Abstract
There is an urgent need for an easily assessable preoperative test to predict postoperative liver function recovery and thereby determine the optimal time point of liver resection, specifically as current markers are often expensive, time consuming, and invasive. Emerging evidence suggests that microRNA (miRNA) signatures represent potent diagnostic, prognostic, and treatment‐response biomarkers for several diseases. Using next‐generation sequencing as an unbiased systematic approach, 554 miRNAs were detected in preoperative plasma of 21 patients suffering from postoperative liver dysfunction (LD) after liver resection and 27 matched controls. Subsequently, we identified a miRNA signature—consisting of miRNAs 151a‐5p, 192‐5p, and 122‐5p—that highly correlated with patients developing postoperative LD after liver resection. The predictive potential for postoperative LD was subsequently confirmed using real‐time PCR in an independent validation cohort of 98 patients. Ultimately, a regression model of the two miRNA ratios 151a‐5p to 192‐5p and 122‐5p to 151a‐5p was found to reliably predict postoperative LD, severe morbidity, prolonged intensive care unit and hospital stays, and even mortality before an operation with a remarkable accuracy, thereby outperforming established markers of postoperative LD. Ultimately, we documented that miRNA ratios closely followed liver function recovery after partial hepatectomy. Conclusion: Our data demonstrate the clinical utility of an miRNA‐based biomarker to support the selection of patients undergoing partial hepatectomy. The dynamical changes during liver function recovery indicate a possible role in individualized patient treatment. Thereby, our data might help to tailor surgical strategies to the specific risk profile of patients.
Abbreviations
- ALPPS
associating liver partition and portal vein ligation
- ALT
alanine transaminase
- AST
aspartate transaminase
- AUC
area under the curve
- CCC
cholangiocellular carcinoma
- CPM
counts per million
- GEO
Gene Expression Omnibus
- GGT
gamma‐glutamyl transpeptidase
- HCC
hepatocellular carcinoma
- ICG
indocyanine green
- ICU
intensive care unit
- ISGLS
International Study Group of Liver Surgery
- LD
liver dysfunction
- LR
logistic regression
- mCRC
metastatic colorectal carcinoma
- miRNA
microRNA
- NGS
next‐generation sequencing
- NPV
negative predictive value
- OR
odds ratio
- POD
postoperative day
- PPV
positive predictive value
- pre‐OP
preoperatively
- qPCR
real‐time quantitative PCR
- ROC
receiver operator characteristic
- SN
sensitivity
- SP
specificity
Liver resection represents the only curative treatment in many liver malignancies. Postresectional hepatic regeneration determines the clinical outcome of patients undergoing liver resection,1 as insufficient hepatic regeneration after liver resection results in postoperative liver dysfunction (LD), which occurs in up to 30% of patients after major hepatic resections.2 Importantly, currently available treatment options for patients with postoperative LD are very limited, mostly symptomatic, and goal‐directed.2, 3 Hence, risk stratification for optimized patient selection before liver operation is key for minimizing the postoperative LD and concomitant complications. This is specifically relevant for patients suffering from extensive disease who require surgical procedures with a high risk of postoperative complications and mortality, such as the associating liver partition and portal vein ligation (ALPPS) procedure, to achieve complete tumor clearance. However, currently available markers are often expensive, time consuming, and invasive, highlighting the need for an easily assessable preoperative test to predict postoperative liver function recovery.
Emerging evidence suggests that microRNA (miRNA) signatures represent potent diagnostic, prognostic, and treatment‐response biomarkers for several diseases.4 As master regulators of expression of multiple genes in different tissues, miRNAs can control virtually every cellular process on a transcriptional level, including cellular development, proliferation, migration, survival, metabolism, homeostasis, and regeneration.5 Estimates based on computational analyses suggest that over 50% of the human transcriptome is regulated by at least one miRNA.6 Hence, it is not surprising that aberrant miRNA expression can have detrimental effects on signaling pathways and indeed has been implicated in a wide range of diseases.7, 8, 9 To date, almost 2,000 miRNAs have been identified, and some have already been validated for in vitro diagnosis and/or prognosis of various malignant diseases, demonstrating their potential as biomarkers in the clinic.10, 11, 12 Moreover, miRNAs are easily accessible from biofluids such as blood, urine, and saliva through noninvasive methods, and they exhibit high stability and relatively low complexity (e.g., no postprocessing modifications) and can be readily assessed by various methods with high specificity (SP), making them superior compared with other classes of biomarkers, including DNA, RNA, and proteins.
Thus, the aim of this study was to systematically elucidate potential differences in the profile of circulating miRNAs between patients with and without postoperative LD and poor postoperative outcome. Thus, we used next‐generation sequencing (NGS) as an unbiased systematic approach to investigate the predictive potential of miRNA profiles in plasma of a discovery cohort of patients undergoing liver resection. We identified a clinically applicable and robust combination of specific miRNAs to efficiently predict postoperative LD as well as complications before operation and verified these findings in an independent prospective validation cohort by real‐time quantitative PCR (qPCR). Importantly, within this analysis, we observed a superior prognostic performance of the established miRNA signature when compared with currently used clinical markers for prediction of postoperative LD. Ultimately, we provide exploratory data suggesting that miRNA signatures can dynamically monitor liver function recovery and might represent a very useful tool to define the most suitable time point for liver resection to avoid postoperative LD and complications.
Materials and Methods
Details on methodology can be found within the Supporting Information for this manuscript.
Study Population
Initially, a discovery cohort of patients was prospectively recruited starting from February 2012. Patients with hepatocellular carcinoma (HCC), cholangiocellular carcinoma (CCC), or metastatic colorectal carcinoma (mCRC) were considered eligible for inclusion. Subsequently, we validated our explorative results in a prospective cohort. Ultimately, we monitored miRNAs of interest in a subset of patients before operation as well as on postoperative day (POD) 1 and POD 5.
The present study was approved by the Institutional Ethics Committee, and written informed consent was obtained.
Definition and Classification of Postoperative LD and Complications
Patients were followed up for a postoperative period of 90 days. Postoperative LD was diagnosed following the criteria issued by the International Study Group of Liver Surgery (ISGLS).13 Accordingly, LD was defined by both abnormal values of serum bilirubin and prothrombin time on or after POD 5. Of note, in case of abnormal preoperative serum bilirubin or prothrombin time, a postoperative deviation and deterioration on 2 subsequent days after POD 5 was identified as postoperative LD. Furthermore, patients reaching normal serum bilirubin or prothrombin time values before POD 5, or who were discharged early because of good clinical performance and hence had no further blood collection, were considered as having “no LD.”
