Abstract
Type 2 diabetes (T2D) is widespread, affecting the health of hundreds of millions worldwide. The disease results from the complex interplay of lifestyle factors acting on a backdrop of inherited DNA risk variants. Detecting and understanding biomarkers, whether genotypes or other downstream biological features that dictate a person’s phenotypic response to different lifestyle exposures, may have tremendous utility in the prevention of T2D. Here, we explore (i) evidence of how human genetic adaptation to diverse local environments might interact with lifestyle factors in T2D, (ii) the key challenges facing the research area of gene × lifestyle interactions in T2D, and (iii) the solutions that might be pursued in future studies. Overall, many preliminary examples of such interactions exist, but none is sufficient to have a major impact on clinical decision making. Future studies, integrating genetics and other biological markers into regulatory networks, are likely to be necessary to facilitate the integration of genomics into lifestyle medicine in T2D.
Introduction
The burden of type 2 diabetes (T2D) is not equally arrayed, as susceptibility to diabetogenic environmental factors tends to segregate by ethnicity and within families [1•,2]. This observation has led many to recognize that diabetogenic environmental factors are likely to convey different effects depending on the genetic background of the individual; it may be necessary to understand this concept, often referred to as ‘gene × lifestyle interaction’, if the etiology of T2D is to be adequately dissected, and lifestyle medicine is to serve its full potential.
The major scientific guidelines for the prevention and treatment of virtually all non-communicable diseases emphasize healthy lifestyle choices as the frontline intervention to prevent disease. However, as is the case with exposure susceptibility, the success of lifestyle medicine in the prevention and treatment of diabetes varies substantially from one person to the next [3••,4]. Much of this may be attributable to adherence to therapeutic recommendations [5]. Nevertheless, there also appears to be a meaningful heritable component to the effects of lifestyle in cardiometabolic disease [6]. Moreover, both gene × gene and gene × environment (G × E) interactions are likely to be important determinants of allelic specific expression, such that between one third and one half of the observed variance in allele specific expression is owing to non-additive genetic effects, including interactions [7]. In combination, these findings raise the possibility that genetic data may be useful in the optimization of lifestyle medicine.
Evidence of genetic adaptation to environmental factors
The first empirical evidence that genes govern phenotypic adaptations to environmental cues came from studies of temperature and eye facet formation in Drosophila [8]. Garrod is often cited as the first to propose mechanistic interactions between genes and food in humans, although none of this early work focused on energy metabolism or diabetes [9]. Indeed, the first commentary about genetic selection in diabetes came decades later, when Neel proposed the “Thrifty genotype hypothesis”, whereby he posited that the high rates of diabetes in contemporary American Indians are the consequence of evolutionary adaptations that favored efficient use and storage of energy during famine and migration [10••]. Although the Thrifty genotype hypothesis is often discussed in the context of genetic adaptations to extreme environmental conditions, there is little empirical evidence to support the existence of thrifty genes [11]. One intriguing exception though, is of an obesity-predisposing variant at CREBRF (p.Arg457Gln), which was discovered in contemporary Samoans [12••]; the variant, which may have protected against starvation during arduous ocean migrations, promotes fat storage and reduces cellular energy metabolism, modestly increasing risk of obesity, but paradoxically reducing the risk of T2D.
