Summary
miRNAs contribute to plant resistance against pathogens. Previously, we found that the function of miR398b in immunity in rice differs from that in Arabidopsis. However, the underlying mechanisms are unclear.
In this study, we characterized the mutants of miR398b target genes and demonstrated that multiple superoxide dismutase genes contribute to miR398b‐regulated rice immunity against the blast fungus Magnaporthe oryzae.
Out of the four target genes of miR398b, mutations in Cu/Zn‐Superoxidase Dismutase1 (CSD1), CSD2 and Os11g09780 (Superoxide DismutaseX,SODX) led to enhanced resistance to M. oryzae and increased hydrogen peroxide (H2O2) accumulation. By contrast, mutations in Copper Chaperone for Superoxide Dismutase (CCSD) resulted in enhanced susceptibility. Biochemical studies revealed that csd1, csd2 and sodx displayed altered expression of CSDs and other superoxide dismutase (SOD) family members, leading to increased total SOD enzyme activity that positively contributed to higher H2O2 production. By contrast, the ccsd mutant showed CSD protein deletion, resulting in decreased CSD and total SOD enzyme activity.
Our results demonstrate the roles of different SODs in miR398b‐regulated resistance to rice blast disease, and uncover an integrative regulatory network in which miR398b boosts total SOD activity to upregulate H2O2 concentration and thereby improve disease resistance.
Keywords: Cu/Zn‐Superoxidase Dismutase, enzyme activity, H2O2, Magnaporthe oryzae, miR398b, rice resistance, Superoxide Dismutase
Introduction
In plants, production of reactive oxygen species (ROS), such as superoxide radicals (O• 2 −), hydroxyl radicals (OH·) and hydrogen peroxide (H2O2), are considered important defense reactions in response to biotic and abiotic stress (Jones & Dangl, 2006; Boller & He, 2009; Saxena et al., 2016). The synthesis and homeostasis of ROS in plants are regulated by a series of enzymes, such as the NADPH oxidases (also known as respiratory burst oxidase homologs (Rbohs)), Superoxidase Dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX). Rbohs are key enzymes in generation of O• 2 − by catalyzing the transfer of electrons from NADPH to oxygen (O2) (Kaur et al., 2014). In Arabidopsis, both RbohD and RbohF are required for ROS production, as atrbohD and atrbohF mutants show lower ROS concentrations during incompatible interactions with bacterial and oomycete pathogens (Torres et al., 2002). In rice, RbohD and RbohF are induced by oxidative stress, leading to increased intracellular ROS accumulation (Jang et al., 2012). RbohB knockdown rice plants display enhanced susceptibility to rice blast fungus Magnaporthe oryzae, suggesting that RbohB is required for resistance to rice blast (Nagano et al., 2016). SODs act as H2O2 synthetases, catalyzing the reduction of O• 2 − into H2O2 (Fridovich, 1995; Gill & Tuteja, 2010). SOD isoenzymes are classified into three types according to their affinity for specific metal ions in plants: the Cu/Zn‐Superoxidase Dismutase (SOD) (CSD), the manganese SOD (SOD‐Mn) and the iron SOD (SOD‐Fe) (Mittler, 2002). Overexpression of SOD‐Mn increases plant tolerance against freezing, water deficit, low temperature (McKersie et al., 1993, 1996, 1999) and methyl viologen‐induced oxidative stress (Bowler et al., 1991; Slooten et al., 1995). Overexpression of SOD‐Fe results in enhanced tolerance against methyl viologen in tobacco (Van Camp et al., 1996) and maize (Van Breusegem et al., 1999). Overexpression of a pea homolog of Arabidopsis CSD2 in tobacco leads to increased tolerance against high light and low‐temperature stresses (Gupta et al., 1993a,b). Overexpression of another pea CSD in tobacco enhances ozone tolerance (Pitcher & Zilinskas, 1996). Overexpression of a CSD in rice results in increased resistance to NaHCO3 and water stress (Guan et al., 2017). Therefore, SODs act as key defense factors against versatile stresses by regulating the H2O2 concentration in plants. H2O2 is a vital ROS in biological processes leading to tolerance against various stresses (Kaur et al., 2016; Saxena et al., 2016).Consistent with the role of ROS in rice immunity, the resistant rice cultivar IRBLKm‐Ts displays higher H2O2 concentrations than the susceptible cultivar Lijiang XinTuan Hegu (LTH) upon M. oryzae infection (Li et al., 2014). In addition, overexpression of l‐ascorbate oxidase (AO) enhanced AO‐mediated H2O2 accumulation, thereby improving resistance to rice stripe virus (Wu et al., 2017). ROS homeostasis can be controlled by CAT, a peroxisome‐located H2O2 scavenging enzyme which catalyzes the reduction of H2O2 into H2O and O2 (Racchi, 2013). In addition, APX and glutathione peroxidase are involved in the removal of H2O2 (Racchi, 2013). Therefore, ROS‐mediated stress tolerance in plants is regulated by the ROS‐homeostasis‐related enzyme system, which, in turn, seems to be fine‐tuned by certain microRNAs (miRNAs) (Baldrich & San Segundo, 2016).
The miRNAs are a category of 20–24‐nucleotide (nt) noncoding RNAs that play important roles in plant development and defense responses by negatively regulating target gene expression (Padmanabhan et al., 2009; Katiyar‐Agarwal & Jin, 2010; Baldrich & San Segundo, 2016; Tang & Chu, 2017). miR398 is a conserved miRNA family that suppresses the expression of the SOD family members in many plants, including Arabidopsis (Guan et al., 2013; Lu et al., 2013), Brassica rapa (Yu et al., 2012), Helianthus annuus (Khaksefidi et al., 2015), wheat (Xin et al., 2010), switchgrass (Hivrale et al., 2016) and Populus (Chen L. et al., 2012; Chen M. et al., 2012). In Arabidopsis, miR398 regulates oxidative stress response by silencing the expression of CSD1 and CSD2 (Sunkar et al., 2006). In wheat, miR398 and its target genes are responsive to multiple stimuli including cold, wounding and salt stresses (Wang et al., 2014). In tomato, miR398a is involved in response to drought stress, and its amounts in sensitive and tolerant genotypes were inversely correlated (Candar‐Cakir et al., 2016). In pea, miR398 accumulation is decreased whereas CSD1 expression is enhanced upon water deficit (Jovanovic et al., 2014). MiR398 and CSDs also regulate plant disease resistance against pathogens. In Arabidopsis, miR398 overexpression lines display enhanced susceptibility to Pseudomonas syringae DC3000 by downregulating CSD1 and CSD2 (Li et al., 2010a). In barley, Mla and Rom1 negatively regulate the miR398 amount, promoting SOD1 accumulation and enhancing resistance against powdery mildew (Xu et al., 2014). In rice, however, miR398b overexpressing lines (OX398b) show enhanced basal defenses against M. oryzae associated with reduced mRNA amounts of CSD1, CSD2, SODX and CCSD (Li et al., 2014), indicating that the miR398‐SOD module may play reverse roles in regulation of resistance against pathogens in rice compared to that in Arabidopsis or in barley.
In rice, the SOD gene family contains 15 members, including eight annotated SOD genes and seven related genes including the chaperone CCSD (Nath et al., 2014). Among the eight annotated SOD genes, four of them encode CSDs, including Os03g22810 (CSD1), Os07g46990 (CSD2), Os03g11960 (CSD3) and Os08g44770 (CSD4). Os04g48410 encodes a copper chaperone for superoxide dismutase (CCSD) that delivers copper to superoxide dismutase, and Os05g25850, Os06g02500 and Os06g05110 encode SOD‐Mn, SOD1‐Fe and SOD2‐Fe, respectively (http://rice.plantbiology.msu.edu). Three of the SOD family members, including CSD1, CSD2 and CCSD, were identified as target genes of miR398b by degradome‐sequencing assays (Wu et al., 2009; Li et al., 2010b). In addition, one uncharacterized gene, Os11g09780 (designated SODX hereafter), is identified as a member of the SOD gene family (Nath et al., 2014) and is predicted to be a miR398b target in rice (Wu et al., 2009). To address why miR398b positively regulates rice immunity against the blast fungus, we analyzed the function of miR398b's target genes in rice blast disease‐resistance by generating mutants and transgenic lines expressing target mimicry of miR398b (MIM398). Our data indicate that CSD1, CSD2 and SODX negatively regulate, whereas CCSD is required for, miR398b‐boosted H2O2 production, leading to resistance to rice blast disease. H2O2 concentration seems to be correlated with total SOD enzyme activity upon pathogen infection, which in turn is regulated by the target genes of miR398b. Mutation of CCSD led to decreased CSD and total SOD enzyme activity upon M. oryzae infection. However, mutation of CSD1 and CSD2 resulted in increased total SOD enzyme activity, and mutation of SODX led to constitutively higher SOD enzyme activity. Therefore, the target genes of miR398b have antagonistic roles in the SOD enzyme system. Our data demonstrate that miR398b boosts H2O2 production via multiple SODs, and explain why the function of miR398b in immunity in rice is different from that in Arabidopsis and barley.
Materials and Methods
Plant materials and growth conditions
The rice (Oryza sativa) indica accession Kasalath, japonica accessions Nipponbare (NPB) and Taipei 309 (TP309) were used for transgenic analysis. For blast‐resistance and resistant response assays, the wild‐type (WT) control and transgenic lines were grown in a greenhouse at 28 ± 2°C and 70% relative humidity under 12 h : 12 h, light : dark cycles.
