Abstract
Silkworm, Bombyx mori, has various advantages as an experimental animal, such as the low cost for rearing and fewer ethical problems. Models utilizing silkworms of infection with pathogenic bacteria have been established for identification of genes encoding virulence factors by large‐scale in vivo screening. In this review, we describe recent progress in the study of silkworm infection models for elucidating the mechanisms of fungi infection. Silkworm infection models have been established for Candida albicans, Candida tropicalis, Candida glabrata and Cryptococcus neoformans, which are yeast type fungi, and Aspergillus fumigatus, Arthroderma vanbreuseghemii, Arthroderma benhamiae, Microsporum canis, Trichophyton rubrum, and Rhizopus oryzae, which are filamentous fungi. Novel genes encoding virulence factors in C. albicans and C. glabrata have been identified by using the silkworm infection models. We here outline the benefits of using silkworm infection models and a strategy for identifying the genes responsible for pathogenicity of microorganisms such as fungi. © 2019 The Authors. Microbiology and Immunology Published by The Societies and John Wiley & Sons Australia, Ltd.
Keywords: human pathogenic fungus, infectious disease, silkworm, virulence factor
Abbreviations
- eGFP
enhanced green fluorescent protein
1. INTRODUCTION
1.1. Animal models for understanding infection systems of pathogens
Pathogenic microorganisms infect humans and cause various infectious diseases. Understanding the pathogenic mechanisms of infectious diseases caused by pathogenic microorganisms is necessary for establishing therapeutic and preventive methods. To achieve this, basic research using animal models mimicking human infectious diseases is indispensable.
Models involving infection of various mammals by pathogenic microorganisms have been proposed 1, 2, 3. However, use of mammalian animals has problems in terms of both cost and ethical considerations. Therefore, studies of screening for identifying virulence factors of pathogenic microorganisms using mammals are not easy, because many animals are required. To overcome these problems, infection models using invertebrates such as fruit flies, nematodes, and greater wax moths have been proposed 4, 5, 6, 7. Invertebrate animals generally have the following benefits compared with mammals: (i) lower cost of breeding the animals, (ii) larger numbers of individuals can be reared in a small space, (iii) fewer ethical problems concerning killing the animals, and (iv) fewer samples needed because of the smaller body sizes (Table 1) 8.
Table 1.
In vivo infection models with invertebrate animals and mice
| Animals | Cost of rearing | Space for rearing | Permission from the Ethics Committee | Requirement for biosafety measures | Quantitative injection of samples with a syringe | Reference |
|---|---|---|---|---|---|---|
| Silkworm (larva) [Bombyx mori] | Low | Small | Not necessary | Low | Easy | 53 |
| Fruit fly (adult) [Drosophila melanogaster] | Low | Small | Not necessary | High | Difficult | 75 |
| Nematode [Caenorhabditis elegans] | Low | Small | Not necessary | Low | Difficult | 76 |
| Greater wax moth (larva) [Galleria mellonella] | Low | Small | Not necessary | Low | Easy | 77 |
| Mouse [Mus musculus] | High | Large | Necessary | High | Easy | 78, 79, 80 |
This table is a modified version from Ishii et al. 12, with permission.
Silkworms have been proposed as an experimental model animal for pathogenic microorganisms that infect humans 9, 10, 11, 12. Various strains of silkworm and rearing methods have been established in the long history of sericulture. Therefore, researchers can easily rear a large number of silkworms in a small space (Table 1). Body temperatures of silkworm can be easily controlled by changing the rearing temperature, whereas this is difficult with mammals. Because silkworms are much larger than fruit flies and nematodes, it is easy to perform experiments requiring injections, for which tuberculin syringes can be used (Figure 1). Researchers can inject accurate volumes of pathogen culture and solutions of drugs into silkworms. Furthermore, in silkworm experiments, researchers can distinguish between intra‐hemolymph and intra‐midgut injections (Figure 1). The former corresponds with intravenous injection in humans and the latter with oral administration. In larva of Galleria mellonella, an invertebrate, approximately 10 μl of sample solution can be injected into hemolymph with using a syringe (Table 1). However, methods for distinguishing between intra‐hemolymph and intra‐midgut injections have not yet been established for these larvae. Given that G. mellonella larvae are smaller than silkworm larvae, accurate injection into the intra‐midgut may be more difficult than with silkworms. Using intra‐midgut injections, silkworms can be used to investigate the pathogenicity of bacteria that infect the intestinal tract. Utilizing the various advantages of the silkworm as an experimental animal, researchers can study molecular mechanisms of infection by pathogenic microorganisms in humans.
Figure 1.
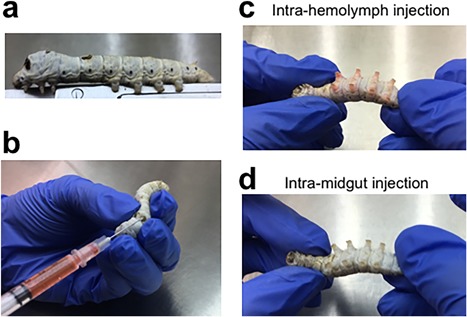
Injection of solution into silkworm. (a) A fifth instar silkworm fed with an artificial diet for one day. (b) Red ink is injected into the silkworm's hemolymph. (c) Red ink has diffused into the silkworm's hemolymph and its legs are stained red (intra‐hemolymph injection). (d) When red ink has been injected into the silkworm's intestinal tract, it stays in the intestinal tract and the silkworm's legs do not stain red (intra‐midgut injection).