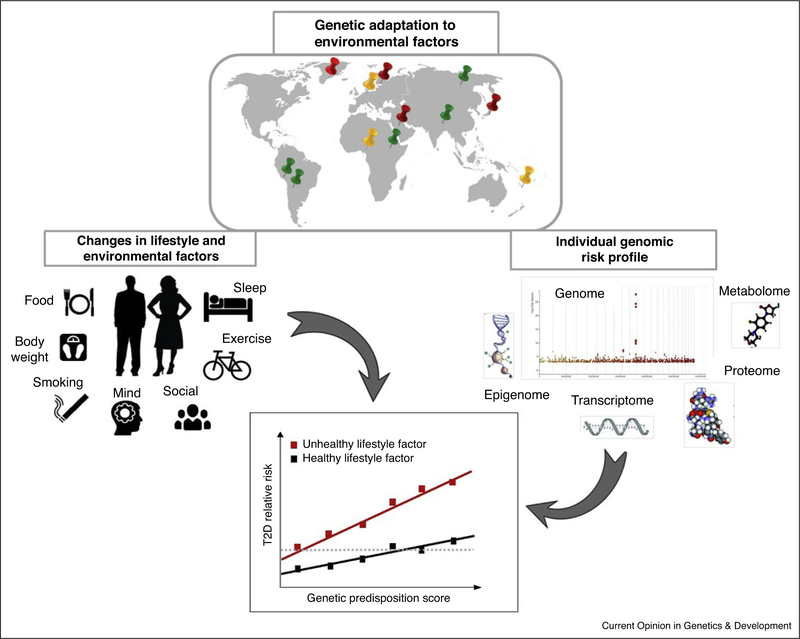
The very rapid emergence of non-autoimmune diabetes in certain indigenous groups, such as Micro-Indonesian Islanders, American Indians, and Greenlandic Inuit, in concert with rapid changes in the environment, has reinforced the belief that T2D is caused by the joint effects of genetic predisposition and environmental triggers [12••,13,14••,15•]. Figure 1 illustrates how genetic adaptation to diverse local environments and downstream genomic information might interact with lifestyle factors in T2D. The Pima Indians of Arizona, for example, have contributed massively to our understanding of diabetes through their decades-long engagement in NIH-led research. The Pima, like many of the indigenous populations of North America were deposed of their land and customs by the Reclamation Act of 1902. The consequential shift away from subsistence farming, toward economically and nutritionally impoverished urban lifestyles, triggered epidemics of obesity and diabetes that persist to this day. The very high lifetime prevalence (>50%) and the very early onset (often in adolescence) of nonautoimmune diabetes in the Pima [13] is in stark contrast to the more favorable disease demographics of European-ancestry populations living in similar circumstances, implicating ethnic-specific catalysts, such as genes. Moreover, the prevalence of diabetes in the closely-related Mexican Pima, who at the time of the study still subsistence farmed, was ~10% in the 1990s [16], suggesting that although the Pima’s susceptibility to diabetes might have a genetic basis, lifestyle is a key modulator of risk. Nevertheless, despite extensive genetic research in this population, little definitive evidence of significant G × E interactions or Thrifty genes has emerged.
Figure 1.
Gene × lifestyle interplay in type 2 diabetes. Schematic representation of G × E interaction in T2D. The spread of humans across the globe has led to genetic adaptation to diverse local environment (red; adaptation to dietary components such as diet rich in fatty acids or cereals, green; genetic adaptation to geographic distribution such as altitude, yellow anthropometric adaptation such as body weight or height. Adapted from Fan S, Hansen ME, Lo Y, Tishkoff SA. Going global by adapting local: A review of recent human adaptation. Science. 2016, 34:54–59). The enriched genome (Manhattan plot for T2D based on the UK Biobank data; available from https://biobankengine.stanford.edu) is unlikely to adapt to rapid changes in environmental factors, therefore genetic susceptible individuals are at higher odds to develop the disease. In addition, downstream genomic information, including epigenomics, transcriptomics, proteomics or metabolomics markers is likely to offer novel insights in the interplay between genomics and lifestyle factors.
By contrast, recent studies of Greenlandic Inuit have yielded convincing examples of how environmental pressures have enriched the human genome with specific loci that affect a person’s response to foods and nutrients [14••,17]. For example, allelic heterogeneity at FADS1 and FADS2 affect fatty acid desaturase activity, which appears to have facilitated selection under varying environmental/dietary conditions [17]. FADS mutations that the Greenlandic Inuit carry are associated with increased levels of linoleic acid and a-linolenic acid (short-chain PUFAs), and decreased levels of arachidonic acid and eicosapentaenoic acid (long-chain PUFAs); accordingly, it appears that these specific FADS variants may be frequent in this population because fatty marine animals were a dominant dietary component throughout the Inuit’s recent evolution [14••]. By contrast, in European populations, selection has favored alleles associated with a decrease in linoleic acid levels and an increase in eicosapentaenoic acid. [17]. A recent multi-cohort analysis, comprised of adults from Europe and North America, showed that Inuit-specific SNPs at the FADS locus do not appear to modify the relationship of omega 6 fatty acids biomarker levels and incident T2D [18].