Plasmid construction and genetic transformation
The miR398b‐insensitive versions of Cu/Zn‐Superoxidase Dismutase2 (CSD2 m) was amplified from NPB cDNA with primers CSD2‐KpnI‐F and CSD2‐SpeI‐R (Supporting Information Table S1). The miR398b‐insensitive version of Copper Chaperone for Superoxidase Dismutase (CCSD)m was generated by site‐directed mutagenesis using the primers CCSD‐KpnI‐F, CCSDm‐R, CCSDm‐F, CCSD‐SpeI‐R (Table S1). The PCR fragments were fused to enhanced‐green fluorescent protein (GFP) and cloned into the KpnI–SpeI site of the binary vector 35S‐pCAMBIA1300 resulting in the overexpression vector. To construct MIM398, a 331‐bp fragment including the endogenous target mimicry sequences of miR398b (chr5:6397583..6397606) (Wu et al., 2013) was amplified from NPB genomic DNA with primers MIM398‐KpnI‐F and MIM398‐SpeI‐R (Table S1), and cloned into the KpnI–SpeI site of the binary vector 35S‐pCAMBIA1300. Agrobacterium strain EHA105 was selected for rice genetic transformation. Hygromycin B was used for screening the genotype of transgenic plants by means of hygromycin resistance analysis.
The CRISPR/Cas9 plasmids construct and mutant screen
The CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 plasmids of target genes were constructed as reported previously (Li et al., 2017b). The guide RNA sequences listed in Table S2 were screened by Cas‐OFFinder system (http://www.rgenome.net/cas-offinder/) to avoid potential off‐target‐sites with the screen parameters to allow <3 bp mismatches and one DNA/RNA bulge. The maize ubiquitin promoter (UBI) upstream of a codon optimized hSpCas9(Cong et al., 2013) was inserted into binary vector pCAMBIA1300 with hygromycin selection (via hygromycin B phosphotransferase). The original BsaI site present in the pCAMBIA1300 backbone was removed using a point mutation kit (Transgen, Beijing, China). A fragment containing an OsU6 promoter(Feng et al., 2013) and a negative selection marker gene ccdB flanked by two BsaI sites and a sgRNA derived from pX260(Cong et al., 2013) was inserted into this vector using In‐fusion cloning kit (Takara, Tokyo, Japan) to produce the CRISPR/Cas9 binary vector pBGK032. Escherichia coli strain DB3.1 was used for maintaining this binary vector. The 23 bp targeting sequences (including PAM) were selected within the target genes and their targeting specificity was confirmed using a Blast search against the rice genome (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Hsu et al., 2013). The designed targeting sequences were synthesized and annealed to form the oligo adaptors. Vector pBGK032 was digested by BsaI and purified using a DNA purification kit (Tiangen, Beijing, China). A ligation reaction (10 μL) containing 10 ng of the digested pBGK032 vector and 0.05 mM oligo adaptor was carried out and directly transformed to E. coli competent cells to produce CRISPR/Cas9 plasmids. Agrobacterium strain EHA105 was selected for rice genetic transformation.
Genomic DNA was extracted from T2 transgenic lines and the primers flanking the designed target site (Table S2) were used for PCR amplification. The PCR products (300–500 bp) were sequenced directly and blasted to the WT genome sequence to identify the mutation sites.
Agrobacterium‐mediated transient expression assay in Nicotiana benthamiana
YFP detection and accumulation was assayed as reported previously (Li et al., 2017a,b). In order to generate miR398 target‐site reporter fusions, we fused yellow fluorescent protein (YFP) with either the target site of Superoxide DismutaseX (SODX) at its N‐terminus (35S:SODX ts ‐YFP) or with a mutated target site (35S:SODX mts ‐YFP) that could not be recognized by miR398b. The sequences of SODXts and SODXmts were synthesized by annealing gene‐specific primers SODX ts‐F/R and SODX mts‐F/R, respectively (Table S1). The isolated fragments were then fused to the N‐terminus of YFP and inserted into KpnI‐SpeI sites of binary vector 35S‐pCAMBIA1300. Agrobacterium strain GV3101 was used for agroinfection assay in N. benthamiana. In brief, Agrobacterium strain GV3101 harboring the respective expression constructs (35S:SODX ts ‐YFP, 35S:miR398b, 35S:MIM398) was incubated at 28°C overnight in LB media containing kanamycin (50 mg ml−1) and carbenicillin (50 mg ml−1) on a table shaking at 250 rpm. The bacteria were collected at 800 g for 5 min and resuspended in an MMA buffer (10 mM MES, 10 mM MgCl2, 100 mM AS). The Agrobacteria harboring the expression constructs were infiltrated into leaves of N. benthamiana for transient expression assay. Leaves were examined at 48 h post‐infiltration (hpi) for image acquisition using a NikonA1 Confocal Laser Scanning Microscope (Nikon Instruments Inc., Shanghai, China). Western blotting analyses were performed to determine the accumulation of YFP. In brief, 15 μg of total extracted protein was electrophoresed on a 10% SDS‐PAGE gel, and then transferred to a membrane. The protein blot was reacted with 3000‐fold‐diluted anti‐GFP sera (Sangon Biotech, Shanghai, D110008, China) and 4000‐fold‐diluted anti‐actin sera (Sangon Biotech, Shanghai, D110007, China), respectively, to detect YFP and actin accumulation.
Pathogen infection and microscopy
Magnaporthe oryzae strains stocked in the laboratory (Guy11, GZ8 and 089) were used in this study. Magnaporthe oryzae strains were incubated on Complete Medium at 28°C with 12 h : 12 h, light : dark cycles for sporulation. After 2 wk, the spores (1 × 105 spores mL−1) were collected for punch‐ and spray‐inoculation. The lesions in infected leaves were observed at 5 d post‐inoculation (dpi) and the fungal biomass was measured as reported previously (Park et al., 2012). In brief, relative fungal biomass was calculated using the DNA amount of M. oryzae Pot2 against the rice genomic ubiquitin DNA amount by quantitative reverse transcription polymerase chain reaction (qRT‐PCR). For observing the infection process of M. oryzae, we inoculated the strain GZ8 on 5‐cm‐long leaf sheaths as described previously (Kankanala et al., 2007). The inoculated epidermal layer was excised and analyzed by fluorescence microscopy (Zeiss Axio Imager A2, Carl Zeiss Co. Ltd, Chengdu, China) during 24–48 hpi.
H2O2 and O·2 − measurement
Hydrogen peroxide (H2O2) accumulations and cell death in infected leaves were observed with the protocol given in Methods S1. The quantification of H2O2 followed the method described in (Wu et al., 2017). Briefly, 50 mg of inoculated fresh leaves were collected and ground with liquid nitrogen. Five hundred microlitres of sodium phosphate buffer (50 mM, pH 7.4) was added to extract H2O2 and the mixture was centrifuged at 12 000 g for 20 min at 4°C to pellet cell debris. The supernatant was used for H2O2 concentration assay with the Amplex Red Hydrogen Peroxide/Peroxidase Assay kit (Invitrogen molecular probe, A22188; Thermo Fisher Scientific Co. Ltd, Shanghai, China) following the manufacturer's directions. Absorbance was measured in a 96‐well microplate reader (Thermo Scientific Microplate Reader, Thermo Fisher Scientific Co. Ltd., Shanghai, China) at 560 nm, and the amount of H2O2 was calculated according to a standard curve. O• 2 − histochemical staining was performed using NBT (Nitrotetrazolium Blue chloride) as in (Wu et al., 2017). Briefly, the leaf samples were spray‐inoculated with Guy11 (1 × 105 mL−1 spores) or water. After 32 h the leaf samples were infiltrated with 50 mM sodium phosphate (pH 7.0) containing 0.05% NBT (Sigma) and 10 mM NaN3, and incubated at 37°C in the dark for 16 h. Leaf samples were then washed with bleaching solution (ethanol : acetic acid, 3 : 1) at 70°C for 30 min to elute the chlorophyll. The leaf sections were observed with a microscope (Zeiss Axio Imager A2), and quantification of O• 2 − concentrations was measured by imagej software.
qRT‐PCR
Four‐ to six‐leaf‐stage rice seedlings were spray‐inoculated or drop‐inoculated with Guy11 and mock at a concentration of 1 × 105 spores ml−1, and samples were collected at 48 hpi. Total RNA was extracted using TRIzol reagent (Invitrogen) and was reverse transcribed to cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser following the manufacturer's instruction (TaKaRa Biotechnology, Dalian, China). qRT‐PCR was performed using SYBR Green mix (QuantiNova SYBR Green PCR Kit, Qiagen) and the indicated primers (Table S1). The rice ubiquitin (UBQ) gene was selected as an internal reference to normalize data.
Enzymatic activity assay
In order to determine the activity of CSD protein in gel, the CSD protein extracts was assayed as reported previously (Davis, 1964; Shah & Nahakpam, 2012). Nondenaturing polyacrylamide gel electrophoresis (PAGE, 7.5% running gel and 3.5% stacking gel (acrylamide : bis‐acrylamide = 29 : 1)) was performed at 4°C using 0.01 M Tris‐glycine (pH 8.3) as electrode buffer. Twenty microliters of protein mixed with glycerol were loaded and electrophoretically run using a current of 25 mA per slab. After electrophoresis, the gels were rinsed with distilled water and incubated for 30 min in 2.5 mM NBT, then immersed in 1.17 × 10−6 M riboflavin for 20 min and removed to a petri dish for irradiance with a fluorescent lamp. Light exposure led to the development of the purple color of insoluble formazan throughout the gel, except for the locations where CSD was localized. For protein visualization, 20 μL of protein mixed with loading buffer were boiled and electrophoresed on 10% SDS‐PAGE gel, and the protein were dyed with Coomassie Brilliant Blue. The enzyme activities of CSD, SOD and CAT were detected using the Activity Assay Kits (A001‐4 for CSD and SOD, Njjcbio, Nanjing, China; BC0205 for CAT, Solarbio, Beijing, China). In brief, six‐leaf‐stage seedlings were spray‐inoculated with Guy11/mock at a concentration of 1 × 105 spore mL−1, and 100 mg rice leaves were collected and powdered in liquid nitrogen and then homogenized with the extracting buffer. The extracts were centrifuged at 15 000 g for 10 min at 4°C and the supernatant were mixed with the provided reagents following the protocols in these kits, respectively, and detected by testing the absorbance of the final reaction mix with a 96‐well microplate reader (Thermo Scientific Microplate Reader) at specific wavelengths (SOD and CSD at 550 nm; CAT at 240 nm). The enzyme activity was calculated following the recommended formulae. For SOD, 1 unit of enzyme activity is defined as the absorbance change ratio when the absorbance is inhibited by 50% per gram fresh sample at 550 nm in 1 mL reaction buffer. In brief, SOD or CSD(U/g FW) = [(A (control) − A (sample)) / A (control)]/50% × [V (total reaction volume) / V (tested sample volume)] × 2(sample fold dilution) /0.1 g mL−1 (sample concentration). For CAT, 1 unit of enzyme activity is defined as the activity of degrading 1 nM H2O2 per gram fresh sample per minute at 240 nm. In brief, CAT(U/g FW) = [ΔA × 1×10−4 L(total volume of the reaction system) / /0.5 cm(96‐well plate light‐diameter) × 109]/[(10 × 10−6 L(the volume of the tested sample) /1 × 10−3 L(the volume of total sample) × 0.1 g(FW)]/1 min(reaction time) = 9180 × ΔA.
