Abstract
Autophagy, a lysosomal degradative pathway in response to nutrient limitation, plays an important regulatory role in lipid homeostasis upon energy demands. Here, we demonstrated that the endoplasmic reticulum–tethered, stress-sensing transcription factor cAMP-responsive element-binding protein, hepatic-specific (CREBH) functions as a major transcriptional regulator of hepatic autophagy and lysosomal biogenesis in response to nutritional or circadian signals. CREBH deficiency led to decreased hepatic autophagic activities and increased hepatic lipid accumulation upon starvation. Under unfed or during energy-demanding phases of the circadian cycle, CREBH is activated to drive expression of the genes encoding the key enzymes or regulators in autophagosome formation or autophagic process, including microtubule-associated protein 1B-light chain 3, autophagy-related protein (ATG)7, ATG2b, and autophagosome formation Unc-51 like kinase 1, and the genes encoding functions in lysosomal biogenesis and homeostasis. Upon nutrient starvation, CREBH regulates and interacts with peroxisome proliferator–activated receptor α (PPARα) and PPARγ coactivator 1α to synergistically drive expression of the key autophagy genes and transcription factor EB, a master regulator of lysosomal biogenesis. Furthermore, CREBH regulates rhythmic expression of the key autophagy genes in the liver in a circadian-dependent manner. In summary, we identified CREBH as a key transcriptional regulator of hepatic autophagy and lysosomal biogenesis for the purpose of maintaining hepatic lipid homeostasis under nutritional stress or circadian oscillation.—Kim, H., Williams, D., Qiu, Y., Song, Z., Yang, Z., Kimler, V., Goldberg, A., Zhang, R., Yang, Z., Chen, X., Wang, L., Fang, D., Lin, J. D., Zhang, K. Regulation of hepatic autophagy by stress-sensing transcription factor CREBH.
Keywords: stress response, transcriptional regulation, hepatic lipid homeostasis
Autophagy, a cellular catabolic process in lysosome, degrades cytoplasmic components, abnormally accumulated proteins, and damaged organelles. Autophagy is started with the formation of phagophore, which expands and forms autophagosome to engulf intracellular cargo, such as cytoplasmic proteins, dysfunctional organelles, and sequestrate cargo proteins (1–3). Autophagosome subsequently fuses with lysosome that promotes the degradation of autophagosomal substrates by lysosomal acid proteases (4). Functionally, autophagy is known to play an important role in maintaining cellular energy homeostasis and mobilizing nutrients, including carbohydrates, lipids, and iron, under starvation or chronic metabolic stress conditions (5–7). Starvation challenge induces transcriptional activities to control the major autophagy steps including autophagosome formation, autophagosome-lysosome fusion, and substrate degradation (8–11). Lipid-specific autophagy, known as lipophagy, was identified in hepatocytes as a major pathway to degrade lipids, and thus plays an important role in hepatic lipid homeostasis (5, 6, 12, 13).
cAMP-responsive element-binding protein, hepatic-specific (CREBH) is an endoplasmic reticulum (ER)-tethered basic-region leucine zipper transcription factor of the cAMP-responsive element-binding protein (CREB)/activating transcription factor family (14, 15). CREBH is selectively expressed in the liver and small intestines in mice (16). Activation of CREBH is mediated through a process called regulated intramembrane proteolysis, in which CREBH transits from the ER to Golgi, where it is cleaved by site-1 and -2 proteases to release the activated CREBH fragment (14). Activated CREBH functions as a potent transcription factor to drive expression of genes encoding functions involved in hepatic acute phase response as well as lipid and glucose metabolism (14, 17–21). Activation of hepatic CREBH is regulated by a variety of stress signals, including ER stress, inflammatory challenge, energy demand, and circadian clock. We revealed that CREBH plays key roles in energy homeostasis by: 1) regulating expression of genes involved in hepatic lipolysis, fatty acid (FA) oxidation, and lipogenesis (17); 2) activating fibroblast growth factor 21 (FGF21) through interaction with peroxisome proliferator–activated receptor α (PPARα) (18, 22); 3) functioning as a circadian metabolic regulator (19); and 4) promoting gluconeogenesis and glycogenolysis by activating expression of the genes encoding phosphoenolpyruvate arboxykinase 1, glucose 6–phosphatase, and glycogen phosphorylase, liver form (20). CREBH deficiency led to profound nonalcoholic steatohepatitis and hyperlipidemia phenotypes in mice fed a high-fat diet (17). In humans, patients with hypertriglyceridemia display high-rate nonsense mutations or rare genetic variant accumulation in the CREBH gene (23–25).
In this study, we demonstrate that CREBH acts as a major driver of energy supplies by promoting the hepatic autophagy process upon energy demands or under the circadian clock. As a transcription factor, CREBH activates expression of the genes encoding the key components of autophagy and lysosome biogenesis in mouse livers or hepatocytes in response to nutrient starvation or across the circadian clock. The findings from this study provide important novel insights into the regulation of both physiologic autophagy and energy homeostasis in the liver.
MATERIALS AND METHODS
Reagents
Glucagon, Oil Red O, and mouse monoclonal anti–β-actin or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were obtained from MilliporeSigma (Burlington, MA, USA). Goat polyclonal anti–autophagy-related protein (Atg) 2B, mouse monoclonal anti-lamin B1, rabbit polyclonal anti–transcription factor EB (TFEB), and anti-CREB antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Rabbit monoclonal anti-Atg7 and monoclonal anti–autophagosome formation Unc-51 like kinase 1 (Ulk1) antibodies were from Cell Signaling Technology (Danvers, MA, USA). Rabbit polyclonal anti–microtubule-associated protein 1B-light chain 3 (LC3b) antibody was from Novus Biologicals (Centennial, CO, USA). Mouse monoclonal anti–sequestosome 1 (p62) antibody was from Abcam (Cambridge, MA, USA). Mouse monoclonal anti-PPARα antibody was from MilliporeSigma. Rabbit anti-CREBH pAb was raised by immunizing rabbits with a mouse CREBH protein fragment spanning N-terminal amino acids 75–250 of mouse CREBH protein as previously described in Zhang et al. (17). Anti-rabbit, anti-mouse, and anti-goat HRP-conjugated secondary antibodies were from Promega (Madison, WI, USA). SlowFade Gold Antifade Mountant with DAPI and Alexa Fluor 488 conjugate chicken anti-rabbit IgG (H + L) secondary antibody were from Thermo Fisher Scientific (Waltham, MA, USA). Bodipy lipid probe was obtained from Molecular Probes (Eugene, OR, USA).
Mouse experiments
CREBH-knockout (KO) mice in which exons 4–7 of the CrebH gene were deleted were previously described in Luebke-Wheeler et al. (16). CREBH-KO and wild-type (WT) control mice∼3 mo on a C57BL/6J background were used for the experiments. Brain and muscle ARNT-like 1 (Bmal1) flox/flox (fl/fl) mice were purchased from The Jackson Laboratory (007668; Bar Harbor, ME, USA) and crossed with Albumin‐Cre transgenic mice (003574) to obtain the liver‐specific Bmal1 KO (Bmal1-LKO) mouse as previously described in Zheng et al. (19). For unfed experiments, the mice were housed in cages containing Pure-o’Cel bedding (Andersons Lab Bedding, Maumee, OH, USA) bedding without food (with water provided) for 6, 12, or 24 h. For circadian studies, the mice were fed ad libitum and maintained in 12-h light/dark cycles. The mice were dissected every 4 h for a period of 24 h. For adenovirus injection experiments, recombinant adenovirus expressing the full-length human CREBH protein or the acetylation-deficient [Lys294Arg (K294R)] mutant protein was injected into CREBH-KO mice through the tail vein (22). Approximately 1 × 1010 plaque-forming units of adenovirus in 0.25 ml PBS was intravenously injected into a mouse with a body weight of ∼20 g. All the animal experiments were approved by the Wayne State University Institutional Animal Care and Use Committee and carried out under the institutional guidelines for ethical animal use.
Cell culture
Primary hepatocytes were isolated from WT or CREBH-KO mice as previously described in Zheng et al. (17). The primary hepatocytes were stimulated with 25 nM glucagon for indicted time point. For in vitro circadian synchronization of mouse primary hepatocytes, the cells were subjected to serum shock (50% horse serum) for 2 h, followed by replacing the shock medium with serum-free medium. Cell lysates were collected at 6-h intervals between 24 (circadian 0 h) and 48 h (circadian 24 h) postserum shock for Western blot analysis. Mouse hepatoma cell line Hepa1-6 was cultured in DMEM containing 10% fetal bovine serum and 100 U/ml penicillin/streptomycin.
Lipid staining
Mouse liver tissues were embedded with optimal cutting temperature compound and stored at −80°C. For Oil Red O staining, frozen sections from liver were fixed with 10% formalin, followed by incubation with Oil Red O solution (0.3% Oil Red O in 60% isopropanol) for 15 min. Slides were mounted in aqueous mounting medium. For boron dipyrromethene (BODIPY) staining of lipid droplet, the cells stimulated with 25 nM glucagon for indicated time were washed with PBS, fixed with 3% formaldehyde, and stained with working Bodipy 493/503 solution (final 1 μg/ml; Thermo Fisher Scientific). Cells were then mounted with Prolong gold antifade reagent containing DAPI (DAPI; Thermo Fisher Scientific).
Immunofluorescence
Frozen liver tissues were sectioned at 8-μm thickness on glass plates. The cryosections were air-dried and fixed with 4% formaldehyde. The cells grown on a glass coverslip were treated with 25 nM glucagon for the indicated time and washed with PBS and then fixed with 4% formaldehyde. The fixed cells were incubated with the primary LC3b or hepatocyte nuclear factor 4α antibody overnight at 4°C, followed by incubation with Alexa Fluor 488- or 594-conjugated anti-rabbit secondary antibody. Cells were then mounted with Prolong gold antifade reagent containing DAPI. The images were observed by using a Zeiss fluorescent microscopy (Carl Zeiss, Oberkochen, Germany).