1.2. Identification of pathogenic genes of bacteria using a silkworm infection model
Silkworm infection models have been established for human pathogenic bacteria such as Staphylococcus aureus 9, Streptococcus pyogenes 10, Pseudomonas aeruginosa 9, pathogenic Escherichia coli 13, Listeria monocytogenes 14, Serratia marcescens 15, and Vibrio cholerae 9. Silkworms are also killed by injection of extracellular toxins, such as α‐toxin and β‐toxin of S. aureus, exotoxin A of P. aeruginosa, diphtheria toxin, and hemolysin of B. cereus 16, 17. These results suggest that the virulence of various pathogenic bacteria infecting humans can be evaluated using silkworm infection models.
Injection of Porphyromonas gingivalis cells causes silkworm death and such death is not prevented by administration of antibiotics 18. Moreover, silkworms die as a result of excessive activation of the innate immune system induced by P. gingivalis peptidoglycans 18. Silkworms recognize not only peptidoglycan and lipopolysaccharide, which are constituents of bacteria, but also β‐glucan, which is a cell wall component of fungi and activates innate immune systems 19, 20, 21, 22. Thus, these studies suggest that death resulting from excessive activation of the innate immune system by bacterial and fungal infection can be evaluated using a silkworm model.
The silkworm infection model is applicable to a variety of pathogenic microorganisms. The strategy for searching for pathogenic genes of microorganisms is shown in Figure 2. The steps include: (i) preparation of gene‐disrupted mutants of the pathogenic microorganism; (ii) screening for mutants with less ability to kill silkworms; (iii) confirming their low killing ability against mice; and (iv) genetic and biochemical analysis of the function of the protein encoded by the identified pathogenic gene. We have succeeded in identifying several pathogenic genes of some bacteria, including S. aureus, by this strategy 10, 23, 24, 25, 26. We constructed a gene‐deficient mutant library of S. aureus and then screened avirulent mutants that showed low pathogenicity against silkworms. Novel genes, cvfA, cvfB, and cvfC, have been identified by screening using a silkworm infection model with S. aureus 10. Mutants of these genes also showed lower pathogenicity against mice 10. Similarly, mechanisms of fungal infection can be clarified by this strategy. In this review focusing on various pathogenic fungi, we outline methods for evaluating their pathogenicity using a silkworm infection model and for discovering novel pathogenic genes using a silkworm infection model.
Figure 2.
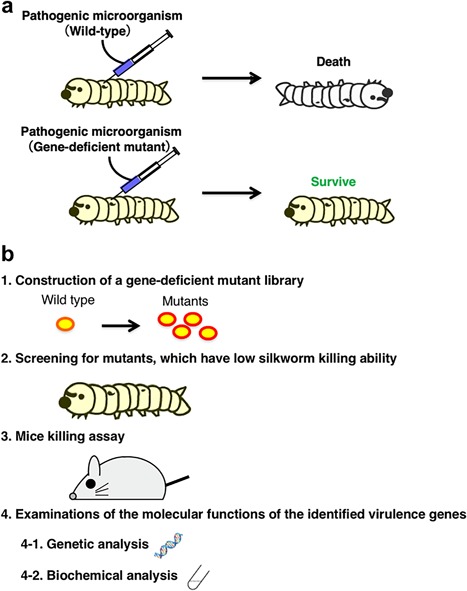
Identification of virulence factors of pathogens using a silkworm infection model. (a) Method for evaluating avirulent mutants from a gene‐deficient mutant library. The wild type strain of a pathogen or gene‐deficient mutants are injected into silkworms and the number that survive measured. (b) Strategies for clarifying the infectious systems of pathogenic microorganisms. A gene‐deficient mutant library of a pathogenic microorganism is prepared and new genes necessary for pathogenicity to silkworms identified. Furthermore, the pathogenicity of the identified mutants against mice is confirmed. Genetic analyses of the identified pathogenic genes and biochemical analysis of the proteins (gene products) are carried out.
2. SILKWORM INFECTION MODELS WITH PATHOGENIC FUNGI
Studies using silkworm infection models with individual pathogenic fungi are described below.
2.1. Candida albicans
C. albicans, a resident fungus on the human body surface and in the gastrointestinal tract and vaginal mucosa, causes candidiasis in patients with reduced immunity 27, 28. C. albicans adheres to tissues and forms hyphae, causing tissue destruction and inflammation 29. Moreover, C. albicans regulates hyphal formation by signal transduction via a protein kinase pathway 30, 31, 32, 33, 34. CMP1 (CNA1), a component of calcineurin complex, which is a serine/threonine protein kinase, is necessary for the pathogenicity of C. albicans mice 30, 31, 34. Protein kinases SIT4 and YVH1 are also required for infection against mice 32. Therefore, these protein kinase pathways are thought to play an important role in regulation of infection against mammals.