For classification of patients with postoperative morbidity, the outline given by Dindo et al.14 was applied. Death within 90 days after operation was classified as postoperative mortality.15
Measurement of Circulating miRNAs
Meticulous preparation of plasma was performed as described.16
RNA Extraction and Small RNA Sequencing
The miRNeasy Mini Kit (Qiagen, Germany) was applied to isolate total RNA, including small RNAs, from plasma. Small RNA sequencing libraries were generated using the CleanTag Small RNA Library Preparation Kit (TriLink) according to the manufacturer's recommendations. Sequencing was performed on an Illumina NextSeq 500, using single‐end reads with 50 cycles (Illumina). Sequencing reads in fastq format were adapter‐trimmed and quality‐checked (generation of fastQC files). Quality filtered reads (phred > 30) were used for alignment against human mature miRNAs (miRBase version 21) using Bowtie 2. Mapped reads were cross‐checked through genome alignments (Bowtie 2, Genome Reference Consortium human genome build 37). Mature miRNA reads were counted (isomiR sequences were summarized) and normalized as counts per million (CPM) to the total number of mapped reads. CPM values were used for statistical analysis (see below). Sequencing raw and normalized data can be accessed under the record GSE123605 from the National Center for Biotechnology Information Gene Expression Omnibus (GEO).
qPCR Analysis
qPCR analyses were performed as described.17 Robustness of RNA extraction, complementary DNA (cDNA) synthesis, and qPCR amplification was assessed using combinations of synthetic spike‐in controls (Exiqon, Denmark). Hemolysis was assessed using the ratios of miRNA‐23a‐3p and miRNA‐451a.18 Of note, no samples failed because of hemolysis or high analytical variance.
Statistical Analyses
Statistical analysis was based on nonparametric tests. Calculations were performed using SPSS (version 23.0; IBM Corp., Armonk, NY) and R (version 3.4.1; R Foundation, Vienna, Austria). P values were adjusted for multiple testing based on the false discovery rate according to the Benjamini‐Hochberg method, and P values < 0.05 were considered significant. For more detail on statistical methods, refer to the Supporting Materials and Methods section.
Results
Patients and Cohorts
A total of 48 patients with mCRC, HCC, or CCC who underwent liver resection between February 2012 and April 2016 were included in the discovery cohort. To achieve a representative cohort, 21 patients suffering from postoperative LD were matched based on basic characteristics, liver function, and extent of liver resection to 27 patients without postoperative LD (Supporting Table S1). Subsequently, an additional 98 patients served as a prospective validation cohort (Table 1). Additionally, the perioperative dynamics of miRNAs were evaluated in a group of 7 patients undergoing regular major liver resection and a group of 8 patients undergoing ALPPS (Supporting Table S2) for whom longitudinal measurements of miRNAs could be performed on the basis of repeatedly collected plasma samples.
Table 1.
Patient Demographics
| Parameter | Entire Cohort (N = 154) | Evaluation Cohort (N = 48) | Validation Cohort (N = 106) | P Value |
|---|---|---|---|---|
| Median (Range) N (%) | Median (Range) N (%) | Median (Range) N (%) | ||
| Sex | 0.696 | |||
| Male | 106 (68.8%) | 32 (66.7%) | 74 (69.8%) | |
| Female | 48 (31.2%) | 16 (33.3%) | 32 (30.2%) | |
| Age (years) | 65 (22‐89) | 65 (36‐89) | 65 (22‐86) | 0.838 |
| Hepatic resection | <0.001 | |||
| Minor (<3 segments) | 55 (35.7%) | 4 (8.3%) | 51 (48.1%) | |
| Major (≥3 segments) | 99 (64.2%) | 44 (91.7%) | 55 (51.9%) | |
| Tumor type | 0.147 | |||
| CRCLM | 62 (40.3%) | 16 (33.3%) | 46 (43.4%) | |
| HCC | 48 (31.2%) | 16 (33.3%) | 32 (30.2%) | |
| CCC | 41 (26.6%) | 16(33.3%) | 25 (23.6%) | |
| Other | 3 (1.9%) | 0 (0.0%) | 3 (2.8%) | |
| Cofactors | ||||
| Neoadjuvant CTx | 55 (35.7%) | 16 (33.3%) | 39 (36.8%) | 0.053 |
| Steatosis (%) | 5 (0‐100) | 5 (0‐40) | 5 (0‐100) | 0.299 |
| Steatohepatitis | 29 (18.8%) | 10 (20.8%) | 19 (17.9%) | 0.341 |
| Fibrosis | 81 (52.6%) | 19 (39.5%) | 61 (57.5%) | 0.411 |
| Cirrhosis | 20 (13.0%) | 4 (8.3%) | 16 (15.1%) | 0.614 |
| Intraoperative RBCs | 13 (8.4%) | 7 (14.6%) | 6 (5.7%) | 0.061 |
| Preoperative parameters | ||||
| PDR (%) | 20.7 (7.6‐40.0) | 20.0 (9.9‐34.8) | 20.9 (7.6‐40.0) | 0.629 |
| R15 (%) | 5.0 (0.3‐32.0) | 5.0 (0.5‐22.7) | 4.8 (0.3‐32.0) | 0.674 |
| Platelets (×103/µL) | 229 (78‐492) | 236 (86‐492) | 227 (78‐470) | 0.535 |
| SB (mg/dL) | 0.57 (0.15‐3.77) | 0.57 (0.15‐3.17) | 0.57 (0.16‐3.77) | 0.515 |
| PT (%) | 100 (40‐150) | 102 (40‐137) | 99 (45‐150) | 0.355 |
| AP (U/L) | 95 (43‐707) | 104 (49‐707) | 90 (43‐418) | 0.190 |
| GGT (U/L) | 74 (13‐576) | 32 (18‐1,576) | 73 (13‐1,335) | 0.120 |
| AST (U/L) | 31 (14‐224) | 33 (17‐224) | 31 (14‐175) | 0.347 |
| ALT (U/L) | 31 (9‐318) | 84 (9‐318) | 28 (8‐196) | 0.279 |
| Albumin (g/L) | 41.8 (30.9‐49.6) | 42.6 (31.5‐47.3) | 41.0 (30.9‐49.6) | 0.175 |
| Morbidity | 0.354 | |||
| No morbidity | 77 (50.0%) | 21 (43.8%) | 56 (52.8%) | |
| Grade I | 14 (9.1%) | 4 (8.3%) | 10 (9.4%) | |
| Grade II | 29 (18.8%) | 7 (14.6%) | 22 (20.8%) | |
| Grade III | 18 (11.6%) | 9 (18.8%) | 9 (8.5%) | |
| Grade IV | 8 (5.1%) | 2 (4.1%) | 6 (5.7%) | |
| Grade V | 8 (5.2%) | 5 (10.4%) | 3 (2.8%) | |
| LD ISGLS | <0.001 | |||
| No LD | 124 (81.1%) | 27 (56.3%) | 98 (92.4%) | |
| ISGLS A | 7 (4.5%) | 5 (10.4%) | 2 (1.9%) | |
| ISGLS B | 8 (5.2%) | 4 (8.3%) | 4 (3.8%) | |
| ISGLS C | 14 (9.1%) | 12 (25.0%) | 2 (1.9%) | |
| Postoperative stay | ||||
| ICU (days) | 1 (0‐25) | 1 (0‐15) | 1 (0‐25) | 0.190 |
| Total hospitalization (days) | 9 (3‐117) | 11 (4‐50) | 9 (3‐117) | 0.096 |
Abbreviations: AP, alkaline phosphatase; CRCLM, colorectal cancer liver metastases; CTx, chemotherapy; PDR, plasma disappearance rate; PT, prothrombin time; R15, retention rate at 15 minutes; RBC, red blood cell; SB, serum bilirubin.