Western blot and coimmunoprecipitation analysis
Equal amounts of denatured mouse liver protein lysates were run on SDS-polyacrylamide gels and then transferred to PVDF membranes. The blots were incubated with the primary antibody (1:1000 dilution) overnight at 4°C and then incubated with secondary antibody conjugated to horseradish peroxidase (1:10,000) for 1 h at room temperature. For coimmunoprecipitation, 300 μg of protein extracts were incubated with 20 µl of protein A agarose beads and 3 μg of CREBH antibody in total volume of 500 µl at 4°C overnight with rotation. After centrifugation, the supernatant containing nonbound protein was removed and 1× sample buffer was added. The eluted proteins were separated by SDS-PAGE for Western blot analysis.
Real-time quantitative PCR
Total RNA from liver was extracted using Trizol by following the manufacturer’s instructions, and cDNA was synthesized from 500 ng of total RNA with High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The abundance of mRNA was measured by real-time quantitative PCR (qPCR) analysis using the SYBR Green PCR Master Mix (Thermo Fisher Scientific). PCR was carried out with 96-well plate using Applied Biosystems 7500 Real-Time PCR Systems (Thermo Fisher Scientific). Mouse β-actin was used as a reference gene. The sequences of the primer sets are listed in Supplemental Table S1.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were carried out with mouse liver tissue or Huh7 cells as previously described in Kim et al. (18). Antibodies used for immunoprecipitation were CREBH and PPARα, and immunoprecipitated DNA was purified using DNA Clean and Concentration Kit (Zymo Research, Irvine, CA, USA). The purified DNA was amplified using primers listed in Supplemental Table S2 to analyze the binding of CREBH or PPARα to target promoter regions, and the PCR products were separated by electrophoresis on a 1% agarose gel and visualized by ethidium bromide staining.
EMSA
EMSAs were performed using a Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific). The 5′ biotin-labeled oligonucleotides were synthesized from Integrated DNA Technologies (Coralville, IA, USA), and sequences are listed in Supplemental Table S2. The protein extracts from hepatic extract or in vitro–translated proteins were incubated with an annealed probe for 30 min at room temperature. The DNA-protein complexes were resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel and detected on X-ray film.
Transmission electron microscopy
Mouse tissues were initially fixed by cardiac perfusion using 2% paraformaldehyde and 2.5% glutaraldehyde in Millonig’s phosphate buffer. Livers were immediately removed, dissected into small cubes, immersed in the same solution, and stored at 4°C overnight. Liver tissue was osmicated, en bloc stained with uranyl acetate, dehydrated with ethanol, infiltrated with Polybed 812, and polymerized overnight at 60°C. Thin sections were poststained with uranyl acetate and lead citrate, visualized on a FEI Morgagni transmission electron microscope (TEM; Thermo Fischer Scientific), and imaged using a side-mount Hamamatsu Orca-HR digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) and AMT Image Capture Engine V601 software (Advanced Microscopy Techniques, Woburn, MA, USA). To estimate the autophagosome content, overlapping images were acquired from randomly selected cells at ×8900 magnification. Montaged images were inspected and scored for the presence of lipid droplets, lipophagic vesicles, and nonlipid autophagic vesicles, using the ImageJ [National Institutes of Health (NIH), Bethesda, MD, USA] software cell counter plugin. The autophagosome number or size was examined manually, and the cytoplasmic area is measured using ImageJ.
Statistical analysis
Experimental results are shown as means ± sem (for variation between animals or experiments). The mean values for biochemical data from the experimental groups were compared by a paired or unpaired, 2-tailed Student’s t test. When more than 2 treatment groups were compared, 1-way ANOVA followed by least significant difference post hoc testing was used. Statistical tests with P ≤ 0.05 were considered significant.
RESULTS
CREBH deficiency increases unfed-induced hepatic lipid accumulation
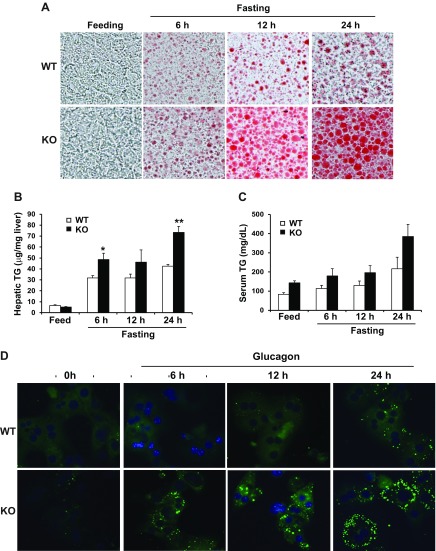
To characterize the functional involvement of CREBH in hepatic lipid homeostasis, we did not feed CREBH-KO and WT control mice for 6, 12, and 24 h, respectively. Oil Red O staining of lipid droplets with the liver tissue sections of CREBH-KO and WT mice showed that CREBH-KO mouse livers displayed significantly increased lipid accumulation in a time-dependent manner, compared to WT mice, upon unfed challenge (Fig. 1A). However, under fed conditions, there was no detectable hepatic lipid accumulation in CREBH-KO or WT mice. These results suggested that CREBH activity prevents exacerbation of unfed-induced hepatic steatosis and that hepatic steatosis caused by CREBH deficiency depends on unfed challenge. Consistent with the results of hepatic lipid staining, quantitative enzymatic analysis confirmed a significant increase in hepatic triglyceride (TG) levels in CREBH-KO mice after being unfed (Fig. 1B). The CREBH-KO mice exhibited increased levels of serum TG compared to the control mice under the unfed conditions (Fig. 1C).
Figure 1.
CREBH deficiency attenuates hepatic lipid degradation and elevates hepatic lipid accumulation in mice under unfed conditions. A) CREBH-KO and WT control mice were fed normal chow or were unfed for 6, 12, and 24 h, respectively. Hepatic lipids were visualized by Oil Red O staining with frozen liver tissue sections. B, C) Enzymatic analysis of hepatic (B) and serum (C) TG levels in CREBH-KO and WT mice under feeding or after a 6, 12, or 24 h nonfeeding. Each bar denotes means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01. Student’s t test was applied. D) Primary hepatocytes isolated from CREBH-KO and WT mice were incubated with Glucagon (25 nM) for indicated time intervals. Hepatic lipids in CREBH-KO and WT mouse primary hepatocytes were stained with BODIPY and visualized by fluorescence microscopy. Cells were mounted with mounting solution containing DAPI for nuclei (blue). Original magnification: ×200 (A); ×400 (D).
To validate the function of CREBH in preventing unfed-induced hepatic lipid accumulation, we examined cytosolic lipids in CREBH-KO and WT mouse primary hepatocytes stimulated with glucagon, a hormone that controls metabolic adaptation in the liver upon starvation (22). Although the control primary hepatocytes showed modest hepatic lipid accumulation, the CREBH-KO primary hepatocytes displayed a time-dependent increase in lipid droplet accumulation in response to glucagon treatments for 6, 12, and 12 h (Fig. 1D). In particular, the CREBH-KO hepatocytes exhibited a high degree of macrosteatosis, which was characterized by augmented cytosolic lipid accumulation, as shown by BODIPY fluorescence staining (Fig. 1D). These results confirmed the functional involvement of CREBH in mitigating hepatic lipid droplet accumulation triggered by energy demands.
CREBH is required for the induction of hepatic autophagy triggered by starvation
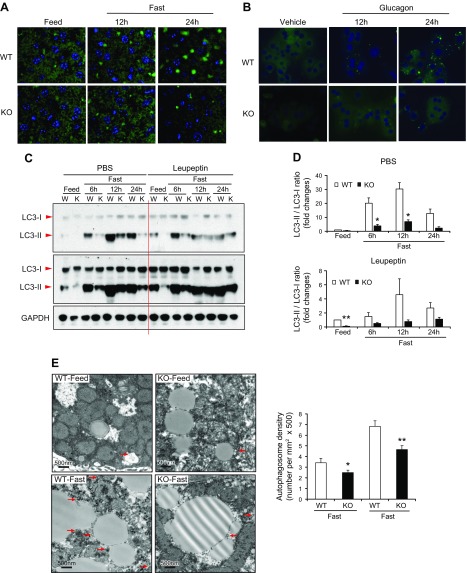
In the liver, hepatic autophagy (or lipophagy) is a cellular process that is activated in response to nutrient limitation, resulting in the degradation of cytoplasmic components, particularly lipid droplets, to provide a source of nutrients and metabolic fuel (26). Given massive cytosolic lipid accumulation observed in unfed-challenged CREBH-null livers or glucagon-stimulated primary hepatocyte, we investigated potential involvement of CREBH in hepatic autophagy in response to starvation. Autophagy can be measured by induction and conversion of LC3b, a central protein in the autophagy pathway that functions in autophagosome membrane expansion and fusion events (27). To test whether CREBH is required for unfed-induced autophagic activity, we examined induction of LC3b in the livers of CREBH-KO and WT control mice upon being unfed. As shown by immunofluorescent staining, the presence of LC3b protein in hepatocytes of WT mice was gradually increased in response to being unfed for 12 and 24 h (Fig. 2A). In contrast, unfed-induced presence of LC3b in the CREBH-KO mouse livers was diminished. Costaining of LC3b and hepatocyte nuclear factor 4α confirmed unfed-induced autophagy activities in the WT mouse livers were from hepatocytes (Supplemental Fig. S1). Together, these data suggested a critical role of CREBH in hepatic autophagy upon starvation. Further, we examined production of LC3b in CREBH-KO and WT mouse primary hepatocytes challenged with glucagon. Whereas WT hepatocytes expressed LC3b protein under glucagon stimulation in a time-dependent manner, CREBH-KO hepatocytes displayed a diminished induction of LC3b upon glucagon challenge (Fig. 2B), thus confirming the requirement of CREBH for hepatic autophagic activity under energy demands.
Figure 2.