Results
miR398b downregulates SODX expression
In order to confirm that SODX was repressed by miR398b, we made constructs expressing YFP fused with the putative target site of SODX at its 5′‐terminus (35S:SODX ts ‐YFP) (Fig. 1a) or with mutated target site (35S:SODX mts ‐YFP) that abolished recognition by miR398b (Fig. 1b). A construct expressing a target mimicry of miR398 (MIM398) also was created to act as a sponge and capture miR398, preventing silencing of its target (Wu et al., 2013; Fig. 1c). The YFP intensity and protein concentration expressed from 35S:SODX ts ‐YFP was obviously lower when coexpressed with miR398b than when expressed alone in N. benthamiana, but recovered when coexpressed with MIM398 (Fig. 1d,f). By contrast, the YFP concentration expressed from 35S:SODX mts ‐YFP was unchanged when coexpressed with miR398b or miR398b plus MIM398 (Fig. 1e,f). These results indicate that miR398b represses SODX expression. Thus, miR398b suppresses its four target genes that belong to the SOD gene family.
Figure 1.
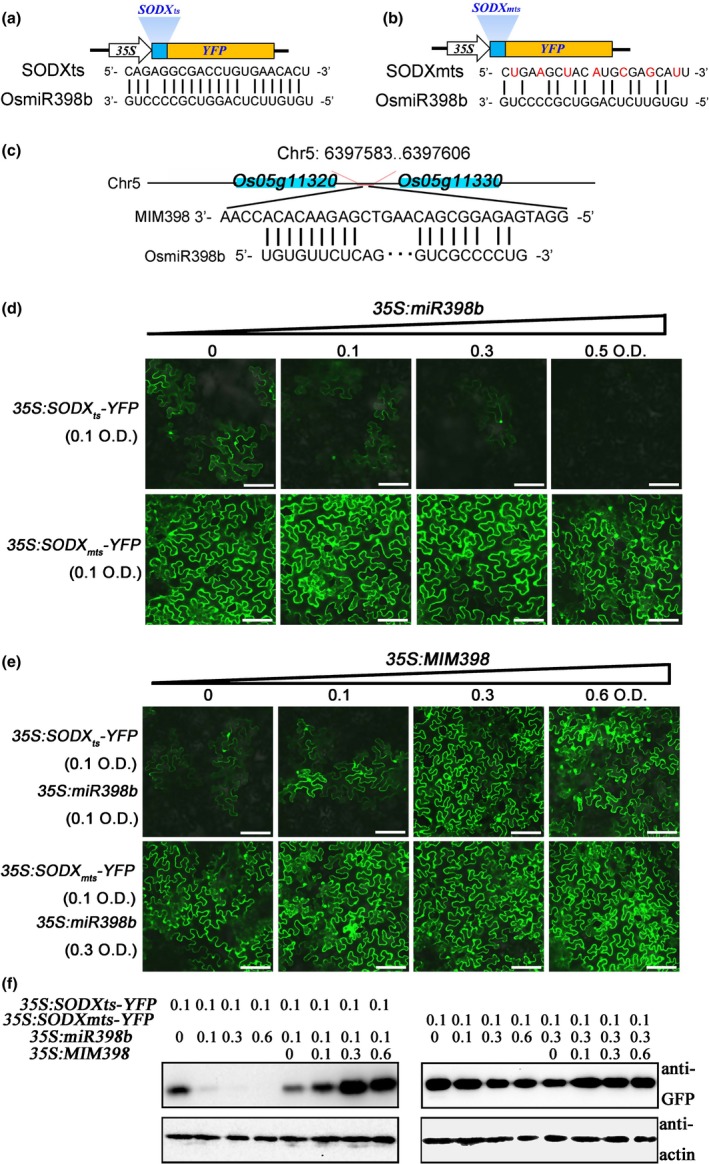
MiR398b represses the expression of superoxidase dismutase (SOD)X. (a) The construct of fused‐yellow fluorescent protein (YFP) with the target site of SODX (SODXts‐YFP) and alignment of miR398b with SODX ts target site sequences. (b) The construct of fused‐YFP with the mutated target site of SODX (SODXmts‐YFP) and alignment of miR398b with SODX mts target site sequences. The red letters indicate the mutated bases. (c) Alignment of miR398b with target mimicry of miR398 (MIM398) target site sequences. (d, e) Confocal images show the protein concentrations of SODXts‐YFP and SODXmts‐YFP. The indicated SODXts‐YFP and SODXmts‐YFP reporter constructs were transiently expressed alone or coexpressed with miR398b (d) or miR398b plus MIM398 (e) in Nicotiana benthamiana leaves using Agrobacterium‐mediated infiltration at the indicated optical density(OD)concentration. Bars, 50 μm. (f) Western blotting analysis shows the protein concentrations of SODXts‐YFP and SODXmts‐YFP. Protein extracts from the same amount of infiltrated leaves were subjected to Western blot analysis using anti‐green fluorescent protein (GFP) sera and anti‐actin sera, respectively. This experiment was repeated two times with similar results.
Mutants of miR398b target genes display different sensitivity to M. oryzae
In a previous report, we demonstrated that rice OX398b lines displayed higher resistance against M. oryzae. Because a miRNA functions through its target genes, we focused on functional characterization of the miR398b target genes and assessed the role of each target gene in blast disease‐resistance. We modified each of the four target genes using the CRISPR/Cas9 technology and identified two homozygous mutants for CSD1 (csd1‐1 AND csd1‐3), two for CSD2 (csd2‐1 and csd2‐33), two for SODX (sodx‐5 and sodx‐14) and one for CCSD (ccsd‐1) (Fig. S1). Although csd1‐1 carried a 3‐bp (CGG) deletion causing a threonine/serine substitution and a glycine deletion (Fig. S1a,b), the other six mutants carried insertions or deletions causing protein truncation (Fig. S1c–n). csd1‐3 carried a 5‐bp deletion resulting in an early stop codon after amino acid residue (aa) 14 (glutamine; Fig. S1c,d); csd2‐1 carried a G/A substitution and a 28‐bp deletion resulting in changes from aa 30 and led to an early stop after aa 76 (Isoleucine, Fig. S1e,f); csd2‐33 carried a 2‐bp deletion resulting in changes starting at aa 34 and led to an early stop after aa 49 (arginine; Fig. S1g,h); ccsd‐1 carried a 1‐bp insertion resulting in an early stop after aa 82 (Valine; Fig. S1i,j); sodx‐5 carried a 5‐bp deletion resulting in changes starting at aa 35 and led to an early stop codon after aa 47 (arginine; Fig. S1k,l); sodx‐14 carried a 1‐bp deletion resulting in changes starting at aa 72 and led to an early stop after aa 75 (histidine; Fig. S1m,n).
We then assessed the sensitivity of these mutants to the rice blast fungus via punch‐ and spray‐inoculation with different virulent strains. Punch‐inoculation results showed that csd1, csd2 and sodx are less susceptible to M. oryzae strains GZ8 and Guy11, displaying smaller disease lesions and significantly lower fungal growth than that of their corresponding WT control (Fig. 2a–c), However, ccsd‐1 displayed enhanced susceptibility to both strains, whereas the ccsd‐1 segregated azygous control line, ccsd‐1(‐), showed similar fungal growth as the WT (Fig. 2a–c). Similarly, spray‐inoculation with another M. oryzae strain 089 showed that OX398b, csd1, csd2 and sodx were less susceptible, but ccsd was more susceptible than WT control (Fig. S2a,b). To understand how csd1, csd2 and sodx were less susceptible and ccsd was more susceptible, we observed the fungal infection progress by quantifying the formation and expansion of invasive hyphae in leaf sheath cells. Consistent with the disease phenotypes, the infection progress is significantly delayed in csd1, csd2 and sodx, but enhanced in ccsd compared with the corresponding WT control (Fig. 3d,e; Table S3). In Nipponbare (NPB), >93.9% of inoculated spores formed invasive hyphae at 24 hpi and > 86.6% of them expanded into neighbor cells at 48 hpi; whereas, in both csd1‐3 and sodx‐5 lines, < 88.1% spores formed invasive hyphae at 24 hpi and < 75.1% of them expanded into neighbor cells at 48 hpi (Fig. 2d,e; Table S3). In TP309, > 74.9% spores formed invasive hyphae at 24 hpi and > 82.4% of them expanded into neighbor cells at 48 hpi; however, in csd2‐33 < 70.2% expanded into neighbor cells at 48 hpi, although no difference was observed at 24 hpi (Fig. 2d,e; Table S3). Conversely, in ccsd‐1, 93.6% spores formed invasive hyphae at 24 hpi and > 88.9% of them extended into neighbor cells at 48 hpi (Fig. 2d,e; Table S3). These observations indicate that different target genes of miR398b play different roles in blast disease‐resistance. Although CSD1, CSD2 and SODX may negatively regulate resistance, CCSD may be required for rice blast disease‐resistance in rice.