Establishing the Predictive miRNA Panel for LD After Operation in the Discovery Cohort
For initial identification of miRNAs associated with postoperative LD, we aimed for an unbiased systematic approach using NGS. miRNAs, small nuclear RNAs, small nucleolar RNAs, transfer RNAs, and Y RNAs were identified in the data set (see Supporting Fig. S1; NGS data were submitted to GEO and can be downloaded using the accession number GSE123605). Subsequently, we focused on miRNA, given the above‐mentioned benefits, and analyzed miRNA profiles in the plasma of all patients within the discovery cohort before operation to determine if miRNA profiles differ between patients who develop LD and those without delayed hepatic recovery. With an average of 3 million miRNA reads per sample, we detected a total of 554 miRNAs across all analyzed plasma samples. To identify potential biomarker candidates, cutoffs for plasma abundance (average log2 CPM [logCPM] > 5), effect size (fold change > 1.3), and significance level (P < 0.2) were implied. As depicted in Fig. 1A, a set of 19 miRNAs remained in the analysis, of which 12 were up‐regulated (red) and 7 were down‐regulated (blue) in the pre‐OP plasma of patients with LD. A clustered expression heat map, comparing expression of these miRNAs in patients with and without LD, is depicted in Fig. 1B.
Figure 1.
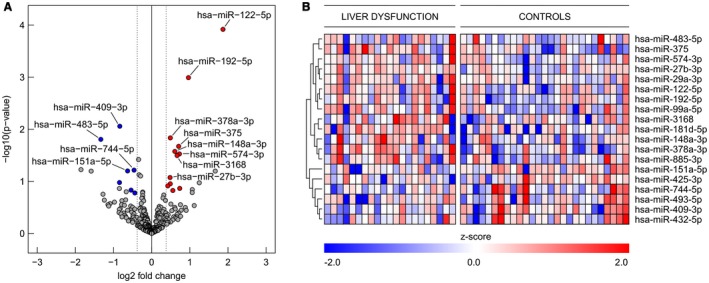
Differences in presurgical microRNA patterns in patients undergoing liver resection. (A) Volcano plot of differentially regulated microRNAs in presurgical plasma of patients with LD. To identify biomarker candidates, cutoffs for plasma concentration (average logCPM > 5), effect size (fold change > 1.3), and significance level (raw P < 0.2) were implied. A set of 19 microRNAs, of which 12 were up‐regulated (red) and 7 were down‐regulated (blue), was identified. (B) Expression heat map of regulated miRNAs in plasma of patients (miR‐wise z score of log2 [CPM + 1]); red indicates higher expression than the mean and blue indicates lower expression than the mean, according to the legend at the bottom.
Subsequently, we evaluated the analytical variability between NGS and qPCR‐based miRNA detection and calculated the relative logarithmic differences between two miRNAs to form self‐normalizing miRNA pairs, thus circumventing the need for a reference miRNA. Six of the top miRNAs, and consequently 15 miRNA pairs, were considered for this analysis. We observed high concordance between data sets (NGS vs. qPCR‐based miRNAs) as illustrated in Supporting Fig. S2. The importance of miRNA pairs for achieving excellent predictive performance of negative postoperative outcomes was analyzed using random forest modelling (Fig. 2A). Two top‐ranked miRNA pairs (151a‐5p_192‐5p, 122‐5p_151a‐5p) were identified with significant pre‐OP differences between individuals developing LD and controls (Fig. 2B). The diagnostic performance of a multivariate model as measured by receiver operator characteristic (ROC) analysis estimated an area under the curve (AUC) of 0.66 for miRNA pair 122‐5p/151a‐5p (Fig. 2C), 0.75 for miRNA pair 151a‐5p/192‐5p (Fig. 2D), and 0.76 for a logistic regression (LR) model using the combination of both miRNA pairs (Fig. 2E).
Figure 2.
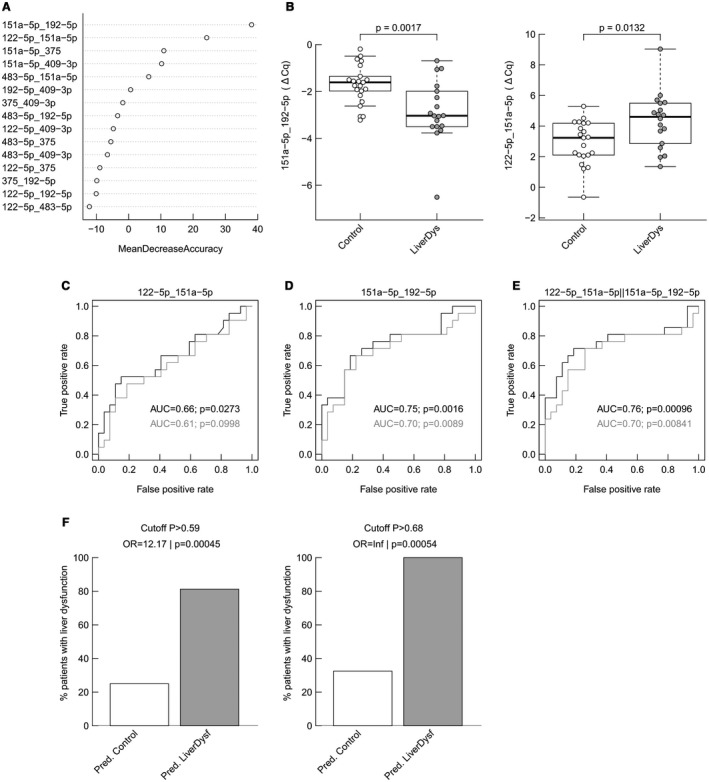
Analysis of the diagnostic performance of miRNA pairs to predict LD. (A) Importance of miRNA pairs in a random forest classification model (the most important ones are at the top with highest mean decrease accuracy). (B) Distribution of ratios measured by qPCR in the discovery cohort for miR151a‐5p/192‐5p (boxplots and P values from two‐sided Wilcoxon rank‐sum test) and for miR122‐5p/151a‐5p. (C) ROC curves for an LR model including miR122‐5p/151a‐5p in the discovery cohort (results from leave‐one‐out cross‐validation are in gray). (D) An LR model including miR151‐5p/192‐5p and (E) an LR model including both miRNA pairs. The performance is described by the AUC, and whether the classification deviates significantly from the random assignment (AUC = 0.5) is indicated by the P value. The percentage of true postoperative LD on predicted controls and predicted LD were analyzed for both model‐defined cutoffs: P > 0.59 and P > 0.68 (F).
Next, two clinically useful cutoffs were defined to predict postoperative LD. Accordingly, a low‐stringency cutoff to identify patients who could undergo liver resection with very low risk (cut‐off P > 0.59) as well as a stringent cutoff to identify patients who should be optimized before surgery or not undergo liver resection (cut‐off P > 0.68) were defined (see Supporting Table S3). For both cutoffs, predictions were found to be significantly associated with postoperative LD (P = 0.00045 and P = 0.00054, respectively; Fig. 2F).
Validation of the Discovered miRNA Ratios in an Independent Validation Cohort
To confirm the clinical utility of our miRNA ratios, we next validated the predictive performance of the two miRNA pairs in an independent prospective validation cohort consisting of 98 patients, 90 without and 8 with postoperative LD, reflecting the natural postsurgical LD incidence of approximately 10%‐20%. The diagnostic performance of a multivariate model as measured by ROC analysis showed an AUC of 0.69 for the two miRNA pairs (Fig. 3A). Further, we validated that the two cutoffs were able to predict postoperative LD for cut‐off P > 0.59 with P = 0.011 (Fig. 3A) and for cut‐off P > 0.68 with P = 0.032 (Fig. 3A), respectively.