Silkworms reportedly die when incubated at 27°C after injection of C. albicans cells into their hemolymph 35. Administration of antifungal drugs to silkworms infected with C. albicans has a therapeutic effect. Thus, killing of silkworms seems to require proliferation of C. albicans within them.
Hanaoka and colleagues constructed gene‐deficient mutants of 21 protein kinases of C. albicans and examined their pathogenicity against silkworms 36. The cmp1, sit4, and yvh1 genes are required for infection of C. albicans against mice 32, 33, 37. Mutants deficient in these genes also showed reduced ability to kill silkworms 36. In addition, deficiency of the ptc1 gene, which has not been reported to be associated with pathogenicity of C. albicans, causes decreased ability to kill silkworms 36. The pathogenicity of a ptc1 gene‐deficient mutant of C. albicans has also been shown to be decreased in mice 36. This study suggests that novel virulence genes of C. albicans can be identified by using a silkworm infection model. Thus, the silkworm infection model is a useful in vivo means of identifying novel virulence factors and exploring the infectious system of C. albicans.
2.2. Candida tropicalis
C. tropicalis infects patients with neutropenia is frequently isolated from patients with leukemia 38, 39, 40. In patients with neutropenia, C. tropicalis is considered more likely to form disseminated lesions than C. albicans. Therefore, C. tropicalis and C. albicans may have different regulatory mechanisms for pathogenicity; however, the molecular mechanisms remain uncertain.
When silkworms are injected with C. tropicalis and incubated at 27°C, they die; however their death under these conditions can be prevented by administration of antifungal drugs 35. Therefore, the silkworm infection model is useful for evaluating the pathogenicity of C. tropicalis. Now that a technique for constructing gene‐deficient mutants of C. tropicalis has been established 41, a gene‐deficient mutant library can be prepared using the technique. It is that new virulence genes of C. tropicalis will be identified by screening avirulent mutants from a gene‐deficient mutant library using a silkworm infection model.
2.3. Candida glabrata
C. glabrata is an opportunistic fungus that is resident in the human intestinal tract and infects individuals with diabetes 42. C. glabrata is rarely isolated alone and is often isolated with other Candida species such as C. albicans 43. Moreover, C. glabrata is resistant to azoles, antifungal drugs; thus, relapse of infections with this organism is a problem clinically 44. Because C. glabrata has low infectivity for mice, establishment of an experimental animal system has been difficult.
Silkworms infected with C. glabrata do not die within 4 days when incubated at 27°C. To establish a silkworm infection model with C. glabrata, we constructed an experimental system for infection by this organism using diabetic silkworms. Feeding a high glucose diet induces diabetes in silkworms 45, 46, 47. When diabetic silkworms injected with C. glabrata are incubated at 37°C, they die within 3 days 48.
Ueno and colleagues have constructed a gene‐deficient mutant library of C. glabrata and searched for genes necessary for infection caused by C. glabrata using a diabetic silkworm infection model 48. Screening for avirulent mutants in this model resulted in identification of lactate dehydrogenase Cyb2 as a virulence‐related factor in C. glabrata 48. A mutant of the gene encoding the lactate dehydrogenase Cyb2 of C. glabrata had decreased ability to kill the diabetic silkworms 48. Furthermore, in an experimental system of intestinal colonization using diabetic mice, the cyb2 gene‐deficient mutant of C. glabrata was found to have decreased ability to adapt in the intestinal tract 48. The CYB2 gene in S. cerevisiae is regulated at the transcriptional stage by the Hap family transcription factor, which is activated by recognizing glucose deprivation 49. Ueno and colleagues consider that cyb2 gene expression is controlled by Hap family transcription factors in C. glabrata and investigated this using gene‐deficient mutants of the transcription factors. The expression of the cyb2 gene was decreased in deficient mutants of hap2 and hap5 genes of C. glabrata 48. However, the deficient mutant of the hap1 gene in C. glabrata was not found to have decreased expression of the cyb2 gene 48. Strains of C. glabrata deficient in the hap2 and the hap5 genes have reduced ability to kill diabetic silkworms, whereas the hap1 gene‐deficient mutant has not been found to have decreased killing ability 48. These results suggest that C. glabrata adapts to environments of diabetic hosts by promoting expression of Cyb2 via transcription factors Hap2 and Hap5. Thus, research using the gene‐deficient mutant library of C. glabrata and a silkworm infection model has revealed a novel mechanism that is necessary for infection of C. glabrata.
2.4. Cryptococcus neoformans
C. neoformans causes cryptococcosis, a fatal fungal disease 50, and is frequently detected in patients with reduced immunity. Patients with AIDS in Africa south of the Sahara Desert have died of infection with C. neoformans 51. C. neoformans meningitis, which has a high mortality rate, is particularly problematic in areas where AIDS is prevalent 52.