Figure 2.
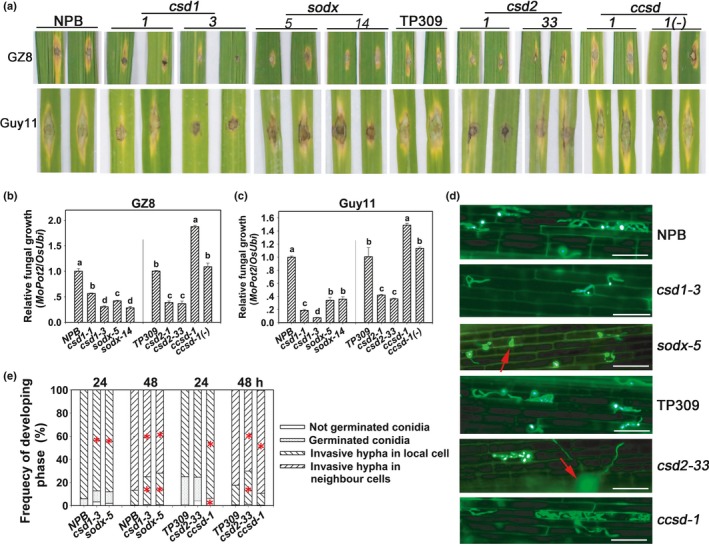
Mutations in miR398b target genes alters sensitivity to Magnaporthe oryzae. (a) Disease phenotypes of the indicated mutant lines (Cu/Zn‐Superoxidase dismutase (csd)1, csd2, copper chaperone for superoxide dismutase (ccsd), superoxidase dismutase (sod)x, ccsd‐1 segregated azygous line (ccsd‐1(‐))) and their corresponding controls (Nipponbare (NPB), Taipei 309 (TP309)) at 5 d post‐inoculation (dpi) by M. oryzae strains GZ8 and Guy11. (b, c) Relative fungal biomass on the inoculated leaves of the inoculated lines at 5 dpi. The relative fungal biomass was measured by using the DNA amount of M. oryzae Pot2 against the rice genomic ubiquitin DNA amount. Values are means ± SD of three replications. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (d) Representative epifluorescent microscopic images show the growth of the eGFP‐tagged M. oryzae strain GZ8 on sheath cells of the indicated mutant lines and the control lines at 36 h post‐inoculation (hpi), respectively. Red arrows indicate germinated but not invaded conidia. Bars, 50 μm. (e) Quantitative analysis of M. oryzae growth. More than 200 conidia in each line were analyzed. Significant differences between the corresponding wild‐type controls and mutant lines as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis are indicated: *, P < 0.05. All of the experiments were repeated two times with similar results.
Figure 3.
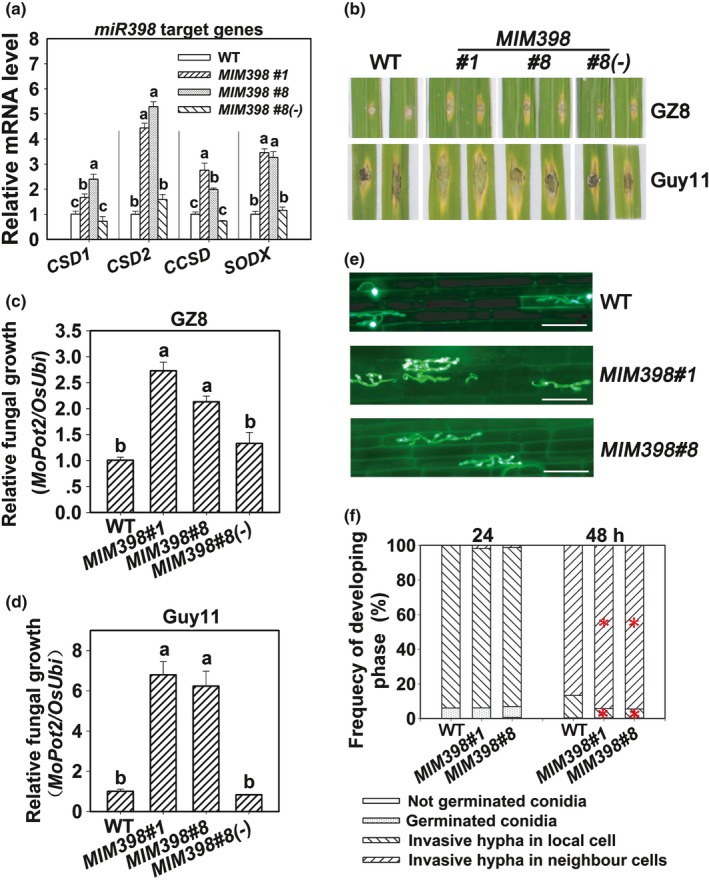
Overexpressing target mimicry of miR398b enhances sensitivity to Magnaporthe oryzae. (a) mRNA amounts of target genes in wild‐type (WT), target mimicry of miR398 (MIM398) and MIM398#8 segregated azygous line (MIM398#8(‐)). The mRNA amounts were normalized to that in WT plants. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (b) Disease phenotypes on leaves of the indicated lines upon M. oryzae strains GZ8 and Guy11 infection at 5 d post‐inoculation (dpi). (c, d) Relative fungal mass of GZ8 and Guy11 on the inoculated leaves of the indicated lines. The relative fungal mass was measured by using the DNA amount of M. oryzae Pot2 against the rice genomic ubiquitin DNA amount. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis. (e) Representative epifluorescent microscopic images show the growth of GZ8 at 36 h post‐inoculation (hpi) on sheath cells of the indicated transgenic lines and the WT plants, respectively. Bars, 50 μm. (f) Quantitative analysis of M. oryzae growth at the indicated time points. More than 200 conidia in each line were analyzed. Significant differences at between the WT and MIM398 lines as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis are indicated: *, P < 0.05. All of the experiments were repeated twice with similar results.
Overexpression of miR398b target genes enhances rice sensitivity to M. oryzae
In order to achieve upregulation of all the target genes simultaneously, we generated transgenic lines expressing MIM398. All of the MIM398 lines express significantly higher mRNA amounts of the four target genes of miR398b, whereas, the segregated azygous control (MIM398#8(‐)) retains WT mRNA amounts (Fig. 3a). Moreover, MIM398 lines are more susceptible to GZ8 and Guy11, with larger disease lesions and significantly more fungal growth than WT control (Fig. 3b–d); whereas MIM398#8(‐) displays the similar susceptibility to both strains (Fig. 3b–d). Consistently with the macroscopic disease phenotypes, the infection progress is significantly enhanced in MIM398 lines compared with WT control (Fig. 3e,f; Table S3). In NPB, > 93.9% spores formed invasive hyphae at 24 hpi and < 86.7% of spore‐formed invasive hypha expanded into neighbor cells at 48 hpi; however, in MIM398 lines, > 94.2% of them expanded into neighbor cells at 48 hpi, although no difference was observed at 24 hpi (Fig. 3e,f; Table S3). These results indicate that upregulation of the miR398b target genes facilitates the growth of M. oryzae presumably via suppressing rice immunity, but miR398b positively contributes to blast resistance by silencing them.
In order to further confirm the roles of target genes in blast disease‐resistance, we generated transgenic lines overexpressing miR398b‐insensitive CCSD m ‐GFP (OXCCSD) and CSD2 m ‐GFP (OXCSD2). CCSDm‐GFP contains six mismatches in the miR398b target site, and CSD2 m ‐GFP excludes the miR398b target site located in the 5′‐UTR of CSD2 (Fig. S3a,b). Higher CSD2 and CCSD mRNA amounts were detected (Fig. S3c,e) and the fusion proteins are present in the nucleus and the cytoplasm (Fig. S3d,f). By contrast to the csd2 mutants that are less susceptible, the OXCSD2 lines are more susceptible to M. oryzae, boasting larger disease lesions and more fungal growth (Fig. 4a,b), and displaying a significantly higher ratio of invasive hyphae from the primary infected cells expanding into neighbor cells at 48 hpi (Fig. 4c,d; Table S3). These observations indicate that CSD2m‐GFP is functional and negatively regulates blast disease‐resistance. The OXCCSD lines display similar susceptibility and disease lesions to WT (Fig. 4a,b), although they show a significantly lower ratio of invasive hyphae at local cells at 24 hpi (Fig. 4c,d; Table S3). Further investigation is required to clarify whether CCSDm‐GFP is functional or over‐accumulation of CCSD does not enhance rice blast resistance.
Figure 4.