Figure 3.
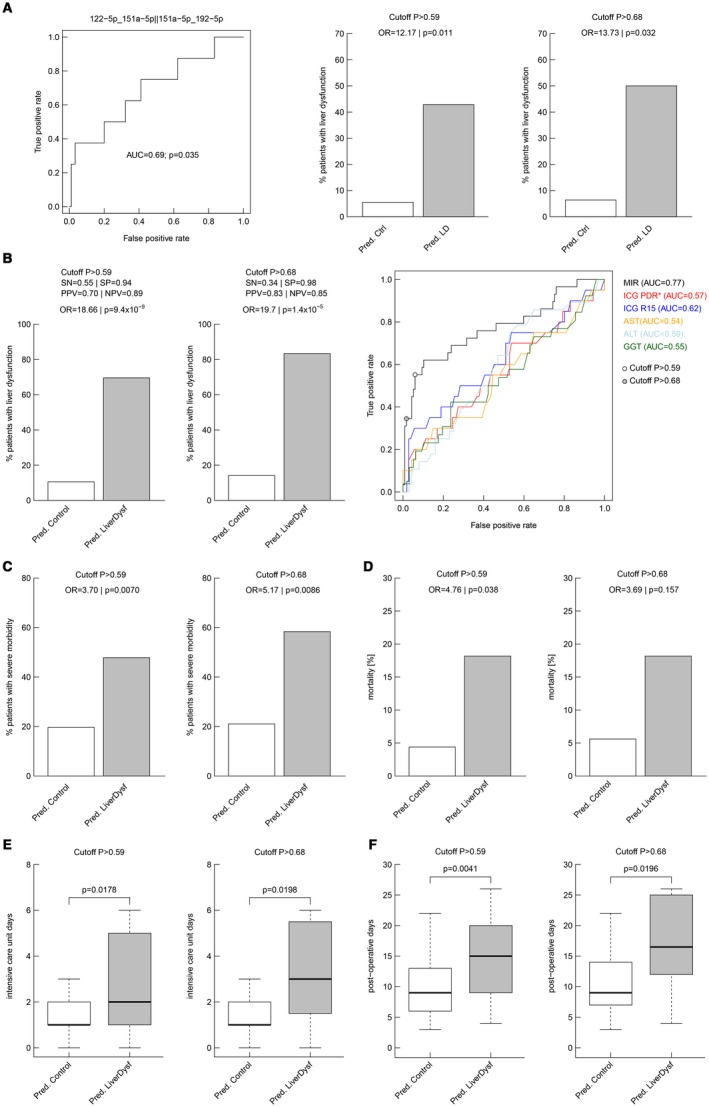
Validation of predictive performance of the top two miRNA ratios in an independent cohort and analyses of the complete data set. miRNAs were analyzed by qPCR in 98 subjects of which 8 (8.1%) experienced the adverse outcome postsurgery. (A) The predictive performance of the previously defined multivariate logistic prediction models was validated using ROC analyses. The performance is described by the AUC and whether the classification deviates significantly from the random assignment (AUC = 0.5) is indicated by the P value. The percentage of true postoperative LD on predicted controls and predicted LD were analyzed for both model‐defined cutoffs: P > 0.59 and P > 0.68. The two cutoffs were further analyzed for their performance to predict postoperative LD in the entire cohort (B). Performance was described using SN, SP, PPV, NPV, and the OR, which is the ratio of odds of suffering from postoperative LD associated with a positive test result compared with a negative test result. The low‐stringency cutoff (P = 0.59) yielded PPV and NPV values of 0.70 and 0.89, respectively, whereas the stringent cutoff (P = 0.68) resulted in a PPV of 0.83, with an NPV of 0.85 (B). This means that 83% of the patients who tested positive suffered from postoperative LD, whereas 85% who tested negative did not suffer from postoperative LD. In contrast, 15% who tested negative did in fact suffer from postoperative LD. The ORs for an adverse event were 18.66 (P < 0.0001) and infinite (P < 0.0001), respectively. ROC curve analysis was performed for the microRNA model to compare its performance against that of standard liver function parameters (B). ORs for other adverse postoperative outcomes were analyzed for both model cutoffs: severe morbidity (C) and mortality (D). Postoperative ICU stay (E) and hospitalization (F) were significantly prolonged in our predicted risk groups (boxplots are shown without outliers; P values from two‐sided Wilcoxon rank‐sum test). Abbreviations: PDR, plasma disappearance rate; R15, retention rate at 15 minutes.
Comparison of the Discovered miRNA Ratios to Other Predictors for Postoperative LD
Next, we evaluated the performance of the miRNA‐based prediction model in the combined data set (N = 146) to increase the power of our analyses. First, to assess independence of the identified miRNA pairs to predict postoperative LD from other known markers of liver damage and function, as well as from clinical parameters, multivariate LR analysis was performed for both pairs independently. Strikingly, both miRNA pairs remained significant on multivariate LR analysis (Table 2).
Table 2.
Univariate and Multivariate LR Models
| Variables | Levels (Unit) | n | Coeff | P Value |
|---|---|---|---|---|
| Univariate LR models | ||||
| 122‐5p/151‐5p | ∆Cq | 56 | 1.098 | 0.0025 |
| 151a‐5p/192‐5p | ∆Cq | 56 | −1.643 | 0.0035 |
| Sex | Male vs. female | 56 | −1.609 | 0.0459 |
| Age | years | 56 | 0.0015 | 0.6731 |
| Extent of hepatic resection | Major vs. minor | 56 | 1.695 | 0.1261 |
| Tumor type | HCC vs. CCC | 56 | 0.223 | 0.8128 |
| CRCLM vs. CCC | −18.180 | 0.9934 | ||
| Steatosis (>10%) | Yes vs. no | 56 | −0.594 | 0.4493 |
| Liver cirrhosis | Yes vs. no | 56 | 0.669 | 0.4638 |
| PDR | % | 56 | 0.000 | 0.9938 |
| R15 | % | 56 | 0.067 | 0.2624 |
| SB | mg/dL | 56 | 0.001 | 0.9981 |
| AP | U/L | 56 | 0.006 | 0.0841 |
| AST | U/L | 56 | 0.017 | 0.0377 |
| ALT | U/L | 56 | 0.015 | 0.0941 |
| GGT | U/L | 56 | 0.004 | 0.0662 |
| Albumin | g/L | 56 | −0.170 | 0.0895 |
| Platelet count | ×103/µL | 56 | 0.003 | 0.6140 |
| PT | % | 56 | −0.016 | 0.3850 |
| Change of platelet count | POD 1 / pre‐OP | 56 | 1.198 | 0.2120 |
| Change of PT | POD 1 / pre‐OP | 56 | 0.421 | 0.8473 |
| Change of albumin | POD 1 / pre‐OP | 50* | −0.955 | 0.8769 |
| Multivariate LR models† | ||||
| Intercept | — | 56 | 6.107 | 0.3693 |
| 122‐5p/151‐5p | ∆Cq | 1.023 | 0.0081 | |
| Sex | Male vs. female | −1.523 | 0.2081 | |
| AP | U/L | −0.019 | 0.2762 | |
| AST | U/L | −0.003 | 0.8796 | |
| ALT | U/L | −0.016 | 0.5406 | |
| GGT | U/L | 0.007 | 0.2979 | |
| Albumin | g/L | −0.211 | 0.1733 | |
| Intercept | — | 56 | 37.646 | 0.0061 |
| 151a‐5p/192‐5p | ∆Cq | 6.622 | 0.0001 | |
| Sex | Male vs. female | −4.140 | 0.0188 | |
| AP | U/L | −0.025 | 0.3829 | |
| AST | U/L | −0.201 | 0.0233 | |
| ALT | U/L | 0.086 | 0.1018 | |
| GGT | U/L | 0.004 | 0.7193 | |
| Albumin | g/L | −1.090 | 0.0022 |
P = the P value from Wald test indicating significant influence in the LR model; n = number of patients included in the model with no missing values in any of the selected parameter.