Cold exposure may also have driven the selection of loci implemented in energy metabolism in the Greenlandic Inuit. For example, a highly-prevalent (minor allele frequency [MAF] of 17%) nonsense variant (p.Arg684ter) at the Tre-2/BUB2/cdc 1 domain family 4 (TBC1D4) locus severely inhibits post-prandial cellular glucose uptake in Greenlandic Inuit [15•]the variant is present in other Inuit populations, but essentially absent in non-Inuit. The protein encoded by TBC1D4 (and TBC1D1) (Rab-GTPase-activating proteins/AS160) regulates the effects of insulin and exercise on skeletal muscle oxidation and transportation of glucose and fatty acids (see [19]). In mice, variants at Tbc1d1 modulate exercise-dependent uptake of glucose into skeletal muscle and body weight, although supportive data in humans has not been published. However, Greenlandic Inuit carrying both copies of p.Arg684ter variant are at relatively high risk of T2D (odds ratio = 10.3) [15•], owing to decreased postprandial glucose transport caused by muscle-selective loss of the long TBC1D4 isoform and corresponding reductions in GLUT4-mediated insulin-stimulated glucose uptake. Furthermore, a thymine for cytosine substitution (at nucleotide 1087 in exon 3, which causes a premature stop at codon 363) has been shown to convey an exaggerated first-phase insulin response to glucose in people with acanthosis nigricans [20]. Whether phosphorylation of AS160 and GLUT4 translocation are affected by TBC1D4 variants or whether such variants interact with exercise in outbred populations is unknown.
Elsewhere, a common risk haplotype in the SLC16A11 locus (MAF ~40–50% among people of Mexican or Latin American descent, but rare among Europeans and absent in Africa) explains ~20% of the increased T2D prevalence in Mexico [21]. A recent study of American Indians found a strong interaction with body mass index (BMI) and rs75493593 in T2D risk, such that the association of the variant and diabetes was strongest in leaner individuals [22]. Although the mechanism of action is not known, lower levels of monocarboxylate transporter 11 (the protein encoded by SLC16A11) in the plasma membrane of primary human hepatocytes affects changes in fatty acid and lipid metabolism that may affect glycemic control in the face of weight gain or weight loss [23].
Evidence of the gene × lifestyle interplay in T2D and obesity
Many of the established T2D-associated loci have been examined for their potential roles in gene × lifestyle interactions, with the largest studies having been performed in European-ancestry populations. The European Prospective Investigation of Cancer-InterAct consortium (EPIC-InterAct), for example, undertook the largest study to date on this topic (n = 12 403 incident cases and 16 154 healthy controls), studying the interactions of 58 established T2D-associated variants with Mediterranean diet and physical activity; these analyses yielded no convincing evidence of interactions at an individual gene variant or burden-score level [24]. The EPIC-Inter-Act cohort has since been used to examine gene interactions with dietary fiber [25], dietary factors influencing body iron status [26], and macronutrient intake [27•], although no compelling evidence has been forthcoming. The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE) has also examined interactions between many of the same loci and quantitative glycemic traits. Those analyses provided tentative evidence of interactions between dietary fiber and zinc intake with variants at GCKR [28] and SLC30A8 [29] respectively. Several reports from single prospective cohorts have also yielded suggestive evidence of gene × lifestyle interactions in incident diabetes (reviewed in [30]), although independent replication analyses are conspicuously absent.
Evidence of gene × lifestyle interactions is considerably stronger for obesity. There are now many large-scale observational studies in which evidence of gene × lifestyle interactions for specific loci and for sets of loci have been explored. Variants with very strong association signals with BMI are especially strong candidates for G × E interactions per se [31], with the single most compelling signal located at FTO [32]. A decade ago, evidence began to emerge of interactions between variants at the FTO locus and physical activity [33], which, within a year, were followed by single-cohort analyses of FTO × lifestyle interaction [34–36]. Subsequently, comprehensive consortia-based analyses confirmed these initial findings, demonstrating that the estimated risk of obesity conferred by FTO may be attenuated by ~30% by a physically active lifestyle [32,37]. The evidence of interactions at the FTO locus have recently been extended to include multiple additional lifestyle exposures: a recent analysis in the UK Biobank cohort (n ~ 120 000 adults), for example, reported statistically robust interactions between an FTO variant (rs1421085) and frequency of alcohol consumption, sleep duration, dietary salt, and diet per se, as well as physical activity [38] Epidemiological analyses focused on genetic risk scores for obesity (usually constructed using BMI-associated risk alleles) have also yielded compelling evidence of gene × lifestyle interactions for physical activity [39], fried food consumption [40], sugar-sweetened beverages [41••], and social deprivation [42]. Importantly though, epidemiological studies of gene × lifestyle interactions are often prone to bias and confounding (examples discussed in [43]), which appear to underlie some of the interactions reported in the UK Biobank dataset [42]; this may explain why, in comprehensive analyses of lifestyle intervention trials, little or no evidence of gene × lifestyle interactions for FTO [44] or other obesity loci [45] has emerged (see [46] for detailed discussion of these issues). The major bottlenecks for translating into clinical trials evidence of gene × lifestyle interactions derived from observational studies, include statistical power (owing to the much smaller sample sizes available in trials and sources of heterogeneity introduced through meta-analysis of diverse trials) and a focus on temporal changes in lifestyle factors instead of cumulative averages of lifestyle exposures (in observational studies this is usually cross-sectionally ascertained BMI, whereas in trials the outcome is usually weight change over months or years).