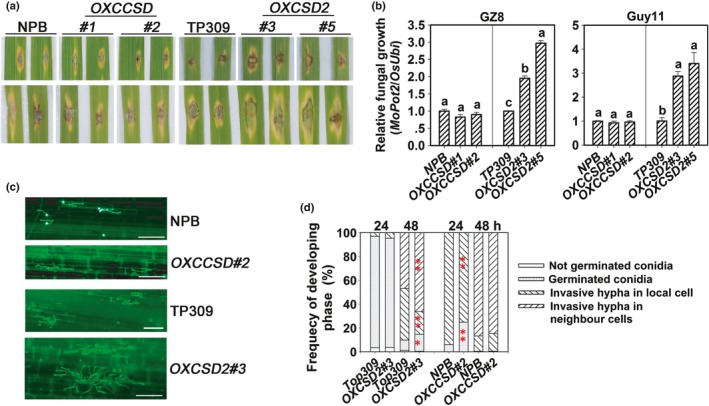
Overexpressing Cu/Zn‐Superoxidase Dismutase (CSD)2 enhances sensitivity to Magnaporthe oryzae. (a) Disease phenotypes on leaves of the inoculated lines 5 d post‐inoculation (dpi) by M. oryzae strains GZ8 and Guy11 infection, separately. (b) The relative fungal mass on the inoculated leaves of the indicated lines at 5 dpi. The relative fungal mass was measured by using the DNA amount of M. oryzae Pot2 against the rice genomic ubiquitin DNA amount. Values are means ± SD of three replications. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (c) Representative epifluorescent microscopic images show the growth of GZ8 at 36 h post‐inoculation (hpi) on sheath cells of the indicated transgenic lines (OXCCSD, Copper Chaperone for Superoxide Dismutase (CCSD) overexpressing line; OXCSD2, CSD2 overexpressing line) and the corresponding wild‐type (WT) controls (NPB, Nipponbare. TP309, Taipei 309), respectively. Bars, 50 μm. (d) The quantitative analysis of M. oryzae growth at indicated time points. More than 200 conidia in each line were analyzed. Significant differences between the corresponding WT controls and overexpression lines as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis: *, P < 0.05; **, P < 0.01. All of the experiments were repeated two times with similar results.
miR398b and its target genes regulate ROS concentration upon M. oryzae infection
Accumulation of H2O2 is a common defense response to M. oryzae in rice (Chen L. et al., 2012; Chen M. et al., 2012; Shimono et al., 2012; Wang et al., 2016). To test whether H2O2 accumulation contributes to the observed resistance phenotypes, we examined H2O2 concentrations in all tested lines. Consistently with the disease phenotypes, upon Guy11 infection, H2O2 concentrations increased significantly higher in OX398b, csd1, csd2 and sodx compared to those in their corresponding WT controls, but decreased and significantly lower in ccsd (Figs 5a, S4). Although MIM398 displayed higher H2O2 concentrations with mock treatment (Fig. S5a,b), upon Guy11 infection, MIM398, OXCSD2 and OXCCSD all showed lower or similar H2O2 concentrations compared to their corresponding WT controls (Fig. S5a,b). These results indicate that miR398b positively regulates H2O2 accumulation by repressing CSD1, CSD2 and SODX in rice infected with M. oryzae, and overexpressing target genes does not positively contribute to blast‐induced H2O2 accumulation.
Figure 5.
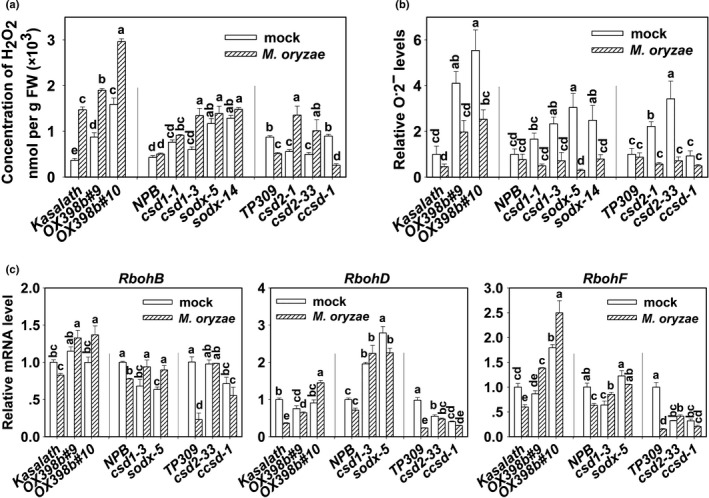
Overexpressing miR398b or mutations in target genes affected reactive oxygen species (ROS) concentration upon Magnaporthe oryzae infection. (a, b) Quantification of hydrogen peroxide (H2O2) (a) and O• 2 − (b) concentrations in leaves of the wild‐type (WT) (Kasalath, Nipponbare (NPB), Taipei 309 (TP309)) and indicated lines (miR398b overexpressing lines (OX398b), mutant of Cu/Zn‐Superoxidase Dismutase (csd)1, csd2, mutant of Copper Chaperone for Superoxide Dismutase (ccsd), mutant of Superoxidase Dismutase (sod)x) with Guy11/mock treatment at 48 h post‐inoculation (hpi), respectively. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (c) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) data showing the expression pattern of the indicated NADPH oxidase genes (Respiratory burst oxidase homologs (Rboh) B, RbohD and RbohF) in the indicated lines upon Guy11/mock treatment at 48 hpi. Relative mRNA amounts were normalized to that in WT mock samples. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis. All of the experiments were repeated two times with similar results.
Because O• 2 − is the precursor of H2O2, we next examined its concentration by Nitroblue tetrazolium (NBT) staining (Wu et al., 2017). Upon mock treatment, O• 2 − concentrations in OX398b, csd1, csd2 and sodx, but not in ccsd, were generally higher compared to those in their corresponding WT controls; in particular, the O• 2 − concentration in OX398b is markedly higher (Figs 5b, S4). Conversely, MIM398, CCSD and CSD2 showed lower or similar O• 2 − concentrations (Fig. S5a,c). These data indicate that silencing of CSD1, CSD2 and SODX by miR398b positively contributes to O• 2 − concentrations, whereas mutation of CCSD or overexpressing target genes do not lead to significant alteration of O• 2 − concentrations. More interestingly, the O• 2 − concentrations markedly decreased upon Guy11 infection in OX398b, csd1, csd2 and sodx lines, but only slightly changed in ccsd and target gene overexpressing lines (Figs 5b, S5a,c), suggesting that overexpression of miR398b, or mutations in CSD1, CSD2 and SODX positively regulated the blast infection‐promoted conversion of O• 2 − to H2O2, whereas overexpression of target genes had little effect on the conversion process.
NADPH oxidase is the key enzyme catalyzing the production of O• 2 − (Kaur et al., 2014). We then monitored the mRNA amounts of three NADPH oxidase genes, RbohB (Nagano et al., 2016), RbohD and RbohF (Jang et al., 2012). Guy11 infection clearly decreased their RNA amounts in all three WT rice varieties – Kasalath, NPB and TP309. By contrast, upon Guy11 infection, the mRNA amounts for those genes in OX398b, csd1, csd2 and sodx were all higher than in WT control plants (Fig. 5c–e). These results indicate that upon M. oryzae infection, overexpression of miR398b or mutations in CSD1, CSD2 and SODX leads to higher ROS concentrations, resulting enhanced blast disease‐resistance.
miR398b and its target genes regulate CSD expression and activity
Reduction of O• 2 − into H2O2 is primarily conducted by SOD isoenzymes (Fridovich, 1995; Gill & Tuteja, 2010). In rice, the SOD gene family contains fifteen members, including four CSDs, four SODs and another seven related chaperones including CCSD (Nath et al., 2014). Among the 15 members, the four CSDs were identified as CSD1, CSD2, CSD3 and CSD4 (Fig. S6). We first tested whether the expression levels of these CSDs were altered in OX398b and the mutants. As shown in Fig. 6a–d, OX398b lines display significantly decreased levels of CSD1, CSD2 and CSD3 with or without blast treatment, indicating that overexpression of miR398b leads to lower CSD expression; upon Guy11 infection, nearly all mutants lines displayed less or unchanged mRNA amounts of the four CSDs, suggesting that M.oryzae suppressed CSD expression.
Figure 6.
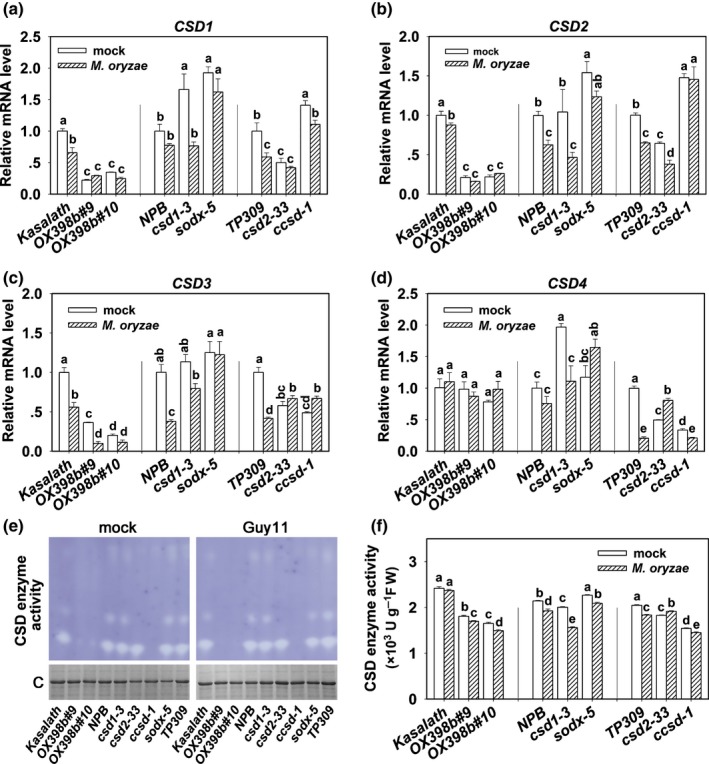
Overexpressing miR398b or mutations in target genes affected Cu/Zn‐Superoxidase Dismutase (CSD) concentrations and enzyme activity. (a–d) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) data showing the expression pattern of the CSD subfamily genes in wild‐type (WT) (Kasalath, Nipponbare (NPB), Taipei 309 (TP309)) and indicated transgenic lines with Magnaporthe oryzae strains Guy11 or mock infection at 48 h post‐inoculation (hpi). Relative mRNA amount was normalized to that in WT mock samples. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (e) The CSD enzyme activity in gel with Guy11/mock treatment. Coomassie (C) dying indicates the protein loading. (f) The enzyme activity of CSD in indicated lines with Guy11/mock treatment. Values are means of four replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis. All of the experiments were repeated two times with similar results.