CREBH is required for starvation-induced hepatic autophagy activity and autophagosome formation. A) Immunofluorescence staining of LC3b protein in the liver sections from CREBH-KO and WT mice fed normal chow or unfed for 12 or 24 h. Liver tissue sections were stained with DAPI for nuclei (blue). Original magnification, ×40. B) Primary hepatocytes isolated from CREBH-KO and WT mice were incubated with 25 nM Glucagon for indicated time periods. LC3b protein was visualized by immunofluorescence staining. Cell nuclei stained by DAPI in blue. Original magnification, ×400. C) Western blot analyses of LC3b (LC3-I and LC3‐II) protein levels in the livers of CREBH-KO and WT control mice subjected to being unfed for 6, 12, and 24 h. The mice were injected with PBS or Leupeptin 3 h before tissue harvest. Pooled samples from 3 mice were used for each lane. Levels of GAPDH were determined as loading controls. D) Quantitation of in vivo autophagy flux based on the Western blot analyses of LC3-II and LC3-I protein levels in the livers of mice injected with Leupeptin as described in C. Following normalization to GAPDH, ratios of Leupeptin‐induced LC3‐II vs. LC3-I protein in mouse livers were determined for each time point. The fold changes of LC3-II vs. LC3-I ratios were shown. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01. E) TEM of liver tissue sections from CREBH-KO and WT control mice under feeding or after being unfed for 14 h. Scale bars, 500 nm. Arrows indicate double‐membraned autophagosomes. The graph shows quantitation of autophagosome abundance. Data represent means ± sd (n = 12 for WT fed; 22 for KO fed; 23 for WT unfed; and 17 for KO unfed). *P < 0.05, **P < 0.01.
During the autophagic process, LC3b is hydrolyzed to a cytosolic form (LC3-I), which is conjugated to phosphatidylethanolamine to form lipid-modified LC3b form (LC3-II) (27). LC3-II is recruited to autophagosomal membranes and promotes autophagosomes fusing with lysosomes to form autolysosomes. Therefore, conversion of LC3-I to LC3-II and lysosomal turnover of LC3-II reflects starvation-induced autophagic activity, and detecting the ratio of LC3-II vs. LC3-I by immunoblotting has been a reliable method for monitoring autophagic activity. To further clarify whether hepatic autophagy activity is regulated by CREBH, we injected a single dose of PBS or leupeptin—a lysosomal protease inhibitor that enables quantitative evaluation of autophagic flux in mouse livers based on LC3b protein turnover (28)—into CREBH-KO and WT control mice. As shown in Fig. 2C, D, in response to being unfed for 6, 12, or 24 h, the rates of LC3-I to LC3-II conversion in the livers of the WT mice were consistently higher than those in the livers of CREBH-KO mice. Following leupeptin injection, the levels of autophagy flux, reflected by conversion and turnover of LC3-II protein during the unfed periods, were significantly lower in the livers of CREBH-KO mice compared to those of WT control mice (Fig. 2C, D). We next performed TEM on liver samples to assess autophagosome formation in the livers of CREBH-KO and WT mice under the feeding or unfed condition. Consistent with the immunostaining and autophagic flux studies, the amount of autophagosomes formed in the livers of WT mice were significantly more than that formed in the livers of CREBH-KO mice under the normally fed condition or after 14 h of being unfed (Fig. 2E). Additionally, we examined the size of autophagosomes in the livers of WT and CREBH-KO mice. The size of autophagosomes present in the CREBH-KO mouse liver was not significantly changed compared to that of WT mouse liver under the feeding or unfed condition (Supplemental Fig. S2A). Together, these results indicate that unfed-induced hepatic autophagy activity, as indicated by the levels of LC3-I to LC3-II conversion and autophagosome formation, is significantly reduced in the liver of CREBH-KO mice.
CREBH regulates expression of the genes encoding key components of autophagy and lysosome biogenesis in the liver
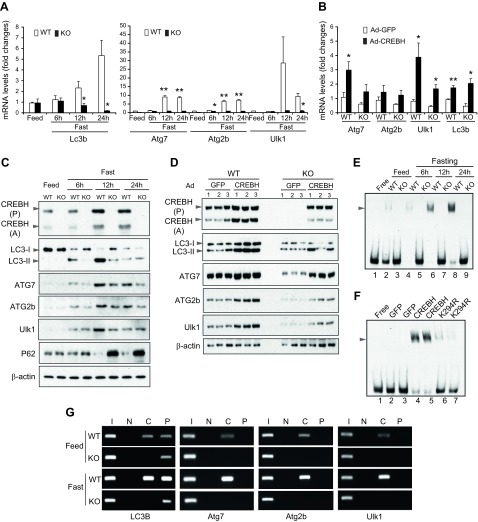
To understand the mechanism by which CREBH regulates hepatic autophagy, we examined whether CREBH, as an unfed-inducible transcriptional activator, regulates expression of the genes encoding the components or regulators of autophagy and lysosomal biogenesis in the liver. Through a microarray analysis with CREBH-KO and WT mouse livers that we previously accomplished (17), we found that the transcripts encoding the key autophagy components were down-regulated in the livers of CREBH-KO mice. qPCR analysis showed that expression levels of the Lc3b gene in WT mouse livers were increased in response to being unfed in a time-dependent manner (Fig. 3A). In contrast, unfed-induced expression of the Lc3b gene was not observed in the livers of CREBH-KO mice. The unfed-induced expression pattern of the Lc3b gene was correlated with the increases in both expression and proteolytic activation of CREBH protein but not CREB, another basic-region leucine zipper transcription factor of the CREB/activating transcription factor family that can potentially bind to the promoters of the CREBH-targeted genes (17, 18), in the livers of WT mice (Supplemental Fig. S2B, C). Further, qPCR analysis indicated that expression levels of the mRNAs encoding major autophagic enzymes or regulators, including ATG7, ATG2b, and ULK1, were significantly reduced in the livers of CREBH-KO mice compared to those of WT control mice under unfed conditions (Fig. 3A). To validate the role of CREBH in driving expression of the autophagy genes in the liver, we overexpressed an activated form of CREBH in mouse livers through tail vein injection of the recombinant adenovirus expressing CREBH into WT mice. qPCR analyses indicated that overexpression of the activated CREBH significantly increased expression of the genes encoding LC3b, ATG7, ATG2b, and ULK1 in the liver (Fig. 3B). The results from the loss- and gain-of-function studies suggested that CREBH is a key transcriptional activator of Lc3, Atg7, Atg2b, and Ulk1 gene expression in mouse livers.
Figure 3.
CREBH directly regulates expression of the key autophagy genes in the liver in response to being unfed. A) Expression levels of Lc3b, Atg7, Atg2b, and Ulk1 mRNAs in the livers of CREBH-KO and WT mice under feeding conditions or after being unfed for 6, 12, or 24 h. mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined by comparison to the mRNA levels in one of the WT control mice under the feeding condition. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01. B) Expression levels of Atg7, Atg2b, Ulk1, and Lc3b mRNAs in the livers of mice infected with Adenovirus (Ad) overexpressing GFP or activated CREBH. mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01. C) Western blot analysis of CREBH, LC3b, ATG7, ATG2b, P62, and Ulk1 proteins in the livers of CREBH-KO and WT mice under feeding conditions or after being unfed for 6, 12, or 24 h. Levels of β-actin were determined as loading controls. CREBH (P), CREBH precursor; CREBH (A), activated/cleaved form of CREBH; LC3-I, a cytosolic form of LC3b; LC3-II, lipid-modified LC3b form. D) Western blot analysis of CREBH, LC3b, ATG7, ATG2b, and Ulk1 proteins in the livers of CREBH-KO and WT mice infected with Ad overexpressing GFP or activated CREBH. E) Binding activities of mouse liver NEs to the CRE-binding motif of the mouse Lc3 gene promoter. NEs were prepared from the livers of CREBH-KO and WT control mice under feeding or after being unfed for 6, 12, or 24 h. EMSA was performed with liver NEs and biotin-labeled Lc3 gene probe containing the CRE-binding motif. F) Binding activities of mouse liver NE expressing GFP, CREBH, or the CREBH mutant K294R to the CRE-binding motif of the mouse Lc3b gene. NE was prepared from the livers of WT mice infected with Ad expressing GFP, CREBH (activated form), or K294R. EMSA was performed using the liver NEs and biotin-labeled Lc3b gene probe containing the CRE-binding motif. G) ChIP analysis of CREBH- and PPARα-binding activities to the endogenous Lc3b, Atg7, Atg2b, and Ulk1 gene promoters in the livers of CREBH-KO and WT mice under feeding or after being unfed for 12 h. For ChIP-PCR, isolated mouse chromatins were immunoprecipitated with the antibody against CREBH (C), PPARα (P), or no antibody (N). Nonimmunoprecipitated chromatins were included as input controls (I). PCR reactions were conducted using the primers amplifying the CRE or PPRE-binding motif present in the Lc3b, Atg7, Atg2b, or Ulk1 gene promoter. The amplified PCR products were visualized on agarose gels.
Next, we verified whether CREBH regulates expression of LC3b, Atg7, Atg2b, and Ulk1 in the liver at the protein level. As shown by the Western blot analyses, levels of expression and proteolytic activation of CRBEH protein were increased in the livers of WT mice in response to being unfed (Fig. 3C and Supplemental Fig. S3A). Along with the increases in hepatic CREBH expression and activation, LC3b expression levels and rates of LC3-I to LC3-II conversion were increased in the livers of WT mice upon being unfed (Fig. 3C). In comparison, unfed-induced expression and conversion of LC3 in the livers of CREBH-KO mice were reduced compared to that of WT control mice. Similarly, unfed-induced expression of ATG7, ATG2b, or ULK1 was also decreased in the livers of CREBH-KO mice. Additionally, we examined the expression of P62, a protein that interacts with LC3 and delivers autophagy cargos for lysosomal degradation (29). P62 is both an essential component and a target of autophagy and is therefore reversely correlated with autophagy activities. Levels of P62 protein were increased in the livers of CREBH-KO mice after being unfed (Fig. 3C), thus confirming the reduction of unfed-induced hepatic autophagy in the absence of CREBH. Furthermore, we examined protein levels of LC3b, ATG7, ATG2b, ULK1, and P62 in the livers of CREBH-KO and WT control mice overexpressing the activated form of CREBH or green fluorescent protein (GFP) control. Supporting the role of CREBH in driving expression of the key autophagic components, overexpression of the activated CREBH significantly increased the levels of expression or conversion of LC3b, ATG7, ATG2b, and ULK1 proteins in the livers of CREBH-KO mice as well as, to a greater extent, WT mice (Fig. 3D and Supplemental Fig. S3B). Additionally, CREBH was also required for unfed-induced expression of the genes encoding the key enzymes or components of lysosome biosynthesis in the liver. qPCR analyses indicated that expression levels of the genes encoding: 1) lysosomal hydrolase and accessory proteins, including cathepsin A (CTSA), glucocerebrosidase (GBA), GLA, and prosaposin (PSAP); 2) lysosomal membrane components, including chloride voltage-gated channel 7 (CLCN7) and mucolipin 1 (MCOLN1); and 3) the lysosomal acidification enzyme ATPase H+ transporting V1 subunit H (ATP6V1H) were significantly reduced in the livers of CREBH-KO mice compared to those of WT control mice in response to being unfed (Supplemental Fig. S4).