Of note, albumin measurement on POD 1 was missing in 6 patients.
Only variables with P < 0.1 in the univariate LR model were included (indicated in italics), all VIFs between explanatory variables < 4; P values < 0.05 are indicated in bold.
Abbreviations: ΔCq, delta quantification cycle; AP, alkaline phosphatase; Coeff, coefficient of the respective variable in the LR model; CRCLM, colorectal cancer liver metastases; PDR, plasma disappearance rate; PT, prothrombin time; R15, retention rate at 15 minutes; SB, serum bilirubin; VIF, variance inflation factor.
Further, the diagnostic performance of the two cutoffs were illustrated using sensitivity (SN), SP, positive predictive value (PPV), negative predictive value (NPV), and the odds ratio (OR). The low‐stringency cutoff (>0.59) yielded PPV and NPV values of 0.70 and 0.89 (Fig. 3B), respectively, whereas the stringent cutoff (>0.68) resulted in a PPV of 0.83 with an NPV of 0.85 (Fig. 3B), indicating that 83% of patients who tested positive suffered from postoperative LD, whereas 85% of patients who tested negative did not suffer from postoperative LD. The ORs for an adverse event were 18.66 (P < 0.0001) and 19.7 (P < 0.0001) for the two cutoffs, respectively. ROC curve analysis was performed for the miRNA model and compared with ROC curves of standard liver function parameters (Fig. 3B). We observed an AUC of 0.77 for the miRNA model, which exceeded the AUC of other parameters, including indocyanine green (ICG) plasma disappearance and retention rates as well as standard blood parameters like alanine transaminase (ALT), aspartate transaminase (AST), and gamma‐glutamyl transpeptidase (GGT). Further, we compared SN, SP, NPV, and PPV in a subset of patients who also had preoperative volumetric analyses (N = 33). miRNA pairs were found to identify patients subsequently suffering from postoperative LD with a higher predictive potential as compared with ICG clearance, but also exceeded the predictive potential of volumetric analyses (cutoff of 30% future liver remnant), as illustrated in Supporting Table S4. Finally, ORs for other adverse postoperative outcomes were analyzed for both cut‐off models. ORs for severe morbidity reached significance for both cutoffs (Fig. 3C). Although OR for mortality was found to be significant for the low‐stringency cutoff, the high‐stringency cutoff was not significant but showed a comparable tendency (Fig. 3D). Further, patients fulfilling our cutoffs were found to stay significantly longer on the intensive care unit (ICU) and remained hospitalized for a prolonged time (Fig. 3E,F).
miRNA Pairs Represent Significant Predictors of Postoperative Outcome in the Respective Tumor Types
As the underlying tumor entity is a known confounding factor for the analysis of postoperative outcomes in this patient cohort, further subgroup analyses were computed. Accordingly, patients were divided into the three largest subgroups—mCRC (n = 56), HCC (n = 46), and CCC (n = 41)—and were analyzed separately (Fig. 4A‐C). Indeed, the preoperative miRNA signature was found to differ significantly between patients with and without postoperative LD for each tumor type. Further, ROC analysis revealed comparably good predictive potentials. Lastly, evaluation of the proposed cutoffs for prediction of LD showed a strong predictive potential, irrespective of tumor entity. (Fig. 4A‐C).
Figure 4.
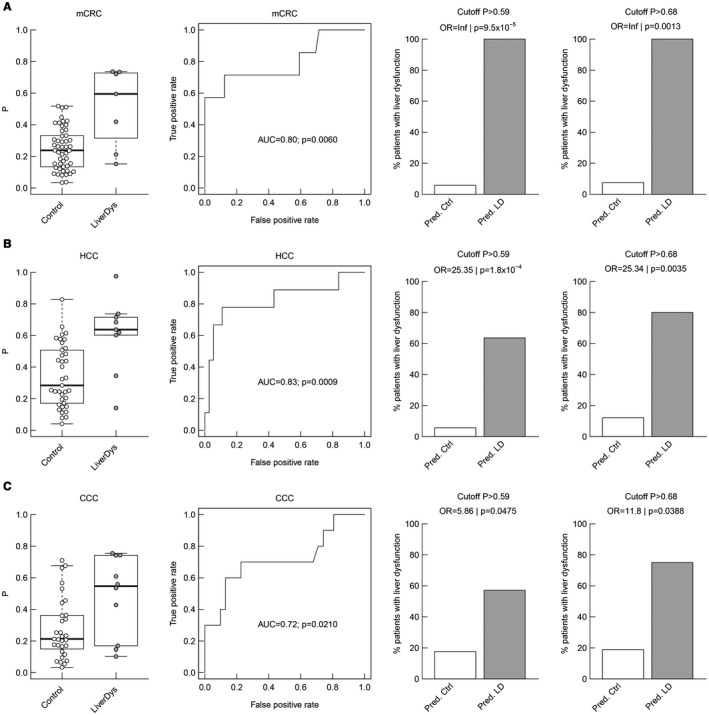
Tumor‐specific subgroup analyses of miRNA predictive potential for postoperative LD. Distribution of the combined ratio (miR151a‐5p/192‐5p//miR122‐5p/151a‐5p), ROC curves for an LR model (the performance is described by the AUC and whether the classification deviates significantly from the random assignment [AUC = 0.5] is indicated by the P value), and the percentage of true postoperative LD on predicted controls and predicted LD with respect to both model‐defined cutoffs (P > 0.59 and P > 0.68) are illustrated for the subgroups of patients with mCRC (A), HCC (B), and CCC (C), respectively.
Discovered miRNA Ratios Allow Dynamical Monitoring of Liver Function Recovery After Liver Resection and Might Enable Identification of an Optimized Time Point for Operation
Next, we aimed to determine dynamical changes of miRNA pairs after operation to evaluate their association with liver function. Accordingly, miRNA ratios were assessed preoperatively (pre‐OP) as well as on POD 1 and POD 5 in a matched cohort of patients with regular major liver resection (n = 7) and patients undergoing the ALPPS procedure (n = 8, see details on the procedure in Fig. 5A and below). Basic characteristics are illustrated in Supporting Table S2. Although miRNA pairs as well as the combined LD probability changed significantly after liver resection (POD 1 vs. pre‐OP) in all patients to a comparable level, most of them recovered until POD 5 in parallel with a regular liver function recovery.
Figure 5.