We next examined CSD enzyme activity in these lines by photochemical method (Shah & Nahakpam, 2012) and spectrophotometry (Wu et al., 2017). Without blast treatment, OX398b displayed a significant reduction of bands in the gels, indicative of less CSD enzyme activity; ccsd lost major bands (Fig. 6e), suggesting that CCSD was required for CSD activity. Consistently, OX398b, csd1, csd2 and ccsd showed reduced CSD enzyme activity compared to their corresponding WT controls, whereas sodx showed slightly higher CSD activity (Fig. 6f). These results indicate that CSD1, CSD2 and CCSD positively, whereas SODX negatively, regulate CSD enzyme activity. Upon Guy11 infection, all tested lines displayed significantly lower CSD activity than the mock inoculation, except csd2 (Fig. 6f), indicating that M. oryzae suppresses CSD enzyme activity. However, the blast‐suppressed CSD activity contradicts the blast‐induced higher H2O2 concentrations in OX398b and mutant lines except ccsd (Fig. 5), indicating that some other SOD family members are possibly involved in miR398b‐regulated H2O2 accumulation upon M. oryzae infection.
We also tested the mRNA amounts of the four CSD genes and CSD enzyme activity in the target gene overexpressing lines. Upon mock treatment, whereas MIM398 displayed higher CSD enzyme activity associated with higher mRNA amount of CSD1 and CSD2 compared to the WT control, OXCSD2 showed higher activity accompanied by a higher CSD2 mRNA amount (Fig. S7a–e). Upon Guy11 infection, although all tested lines displayed decreased CSD activity, MIM398 and OXCCSD also showed higher activity compared to the WT control (Fig. S7e). These results confirm that rice blast suppresses CSD expression and enzyme activity, and imply that overexpression of miR398b target genes or overexpression of CSD2 alone enhances CSD enzyme activity.
miR398b and its target genes regulate SOD concentration and enzyme activity
In order to assess whether other SOD family genes contribute to the elevated H2O2 concentrations in OX398b, csd1, csd2 and sodx, we examined the mRNA amount of the other SOD family members and total SOD enzyme activity. Our data showed that upon M. oryzae infection, the mRNA amounts of SOD1‐Fe, SOD2‐Fe, SOD‐Mn, and a SOD family chaperone, Os02g54060, were significantly decreased in WT and ccsd, but unchanged or even increased in OX398b, csd1, csd2 and sodx, although SOD‐Mn was decreased in csd2 (Fig. 7a–d). Consistently, total SOD enzyme activity was upregulated in OX398b, csd1, csd2 and sodx, but was downregulated or remained unchanged in WT control plants and the ccsd mutant upon Guy11 infection (Fig. 7e). By contrast, the MIM398, OXCSD2 and OXCCSD lines showed lower or unchanged mRNA amount of these genes and unchanged or decreased SOD enzyme activity upon Guy11 infection compared to their corresponding WT controls (Fig. S8a–e). These data indicate that upon M. oryzae infection, miR398b silences target genes CSD1, CSD2 and SODX, which in turn triggers upregulation of other SOD family members resulting in higher total SOD enzyme activity, leading to higher H2O2 production and enhanced blast disease‐resistance.
Figure 7.
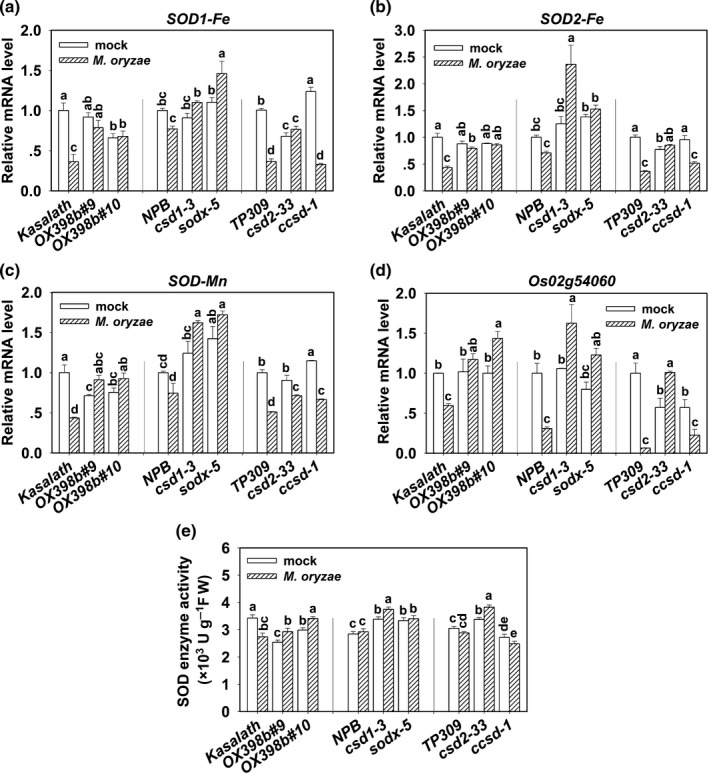
Overexpressing miR398b or mutations in target genes affected total Superoxidase Dismutase (SOD) concentrations and enzyme activity. (a–d) Quantitative reverse transcription polymerase chain reaction (qRT‐PCR) data showing the expression pattern of the SOD family genes in wild‐type (WT) (Kasalath, Nipponbare (NPB), Taipei 309 (TP309)) and indicated transgenic lines upon Magnaporthe oryzae strains Guy11 or mock treatment at 48 h post‐inoculation (hpi). Relative mRNA amount was normalized to that in WT mock samples at 48 hpi. Values are means of three replications. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey honest significant difference (HSD) analysis. (e) The total SOD enzyme activity in transgenic lines and WT plants with Guy11 or mock treatment at 48 hpi. Results are the means of four replicates. Error bars indicate SD. Different letters above the bars indicate significant differences at P < 0.01 as determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis. All of the experiments were repeated two times with similar results.
Because the observed H2O2 accumulation could be due to reduced H2O2 degradation in OX398b and the target gene mutants, we tested the activity of the main H2O2 degradation enzyme, catalase (CAT) (Kim et al., 2017). We found no obvious differences between the transgenic lines and WT upon M. oryzae infection (Fig. S9), indicating that CAT was not involved in miR398b‐mediated resistance to M. oryzae by catalyzing the conversion of H2O2 into H2O and O2.
Discussion
In a previous paper, miR398b is reported to positively regulate rice basal defense against M. oryzae, which is different from its negative role in defense in Arabidopsis (Li et al., 2010a, 2014). Here, we further demonstrated that miR398b coordinates multiple pathways to boost hydrogen peroxide (H2O2) production by regulating the expression of Cu/Zn Superoxidase Dismutase (CSD)1, CSD2, Copper Chaperone for Superoxide Dismutase (CCSD) and Superoxidase DismutaseX (SODX) upon M. oryzae infection. Functional analysis of these mutants indicates that CSD1, CSD2, CCSD and SODX play different roles in regulation of H2O2 concentrations and host resistance (Figs 2, 3, 4, 5). Mutations in SODX lead to higher CSD and total SOD activity, whereas mutations in CCSD compromise CSD activity (Figs 6, 7). Furthermore, mutations in CSD1 and CSD2 compromise CSD activity but lead to increased expression of other SODs and total SOD activity (Figs 6, 7), indicating that some compensatory regulation mechanism exists between CSDs and other SOD family members. Based on these data, we propose a working model to explain how miR398b boosts H2O2 production via multiple SODs (Fig. 8). Although CCSD is required for CSD activity to transfer copper to CSDs, SODX negatively regulates the activity of CSD and other SODs, such as SOD‐Fe and SOD‐Mn, by unknown mechanisms. These might be certain compensatory regulation or balance between CSDs and SODs. Without M. oryzae infection, miR398b regulates the expression of CSD1, CSD2, SODX and CCSD to maintain a basal CSD and total SOD activity level for normal H2O2 concentrations and related metabolic activity in rice cells (Fig. 8). Upon M. oryzae infection, the increased accumulation of miR398b represses the expression of all target genes. On the one hand, reduced accumulation of CSDs and CCSD leads to decreased CSD activity, which in turn results in increased expression of other SODs and higher total SOD activity by certain compensatory regulation mechanisms to enhance conversion of O• 2 − into H2O2; on the other, the reduction of SODX compromises the repression on CSD and SODs, resulting in higher total SOD activity (Fig. 8). In summary, upon M. oryzae infection, miR398b overexpression enhances resistance by repressing target genes to upregulate total SOD concentrations and enzyme activity to generate more H2O2 (Fig. 8). Therefore, our data provide new insight into the roles of miR398b in regulation of rice disease resistance.
Figure 8.
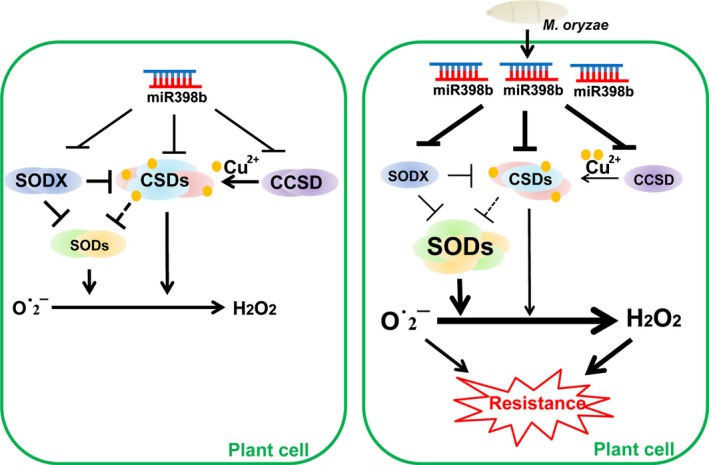
A model of miRNA398b functions in plant immunity to Magnaporthe oryzae. Without pathogen infection, miR398b‐regulated Cu/Zn‐Superoxidase Dismutase (CSD)1, CSD2, Superoxidase Dismutase (SOD)X and Copper Chaperone for Superoxide Dismutase (CCSD) maintain a basal CSD and total SOD enzyme activity level. CCSD positively contributes to CSD activity by transferring coppers to CSD protein. By contrast, SODX negatively regulates CSD and SOD activity by unknown mechanisms. A compensatory regulation mechanism exists between CSD and other SODs to maintain the hydrogen peroxide (H2O2) concentrations for normal metabolism in cells. Upon M. oryzae infection, the elevated miR398b accumulation represses the expression of CSDs/SODX/CCSD. On the one hand, the decreased CSDs and CCSD leads to lower CSD enzyme activity, which in turn results in higher expression and activity of other SODs by the unknown compensatory regulation mechanism. On the other, the decreased SODX compromises the repression on CSDs and SODs, and results in higher total SOD activity. As a result, the enhanced total SOD enzyme activity generates more H2O2 synthesis and results in enhanced resistance. Arrows indicate positive regulation and blunt‐ended bars indicate inhibition. The dotted lines indicate unidentified regulation between CSDs and SODs.