CREBH activates the transcription of autophagy genes through direct promoter occupancy
To understand the molecular basis underlying CREBH-mediated transcriptional regulation of hepatic autophagy, we studied cis-regulatory elements in the promoter regions of the CREBH-regulated autophagy genes. Highly conserved CRE-binding DNA sequences were identified in the promoter regions of the Lc3, Atg7, Atg2b, and Ulk1 genes of both human and mouse species (Supplemental Fig. S5). To test whether CREBH can bind to the promoter region of the key autophagy gene, we first performed EMSA with a mouse Lc3 gene promoter oligo containing the CRE-binding consensus sequence (nt −14 to −53) and the liver nuclear extract (NE) from CREBH-KO and WT mice under feeding or after being unfed for 6, 12, or 24 h. Binding activities to the Lc3 gene promoter were detected with the liver NE from the WT mice, and these activities were increased in a time-dependent manner in response to being unfed (Fig. 3E). In contrast, binding activity of the liver NE of CREBH-KO mice to the Lc3 gene promoter oligo was not detectable, suggesting that CREBH indeed binds to the Lc3 gene promoter. Further, we performed EMSA with the Lc3 gene promoter oligo and the liver NE from CREBH-KO mice expressing the activated form of CREBH, an acetylation-deficient CREBH isoform (K294R), or GFP control. When CREBH, but not GFP, was overexpressed, shifted binding signals, which reflected the binding activities to the Lc3 gene promoter, were detected (Fig. 3F). However, the NE from the mouse livers expressing K294R, a CREBH isoform containing a site mutation that prevents lysine (K)-linked acetylation and thus disrupts CREBH transcriptional activity (22), displayed diminished binding activities to the Lc3 gene promoter (Fig. 3F). Together, these results confirmed the capability of the functional CREBH to directly bind to the Lc3 gene promoter.
It was also reported that PPARα regulates autophagy by activating expression of LC3b (10). We examined the presence of both CREBH- and PPARα-binding sequences in the promoter regions of the major autophagy genes. The Lc3 gene contains both the cAMP-response element (CRE)- and PPRE-binding sequences in the proximal promoter region (Supplemental Fig. S5). To evaluate the binding activities of CREBH and PPARα to the Lc3b gene promoter, we carried out ChIP analysis with the liver tissue samples from CREBH-KO and WT mice under feeding or after 12 h of being unfed. Binding activities of CREBH or PPARα to the mouse endogenous Lc3 gene promoter region containing the CRE- and PPRE-binding motifs were detected in the livers of WT mice, and these activities were increased in response to being unfed (Fig. 3G). Interestingly, although binding activities of PPARα to the Lc3 gene promoter can be detected in the CREBH-KO mouse livers, CREBH deficiency eliminated unfed-induced increase in PPARα binding activities to the Lc3 gene promoter in the liver (Fig. 3G), suggesting that CREBH may synergize PPARα binding to the Lc3 gene promoter in the liver upon being unfed. Additionally, we examined the binding activities of CREBH or PPARα to the Atg7, Atg2b, and Ulk1 gene promoters in the liver through ChIP analysis. We were able to detect unfed-induced CREBH binding activities to the endogenous Atg7, Atg2b, or Ulk1 gene promoter in the livers of WT mice (Fig. 3G). However, no PPARα binding activity to the Atg7, Atg2b, or Ulk1 gene promoter was detected in the mouse livers, suggesting that PPARα may not directly activate expression of these genes.
CREBH interacts with PPARα to regulate expression of hepatic autophagy genes in response to being unfed
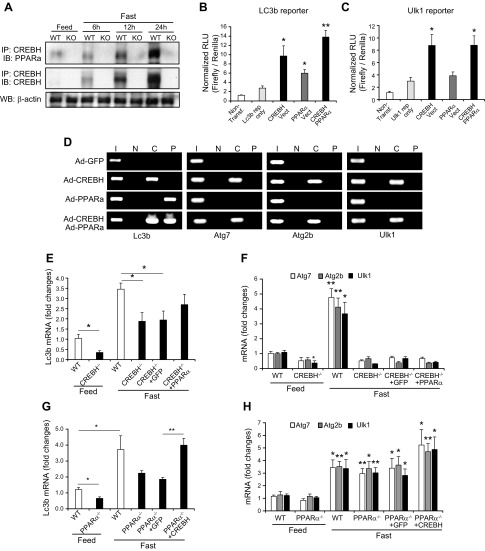
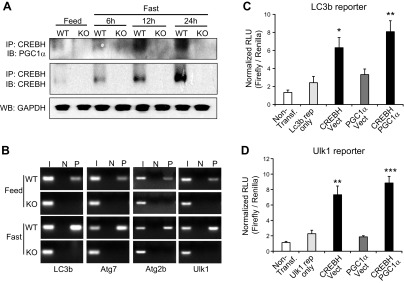
Our ChIP analyses indicated that CREBH and PPARα act in synergy to bind to the promoter region of the Lc3b gene but not the Atg7, Atg2b or Ulk1 genes (Fig. 3G). To validate the interactive relationship between CREBH and PPARα in driving expression of the autophagy genes, we examined the interaction between CREBH and PPAR in the liver of mice upon being unfed. IP-Western blot analysis showed that the levels of CREBH protein associated with PPARα were increased in a time-dependent manner in the livers of WT mice subjected to being unfed for 6, 12, and 24 h (Fig. 4A). To test the synergistic effect of CREBH and PPARα on activating the autophagy genes, we performed autophagy gene reporter analysis with overexpression of CREBH or PPARα in the murine hepatocellular carcinoma cell line Hepa1-6. Reporter analysis showed that either CREBH or PPARα can significantly increase expression of luciferase reporter under the control of the human Lc3b gene prompter (Fig. 4B). Importantly, coexpression of CREBH and PPARα drove the expression of the Lc3b promoter reporter to a level higher than that driven by expression of CREBH or PPARα alone, suggesting an additive effect of CREBH and PPARα on promoting Lc3b expression. However, CREBH, but not PPARα, can increase expression of the Ulk1 gene promoter reporter (Fig. 4C). Coexpression of CREBH and PPARα failed to increase expression levels of the Ulk1 promoter reporter compared with expression of CREBH alone. This is consistent with the observation that CREBH, but not PPARα, can bind to the promoter of the Atg7, Atg2b, and Ulk1 genes (Fig. 3G). Furthermore, we performed ChIP analysis with the human hepatocellular cell line Huh-7 expressing activated CREBH or PPARα protein. Binding activity of CREBH or PPARα to the human LC3b gene promoter region containing the CRE or PPRE motifs can be detected with the NEs from Huh-7 cells overexpressing CREBH or PPARα but not GFP control (Fig. 4D). Importantly, binding activity of CREBH or PPARα to the LC3b gene promoter in the cells expressing both CREBH and PPARα was significantly increased, compared with that in the cells expressing either CREBH or PPARα alone (Fig. 4D). These results suggest that both CREBH and PPARα may be recruited to the LC3b gene promoter, and both factors are likely interactive to enhance their binding activities. This is consistent with the observation that CREBH and PPARα can interact and act in synergy to activate the LC3b gene promoter (Fig. 4A, B). Additionally, we examined the binding activities of CREBH or PPARα to the ATG7, ATG2b, and ULK1 gene promoters in Huh7 cells expressing PPARα or CREBH. ChIP assay confirmed that CREBH, but not PPARα, can directly bind to the human ATG7, ATG2b, and ULK1 gene promoters in Huh-7 cells (Fig. 4D).
Figure 4.
CREBH interacts with PPARα to regulate expression of LC3b in the liver upon being unfed. A) IP and Western blot analysis of interactions between CREBH and PPARα in the livers of mice under feeding or after being unfed for 6, 12, or 24 h. Total liver protein lysates pooled from 3 mice per group were immunoprecipitated with the anti-CREBH antibody, followed by immunoblotting with the anti-PPARα or anti-CREBH antibody. Levels of β-actin were determined by Western blot analysis as input controls. B, C) Luciferase reporter (Rep) analyses of transcriptional activation of the human LC3b (B) or Ulk1 (C) gene promoter by CREBH alone or in combination with PPARα. Hepa1-6 cells were transduced with the reporter vector or a vehicle. After 24 h, the transfected cells were infected with Ad expressing GFP (control), CREBH, or PPARα. A Renilla reporter plasmid was included in the cotransfection for the normalization of luciferase reporter activities. The same adenovirus titers were used for individual infections. Data represent means ± sem (n = 3 biologic repeats). Non-transf, nontransfected cell control. *P ≤ 0.05, **P ≤ 0.01. D) ChIP analysis of CREBH and PPARα binding activities to the LC3b, Atg7, Atg2b, and Ulk1 gene promoters in Huh-7 cells infected with Ad expressing GFP, the activated CREBH, PPARα, or the same titer combination of CREBH-expressing and PPARα-expressing Ad. An anti-CREBH (C) or anti-PPARα (P) antibody was used to pull down the exogenously expressed activated CREBH or PPARα binding complex from the chromatins isolated from the Huh-7 cells. PCR reactions were conducted using the primers amplifying the CRE or PPRE-binding motif present in the Lc3b, Atg7, Atg2b, or Ulk1 gene promoter. N, no antibody; I, input controls (nonimmunoprecipitated chromatin). E, F) Levels of the Lc3b (E), Atg7, Atg2b, and Ulk1 mRNAs (F) in livers of WT mice, CREBH-KO mice, or CREBH-KO mice infected with Ad expressing GFP or PPARα after being unfed for 14 h. G, H) Levels of the Lc3b (G), Atg7, Atg2b, and Ulk1 mRNAs (H) in livers of WT mice, PPARα-KO mice, or PPARα-KO mice infected with Ad expressing GFP or the activated form of CREBH after being unfed for 14 h. E–H) mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01.