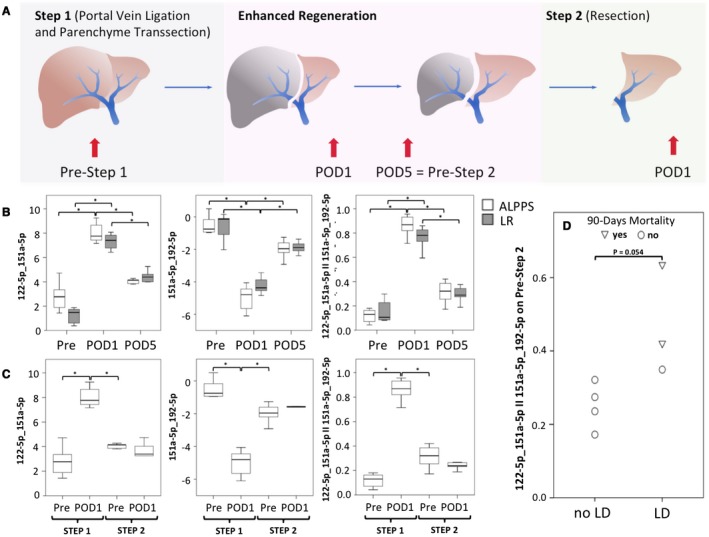
miRNA pairs follow liver function recovery after partial hepatectomy and predict postoperative LD after the second step of ALPPS. Panel (A) illustrates the study design of this additional exploratory study as well as summarizes the procedural algorithm of ALPPS. The ALPPS procedure was described by Schnitzbauer et al. and has been developed to allow for rapid liver regeneration in borderline‐resectable patients with an insufficient liver remnant.50 Briefly, during the first step of the ALPPS procedure, the portal vein branch, feeding the tumor‐bearing liver lobe, is selectively ligated, whereas the arterial as well as bile structures are preserved and the liver parenchyma is further transected during this initial step of operation. This procedure leads to an improved liver regeneration within the time frame of days. Still, after this substantial gain of liver regeneration, a second surgical procedure has to be performed in which the ligated remaining liver lobes need to be removed. Perioperative dynamics of miRNAs were evaluated in a group of 7 patients with regular partial hepatectomy and 8 patients undergoing ALPPS (details are listed in Supporting Table S2) for which longitudinal measurement of miRNAs could be performed on the basis of repeatedly collected plasma samples. Time points of blood collection are given in (A). Perioperative dynamics of miRNA pairs as well as combined pairs are illustrated in (B) (POD 1 vs. pre‐OP, 151a‐5p_192‐5p, ALPPS, P = 0.012, regular major liver resection, P = 0.018; 122‐5p_151a‐5p, ALPPS, P = 0.012, regular major liver resection, P = 0.018, 122‐5p_151a‐5p II 151a‐5p_192‐5p, ALPPS, P = 0.012, regular major liver resection, P = 0.018; POD 5 vs. POD 1, 151a‐5p_192‐5p, ALPPS, P = 0.018, regular major liver resection, P = 0.018; 122‐5p_151a‐5p, ALPPS, P = 0.018, regular major liver resection, P = 0.018, 122‐5p_151a‐5p II 151a‐5p_192‐5p, ALPPS, P = 0.018, regular major liver resection, P = 0.018). As during the second step of ALPPS, only the atrophic liver lobe is removed; we further analyzed miRNA pairs after the second step of ALPPS as illustrated in (C), showing an almost vanished increase in miRNA ratios after this second operation. Ultimately, (D) illustrates the predictive potential of the combined miRNA pairs before the second step of ALPPS as stratified according to postoperative LD and mortality after the removal of the atrophic lobe. *P < 0.05, **P < 0.005.
As timing of the second stage of the ALPPS procedure is still a challenge for clinicians, the usefulness of miRNA pairs for this task was evaluated. Interestingly, patients undergoing the ALPPS procedure were found to display unaltered miRNA ratios after the second step of the procedure (regeneration grossly completed), whereas they behaved similarly to patients undergoing regular liver resection after the first step of the procedure (Fig. 5C). Of note, all patients who developed postoperative LD after the second step of the ALPPS procedure showed a worsened pattern of miRNA profile when compared with the remaining patients (P = 0.054, Fig. 5D). More importantly, the 2 patients with highest detected miRNA ratios before the second operation died after the second step of the procedure (Fig. 5D), indicating that the miRNA ratios are indeed predictive of functional liver recovery.
Discussion
Using NGS as an unbiased systematic approach, we were able to detect 554 miRNAs in the plasma of patients before liver resection. Of those, a signature was identified—consisting of three miRNAs 151a‐5p, 192‐5p, and 122‐5p—that specifically detected, before the operation, the patients who developed postoperative LD. In particular, an LR model of the two miRNA ratios 151a‐5p/192‐5p and 122‐5p/151a‐5p reliably predicted postoperative LD, severe morbidity, prolonged ICU as well as hospital stay, and even mortality before the operation with a remarkable accuracy. Of note, multivariate LR of the entire cohort found both miRNA ratios to be independent predictors of postoperative LD (study design summarized in Fig. 6). Given the clinical relevance of predicting potentially fatal postoperative clinical outcomes after liver resection, our data demonstrate the clinical utility of an miRNA‐based biomarker to support the selection of patients undergoing partial hepatectomy.
Figure 6.

Study overview. In the discovery phase, plasma of 48 patients (21 with LD matched with 27 patients without LD after liver resection) was analyzed before operation to determine if miRNA signatures can serve as predictive markers. Using RNA sequencing, 19 microRNAs of interest were discovered and subjected to qPCR confirmation. In‐depth analysis predicted a marker potential for the miRNA pairs miR‐122‐5p/miR151a and miR‐151a/miR192‐5p (silver box). In the validation phase, the two miRNA pairs were reevaluated in an independent cohort consisting of 98 patients (8 with LD and 90 without LD after liver resection) (golden box). For performance evaluation, the combined data set was used to evaluate SN, SP, PPV, NPV, and ORs and compare them to previously used clinical markers (platinum box).
Specifically in early stages of liver disease, clinical evaluation and quantification of liver function remains challenging. However, even slightly diminished liver function can be of major relevance if certain stressors, such as extensive liver resection, come into play. Although several tests have been developed, only a few have found their way into routine clinical application. Major drawbacks of available predictors are availability, high costs, and invasiveness.19 Although hepatic venous pressure gradient predicts postoperative clinical outcomes in HCC patients,20, 21, 22 it remains reserved for high‐risk patients because of its invasiveness. Other less invasive and well‐established markers to assess liver function rely on dynamic functional assessment of the liver. In this context, multiple groups have documented that ICG clearance is vital to predict postoperative LD and morbidity.23 However, we here document that miRNA signatures outperform ICG in terms of diagnostic accuracy by far. Furthermore, miRNA signatures even exceeded volumetric analyses in terms of prediction of postoperative LD (see Supporting Table S4).