It is possible that miR398b activates a coordinated regulation network of CSD and SOD to positively regulate host resistance to rice blast disease. Without pathogen infection, OX398b displayed significantly lower CSD (Fig. 6) and total SOD activity compared to WT control (Fig. 7). However, upon M. oryzae infection, OX398b showed higher expression of other SOD genes and total SOD activity (Fig. 7) associated with higher H2O2 accumulation (Fig. 5), suggesting that miR398b upregulates total SOD activity to fight against M. oryzae invasion via upregulation of H2O2 production. By contrast, mutations of CSD1 and CSD2, respectively, resulted in lower CSD enzyme activity (Fig. 6e,f), whereas overexpression of CSD2 led to higher CSD enzyme activity (Fig. S7e), suggesting CSD1 and CSD2 are important CSD enzymes and positively contribute to CSD activity. However, upon M. oryzae infection, both csd1 and csd2 showed increased SOD‐Fe and SOD‐Mn expression, and total enzyme activity (Fig. 7a–e), resulting in increased H2O2 concentration (Fig. 5) and enhanced host resistance (Figs 2, S2), suggesting the existence of compensatory regulation mechanisms that perceive ROS concentrations upon M. oryzae infection and result in increased expression and activity of other SOD family members. However, how the decreased expression and activity of CSD triggers the compensatory regulation mechanisms needs further investigation. Interestingly, similar compensatory regulation mechanism possibly exists in other plants. For example, miR398 is upregulated, whereas CSD1 is downregulated in response to water deficit‐tolerance in Medicago truncatula, (Trindade et al., 2010), and miR398 positively contributes to heat tolerance by reducing transcripts of CSD1, CSD2 and CCS in Arabidopsis (Guan et al., 2013; Lu et al., 2013).
The two target genes of miR398b, CCSD and SODX, act antagonistically in regulating CSD and total SOD activity. CCSD is a chaperone of CSD, and ccsd showed lower CSD (Fig. 6e,f) and total SOD enzyme activity (Fig. 7e), indicating that CCSD is important for the CSD activity. However, OXCCSD displayed unaffected fungal growth, CSD and total SOD activity compared to wild‐type plants (Figs 3, 8, S7). One possible explanation is that CCSD is a CSD chaperone protein, not a CSD or SOD protein, and overexpression of CCSD cannot enhance CSD protein concentrations, or activity. The other possible explanation is that the GFP fusion disturbs the normal function of CCSD in the transgenic lines. SODX, an uncharacterized protein, is located near SOD‐Fe and SOD‐Mn in the phylogenetic tree (Fig. S6). We confirmed that its expression was repressed by miR398b (Fig. 1). Mutation of this gene in two different sites resulted in increased host resistance and higher ROS concentrations (Figs 2, 5, S2), indicating that SODX negatively regulates rice resistance. sodx displayed higher CSD concentration and activity (Fig. 7), as well as higher expression of SOD‐Fe and SOD‐Mn, and higher total SOD activity (Fig. 7), implying that SODX suppresses both CSDs and other SODs. However, the biochemical function of SODX, and how SODX negatively regulates CSD and total SOD activity needs further investigation.
miR398b mediates a coordinated and balanced regulatory module via CCSD, CSD and SODX. Overexpression of OX398b led to a significant reduction of CCSD concentration, but enhanced resistance to the blast disease, which appears to contradict the susceptibility of the ccsd mutant. There are several possible reasons for this paradox. First of all, OX398b reduced, but did not completely abolish the expression of CCSD (Li et al., 2014), CSD1 and CSD2 (Fig. 6a,b), indicating that OX398b retained certain CCSD‐dependent CSD activity. Second, miR398b also repressed SODX, releasing the SODX‐mediated suppression of CSD and SOD, resulting in higher CSD and SOD enzyme activity. Third, upon M. oryzae infection, the lower accumulation of CSD1 and CSD2 triggered a compensatory regulation mechanism resulting in higher expression of the other SOD family members and higher SOD activity. Thus, miR398b integratively upregulates the total SOD activity upon M. oryzae infection.
The SODs catalyze the conversion of O• 2 − into H2O2, which acts as a signal molecule to trigger resistance to various biotic and abiotic stresses (Quan et al., 2008; Kaur et al., 2016; Saxena et al., 2016). However, the excess H2O2 results in the occurrence of oxidative stress and leads to programmed cell death (PCD), which is harmful to plant development (Quan et al., 2008). Under this scenario, miR398b integratively regulates the activity of total SOD at certain levels to control moderate H2O2 accumulation. Upon M. oryzae infection, increased miR398b levels result in lower CSD1, CSD2 and CCSD concentrations and lower CSD activity, which is disadvantageous for rice to accumulate more H2O2 and fight against blast disease. In turn, the lower CSD activity triggers higher expression of other SOD family members, leading to higher total SOD activity. Such moderate compensatory regulation is based on the decreased CSD concentrations and avoids overaccumulation of H2O2, thereby boosting resistance in as well as avoiding unnecessary harm to rice.
In conclusion, upon M. oryzae infection, silencing of CSD1 and CSD2 by miR398b results in higher H2O2 concentrations associated with higher total SOD activity and greater resistance to infection; silencing of SODX leads to higher H2O2 concentrations accompanied by higher activity of both CSDs and SODs, whereas silencing of CCSD leads to lower H2O2 concentrations associated with lower activity of CSDs, which in turn triggers higher expression of other SODs. Therefore, the output of multiple SODs contributes to the miR398b‐regulated rice immunity against the blast fungus M. oryzae. Future research is needed to dissect the function of each member of the SOD family and their genetic interactions.
Author contributions
YL, X‐LC and W‐MW designed the experiments; YL, X‐LC, YZ, K‐NZ, X‐MY, HW, Z‐YX, Y‐PZ, J‐QZ and W‐MW performed the experiments; YL, X‐LC, JW, O‐XD, X‐WC and W‐MW analyzed the data; YL, MC and W‐MW wrote the paper. J‐HZ, L‐LZ, G‐BL, JF, X‐QC and X‐JWu discussed the results and commented on the manuscript; and YL and X‐LC contributed equally to this work.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Mutation sites of miR398b target genes in mutant lines.
Fig. S2 Overexpressing miR398b or mutations in target genes alters sensitivity to M. oryzae.
Fig. S3 The mRNA and protein concentrations of CSD2 and CCSD increases in overexpressing lines.
Fig. S4 Representative leaf sections from the OX398b and mutant lines show the accumulation of H2O2 and O• 2 −.
Fig. S5 Overexpressing MIM398 or overexpression of CSD2 does not upregulate ROS accumulation upon M. oryzae infection.
Fig. S6 Phylogenetic tree of SOD family members in rice.
Fig. S7 Overexpressing MIM398 or overexpression of miR398b target genes upregulate CSD accumulation and enzyme activity upon M. oryzae infection.
Fig. S8 Overexpressing MIM398 or overexpression of miR398b target genes does not upregulate total SOD enzyme activity upon M. oryzae infection.
Fig. S9 Overexpressing miR398b or mutations in target genes does not affect CAT enzyme activity.
Methods S1 H2O2 measurement.
Table S1 Primers used in this study.
Table S2 Guide RNAs and target sites used in CRISPR/Cas9 technology.
Table S3 Quantitative analysis of M. oryzae growth at the indicated time points.
Acknowledgements
We thank Dr Cai‐Lin Lei (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the monogenic resistant lines IRBLKm‐Ts, Dr Michael Steinwand (Department of Plant Pathology, University of California Davis) edited the manucript. This work was supported by the National Natural Science Foundation of China (grants 31471761 to YL and 31430072 to W‐MW).
Contributor Information
Yan Li, Email: jiazaihy@163.com.
Wen‐Ming Wang, Email: j316wenmingwang@sicau.edu.cn.