Next, we evaluated the regulation of the key autophagy gene expression by CREBH or PPARα in CREBH-KO and PPARα-KO mice reconstituted by PPARα or CREBH overexpression. Expression level of the Lc3b gene in the livers of CREBH-KO mice were reduced, compared to that of WT control mice, under the feeding or 14-h unfed condition (Fig. 4E). Overexpression of PPARα can partially restore expression levels of the Lc3b transcripts in the CREBH-KO livers under unfed conditions. Moreover, although expression levels of the Atg7, Atg2b, and Ulk1 genes in the livers of CREBH-KO mice were significantly reduced (Fig. 4F), overexpression of PPARα failed to rescue expression of the Atg7, Atg2b, and Ulk1 genes in the CREBH-null livers, which is consistent with the observation that PPARα did not bind to or activate the promoter of the Atg7, Atg2b, and Ulk1 genes (Fig. 4C, D). On the other hand, expression levels of the Lc3b gene were reduced in the livers of PPARα-KO mice under the feeding or after being unfed (Fig. 4G). Overexpression of the activated form of CREBH significantly increased expression of the Lc3b gene to a level comparable to that in WT mice under unfed conditions. Additionally, expression of the Atg7, Atg2b, or Ulk1 gene in the livers of PPARα-KO mice was not reduced, compared to those of WT mice (Fig. 4H). Overexpression of CREBH increased expression levels of the Atg7, Atg2b, or Ulk1 transcripts in the livers of PPARα-KO mice (Fig. 4H). Taken together, these results suggested that CREBH and PAPRα interact and act in synergy to regulate expression of Lc3b but not Atg7, Atg2b, or Ulk1.
PPARγ coactivator 1α acts as a coactivator of CREBH to regulate expression of hepatic autophagy genes upon nutrient starvation
In response to energy demands or nutritional cues, PPARγ coactivator 1α (PGC1α) functions as a major transcriptional coactivator by docking to specific nuclear receptors or transcription factors to provide a platform for the recruitment of regulatory transcriptional complexes (30). To test the interactive relationship between CREBH and PGC1α in driving expression of the autophagy genes, we first examined the interaction between CREBH and PGC1α in the liver of mice upon not being fed. IP-Western blot analysis showed that the levels of CREBH protein associated with PGC1α were increased in a time-dependent manner in the livers of WT mice subjected to being unfed for 6, 12, and 24 h (Fig. 5A). Next, we tested whether PGC1α is in complex with CREBH that binds to the CREBH-target autophagy genes. The promoter regions of the Lc3, Atg7, Atg2b, and Ulk1 genes of both human and mouse species possess highly conserved DNA sequences of CREBH but not PGC1α (Supplemental Fig. S5). Because our ChIP analyses have revealed unfed-induced CREBH binding activities to the Lc3b, Atg7, Atg2b, and Ulk1 gene promoter in mouse livers (Fig. 3G), we performed ChIP analyses to examine whether CREBH can bind to the mouse liver chromatins pulled down by the PGC1α antibody. Binding activities of CREBH to the PGC1α-precipitated Lc3b, Atg7, Atg2b or Ulk1 gene promoter were detected in the livers of WT mice, and these activities were increased after 12 h of being unfed (Fig. 5B). These results confirmed PGC1α’s function as a coactivator to complex with CREBH to bind to the autophagy gene promoters.
Figure 5.
PGC1α interacts with CREBH to regulate expression of hepatic autophagy genes upon nutrient starvation. A) IP and Western blot analysis of interactions between CREBH and PGC1α in the livers of mice under feeding or after being unfed for 6, 12, or 24 h. Total liver protein lysates pooled from 3 mice per group were immunoprecipitated with the anti-CREBH antibody, followed by immunoblotting with the anti-PGC1α or anti-CREBH antibody. Levels of GAPDH were determined by Western blot analysis as input controls. B) ChIP analysis of CREBH binding activities to the endogenous Lc3b, Atg7, Atg2b, and Ulk1 gene promoters in the liver chromatins (pulled down by the anti-PGC1α antibody) of CREBH-KO and WT mice under feeding or after being unfed for 12 h. For ChIP-PCR, isolated mouse chromatins were immunoprecipitated with the antibody against PGC1α (P) or no antibody (N). Nonimmunoprecipitated chromatins were included as input controls (I). PCR reactions were conducted using the primers amplifying the CRE-binding motif present in the Lc3b, Atg7, Atg2b, or Ulk1 gene promoter (sequences provided in Supplemental Fig. S5). The amplified PCR products were visualized on agarose gels. C, D) Luciferase reporter (Rep) analyses of transcriptional activation of the human LC3b (C) or Ulk1 (D) gene promoter by CREBH alone or in combination with PGC1α. Hepa1-6 cells were transduced with the vectors expressing the reporter, CREBH, or PGC1α. A Renilla reporter plasmid was included in the cotransfection for the normalization of luciferase reporter activities. Data represent means ± sem (n = 3 biologic repeats). Non-transf, nontransfected cell control. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
To evaluate the potential synergistic effect of CREBH and PGC1α on activating the autophagy genes, we performed autophagy gene reporter analysis with overexpression of CREBH or PGC1α in the murine hepatocellular carcinoma cell line Hepa1-6. Reporter analysis showed that CREBH, but not PGC1α, can significantly increase expression of luciferase reporter under the control of the human Lc3b or Ulk1 gene prompter (Fig. 5C, D). Coexpression of CREBH and PGC1α drove the expression of the Lc3b or Ulk1 gene promoter reporter to levels higher than those driven by expression of CREBH alone, suggesting additive effects of CREBH and PGC1α on promoting Lc3b or Ulk1 gene expression. These results are consistent with the function of PGC1α as a coactivator of CREBH in driving expression of the Atg7, Atg2b, and Ulk1 genes in response to nutrient starvation.
CREBH promotes starvation-induced expression and subcellular translocation of TFEB through regulating PPARα
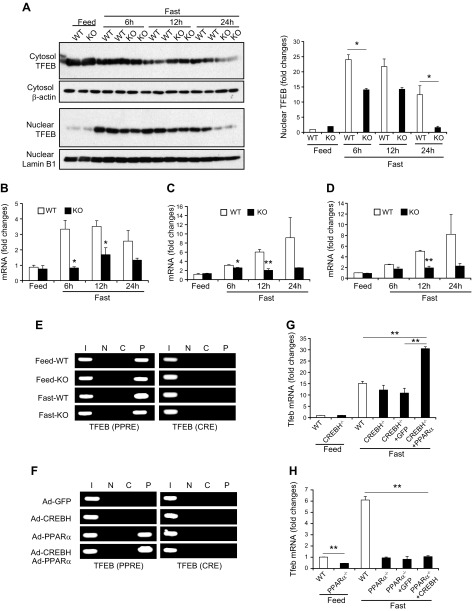
Next, we examined whether CREBH is functionally associated with TFEB, a master regulator of lysosomal biogenesis and autophagy (31), in unfed-induced hepatic autophagy. Upon nutrient starvation, TFEB is induced and rapidly translocated into nucleus to activate a transcriptional program that controls major autophagy steps, including autophagosome formation, autophagosome-lysosome fusion, and substrate degradation (32). We examined the presence of TFEB protein in the livers of CREBH-KO and WT mice under the feeding or unfed condition. Western blot analyses with liver subcellular protein fractions showed that the levels of nuclear TFEB or total (cytosolic and nuclear) TFEB protein were increased in WT mouse livers in response to being unfed (Fig. 6A). In comparison, the levels of nuclear TFEB protein in the livers of CREBH-KO mice were significantly reduced, compared to those of WT mice, under unfed conditions (Fig. 6A). Further, qPCR analyses indicated that expression levels of the Tfeb transcript were significantly decreased in the livers of CREBH-null mice upon not being fed (Fig. 6B). It was known that TFEB is induced by starvation through an autoregulatory feedback loop via PPARα and PGC1α to exert a broad transcriptional control on autophagy process and lipid catabolism (31, 33). We evaluated whether CREBH regulates TFEB expression in the liver through regulating PPARα and PGC1α. Similar to the expression profile of the Tfeb gene, expression levels of the Pparα and Pgc1α genes were decreased in the liver of CREBH-KO mice under unfed conditions (Fig. 6C, D). Based on the promoter sequence analysis of the Tfeb gene, both CRE- and PPRE-binding motifs were identified in the promoter region of mouse Tfeb gene (Supplemental Fig. S5). However, ChIP analysis showed that CREBH did not bind to the CRE-binding motifs present in the Tfeb gene promoter in the mouse liver after being unfed (Fig. 6E). In contrast, PPARα exhibited strong binding activity to the PPRE-binding motif in the Tfeb gene promoter in the unfed mouse liver. Further, NE from Huh7 cells overexpressing the activated form of CREBH did not exhibit binding activity to the human TFEB gene promoter, whereas NE from Huh7 cells overexpressing PPARα displayed strong binding activity to the human TFEB promoter (Fig. 6F). Interestingly, when PPARα and CREBH were coexpressed, the binding activity of PPARα to the Tfeb promoter was increased compared to expression of PPARα alone, suggesting that CREBH may interact with PPARα to enhance PPARα activity to activate the Tfeb gene promoter. Together, these results suggested that CREBH promoted TFEB expression by regulating and interacting with PPARα and PGC1α, although CREBH does not directly drive expression of the Tfeb gene. To validate PPARα-dependent regulation of TFEB by CREBH, we examined expression of the Tfeb gene in CREBH-KO or PPARα-KO mice reconstituted by PPARα or CREBH overexpression. The expression level of the Tfeb gene in the livers of CREBH-KO mice was reduced under feeding or after 14 h of being unfed (Fig. 6G). Overexpression of PPARα restored, and even increased, unfed-induced expression of the Tfeb gene in the liver of CREBH-KO mice, compared to that of CREBH-KO mice overexpressing GFP or WT mice. Moreover, expression of the Tfeb gene was reduced in the livers of PPARα-KO mice (Fig. 6H). Overexpression of the activated form of CREBH failed to restore unfed-induced expression of the Tfeb gene in the liver of PPARα-KO mice. These results confirmed that CREBH regulates starvation-induced expression of the Tfeb gene through PPARα in the liver.