Meta-analysis of data on G × E interactions can also lead to the under-or over-estimation of interaction effects. In order to achieve a standardized set of meta-data, metaanalyses often require that key variables within each contributing cohort are reduced to the lowest common form: in trials, this might include collapsing treatment arms (e.g., by combining drug and lifestyle interventions), focusing on the shortest period of follow-up, or by selecting the most rudimentary outcome variables. In G × E interaction meta-analyses of data from observational studies, the environmental exposure variables are frequently dichotomized, typically by combining two or more categories and comparing this against a referent category or by stratifying continuous variables above and below fairly arbitrary cut-points to create binary variables. Combining data from multiple cohorts can also increase the variance in the outcome variable, which further reduces statistical power. Furthermore, Winner’s curse can lead the marginal and interaction effects generated by a discovery study to be inflated, thereby requiring replication studies to be considerably larger than the primary study. As highlighted above, for FTO’s interaction with physical activity, the interaction effect estimate derived from the major replication meta-analysis [44], yielded an interaction effect estimate considerably smaller in magnitude than the discovery study had reported [32]. In combination, these issues can demand that, to be adequately powered, G × E replication studies are an order of magnitude larger than the original discovery studies [32,37,42,47], which is often unachievable when discoveries are made in large epidemiological cohorts, and replication is sought in clinical trials [46].
Despite the much greater interrogation of how genetic variants, rather than other types of biological variants (e. g., transcripts, epigenomic marks, proteins, metabolites, bacteria) interact with lifestyle, the most compelling evidence that variations in personal biology affect the relationships of food and metabolic health has come from a handful of relatively small studies focused on the gut microbiome. These studies have helped determine that postprandial glycemic responses to specific foods and degree of weight regain after dieting varies from one person to the next and can be predicted at an individual level using biomarker data [3••,4,48].
Future key challenges and concluding remarks
Detecting and understanding biomarkers, whether geno-types or other biological variants, that dictate a person’s phenotypic responses to lifestyle exposures is being facilitated by major technological developments in bio-marker assays and monitoring devices, as well as advances in data processing [49•]. As these advances continue, some of the key challenges that have inhibited the detection of gene × lifestyle interactions in T2D will be overcome.
A significant limitation of many published gene × lifestyle interaction studies is that the self-report data on diet and physical activity upon which results are based are likely to be biased or confounded. Much of the “objective” data, particularly for physical activity, is also likely to be error-prone (because data are generally collected for relatively short durations and at relatively low resolution) and may be biased (owing to Hawthorne effect—i.e., change in a participant’s behavior owing to their awareness of being under observation). Thus, there is a need for new technologies that allow unobtrusive assessments of lifestyle and over long durations. The development of popular wearable technologies or mobile phones that monitor lifestyle [50] is likely to address much of this need; so too may innovations in the use of metabolomics to assess dietary exposures [51••]. Furthermore, the careful integration of these more detailed exposure measures with multi-omics data (epigenomics, transcriptomics, proteomics metabolomics or metagenomics) [52••], is likely to be necessary if we are to understand the complexities of gene × lifestyle interactions, and translate this knowledge into clinical action. Moreover, the application of cutting-edge technologies to biomaterials from historical cohorts, whilst powerful in many ways, requires the design and implementation of new studies, intended specifically for the investigation of gene × lifestyle interactions; such studies are likely to include genotype-based recall clinical trials [53].
In summary, there is an extensive body of literature on the interplay of genomics and lifestyle in T2D and related traits. The most compelling evidence comes from studies of gene × lifestyle interactions in obesity and of micro-biome variation in postprandial glycemic response; however, at this time, there is very little compelling data supporting the integration of genetics and lifestyle into the prevention or management of T2D.