Despite using an unbiased approach, the miRNAs predicting LD all have very distinct roles in liver (patho)physiology and therefore could potentially serve not only as predictive markers but also as therapeutic targets. Although various miRNAs regulate liver functions in health and disease,24 miRNA‐122 is the most prominent one, as it constitutes 70% of miRNA copies in hepatocytes.25, 26 Altered miRNA‐122 expression has been implicated in a variety of liver diseases, including viral hepatitis, alcohol‐induced liver disease, nonalcoholic fatty liver disease, and HCC,27, 28, 29, 30, 31 reflecting its critical and pleiotropic role in regulating liver function. Specifically, miRNA‐122 negatively controls the expression of genes involved in cell cycle–associated processes, hepatic metabolism, and extracellular matrix receptor interactions,32, 33, 34, 35 all of which may contribute to liver regeneration. Furthermore, circulating levels of miRNA‐122 are considered as an early and sensitive biomarker for liver injury,36, 37, 38 and pharmacological inhibition or genetic depletion of miRNA‐122 affects hepatocyte differentiation and spontaneous progression to fibrosis and carcinogenesis.27, 39
Aside from miRNA‐122, miRNA‐192 was also enriched in liver tissue. Further, blood levels have been reported to correlate with the histopathology of liver degeneration and HCC.31 However, aside from its direct marker potential, miRNA‐192 might influence liver regeneration through its antiangiogenic properties, as it has been shown on delivery in various tumor models by targeting two key transcription factors, early growth response 1 and homeobox B9.40 These transcription factors further promote the expression of several angiogenic factors.40, 41, 42, 43, 44, 45
Ultimately, miRNA‐151 has also recently been identified as an oncogenic miRNA in HCC, in which increased levels in patients correlated with poor prognosis. It has been suggested that miRNA‐151 promotes metastasis in liver cancer in synergy with its host gene, focal adhesion kinase, through down‐regulating the metastasis suppressor Rho guanosine diphosphate dissociation inhibitor alpha and thereby inducing Rac family small, cell division cycle 42, and Rho guanosine triphosphatases.46 Furthermore, in lung cancer, miRNA‐151a has been reported to target E‐cadherin and promote cell proliferation and motility of cancer cells, while not affecting normal lung endothelial growth,47 suggesting a pro‐proliferative effect of miRNA‐151a. In summary, the existing data on physiologic interactions of the selected miRNAs suggest an additional deleterious effect on liver regeneration, next to its highly sensitive prognostic potential for LD. If miRNAs are setting the scene for sufficient liver regeneration, they might also represent an attractive therapeutic target. However, this aspect will need more detailed evaluation.
It should be pointed out that we used miRNA pairs to allow for self‐normalization of circulating plasma miRNAs. This is important because robust endogenous references, which are commonly applied for normalization of cell‐based or tissue‐based expression data, are not available for plasma. Therefore, Sheinerman et al. introduced the concept of “self‐normalizing” miRNA pairs (i.e., the combination of two miRNAs) to improve both the robustness and performance of miRNA biomarkers.48, 49 This self‐normalization of miRNAs was essential to allow for the precise prediction of postoperative outcome after liver resection, which was not achieved by individual miRNAs.
Of note, perioperative time course of miRNA pairs was also evaluated in an additional exploratory study. As an acute reaction to the operative trauma (on POD 1), miRNA pairs worsen excessively in all patients. However, as liver function recovers after operation, miRNA pairs seem to closely follow and normalized in most patients until POD 5. In this context, the data of the ALPPS model are of specific interest. miRNA pairs significantly worsened after the first step of this operation, which comprises the main part of tissue destruction. Intriguingly however, there was almost no dynamic in the level of the evaluated miRNA ratios after accomplishment of the second step, when the atrophic liver lobe was removed. This parallels the regeneratory pattern of the ALPPS procedure, which consists of augmented induction after step 1 and only little alternation after the second operation. Hence, the analyzed miRNA pairs seem to reflect liver function and its recovery. Notably, these data show a fast adaptation of miRNA profiles in accordance with the clinical setting, indicating that the proposed miRNA‐based marker represents a dynamic tool for estimation of hepatic reserve. However, these data are only of exploratory nature and have to be validated in larger cohorts. The analyses of miRNA pairs in the ALPPS model allowed for generation of the presumably most interesting and clinically relevant data of this manuscript. In particular, we observed that patients who did not recover after the first step of ALPPS in terms of combined miRNA pairs were indeed those who developed very poorly after the second step of ALPPS. Indeed, all 3 patients who suffered from LD after the second step had the highest miRNA pair values before the second step. More importantly, the 2 patients who died as a result of small‐for‐size syndrome showed the highest values of miRNA ratios. These exploratory data are highly indicative of the crucial relevance of miRNA in dynamically monitoring liver function and in potentially determining the optimal time point for liver resection. This could also be relevant for patients treated with portal vein embolization/ligation or following extensive neoadjuvant chemotherapy.
Taken together, the data indicate that this marker could help to provide an improved strategy to identify patients who will not benefit from operation or may even suffer from potentially lethal complications. Thereby, our data should help to tailor surgical strategies to the specific risk profile of individual patients in an easy, cost‐effective, and noninvasive manner. This could pave the way to personalizing liver operation in patients with liver tumors, thereby increasing therapeutic effectiveness and the quality of a patient's life and dramatically reducing health care costs.
Supporting information
Supported by the Association of Research on the Biology of Liver Tumors, Vienna, Austria. Further, the Austrian Research Promotion Agency has supported this work (855576).
Potential conflict of interest: Dr. Skalicky is employed by TAmiRNA GmbH. Dr. M. Hackl is employed by and owns stock in TAmiRNA GmbH.
Contributor Information
Patrick Starlinger, Email: patrick.starlinger@meduniwien.ac.at.
Alice Assinger, Email: alice.assinger@meduniwien.ac.at.
References
- 1. Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473‐485. [DOI] [PubMed] [Google Scholar]
- 2. Lafaro K, Buettner S, Maqsood H, Wagner D, Bagante F, Spolverato G, et al. Defining post hepatectomy liver insufficiency: where do we stand? J Gastrointest Surg 2015;19:2079‐2092. [DOI] [PubMed] [Google Scholar]
- 3. Qadan M, Garden OJ, Corvera CU, Visser BC. Management of postoperative hepatic failure. J Am Coll Surg 2016;222:195‐208. [DOI] [PubMed] [Google Scholar]
- 4. Hackl M, Heilmeier U, Weilner S, Grillari J. Circulating microRNAs as novel biomarkers for bone diseases—complex signatures for multifactorial diseases? Mol Cell Endocrinol 2016;432:83‐95. [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 6. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92‐105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lecellier CH, Dunoyer P, Arar K, Lehmann‐Che J, Eyquem S, Himber C, et al. A cellular microRNA mediates antiviral defense in human cells. Science 2005;308:557‐560. [DOI] [PubMed] [Google Scholar]
- 8. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova‐Agadjanyan EL, et al. Circulating microRNAs as stable blood‐based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513‐10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress‐responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A 2006;103:18255‐18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, et al. miR‐Test: a blood test for lung cancer early detection. J Natl Cancer Inst 2015;107:djv063 10.1093/jnci/djv063 https://academic.oup.com/jnci. [DOI] [PubMed] [Google Scholar]