Figure 6.
CREBH regulates TFEB through PPARα in the liver under being unfed. A) Western blot analysis of TFEB proteins in the cytosolic and nuclear fractions from the livers of mice under feeding or after being unfed for 6, 12, or 24 h. Levels of β-actin and Lamin B1 were determined as loading controls for cytosolic and nuclear proteins, respectively. The graph shows the quantification of TFEB protein signals based on Western blot analysis. The intensity of TFEB protein signals, determined by Western blot densitometry, was normalized to that of input controls. Fold changes in protein levels were determined by comparison to one of WT control under the feeding condition. B–D) Expression levels of Tfeb (B), Pparα (C), and Pgc1α (D) mRNAs in the livers of CREBH-KO and WT mice under feeding conditions or after being unfed for 6, 12, or 24 h. mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined by comparison to the mRNA levels in one of the WT control mice under the feeding conditions. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01. E) ChIP analysis of CREBH and PPARα binding activities to the endogenous Tfeb gene promoter in the livers of CREBH-KO and WT mice under feeding or after being unfed for 12 h. For PCR, isolated mouse chromatins were immunoprecipitated with the antibody against CREBH (C), PPARα (P), or no antibody (N). Nonimmunoprecipitated chromatins were included as input controls (I). PCR reactions were conducted using the primers amplifying the CRE or PPRE-binding motif present in the Tfeb gene promoter. F) ChIP analysis of CREBH and PPARα binding activities to the Tfeb gene promoter in Huh-7 cells infected with Ad expressing GFP, the activated CREBH, PPARα, or the same titer of combination of CREBH-expressing and PPARα-expressing Ad. An anti-CREBH (C) or anti-PPARα (P) antibody was used to pull down the exogenously expressed activated CREBH or PPARα binding complex from the chromatins. PCR reactions were conducted using the primers amplifying the CRE or PPRE-binding motif present in the Tfeb gene promoter. N, no antibody; I, input controls. G) Levels of the Tfeb mRNA in livers of WT mice, CREBH-KO mice, or CREBH-KO mice infected with Ad expressing GFP or PPARα after being unfed for 14 h. H) Levels of the Tfeb mRNAs in livers of WT mice, PPARα-KO mice, or PPARα-KO mice infected with Ad expressing GFP or the activated form of CREBH after being unfed for 14 h. For (G, H) mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined. Data represent means ± sem (n = 3). *P ≤ 0.05, **P ≤ 0.01.
CREBH regulates circadian rhythmic expression of the key genes involved in hepatic autophagy process
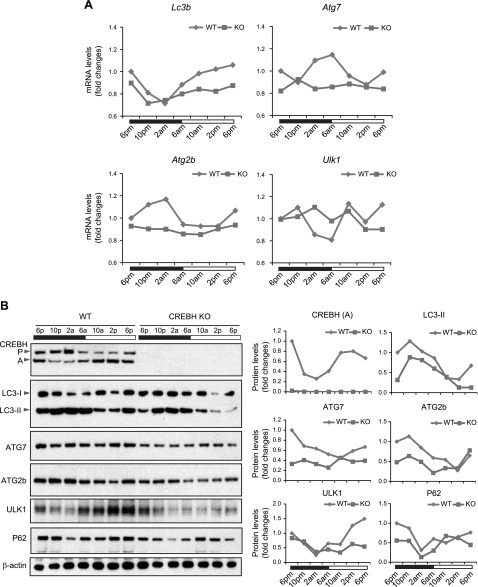
We recently revealed that CREBH is a circadian-regulated transcription factor of hepatic energy metabolism (19). We asked whether rhythmic activation of hepatic autophagy is regulated by CREBH across the circadian cycle. To address this question, we examined the rhythmic expression of the key autophagy genes in the livers of CREBH-KO and WT control mice under the 12-h light/dark circadian cycle. qPCR analyses revealed that expression of the genes encoding the key enzymes or regulators in autophagy, including LC3b, ATG7, ATG2b, and ULK1, displayed robust oscillation in mouse livers throughout the 24-h circadian cycle (Fig. 7A). In contrast, the CREBH-KO livers displayed impaired rhythmicity and decreased amplitudes in expression of the Lc3b, Atg7, Atg2b, and Ulk1 genes across the circadian cycle. Additionally, rhythmic expression of the genes encoding the enzymes or regulators in lysosomal homeostasis, including CTSA, GBA, GLA, PSAP, CLCN7, MCOLN1, and ATP6V1H, were also altered in the livers of CREBH-KO mice (Supplemental Fig. S6), albeit these alterations were not as dramatic as those of Lc3b, Atg7, Atg2b, and Ulk1 in CREBH-KO livers (Fig. 7A). Consistent with the mRNA expression profile, circadian rhythmicity and amplitude of the autophagy proteins were impaired in the livers of CREBH-KO mice. In particular, rhythmic conversion of LC3-I to LC3-II and expression of ATG7, ATG2b, and ULK1 proteins were reduced in the livers of CREBH-KO mice throughout the 24-h circadian cycle (Fig. 7B).
Figure 7.
CREBH regulates rhythmic expression of the key autophagy genes in the liver of mice under the circadian clock. A) Rhythmic expression levels of Lc3b, Atg7, Atg2b, and Ulk1 genes in the livers of CREBH-KO and WT control mice across a 24-h circadian cycle. mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined by comparison to levels in one of the WT control mice at the starting circadian time point. Each expression point represents a pooled liver sample from 3 to 5 mice per group per time point. Black and white bars represent circadian light/dark phases. B) Western blot analysis of rhythmic levels of CREBH, LC3b, ATG7, ATG2b, ULK1, P62, and β-actin proteins in the livers of CREBH-KO and WT mice, collected every 4 h in a 24-h circadian period. Pooled liver protein lysates from 3 to 5 mice per time point per genotype group were used. The graphs show the quantification of the protein levels in mouse livers over the circadian cycle. The intensity of the protein signals, determined by Western blot densitometry, was normalized to that of β-actin. Fold changes in protein levels were determined by comparison to the protein level at 6 pm.
CREBH is regulated by BMAL1 in activating hepatic autophagy genes under the circadian clock
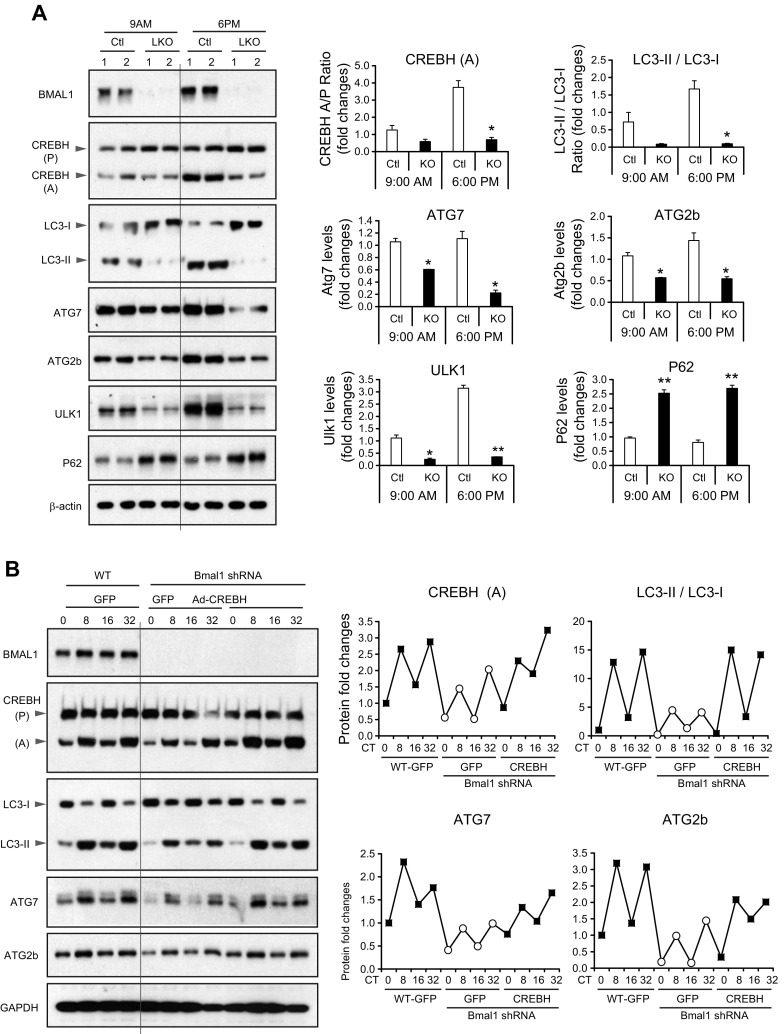
BMAL1, the core circadian oscillator, plays a central role in regulating expression or activities of circadian output regulators (34). To evaluate whether BMAL1 controls CREBH activity in regulating rhythmic autophagy in the liver under the circadian clock, we examined proteolytic activation of CREBH and expression of LC3b, ATG7, ATG2b, ULK1, and P62 proteins in the livers of liver-specific Bmal1 conditional KO (Bmal1-LKO) and WT mice during the resting and active circadian phases. Western blot analysis showed that the levels of the cleaved/activated CREBH protein, but not the CREBH precursor, were reduced in the livers of Bmal1-LKO mice across the resting (9:00 am) and active (6:00 pm) periods compared with those in the control fl/fl mouse livers (Fig. 8A). Correlated with the rhythm of CREBH activation, the levels of the activated LC3b (LC3-II), ATG7, ATG2b, and ULK1 proteins were reduced, whereas the level of P62 protein was increased, in the livers of Bmal1-LKO mice at both 9:00 am and 6:00 pm. These results suggested that CREBH may function as a circadian transcriptional regulator of hepatic autophagy under the control of the circadian oscillator BMAL1.
Figure 8.