References
- Baldrich P, San Segundo B. 2016. MicroRNAs in rice innate immunity. Rice 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, He SY. 2009. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Slooten L, Vandenbranden S, De Rycke R, Botterman J, Sybesma C, Van Montagu M, Inze D. 1991. Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO Journal 10: 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candar‐Cakir B, Arican E, Zhang B. 2016. Small RNA and degradome deep sequencing reveals drought and tissue‐specific microRNAs and their important roles in drought‐sensitive and drought‐tolerant tomato genotypes. Plant Biotechnology Journal 14: 1727–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ren Y, Zhang Y, Xu J, Sun F, Zhang Z, Wang Y. 2012. Genome‐wide identification and expression analysis of heat‐responsive and novel microRNAs in populus tomentosa . Gene 504: 160–165. [DOI] [PubMed] [Google Scholar]
- Chen M, Zeng H, Qiu D, Guo L, Yang X, Shi H, Zhou T, Zhao J. 2012. Purification and characterization of a novel hypersensitive response‐inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 7: e37654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BJ. 1964. Disc electrophoresis. II. Method and application to human serum protein. Annals of the New York Academy of Sciences 121: 404–427. [DOI] [PubMed] [Google Scholar]
- Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y. 2013. Efficient genome editing in plants using a CRISPR/Cas system. Cell Research 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridovich I. 1995. Superoxide radical and superoxide dismutases. Annual Review of Biochemistry 64: 97–112. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiology and Biochemistry 48: 909–930. [DOI] [PubMed] [Google Scholar]
- Guan Q, Liao X, He M, Li X, Wang Z, Ma H, Yu S, Liu S. 2017. Tolerance analysis of chloroplast OsCu/Zn‐SOD overexpressing rice under NaCl and NaHCO3 stress. PLoS ONE 12: e0186052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. 2013. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. The Plant Journal 74: 840–851. [DOI] [PubMed] [Google Scholar]
- Gupta AS, Heinen JL, Holaday AS, Burke JJ, Allen RD. 1993a. Increased resistance to oxidative stress in transgenic plants that overexpress chloroplastic Cu/Zn superoxide‐dismutase. Proceedings of the National Academy of Sciences, USA 90: 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta AS, Webb RP, Holaday AS, Allen RD. 1993b. Overexpression of superoxide dismutase protects plants from oxidative stress. Plant Physiology 103: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hivrale V, Zheng Y, Puli COR, Jagadeeswaran G, Gowdu K, Kakani VG, Barakat A, Sunkar R. 2016. Characterization of drought‐ and heat‐responsive microRNAs in switchgrass. Plant Science 242: 214–223. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O et al 2013. DNA targeting specificity of RNA‐guided Cas9 nucleases. Nature Biotechnology 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SJ, Wi SJ, Choi YJ, An G, Park KY. 2012. Increased polyamine biosynthesis enhances stress tolerance by preventing the accumulation of reactive oxygen species: T‐DNA mutational analysis of Oryza sativa lysine decarboxylase‐like protein 1. Molecules and Cells 34: 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Jovanovic Z, Stanisavljevic N, Mikic A, Radovic S, Maksimovic V. 2014. Water deficit down‐regulates miR398 and miR408 in pea (Pisum sativum L.). Plant Physiology and Biochemistry 83: 26–31. [DOI] [PubMed] [Google Scholar]
- Kankanala P, Czymmek K, Valent B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar‐Agarwal S, Jin H. 2010. Role of small RNAs in host–microbe interactions. Annual Review of Phytopathology 48: 225–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur G, Sharma A, Guruprasad K, Pati PK. 2014. Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnology Advances 32: 551–563. [DOI] [PubMed] [Google Scholar]
- Kaur N, Dhawan M, Sharma I, Pati PK. 2016. Interdependency of reactive oxygen species generating and scavenging system in salt sensitive and salt tolerant cultivars of rice. BMC Plant Biology 16: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaksefidi R, Mirlohi S, Khalaji F, Fakhari Z, Shiran B, Fallahi H, Rafiei F, Budak H, Ebrahimie E. 2015. Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus . Frontiers in Plant Science 6: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Khan AL, Waqas M, Lee IJ. 2017. Silicon regulates antioxidant activities of crop plants under abiotic‐induced oxidative stress: a review. Frontiers in Plant Science 8: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J et al 2017b. A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell 170: e115. [DOI] [PubMed] [Google Scholar]
- Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ et al 2014. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae . Plant Physiology 164: 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. 2010a. Identification of microRNAs involved in pathogen‐associated molecular pattern‐triggered plant innate immunity. Plant Physiology 152: 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL, Xu YJ, Zhao ZX, Xiao ZY, Yang N et al 2017a. Osa‐miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae . Frontiers in Plant Science 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zheng Y, Addo‐Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. 2010b. Transcriptome‐wide identification of microRNA targets in rice. The Plant Journal 62: 742–759. [DOI] [PubMed] [Google Scholar]
- Lu X, Guan Q, Zhu J. 2013. Downregulation of CSD2 by a heat‐inducible miR398 is required for thermotolerance in Arabidopsis. Plant Signaling & Behavior 8: pii: e24952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Harjanto E, Leprince O. 1996. Water‐deficit tolerance and field performance of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiology 111: 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Bowley SR, Jones KS. 1999. Winter survival of transgenic alfalfa overexpressing superoxide dismutase. Plant Physiology 119: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKersie BD, Chen Y, de Beus M, Bowley SR, Bowler C, Inze D, D'Halluin K, Botterman J. 1993. Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.). Plant Physiology 103: 1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7: 405–410. [DOI] [PubMed] [Google Scholar]
- Nagano M, Ishikawa T, Fujiwara M, Fukao Y, Kawano Y, Kawai‐Yamada M, Shimamoto K. 2016. Plasma membrane microdomains are essential for RAC1‐RbohB/H‐mediated immunity in rice. Plant Cell 28: 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath K, Kumar S, Poudyal RS, Yang YN, Timilsina R, Park YS, Nath J, Chauhan PS, Pant B, Lee CH. 2014. Developmental stage‐dependent differential gene expression of superoxide dismutase isoenzymes and their localization and physical interaction network in rice (Oryza sativa L.). Genes & Genomics 36: 45–55. [Google Scholar]
- Padmanabhan C, Zhang X, Jin H. 2009. Host small RNAs are big contributors to plant innate immunity. Current Opinion in Plant Biology 12: 465–472. [DOI] [PubMed] [Google Scholar]
- Park CH, Chen S, Shirsekar G, Zhou B, Khang CH, Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M et al 2012. The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. Plant Cell 24: 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher LH, Zilinskas BA. 1996. Overexpression of Copper/Zinc superoxide dismutase in the cytosol of transgenic tobacco confers partial resistance to ozone‐induced foliar necrosis. Plant Physiology 110: 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan LJ, Zhang B, Shi WW, Li HY. 2008. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. Journal of Integrative Plant Biology 50: 2–18. [DOI] [PubMed] [Google Scholar]
- Racchi ML. 2013. Antioxidant defenses in plants with attention to Prunus and Citrus spp. Antioxidants 2: 340–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena I, Srikanth S, Chen Z. 2016. Cross talk between H2O2 and interacting signal molecules under plant stress response. Frontiers in Plant Science 7: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Nahakpam S. 2012. Heat exposure alters the expression of SOD, POD, APX and CAT isozymes and mitigates low cadmium toxicity in seedlings of sensitive and tolerant rice cultivars. Plant Physiology and Biochemistry 57: 106–113. [DOI] [PubMed] [Google Scholar]
- Shimono M, Koga H, Akagi A, Hayashi N, Goto S, Sawada M, Kurihara T, Matsushita A, Sugano S, Jiang CJ et al 2012. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathology 13: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slooten L, Capiau K, Van Camp W, Van Montagu M, Sybesma C, Inze D. 1995. Factors affecting the enhancement of oxidative stress tolerance in transgenic tobacco overexpressing manganese superoxide dismutase in the chloroplasts. Plant Physiology 107: 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. 2006. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Chu C. 2017. MicroRNAs in crop improvement: fine‐tuners for complex traits. Nature Plants 3: 17077. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JD. 2002. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proceedings of the National Academy of Sciences, USA 99: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I, Capitao C, Dalmay T, Fevereiro MP, Santos DM. 2010. MiR398 and miR408 are up‐regulated in response to water deficit in Medicago truncatula . Planta 231: 705–716. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Slooten L, Stassart JM, Moens T, Botterman J, Van Montagu M, Inze D. 1999. Overproduction of Arabidopsis thaliana FeSOD confers oxidative stress tolerance to transgenic maize. Plant and Cell Physiology 40: 515–523. [DOI] [PubMed] [Google Scholar]
- Van Camp W, Capiau K, Van Montagu M, Inze D, Slooten L. 1996. Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe‐superoxide dismutase in chloroplasts. Plant Physiology 112: 1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Sun YF, Song N, Wei JP, Wang XJ, Feng H, Yin ZY, Kang ZS. 2014. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L.. Plant Physiology and Biochemistry 80: 90–96. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu J, Kim SG, Tsuda K, Gupta R, Park SY, Kim ST, Kang KY. 2016. Magnaporthe oryzae‐secreted protein MSP1 induces cell death and elicits defense responses in rice. Molecular Plant–Microbe Interactions 29: 299–312. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Wang ZM, Wang M, Wang XJ. 2013. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiology 161: 1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang R, Yang Z, Yao S, Zhao S, Wang Y, Li P, Song X, Jin L, Zhou T et al 2017. ROS accumulation and antiviral defence control by microRNA528 in rice. Nature Plants 3: 16203. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. 2009. Rice microRNA effector complexes and targets. Plant Cell 21: 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. 2010. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biology 10: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Meng Y, Wise RP. 2014. Mla‐ and Rom1‐mediated control of microR398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytologist 201: 1396–1412. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang H, Lu YZ, de Ruiter M, Cariaso M, Prins M, van Tunen A, He YK. 2012. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa . Journal of Experimental Botany 63: 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Mutation sites of miR398b target genes in mutant lines.
Fig. S2 Overexpressing miR398b or mutations in target genes alters sensitivity to M. oryzae.
Fig. S3 The mRNA and protein concentrations of CSD2 and CCSD increases in overexpressing lines.
Fig. S4 Representative leaf sections from the OX398b and mutant lines show the accumulation of H2O2 and O• 2 −.
Fig. S5 Overexpressing MIM398 or overexpression of CSD2 does not upregulate ROS accumulation upon M. oryzae infection.
Fig. S6 Phylogenetic tree of SOD family members in rice.
Fig. S7 Overexpressing MIM398 or overexpression of miR398b target genes upregulate CSD accumulation and enzyme activity upon M. oryzae infection.
Fig. S8 Overexpressing MIM398 or overexpression of miR398b target genes does not upregulate total SOD enzyme activity upon M. oryzae infection.
Fig. S9 Overexpressing miR398b or mutations in target genes does not affect CAT enzyme activity.
Methods S1 H2O2 measurement.
Table S1 Primers used in this study.
Table S2 Guide RNAs and target sites used in CRISPR/Cas9 technology.
Table S3 Quantitative analysis of M. oryzae growth at the indicated time points.