Although silkworms infected with C. neoformans do not die after incubation at 27°C for 4 days, they die within 3 days when incubated at 37°C 53. C. neoformans strains of serotype A have a higher infectivity in mammals than those of Serotype D 54. In infection experiments using silkworms, C. neoformans strains of Serotype A had a greater ability to kill silkworms than those of Serotype D 53, suggesting that a silkworm infection model is useful for distinguishing between weakly pathogenic and highly pathogenic strains of C. neoformans. C. neoformans cells translocate to the brain by evading host immunity as a result of capsular formation and melanin production 55. In C. neoformans, capsule formation and melanin production are regulated by GPA1, an α‐subunit of G protein, PKA1, a cyclic AMP‐dependent protein kinase, and CNA1, a catalytic subunit of calcineurin 56, 57, 58. Thus, the pathogenicity of C. neoformans is controlled by various signaling pathways, including the GPA‐PKA and calcineurin pathways 59. Deficient mutants of gpa1, pka1 and cna1 genes, which are necessary for the pathogenicity of C. neoformans against mammals, are less able to kill silkworms than the parent strain 53. On the basis of these results, it is expected that silkworm infection models will be used to clarify the mechanism of pathogenicity of C. neoformans via the GPA‐PKA and calcineurin pathways.
2.5. Aspergillus fumigatus
A. fumigatus, a type of environmental filamentous fungus that is widely present in nature, causes opportunistic infections 60. A. fumigatus has low infectivity against healthy individuals, but causes pulmonary infections in humans with reduced immunity 61. Given that the number of patients with invasive aspergillosis caused by A. fumigatus is increasing and this disease is fatal unless treatment is started early, this pathogen is a clinical problem 62, 63, 64.
When silkworms are injected with A. fumigatus and incubated at 27°C, they die 65. Amphotericin B and voriconazole, anti‐fungal drugs that are used to treat aspergillosis show therapeutic effects against silkworms infected with A. fumigatus 65. Using a silkworm infection model, Nakamura and colleagues succeeded in identifying a novel antifungal agent, ASP2397, for treating infections caused by A. fumigatus 65. Whether it will be possible to identify pathogenic genes of A. fumigatus by using a silkworm infection model is yet to be determined.
2.6. Arthroderma vanbreuseghemii, Arthroderma benhamiae, Microsporum canis and Trichophyton rubrum
A. vanbreuseghemii, A. benhamiae, M. canis and T. rubrum are all fungi that can cause dermatophytoses 66. Given that a quarter of the world's population contracts superficial cutaneous fungal infections caused by these dermatophytes, they are a problem worldwide 67. Dermatophytes grow as hyphae, forming filamentous structures that are referred to as a mycelium. The hyphae can damage host tissues and cause inflammation, thus causing superficial mycoses 68. Yamada and colleagues succeeded in establishing a genetic method for constructing recombinant strains of the dermatophyte A. vanbreuseghemii 69. Therefore, it is considered that it will be possible to evaluate pathogenic factors of dermatophytes by studying their gene‐deficient mutants in an infected animal model. A skin infection model using guinea pigs has been established; however, this model is costly and associated with ethical problems. Therefore, it has been difficult to conduct large‐scale in vivo screening to isolate avirulent mutants from a gene‐deficient mutant library.
A silkworm infection model for dermatophytes has been established 8. After injection of conidia of A. vanbreuseghemii, silkworms die within 100 hours of incubation at 30°C 8. Moreover, germination of conidia of A. vanbreuseghemii in a liquid medium is strongly pathogenic to silkworms. Furthermore, microscopic analysis of a strain of A. vanbreuseghemii expressing eGFP revealed that this fungus forms hyphae in the organs of silkworms (Figure 3). Terbinafine is an antifungal agent that inhibits hyphal formation by dermatophytes. Administration of terbinafine to silkworms injected with A. vanbreuseghemii suppresses hyphal formation in their organs and the infected silkworms administrated with terbinafine live longer than those that do not receive terbinafine 8. These results suggest that hyphal formation of A. vanbreuseghemii plays an important role in its pathogenicity to silkworms. Therefore, it is expected that the pathogenic factors related to hyphal formation by A. vanbreuseghemii will be identified by using a silkworm infection model.
Figure 3.
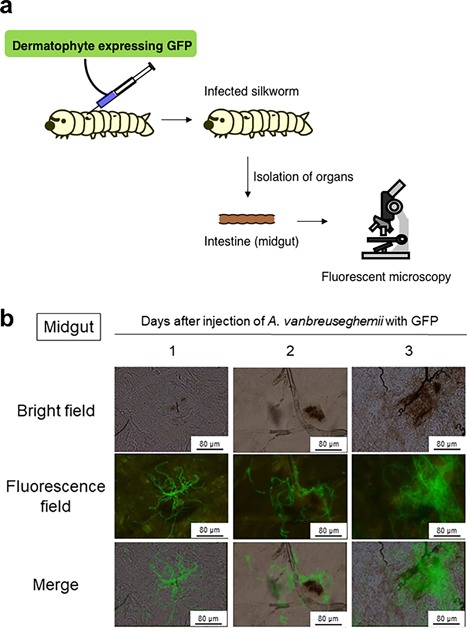
Visualization of infection in organs of silkworm using a dermatophyte expressing eGFP. (a) Method for evaluation of hyphal growth of dermatophytes in organs of silkworm by fluorescent imaging. (b) Fluorescence microscope images of midgut of silkworm infected with A. vanbreuseghemii expressing eGFP . Figure 3b was reproduced from Ishii et al., (8) with permission.