CREBH is regulated by BMAL1 in activating hepatic autophagic genes under the circadian clock. A) Western blot analysis of CREBH, LC3b, ATG7, ATG2b, ULK1, and P62 in the livers of Bmal1-LKO and fl/fl control mice during representative day and night circadian periods. The liver protein lysates were prepared from pooled liver tissues of Bmal1-LKO and fl/fl mice collected at 9 am or 6 pm (n = 3–4 mice/genotype/time point). Levels of BMAL1 and β-actin were determined as controls. The graphs show rhythmic fold changes of protein intensities or ratios (LC3-II/LC3-I) in the livers of Bmal1-LKO and fl/fl mice during the circadian cycle. The protein signals, determined by Western blot densitometry, were normalized to that of β-actin. Fold change of the protein levels was determined by comparing with that at 9 am. B) Western blot analyses of BMAL1, CREBH, LC3b, ATG7, and ATG2b and GAPDH in Bmal1-knockdown and WT control mouse primary hepatocytes expressing the activated CREBH or GFP control under the circadian clock. Mouse primary hepatocytes were infected with Ad expressing Bmal1 short hairpin RNA, CREBH, or GFP before being subjected to horse serum shock for circadian synchronization. Cell lysates were collected throughout a 32-h circadian cycle for Western blot analyses. The graphs show the rhythmic fold changes of activated CREBH, LC3-II/LC3-I ratios, ATG7, and ATG2b protein levels, determined by Western blot densitometry, in Bmal1-knockdown and WT control hepatocytes expressing CREBH or GFP control. Fold change of the normalized protein levels at each circadian time point was compared with that of the starting circadian time.
To test whether CREBH is regulated by BMAL1 to regulate rhythmic expression of the hepatic autophagy genes, we performed reconstitution experiments by overexpressing the activated form of CRBEH or GFP control in Bmal1-knockdown primary hepatocytes. BMAL1 was knocked down in mouse primary hepatocytes using adenoviral-based expression of Bmal1 short hairpin RNA (19) and then subjected to horse serum shock for circadian synchronization (35). Western blot analysis showed that the amplitudes of the cleaved/activated CREBH protein in mouse primary hepatocytes across a 32-h circadian period were significantly repressed by knockdown of BMAL1 (Fig. 8B). In parallel with the reduced levels of the activated CREBH protein, rhythmic expression levels of LC3b, ATG7, and ATG2b were decreased in Bmal1-knockdown primary hepatocytes during circadian oscillation. However, when the activated form of CREBH was expressed in the Bmal1-knockdown mouse primary hepatocytes, circadian rhythmic expression of LC3-II, ATG7, and ATG2b in the Bmal1-knockdown hepatocytes was restored to the levels comparable to those in the WT mouse primary hepatocytes expressing GFP control (Fig. 8B). These results supported the conclusion that CREBH is controlled by BMAL1 in regulating rhythmic expression of the key autophagy genes in the liver under the circadian clock.
DISCUSSION
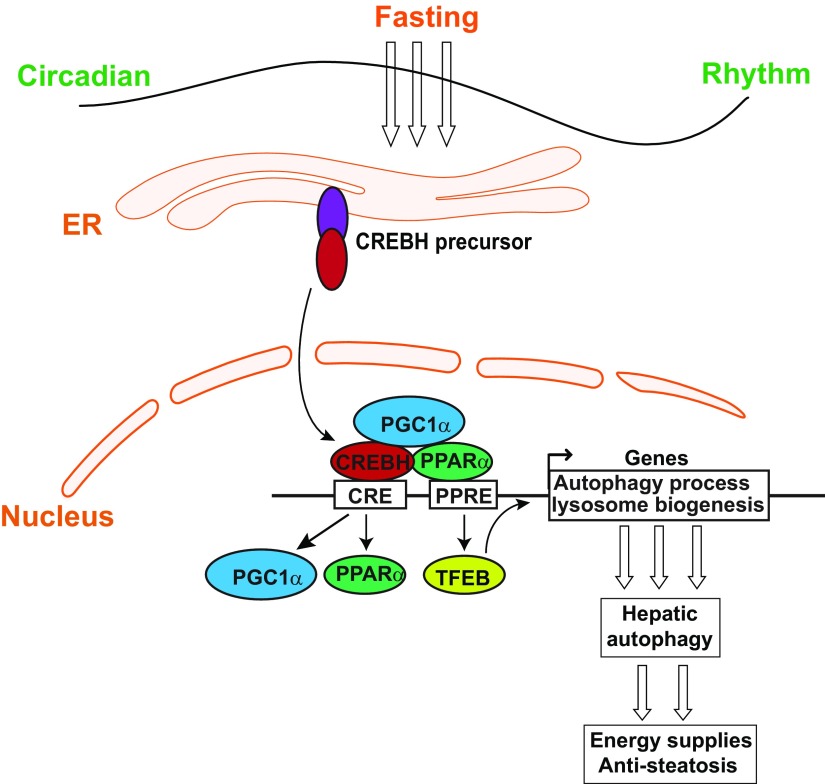
In this study, we demonstrated that CREBH functions as a major transcriptional regulator of hepatic autophagy by activating expression of the genes encoding the key enzymes or regulators involved in autophagic process and lysosomal biogenesis in response to being unfed or under the circadian clock (Fig. 9). CREBH-regulated hepatic autophagy likely plays an important role in preventing hepatic lipid accumulation and the associated metabolic disorders. Specifically, our major findings include the following: 1) CREBH activity prevents unfed-induced hepatic lipid accumulation by activating hepatic autophagy; 2) CREBH is required for the activation of starvation-induced hepatic autophagy by directly activating expression of the genes encoding key enzymes or regulators in autophagic process and lysosome biogenesis in the liver; 3) CREBH regulates and interacts with PPARα and the transcriptional coactivator PGC1α to synergistically drive expression of the key autophagy genes in response to nutrient starvation; 4) CREBH regulates rhythmic expression of the key genes involved in hepatic autophagy process under the circadian clock; and 5) the central circadian oscillator BMAL1 regulates CREBH activity to drive rhythmic expression of hepatic autophagy genes under the circadian clock. These findings demonstrate that CREBH functions as a major transcriptional regulator for autophagy activation in the liver upon energy demands. CREBH-mediated hepatic autophagy may play a protective role in preventing hepatic lipid accumulation in response to energy fluctuation triggered by nutrient starvation or under the circadian clock, and its blockade may accelerate nonalcoholic fatty liver disease progression.
Figure 9.
Illustration of transcriptional regulation of hepatic autophagy by CREBH.
CREBH is a stress-inducible, liver-enriched transcription regulator that plays critical roles in maintaining energy homeostasis. We previously demonstrated that CREBH deficiency leads to hepatic lipid accumulation, which is partially due to impaired FA oxidation and TG lipolysis (17). In this study, however, we established a link of CREBH to hepatic autophagy activation in regulating lipid homeostasis. Impaired hepatic autophagy contributes to hepatic steatosis in CREBH-KO mice. As a conserved cellular process that contributes to nutrient and organelle homeostasis, hepatic autophagy machinery has been found to associate with lipid droplets and participate in the hydrolysis and oxidation of lipids (5). Transcriptional regulation of autophagy genes is an important mechanism in the control of cellular autophagy activity (10, 11, 36). Through gain- and loss-of-function analyses, we demonstrated that CREBH is a major transcription activator of the genes encoding the key enzymes or regulators in autophagosome formation (LC3b, ATG7, ATG2b), autophagic process (ULK1), and lysosome biogenesis (CTSA, GBA, GLA, PSAP, CLCN7, MCOLN1, and ATP6V1H) (Fig. 9). CREBH appears to be a direct transcriptional activator of autophagy gene expression because CREBH was found to bind to and activate the promoters of endogenous Lc3b, Atg7, Atg2b, and Ulk1 genes in the liver (Figs. 3 and 4). In particular, we have identified the consensus CREBH and PPARα binding sites in the promoter region of the human or mouse LC3b gene. ChIP and reporter analyses indicated that both CREBH and PPARα bind to the Lc3b gene promoter and synergistically activate expression of the gene in the liver hepatocytes in response to being unfed (Fig. 4). The CREBH-mediated transcriptional regulation of LC3b is similar to the regulation of metabolic hormone FGF21, where CREBH coordinates with PPARα to synergistically drive an optimized production of FGF21 from the liver upon being unfed or under the circadian clock (19, 20).
Our work depicted a transcriptional network through which CREBH regulates and interacts with 3 stress-inducible transcriptional regulators, PPARα, PGC1α, and TFEB, to promote expression of the genes encoding functions involved in lysosomal biogenesis and autophagy process (Figs. 3–6 and 9). Although CREBH interacts with PPARα and binds to the autophagy gene promoters, PGC1α (as a transcriptional coactivator) interacts with CREBH but does not directly bind to the autophagy gene promoters. However, PGC1α interaction with CREBH provides a platform for the recruitment of transcriptional complexes and synergizes CREBH activity in driving autophagy gene transcription (Fig. 5). Furthermore, CREBH functions as a key driver of PPARα-induced expression and subcellular localization of TFEB, a master transcriptional regulator of lysosomal biogenesis during autophagy triggered by starvation (Fig. 6). However, CREBH does not bind to the Tfeb gene promoter, although it directly regulates expression of PPARα and PGC1α in the livers of mice under being unfed, implicating that the regulation of TFEB by CREBH is likely through PPARα and PGC1α, the known transcriptional drivers of TFEB expression (31, 33). Taken together, the interactive and interdependent transcriptional network formed by CREBH, PPARα, PGC1α, and TFEB likely represents a major regulatory loop that drives the activation and process of the autophagy program in the liver induced by nutrient starvation (Fig. 9).