- 11. Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, et al. Clinical utility of a plasma‐based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol 2016;231:25‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahbari NN, Garden OJ, Padbury R, Brooke‐Smith M, Crawford M, Adam R, et al. Posthepatectomy liver failure: a definition and grading by the International Study Group of Liver Surgery (ISGLS). Surgery 2011;149:713‐724. [DOI] [PubMed] [Google Scholar]
- 14. Dindo D, Demartines N, Clavien PA. Classification of surgical complications: a new proposal with evaluation in a cohort of 6336 patients and results of a survey. Ann Surg 2004;240:205‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schiergens TS, Dorsch M, Mittermeier L, Brand K, Kuchenhoff H, Lee SM, et al. Thirty‐day mortality leads to underestimation of postoperative death after liver resection: a novel method to define the acute postoperative period. Surgery 2015;158:1530‐1537. [DOI] [PubMed] [Google Scholar]
- 16. Starlinger P, Alidzanovic L, Schauer D, Brugger P, Sommerfeldt S, Kuehrer I, et al. Platelet‐stored angiogenesis factors: clinical monitoring is prone to artifacts. Dis Markers 2011;31:55‐65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kocijan R, Muschitz C, Geiger E, Skalicky S, Baierl A, Dormann R, et al. Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab 2016;101:4125‐4134. [DOI] [PubMed] [Google Scholar]
- 18. Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013;59:S1‐S6. [DOI] [PubMed] [Google Scholar]
- 19. La Mura V, Nicolini A, Tosetti G, Primignani M. Cirrhosis and portal hypertension: the importance of risk stratification, the role of hepatic venous pressure gradient measurement. World J Hepatol 2015;7:688‐695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stremitzer S, Tamandl D, Kaczirek K, Maresch J, Abbasov B, Payer BA, et al. Value of hepatic venous pressure gradient measurement before liver resection for hepatocellular carcinoma. Br J Surg 2011;98:1752‐1758. [DOI] [PubMed] [Google Scholar]
- 21. Boleslawski E, Petrovai G, Truant S, Dharancy S, Duhamel A, Salleron J, et al. Hepatic venous pressure gradient in the assessment of portal hypertension before liver resection in patients with cirrhosis. Br J Surg 2012;99:855‐863. [DOI] [PubMed] [Google Scholar]
- 22. Hidaka M, Takatsuki M, Soyama A, Tanaka T, Muraoka I, Hara T, et al. Intraoperative portal venous pressure and long‐term outcome after curative resection for hepatocellular carcinoma. Br J Surg 2012;99:1284‐1289. [DOI] [PubMed] [Google Scholar]
- 23. Haegele S, Reiter S, Wanek D, Offensperger F, Pereyra D, Stremitzer S, et al. Perioperative non‐invasive indocyanine green‐clearance testing to predict postoperative outcome after liver resection. PLoS One 2016;11:e0165481 10.1371/journal.pone.0165481. https://journals.plos.org/plosone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 2013;10:542‐552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR‐122, a mammalian liver‐specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT‐1. RNA Biol 2004;1:106‐113. [DOI] [PubMed] [Google Scholar]
- 26. Lagos‐Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue‐specific microRNAs from mouse. Curr Biol 2002;12:735‐739. [DOI] [PubMed] [Google Scholar]
- 27. Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti‐inflammatory, and anti‐tumorigenic functions of miR‐122 in liver. J Clin Invest 2012;122:2871‐2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta‐Vellve A, Tornai D, et al. MicroRNA 122, regulated by grlh2, protects livers of mice and patients from ethanol‐induced liver disease. Gastroenterology 2018;154:238‐252.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA‐122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012;122:2884‐2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA‐122 as a biomarker for viral‐, alcohol‐, and chemical‐related hepatic diseases. Clin Chem 2010;56:1830‐1838. [DOI] [PubMed] [Google Scholar]
- 31. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus‐related hepatocellular carcinoma. J Clin Oncol 2011;29:4781‐4788. [DOI] [PubMed] [Google Scholar]
- 32. He B, He Y, Shi W, Gong S, Chen X, Xiao J, et al. Bioinformatics analysis of gene expression alterations in microRNA122 knockout mice with hepatocellular carcinoma. Mol Med Rep 2017;15:3681‐3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR‐122 targets an anti‐apoptotic gene, Bcl‐w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun 2008;375:315‐320. [DOI] [PubMed] [Google Scholar]
- 34. Rivkin M, Simerzin A, Zorde‐Khvalevsky E, Chai C, Yuval JB, Rosenberg N, et al. Inflammation‐induced expression and secretion of microRNA 122 leads to reduced blood levels of kidney‐derived erythropoietin and anemia. Gastroenterology 2016;151:999‐1010.e3. [DOI] [PubMed] [Google Scholar]
- 35. Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, et al. MicroRNA‐122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/beta‐catenin pathway. Liver Int 2012;32:752‐760. [DOI] [PubMed] [Google Scholar]
- 36. Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen‐induced acute liver injury at first presentation to hospital. Hepatology 2013;58:777‐787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dubin PH, Yuan H, Devine RK, Hynan LS, Jain MK, Lee WM. Micro‐RNA‐122 levels in acute liver failure and chronic hepatitis C. J Med Virol 2014;86:1507‐1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ward J, Kanchagar C, Veksler‐Lublinsky I, Lee RC, McGill MR, Jaeschke H, et al. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci U S A 2014;111:12169‐12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR‐122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87‐98. [DOI] [PubMed] [Google Scholar]
- 40. Wu SY, Rupaimoole R, Shen F, Pradeep S, Pecot CV, Ivan C, et al. A miR‐192‐EGR1‐HOXB9 regulatory network controls the angiogenic switch in cancer. Nat Commun 2016;7:11169 10.1038/ncomms11169. https://www.nature.com/ncomms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kundumani‐Sridharan V, Niu J, Wang D, Van Quyen D, Zhang Q, Singh NK, et al. 15(S)‐hydroxyeicosatetraenoic acid‐induced angiogenesis requires Src‐mediated Egr‐1‐dependent rapid induction of FGF‐2 expression. Blood 2010;115:2105‐2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee KH, Kim JR. Hepatocyte growth factor induced up‐regulations of VEGF through Egr‐1 in hepatocellular carcinoma cells. Clin Exp Metastasis 2009;26:685‐692. [DOI] [PubMed] [Google Scholar]
- 43. Shimoyamada H, Yazawa T, Sato H, Okudela K, Ishii J, Sakaeda M, et al. Early growth response‐1 induces and enhances vascular endothelial growth factor‐A expression in lung cancer cells. Am J Pathol 2010;177:70‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain‐containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J 2012;279:3715‐3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singha B, Gatla HR, Manna S, Chang TP, Sanacora S, Poltoratsky V, et al. Proteasome inhibition increases recruitment of IkappaB kinase beta (IKKbeta), S536P–p65, and transcription factor EGR1 to interleukin‐8 (IL‐8) promoter, resulting in increased IL‐8 production in ovarian cancer cells. J Biol Chem 2014;289:2687‐2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, et al. Gain of miR‐151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol 2010;12:390‐399. [DOI] [PubMed] [Google Scholar]
- 47. Daugaard I, Sanders KJ, Idica A, Vittayarukskul K, Hamdorf M, Krog JD, et al. miR‐151a induces partial EMT by regulating E‐cadherin in NSCLC cells. Oncogenesis 2017;6:e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sheinerman KS, Tsivinsky VG, Abdullah L, Crawford F, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging (Albany NY) 2013;5:925‐938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sheinerman KS, Tsivinsky VG, Umansky SR. Analysis of organ‐enriched microRNAs in plasma as an approach to development of Universal Screening Test: feasibility study. J Transl Med 2013;11:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schnitzbauer AA, Lang SA, Goessmann H, Nadalin S, Baumgart J, Farkas SA, et al. Right portal vein ligation combined with in situ splitting induces rapid left lateral liver lobe hypertrophy enabling 2‐staged extended right hepatic resection in small‐for‐size settings. Ann Surg 2012;255:405‐414. 10.1186/1479-5876-11-304. https://journals.lww.com/annalsofsurgery/pages/default.aspx. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