Because a single hypha may comprise numerous cells, quantification of hyphal formation in animals by a colony counting method is difficult 70. To overcome this problem, a fluorescence imaging system has been developed for quantitatively evaluating dermatophyte growth in vivo. Hyphal formation can be evaluated on the basis of detection of the fluorescence of eGFP‐expressing dermatophytes in silkworms 8. Thus, a silkworm infection model with dermatophytes expressing eGFP is useful for evaluating hyphal formation in vivo.
In addition, other dermatophytes such as T. rubrum, A. benhamiae and M. canis kill silkworms under the same experimental conditions as A. vanbreuseghemii 8. In particular, silkworms are a useful animal for investigating infection with T. rubrum, which is the most frequent cause of dermatophytosis in humans 71. Thus far, silkworm models can only determine the pathogenicity of dermatophytes by monitoring the death of individual animals. Research on identification of pathogenic factors using a gene‐deficient mutant library of dermatophytes and silkworm infection models is expected to progress.
2.7. Rhizopus oryzae
Rhizopus oryzae, which infects from the nasal cavities of severely immunocompromised patients, causes deep mycosis 72. There are many clinical reports but few basic studies on the pathogenic factors of R. oryzae.
Tominaga and colleagues found that silkworms die when incubated at 27°C after being injected with R. oryzae 73. Furthermore, administration of amphotericin B increases the survival time of these silkworms 73. A silkworm infection model with R. oryzae is expected to contribute to the understanding of zygomycosis.
3. SILKWORM DEATH CAUSED BY FUNGAL INFECTION
The fact that antifungal drugs can be successfully used to treat silkworms in silkworm infection models with pathogenic fungi indicates that growth of fungi contributes to the silkworms’ death. Calcineurin is involved in the hyphal formation that enables C. albicans to kill silkworms; tissue damage and inflammation caused by hyphal formation may cause silkworm death. In comparison, in C. neoformans, calcineurin contributes to cell wall synthesis at 37°C 74. Cell wall components of fungi activate silkworms’ immune systems 19. The cell wall components of C. neoformans, which grows in the bodies of silkworms, may induce excessive immunity that causes silkworm death. Elucidation of the molecular mechanisms of silkworm death caused by fungal infection is an important subject for future study.
4. CONCLUSIONS
In this review, we have stated that silkworm infection models for several pathogenic fungi have been established and that genes necessary for their pathogenicity can be identified by screening of gene‐deficient mutant libraries. Being relatively cheap and lacking ethical problems, silkworms are suitable for large‐scale in vivo screening. In silkworm infection models, it is easy to quantitatively measure pathogenicity of fungi on the basis of determination of the lethal dose for 50% of these animals. Moreover, it is possible to establish an infectious system for investigating weakly pathogenic fungi, which are difficult to investigate in mammals, by changing the incubation temperature (Table 2). Silkworm infection models will contribute to establishing novel preventive and therapeutic strategies by elucidating the infectious systems of pathogenic fungi.
Table 2.
Assay conditions in silkworm infection models with pathogenic fungi
| Species | Incubation temperature | Diabetic state | Reference |
|---|---|---|---|
| Candida albicans | 27°C | − | 35 |
| Candida tropicalis | 27°C | − | 35 |
| Candida glabrata | 37°C | + | 48 |
| Cryptococcus neoformans | 37°C | − | 53 |
| Aspergillus fumigatus | 27°C | − | 65 |
| Arthroderma vanbreuseghemii | 30°C | − | 8 |
| Arthroderma benhamiae | 30°C | − | 8 |
| Microsporum canis | 30°C | − | 8 |
| Trichophyton rubrum | 30°C | − | 8 |
| Rizopus oryzae | 27°C | − | 73 |
Acknowledgment
This work was supported by JSPS KAKENHI (Grant No. JP15H05783 for scientific research to KS and Grant No. JP17K08288 for scientific research to YM). The authors declare that they have no conflicts of interest regarding this work.
Matsumoto Y, Sekimizu K. Silkworm as an experimental animal for research on fungal infections. Microbiol Immunol. 2019;63:41–50. 10.1111/1348-0421.12668
Contributor Information
Yasuhiko Matsumoto, Email: ymatsumoto@main.teikyo-u.ac.jp.
Kazuhisa Sekimizu, Email: sekimizu@main.teikyo-u.ac.jp.