Footnotes
Conflict of interest
P.W.F. is a paid consultant for the PREDICT trial and has received funding from diabetes drug companies as part of the Innovative Medicines Initiative of the European Union.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Menke A, Casagrande S, Geiss L, Cowie CC: Prevalence of and trends in diabetes among adults in the United States, 1988–2012. JAMA 2015, 314:1021. [DOI] [PubMed] [Google Scholar]; • Shows staggering proportion of diabetes among adults in the United States.
- 2.Mayer-Davis EJ, Lawrence JM, Dabelea D, Divers J, Isom S, Dolan L, Imperatore G, Linder B, Marcovina S, Pettitt DJ et al. : Incidence trends of type 1 and type 2 diabetes among youths, 2002–2012. NEnglJ Med 2017, 376:1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M et al. : Personalized nutrition by prediction of glycemic responses. Cell 2015, 163:1079–1094. [DOI] [PubMed] [Google Scholar]; •• Relevant study in which a machine learning algorithm integrating variations in dietary intake, lifestyle, host phenotype, and the gut microbiome accurately predicted the individual glycemic response to meals. This study provides an important first step toward personalized nutrition.
- 4.Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan M, Avnit-Sagi T, Kosower N, Malka G, Rein M et al. : Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab 2017, 25:1243–1253.e5. [DOI] [PubMed] [Google Scholar]
- 5.García-Perez L-E, Alvarez M, Dilla T, Gil-Guillen V, Orozco-Beltran D: Adherence to therapies in patients with type 2 diabetes. Diabetes Ther 2013, 4:175–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poveda A, Chen Y, Brändström A, Engberg E, Hallmans G, Johansson I, Renstrom F, Kurbasic A, Franks PW: The heritable basis of gene-environment interactions in cardiometabolic traits. Diabetologia 2017, 60:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buil A, Brown AA, Lappalainen T, Viñuela A, Davies MN, Zheng H-F, Richards JB, Glass D, Small kS, Durbin R et al. : Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat Genet 2015, 47:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krafka J: The effect of temperature upon facet number in the bar-eyed mutant of Drosophila: Part I. J Gen Physiol 1920, 2:409–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrod AE: Inborn errors of metabolism; the Croonian lectures delivered before the Royal College of Physicians of London, in June, 1908. 1909. [Google Scholar]
- 10.NeeL JV: Diabetes mellitus: a ‘thrifty’ genotype rendered detrimental by ‘progress’? Am J Hum Genet 1962, 14:353–362. [PMC free article] [PubMed] [Google Scholar]; •• Formulation of the Thrifty genotype hypothesis.
- 11.Ayub Q, Moutsianas L, Chen Y, Panoutsopoulou K, Colonna V, Pagani L, Prokopenko I, Ritchie GRS, Tyler-Smith C, McCarthy Ml et al. : Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes. Am J Hum Genet 2014, 94:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minster RL, Hawley NL, Su C-T, Sun G, Kershaw EE, Cheng H, Buhule OD, Lin J, Reupena MS, Viali S et al. : A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat Genet 2016, 48:1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Relevant study demonstrating that CREBRF can be a genetic variant that confluences protection against type 2 diabetes despite the association with higher BMI.
- 13.Bennett PH, Burch TA, Miller M: Diabetes mellitus in American (Pima) Indians. Lancet (London, England) 1971, 2:125–128. [DOI] [PubMed] [Google Scholar]
- 14.Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A et al. : Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science (80-) 2015, 349:1343–1347. [DOI] [PubMed] [Google Scholar]; •• Study showing genetic adaptation to adverse diet and climate conditions.
- 15.Moltke I, Grarup N, Jørgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT et al. : A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 2014, 512:190–193. [DOI] [PubMed] [Google Scholar]; • Study suggesting ethnic-specific genetic adaptation.
