Jensfelt B, Persson T, Lofberg R. A reproducible grading scale for histological assessment of inflammation in ulcerative colitis. Gut. 2000;47:404‐409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Magro F, Lopes J, Borralho P, et al. Comparison of different histological indexes in the assessment of UC activity and their accuracy regarding endoscopic outcomes and faecal calprotectin levels. Gut. 2018:1301-10. [DOI] [PubMed] [Google Scholar]
- 37. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high‐throughput community sequencing data. Nat Methods. 2010;7:335‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haddish‐Berhane N, Farhadi A, Nyquist C, Haghighi K, Keshavarzian A. Biological variability and targeted delivery of therapeutics for inflammatory bowel diseases: an in silico approach. Inflamm Allergy Drug Targets. 2007;6:47‐55. [DOI] [PubMed] [Google Scholar]
- 40. Greenstein RJ, Su L, Shahidi A, Brown ST. On the action of 5‐amino‐salicylic acid and sulfapyridine on M. avium including subspecies paratuberculosis. PLoS ONE. 2007;2:e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mukhopadhya I, Hansen R, El‐Omar EM, Hold GL. IBD‐what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219‐230. [DOI] [PubMed] [Google Scholar]
- 43. Michail S, Durbin M, Turner D, et al. Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis. 2012;18:1799‐1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate‐producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275‐1283. [DOI] [PubMed] [Google Scholar]
- 45. Tyler AD, Knox N, Kabakchiev B, et al. Characterization of the gut‐associated microbiome in inflammatory pouch complications following ileal pouch‐anal anastomosis. PLoS ONE. 2013;8:e66934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kump P, Wurm P, Gröchenig Hp, et al. The taxonomic composition of the donor intestinal microbiota is a major factor influencing the efficacy of faecal microbiota transplantation in therapy refractory ulcerative colitis. Aliment Pharmacol Ther. 2018;47:67‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Varela E, Manichanh C, Gallart M, et al. Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment Pharmacol Ther. 2013;38:151‐161. [DOI] [PubMed] [Google Scholar]
- 48. Ricart E, Taylor WR, Loftus EV, et al. N‐acetyltransferase 1 and 2 genotypes do not predict response or toxicity to treatment with mesalamine and sulfasalazine in patients with ulcerative colitis. Am J Gastroenterol. 2002;97:1763‐1768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