REFERENCES
- 1. Cambier L, Heinen M‐P, Mignon B. Relevant animal models in dermatophyte research. Mycopathologia. 2017; 182:229–240. [DOI] [PubMed] [Google Scholar]
- 2. Lewis RE, Verweij PE. Animal models for studying triazole resistance in Aspergillus fumigatus . J Infect Dis. 2017; 216:S466–S473. [DOI] [PubMed] [Google Scholar]
- 3. Segal E, Frenkel M. Experimental in vivo models of candidiasis. J Fungi (Basel). 2018; 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuo C‐J, Hansen M, Troemel E. Autophagy and innate immunity: Insights from invertebrate model organisms. Autophagy. 2018; 14:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singulani JL, Scorzoni L, de Oliveira HC, et al. Applications of invertebrate animal models to dimorphic fungal infections. J Fungi (Basel). 2018; 4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. AL‐Maliki HS, Martinez S, Piszczatowski P, Bennett JW. Drosophila melanogasteras as a model for studying Aspergillus fumigatus . Mycobiology. 2018; 45:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elkabti A, Issi L, Rao R. Caenorhabditis elegans as a model host to monitor the Candida infection processes. J Fungi (Basel). 2018; 4:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ishii M, Matsumoto Y, Yamada T, Abe S, Sekimizu K. An invertebrate infection model for evaluating anti‐fungal agents against dermatophytosis. Sci Rep. 2017; 7:12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaito C, Akimitsu N, Watanabe H, Sekimizu K. Silkworm larvae as an animal model of bacterial infection pathogenic to humans. Microb Pathog. 2002; 32:183–190. [DOI] [PubMed] [Google Scholar]
- 10. Kaito C, Kurokawa K, Matsumoto Y, et al. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol. 2005; 56:934–944. [DOI] [PubMed] [Google Scholar]
- 11. Miyazaki S, Matsumoto Y, Sekimizu K, Kaito C. Evaluation of Staphylococcus aureus virulence factors using a silkworm model. FEMS Microbiol Lett. 2012; 326:116–124. [DOI] [PubMed] [Google Scholar]
- 12. Ishii M, Matsumoto Y, Sekimizu K. Usefulness of silkworm as a host animal for understanding pathogenicity of Cryptococcus neoformans . Drug Discov Ther. 2016; 10:9–13. [DOI] [PubMed] [Google Scholar]
- 13. Miyashita A, Iyoda S, Ishii K, Hamamoto H, Sekimizu K, Kaito C. Lipopolysaccharide O‐antigen of enterohemorrhagic Escherichia coli O157:H7 is required for killing both insects and mammals. FEMS Microbiol Lett. 2012; 333:59–68. [DOI] [PubMed] [Google Scholar]
- 14. Castillo Y, Suzuki J, Watanabe K, Shimizu T, Watarai M. Effect of vitamin A on Listeria monocytogenes infection in a silkworm model. PLoS ONE 2016; 11:e0163747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishii K, Adachi T, Imamura K, et al. Serratia marcescens induces apoptotic cell death in host immune cells via a lipopolysaccharide‐ and flagella‐dependent mechanism. J Biol Chem. 2012; 287:36582–36592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hossain MS, Hamamoto H, Matsumoto Y, et al. Use of silkworm larvae to study pathogenic bacterial toxins. J Biochem. 2006; 140:439–444. [DOI] [PubMed] [Google Scholar]
- 17. Usui K, Miyazaki S, Kaito C, Sekimizu K. Purification of a soil bacteria exotoxin using silkworm toxicity to measure specific activity. Microb Pathog. 2009; 46:59–62. [DOI] [PubMed] [Google Scholar]
- 18. Ishii K, Hamamoto H, Imamura K, et al. Porphyromonas gingivalis peptidoglycans induce excessive activation of the innate immune system in silkworm larvae. J Biol Chem. 2010; 285:33338–33347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohta M, Watanabe A, Mikami T, et al. Mechanism by which Bombyx mori hemocytes recognize microorganisms: direct and indirect recognition systems for PAMPs. Dev Comp Immunol. 2006; 30:867–877. [DOI] [PubMed] [Google Scholar]
- 20. Ha Lee J, Hee Lee I, Noda H, Mita K, Taniai K. Verification of elicitor efficacy of lipopolysaccharides and peptidoglycans on antibacterial peptide gene expression in Bombyx mori . Insect Biochem Mol Biol. 2007; 37:1338–1347. [DOI] [PubMed] [Google Scholar]
- 21. Miyashita A, Kizaki H, Kawasaki K, Sekimizu K, Kaito C. Primed immune responses to gram‐negative peptidoglycans confer infection resistance in silkworms. J Biol Chem 2014; 289:14412–14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyashita A, Takahashi S, Ishii K, Sekimizu K, Kaito C. Primed immune responses triggered by ingested bacteria lead to systemic infection tolerance in silkworms. PLoS ONE. 2015; 10:e0130486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus . Mol Microbiol. 2006; 62:1601–1617. [DOI] [PubMed] [Google Scholar]
- 24. Kyuma T, Kimura S, Hanada Y, Suzuki T, Sekimizu K, Kaito C. Ribosomal RNA methyltransferases contribute to Staphylococcus aureus virulence. FEBS J. 2015; 282:2570–2584. [DOI] [PubMed] [Google Scholar]
- 25. Kyuma T, Kizaki H, Ryuno H, Sekimizu K, Kaito C. 16S rRNA methyltransferase KsgA contributes to oxidative stress resistance and virulence in Staphylococcus aureus . Biochimie. 2015; 119:166–174. [DOI] [PubMed] [Google Scholar]