In this study, we observed the expression of the core autophagy genes, including Lc3b, Atg7, Atg2b, and Ulk1, exhibited robust circadian rhythmicity in mouse livers and that CREBH functions as a key circadian transcription regulator that orchestrates a rhythmic expression of these genes. Rhythmic activation of autophagy in mammalian tissues provides a steady supply of nutrients throughout the circadian cycles (36). The nutrients released through autophagy enter systemic circulation and supply metabolites for organismal energy homeostasis. Previously, we demonstrated that proteolytic cleavage/activation of CREBH is rhythmically regulated through the BMAL1-AKT/PKB-glycogen synthase kinase (GSK)3β circadian regulatory axis (19). In mouse livers, suppression of BMAL1 abolished diurnal autophagy rhythm (Fig. 8A). The circadian autophagy phenotype caused by BMAL1 deficiency was consistent with the observation that CREBH deficiency disrupted circadian rhythmicity of hepatic autophagy. Importantly, the reconstitution of activated CREBH in BMAL1-defective mouse primary hepatocytes rescued impaired circadian autophagy rhythm (Fig. 8B), suggesting CREBH acts as a key factor that links hepatic autophagy to biologic clock and maintains nutrient homeostasis throughout the circadian cycle. Additionally, the level of cAMP in the liver of CREBH-KO or Bmal1-LKO mice was significantly lower than that of WT control mice (Supplemental Fig. S7A). Because cAMP regulates hepatic autophagy (37) and decreased cAMP in hepatocytes contributes to hepatic steatosis (38), the reduced cAMP level in CREBH-KO or Bmal1-LKO mouse liver supports our finding that BMAL1-CREBH functions as a regulatory axis of hepatic lipid homeostasis by promoting physiologic autophagy in the liver. Impaired autophagy reduces TG hydrolysis and insulin sensitivity and therefore participates in the pathogenesis of hepatic steatosis, obesity, and type 2 diabetes (5, 6). Indeed, decreased expression of CREBH or nonsense mutation in human CREBH gene was associated nonalcoholic fatty liver disease, hyperlipidemia, and atherosclerosis of human patients or animal models (Supplemental Fig. S7B) (17, 23–25). We previously defined that CREBH functions as a key circadian regulator of TG lipolysis, FA oxidation, lipogenesis, and glucose homeostasis and that CREBH deficiency causes hepatic steatosis, nonalcoholic steatohepatitis, hyperlipidemia, or insulin resistance (17–20). In the present study, we revealed the link of CREBH deficiency to impaired hepatic autophagy and subsequent hepatic lipid accumulation. Given that CREBH is critically involved in hepatic FA oxidation and TG lipolysis, the extent by which CREBH-driven hepatic autophagy contributes to hepatic lipid homeostasis remains to be further elucidated.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
Portions of this work were supported by U.S. National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases Grants DK090313 (to K.Z), DK110314 (to X.C.), and DK102456 (to J.D.L.); NIH National Institute of Environmental Health Sciences Grant ES017829 (to K.Z.); NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR066634 (to D.F. and K.Z.); NIH National Eye Institute Grant EY025291 (to A.G.); and American Heart Association Grants 0635423Z and 09GRNT2280479 (to K.Z.). The authors declare no conflicts of interest.
Glossary
- ATG
autophagy-related protein
- ATP6V1H
ATPase H+ transporting V1 subunit H
- BMAL1
brain and muscle ARNT-like 1
- Bodipy
boron dipyrromethene
- ChIP
chromatin immunoprecipitation
- CLCN7
chloride voltage-gated channel 7
- CRE
cAMP-response element
- CREB
cAMP-responsive element-binding protein
- CREBH
cAMP-responsive element-binding protein, hepatic-specific
- CTSA
cathepsin A
- ER
endoplasmic reticulum
- FA
fatty acid
- FGF21
fibroblast growth factor 21
- fl/fl
flox/flox
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GBA
glucocerebrosidase
- GFP
green fluorescent protein
- KO
knockout
- K294R
Lys294Arg
- LC3b
microtubule-associated protein 1B-light chain 3
- MCOLN1
mucolipin 1
- NE
nuclear extract
- P62
sequestosome 1
- PGC1α
PPARγ coactivator 1α
- PPARα
peroxisome proliferator–activated receptor α
- PSAP
prosaposin
- qPCR
quantitative PCR
- TEM
transmission electron microscope
- TFEB
transcription factor EB
- TG
triglyceride
- ULK1
autophagosome formation Unc‐51 like kinase 1
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
H. Kim and K. Zhang designed and conducted the experiments, analyzed the data, and wrote the manuscript; D. Williams, Y. Qiu, Z. Song, Z. Yang, V. Kimler, A. Goldberg, R. Zhang, Z. Yang, and X. Chen performed the experiments and acquired the data; and L. Wang, D. Fang, and J. D. Lin provided key reagents or critical comments.
REFERENCES
- 1.Lamb C. A., Yoshimori T., Tooze S. A. (2013) The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759–774 [DOI] [PubMed] [Google Scholar]
- 2.Ylä-Anttila P., Vihinen H., Jokitalo E., Eskelinen E. L. (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5, 1180–1185 [DOI] [PubMed] [Google Scholar]
- 3.Hailey D. W., Rambold A. S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P. K., Lippincott-Schwartz J. (2010) Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W. W., Li J., Bao J. K. (2012) Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 69, 1125–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L., Li P., Fu S., Calay E. S., Hotamisligil G. S. (2010) Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 11, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto S., Kuramoto K., Wang N., Situ X., Priyadarshini M., Zhang W., Cordoba-Chacon J., Layden B. T., He C. (2018) Autophagy differentially regulates insulin production and insulin sensitivity. Cell Rep. 23, 3286–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M. C., Kroemer G. (2013) Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 23, 310–322 [DOI] [PubMed] [Google Scholar]
- 9.Füllgrabe J., Klionsky D. J., Joseph B. (2014) The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 15, 65–74 [DOI] [PubMed] [Google Scholar]
- 10.Lee J. M., Wagner M., Xiao R., Kim K. H., Feng D., Lazar M. A., Moore D. D. (2014) Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seok S., Fu T., Choi S. E., Li Y., Zhu R., Kumar S., Sun X., Yoon G., Kang Y., Zhong W., Ma J., Kemper B., Kemper J. K. (2014) Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu K., Czaja M. J. (2013) Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha R. A., Farah B. L., Singh B. K., Siddique M. M., Li Y., Wu Y., Ilkayeva O. R., Gooding J., Ching J., Zhou J., Martinez L., Xie S., Bay B. H., Summers S. A., Newgard C. B., Yen P. M. (2014) Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology 59, 1366–1380 [DOI] [PubMed] [Google Scholar]
- 14.Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 15.Omori Y., Imai J., Watanabe M., Komatsu T., Suzuki Y., Kataoka K., Watanabe S., Tanigami A., Sugano S. (2001) CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29, 2154–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luebke-Wheeler J., Zhang K., Battle M., Si-Tayeb K., Garrison W., Chhinder S., Li J., Kaufman R. J., Duncan S. A. (2008) Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology 48, 1242–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C., Wang G., Zheng Z., Maddipati K. R., Zhang X., Dyson G., Williams P., Duncan S. A., Kaufman R. J., Zhang K. (2012) Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 55, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., Mendez R., Zheng Z., Chang L., Cai J., Zhang R., Zhang K. (2014) Liver-enriched transcription factor CREBH interacts with peroxisome proliferator-activated receptor α to regulate metabolic hormone FGF21. Endocrinology 155, 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Z., Kim H., Qiu Y., Chen X., Mendez R., Dandekar A., Zhang X., Zhang C., Liu A. C., Yin L., Lin J. D., Walker P. D., Kapatos G., Zhang K. (2016) CREBH couples circadian clock with hepatic lipid metabolism. Diabetes 65, 3369–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H., Zheng Z., Walker P. D., Kapatos G., Zhang K. (2017) CREBH maintains circadian glucose homeostasis by regulating hepatic glycogenolysis and gluconeogenesis. Mol. Cell. Biol. 37, e00048-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., Hanson R. W., Choi H. S., Koo S. H. (2010) Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 11, 331–339 [DOI] [PubMed] [Google Scholar]
- 22.Kim H., Mendez R., Chen X., Fang D., Zhang K. (2015) Lysine acetylation of CREBH regulates fasting-induced hepatic lipid metabolism. Mol. Cell. Biol. 35, 4121–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J. H., Giannikopoulos P., Duncan S. A., Wang J., Johansen C. T., Brown J. D., Plutzky J., Hegele R. A., Glimcher L. H., Lee A. H. (2011) The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med. 17, 812–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen C. T., Wang J., McIntyre A. D., Martins R. A., Ban M. R., Lanktree M. B., Huff M. W., Péterfy M., Mehrabian M., Lusis A. J., Kathiresan S., Anand S. S., Yusuf S., Lee A. H., Glimcher L. H., Cao H., Hegele R. A. (2012) Excess of rare variants in non-genome-wide association study candidate genes in patients with hypertriglyceridemia. Circ. Cardiovasc. Genet. 5, 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cefalù A. B., Spina R., Noto D., Valenti V., Ingrassia V., Giammanco A., Panno M. D., Ganci A., Barbagallo C. M., Averna M. R. (2015) Novel CREB3L3 nonsense mutation in a family with dominant hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 35, 2694–2699 [DOI] [PubMed] [Google Scholar]
- 26.Rabinowitz J. D., White E. (2010) Autophagy and metabolism. Science 330, 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanida I., Ueno T., Kominami E. (2008) LC3 and autophagy. Methods Mol. Biol. 445, 77–88 [DOI] [PubMed] [Google Scholar]
- 28.Haspel J., Shaik R. S., Ifedigbo E., Nakahira K., Dolinay T., Englert J. A., Choi A. M. (2011) Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy 7, 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 30.Finck B. N., Kelly D. P. (2006) PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A., Jana M., Modi K., Gonzalez F. J., Sims K. B., Berry-Kravis E., Pahan K. (2015) Activation of peroxisome proliferator-activated receptor α induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J. Biol. Chem. 290, 10309–10324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T. J., Wollenberg A. C., Di Bernardo D., Chan L., Irazoqui J. E., Ballabio A. (2013) TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647–658; erratum: 1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowrey P. L., Takahashi J. S. (2004) Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5, 407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balsalobre A., Damiola F., Schibler U. (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 36.Ma D., Panda S., Lin J. D. (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 30, 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holen I., Gordon P. B., Strømhaug P. E., Seglen P. O. (1996) Role of cAMP in the regulation of hepatocytic autophagy. Eur. J. Biochem. 236, 163–170 [DOI] [PubMed] [Google Scholar]
- 38.Avila D. V., Barker D. F., Zhang J., McClain C. J., Barve S., Gobejishvili L. (2016) Dysregulation of hepatic cAMP levels via altered Pde4b expression plays a critical role in alcohol-induced steatosis. J. Pathol. 240, 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.