- 26. Imae K, Saito Y, Kizaki H, et al. Novel nucleoside diphosphatase contributes to Staphylococcus aureus virulence. J Biol Chem. 2016; 291:18608–18619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pappas PG, Lionakis MS, Arendrup MC, Ostrosky‐Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. 2018; 4:18026. [DOI] [PubMed] [Google Scholar]
- 28. Naglik JR, König A, Hube B, Gaffen SL. Candida albicans–epithelial interactions and induction of mucosal innate immunity. Curr Opin Microbiol. 2017; 40:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Desai JV. Candida albicans hyphae: from growth initiation to invasion. J Fungi (Basel). 2018; 4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu S‐J, Chang Y‐L, Chen Y‐L. Calcineurin signaling: lessons from Candida species. FEMS Yeast Res. 2015; 15:fov016. [DOI] [PubMed] [Google Scholar]
- 31. Blankenship JR, Wormley FL, Boyce MK, et al. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryotic Cell. 2003; 2:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee C‐M, Nantel A, Jiang L, Whiteway M, Shen S‐H. The serine/threonine protein phosphatase SIT4 modulates yeast‐to‐hypha morphogenesis and virulence in Candida albicans . Mol Microbiol. 2004; 51:691–709. [DOI] [PubMed] [Google Scholar]
- 33. Hanaoka N, Umeyama T, Ueno K, et al. A putative dual‐specific protein phosphatase encoded by YVH1 controls growth, filamentation and virulence in Candida albicans . Microbiology. 2005; 151:2223–2232. [DOI] [PubMed] [Google Scholar]
- 34. Bader T, Schröppel K, Bentink S, Agabian N, Köhler G, Morschhäuser J. Role of calcineurin in stress resistance, morphogenesis, and virulence of a Candida albicans wild‐type strain. Infect Immun. 2006; 74:4366–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamamoto H, Kurokawa K, Kaito C, et al. Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob Agents Chemother. 2004; 48:774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanaoka N, Takano Y, Shibuya K, Fugo H, Uehara Y, Niimi M. Identification of the putative protein phosphatase gene PTC1 as a virulence‐related gene using a silkworm model of Candida albicans infection. Eukaryotic Cell. 2008; 7:1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bader T, Bodendorfer B, Schröppel K, Morschhäuser J. Calcineurin is essential for virulence in Candida albicans . Infect Immun. 2003; 71:5344–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kontoyiannis DP, Vaziri I, Hanna HA, et al. Risk factors for Candida tropicalis fungemia in patients with cancer. Clin Infect Dis. 2001; 33:1676–1681. [DOI] [PubMed] [Google Scholar]
- 39. Goldani LZ, Mário PSS. Candida tropicalis fungemia in a tertiary care hospital. J Infect. 2003; 46:155–160. [DOI] [PubMed] [Google Scholar]
- 40. Pappas PG. Invasive candidiasis. Infect Dis Clin North Am. 2006; 20:485–506. [DOI] [PubMed] [Google Scholar]
- 41. Lin C‐J, Wu C‐Y, Yu S‐J, Chen Y‐L. Protein kinase A governs growth and virulence in Candida tropicalis . Virulence. 2018; 9:331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Segireddy M, Johnson LB, Szpunar SM, Khatib R. Differences in patient risk factors and source of candidaemia caused by Candida albicans and Candida glabrata . Mycoses. 2011; 54:e39–43. [DOI] [PubMed] [Google Scholar]
- 43. Rodrigues CF, Silva S, Henriques M. Candida glabrata: a review of its features and resistance. Eur J Clin Microbiol Infect Dis. 2014; 33:673–688. [DOI] [PubMed] [Google Scholar]
- 44. Kamikawa Y, Mori Y, Nagayama T, et al. Frequency of clinically isolated strains of oral Candida species at Kagoshima University Hospital, Japan, and their susceptibility to antifungal drugs in 2006–2007 and 2012–2013. BMC Oral Health. 2014; 14:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Matsumoto Y, Sumiya E, Sugita T, Sekimizu K. An invertebrate hyperglycemic model for the identification of anti‐diabetic drugs. PLoS ONE. 2011; 6:e18292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsumoto Y, Ishii M, Hayashi Y, et al. Diabetic silkworms for evaluation of therapeutically effective drugs against type II diabetes. Sci Rep. 2015; 5:10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsumoto Y, Sekimizu K. Evaluation of anti‐diabetic drugs by using silkworm, Bombyx mori . Drug Discov Ther 2016; 10:19–23. [DOI] [PubMed] [Google Scholar]
- 48. Ueno K, Matsumoto Y, Uno J, et al. Intestinal resident yeast Candida glabrata requires Cyb2p‐mediated lactate assimilation to adapt in mouse intestine. PLoS ONE. 2011; 6:e24759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ramil E, Agrimonti C, Shechter E, Gervais M, Guiard B. Regulation of the CYB2 gene expression: transcriptional co‐ordination by the Hap1p, Hap2/3/4/5p and Adr1p transcription factors. Mol Microbiol. 2000; 37:1116–1132. [DOI] [PubMed] [Google Scholar]
- 50. Srikanta D, Santiago‐Tirado FH, Doering TL. Cryptococcus neoformans: historical curiosity to modern pathogen. Yeast. 2014; 31:47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009; 23:525–530. [DOI] [PubMed] [Google Scholar]
- 52. Limper AH, Adenis A, Le T, Harrison TS. Fungal infections in HIV/AIDS. Lancet Infect. Dis 2017; 17:e334–e343. [DOI] [PubMed] [Google Scholar]
- 53. Matsumoto Y, Miyazaki S, Fukunaga DH, Shimizu K, Kawamoto S, Sekimizu K. Quantitative evaluation of cryptococcal pathogenesis and antifungal drugs using a silkworm infection model with Cryptococcus neoformans . J Appl Microbiol. 2012; 112:138–146. [DOI] [PubMed] [Google Scholar]
- 54. Lin X, Nielsen K, Patel S, Heitman J. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans . Infect Immun. 2008; 76:2923–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi M, Mody CH. Fungal infection in the brain: what we learned from intravital imaging. Front Immunol. 2016; 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G‐protein alpha subunit GPA1 and cAMP. Genes Dev. 1997; 11:3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. D'Souza CA, Alspaugh JA, Yue C, et al. Cyclic AMP‐dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans . Mol Cell Biol. 2001; 21:3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans . EMBO J. 1997; 16:2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus . Cell Microbiol. 2009; 11:370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé J‐P. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol. 2017; 15:661–674. [DOI] [PubMed] [Google Scholar]
- 61. Latgé JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 1999; 12:310–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Denning DW. Invasive aspergillosis. Clin Infect Dis. 1998; 26:781–803. [DOI] [PubMed] [Google Scholar]
- 63. Patterson TF, Kirkpatrick WR, White M, et al. Invasive aspergillosis. Disease spectrum, treatment practices, and outcomes. I3 Aspergillus study group. Medicine (Baltimore). 2000; 79:250–260. [DOI] [PubMed] [Google Scholar]
- 64. Marr KA, Carter RA, Boeckh M, Martin P, Corey L. Invasive aspergillosis in allogeneic stem cell transplant recipients: changes in epidemiology and risk factors. Blood. 2002; 100:4358–4366. [DOI] [PubMed] [Google Scholar]
- 65. Nakamura I, Kanasaki R, Yoshikawa K, et al. Discovery of a new antifungal agent ASP2397 using a silkworm model of Aspergillus fumigatus infection. J Antibiot. 2017; 70:41–44. [DOI] [PubMed] [Google Scholar]
- 66. Garber G. An overview of fungal infections. Drugs. 2001; 61:1–12. [DOI] [PubMed] [Google Scholar]
- 67. Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008; 51:2–15. [DOI] [PubMed] [Google Scholar]
- 68. Rouzaud C, Hay R, Chosidow O, et al. Severe dermatophytosis and acquired or innate immunodeficiency: a review. J Fungi (Basel). 2016; 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yamada T, Makimura K, Uchida K, Yamaguchi H. Reproducible genetic transformation system for two dermatophytes, Microsporum canis and Trichophyton mentagrophytes . Med Mycol. 2005; 43:533–544. [DOI] [PubMed] [Google Scholar]
- 70. Clemons KV, Stevens DA. Conventional or molecular measurement of Aspergillus load. Med Mycol. 2009; 47:S132–S137. [DOI] [PubMed] [Google Scholar]
- 71. Zhan P, Liu W. The changing face of dermatophytic infections worldwide. Mycopathologia. 2017; 182:77–86. [DOI] [PubMed] [Google Scholar]
- 72. Mohammadi R, Nazeri M, Sayedayn SMA, Ehteram H. A successful treatment of rhinocerebral mucormycosis due to Rhizopus oryzae . J Res Med Sci. 2014; 19:72–74. [PMC free article] [PubMed] [Google Scholar]
- 73. Tominaga T, Uchida R, Koyama N, Tomoda H. Anti‐Rhizopus activity of tanzawaic acids produced by the hot spring‐derived fungus Penicillium sp. BF‐0005. J Antibiot. 2018; 71:626–632. [DOI] [PubMed] [Google Scholar]
- 74. Chow EWL, Clancey SA, Billmyre RB, et al. Elucidation of the calcineurin‐Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans . PLOS Genetics. 2017; 13:e1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryotic Cell. 2004; 3:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA. 2002; 99:15675–15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mylonakis E, Moreno R, Khoury El JB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005; 73:3842–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zaragoza O, Alvarez M, Telzak A, Rivera J, Casadevall A. The relative susceptibility of mouse strains to pulmonary Cryptococcus neoformans infection is associated with pleiotropic differences in the immune response. Infect Immun. 2007; 75:2729–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Desalermos A, Tan X, Rajamuthiah R, et al. A multi‐host approach for the systematic analysis of virulence factors in Cryptococcus neoformans . J Infect Dis. 2015; 211:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McClelland EE, Hobbs LM, Rivera J, et al. The role of host gender in the pathogenesis of Cryptococcus neoformans infections. PLoS ONE. 2013; 8:e63632. [DOI] [PMC free article] [PubMed] [Google Scholar]