- 16.Ravussin E, Valencia ME, Esparza J, Bennett PH, Schulz LO: Effects of a traditional lifestyle on obesity in Pima Indians. Diabetes Care 1994, 17:1067–1074. [DOI] [PubMed] [Google Scholar]
- 17.Buckley MT, Racimo F, Allentoft ME, Jensen MK, Jonsson A, Huang H, Hormozdiari F, Sikora M, Marnetto D, Eskin E et al. : Selection in Europeans on fatty acid desaturases associated with dietary changes. Mol Biol Evol 2017, 34:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu JHY, Marklund M, Imamura F, Tintle N, Ardisson Korat AV, de Goede J, Zhou X, Yang W-S, de Oliveira Otto MC, Kroger J et al. : Omega-6 fatty acid biomarkers and incident type 2 diabetes: pooled analysis of individual-level data for 39 740 adults from 20 prospective cohort studies. Lancet Diabetes Endocrinol 2017, 5:965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartee GD: Roles of TBC1D1 and TBC1D4 in insulin-and exercise-stimulated glucose transport of skeletal muscle. Diabetologia 2015, 58:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dash S, Sano H, Rochford JJ, Semple RK, Yeo G, Hyden CSS, Soos MA, Clark J, Rodin A, Langenberg C et al. : A truncation mutation in TBC1D4 in a family with acanthosis nigricans and postprandial hyperinsulinemia. Proc Natl Acad Sci USA 2009, 106:9350–9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.SIGMA Type 2 Diabetes Consortium AL, Williams AL, Jacobs SBR, Moreno-Macías H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, García-Ortíz H, Gomez-Vazquez MJ, Burtt nP et al. : Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 2014, 506:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traurig M, Hanson RL, Marinelarena A, Kobes S, Piaggi P, Cole S, Curran JE, Blangero J, Goring H, Kumar S et al. : Analysis of SLC16A11 variants in 12,811 American Indians: genotype-obesity interaction for type 2 diabetes and an association with RNASEK expression. Diabetes 2016, 65:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusu V, Hoch E, Mercader JM, Tenen DE, Gymrek M, Hartigan CR, DeRan M, von Grotthuss M, Fontanillas P, Spooner A et al. : Type 2 diabetes variants disrupt function of SlC16A11 through two distinct mechanisms. Ce// 2017, 170:199–212.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langenberg C, Sharp SJ, Franks PW, Scott RA, Deloukas P, Forouhi NG, Froguel P, Groop LC, Hansen T, Palla L et al. : Gene-lifestyle interaction and type 2 diabetes: the EPIC InterAct case-cohort study. PLoS Med 2014, 11:e1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.InterAct Consortium: Investigation of gene-diet interactions in the incretin system and risk of type 2 diabetes: the EPIC-InterAct study. Diabeto/ogia 2016, 59:2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meidtner K, Podmore C, Kröger J, van der Schouw YT, Bendinelli B, Agnoli C, Arriola L, Barricarte A, Boeing H, Cross AJ et al. : Interaction of dietary and genetic factors influencing body iron status and risk of type 2 diabetes within the EPIC-InterAct study. Diabetes Care 2017. 10.2337/dc17-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SX, Imamura F, Ye Z, Schulze MB, Zheng J, Ardanaz E, Arriola L, Boeing H, Dow C, Fagherazzi G et al. : Interaction between genes and macronutrient intake on the risk of developing type 2 diabetes: systematic review and findings from European Prospective Investigation into Cancer (EPIC)-InterAct. Am J C/in Nutr 2017. 10.3945/ajcn.116.150094. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study attempted to replicate previous gene × diet studies and showed no evidence of significant interactions.
- 28.Nettleton JA, McKeown NM, Kanoni S, Lemaitre RN, Hivert M-F, Ngwa J, van Rooij FJA, Sonestedt E, Wojczynski MK, Ye Z et al. : Interactions of dietary whole-grain intake with fasting glucose-and insulin-related genetic loci in individuals of European descent: a meta-analysis of 14 cohort studies. Diabetes Care 2010, 33:2684–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanoni S, Nettleton JA, Hivert M-F, Ye Z, van Rooij FJA, Shungin D, Sonestedt E, Ngwa JS, Wojczynski MK, Lemaitre RN et al. : Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant: a 14-cohort meta-analysis. Diabetes 2011, 60:2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mutie PM, Giordano GN, Franks PW: Lifestyle precision medicine: the next generation in type 2 diabetes prevention? BMC Med 2017, 15:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shungin D, Deng WQ, Varga TV, Luan J, Mihailov E, Metspalu A, GIANT Consortium aP, Morris AP, Forouhi NG, Lindgren C et al. : Ranking and characterization of established BMI and lipid associated loci as candidates for gene-environment interactions. PLoS Genet 2017, 13:e1006812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpeläinen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, Ahmad T, Mora S, Kaakinen M, Sandholt CH et al. : Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med 2011, 8:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilpeläinen TO, Franks PW: Gene-physical activity interactions and their impact on diabetes. Med Sport Sci 2014:94–103. [DOI] [PubMed] [Google Scholar]
- 34.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, Nielsen AL, Albrechtsen A, Borch-Johnsen K, Rasmussen SS et al. : Low physical activity accentuates the effect of the FTO rs9939609 polymorphism body fat accumulation. Diabetes 2008, 57:95–101. [DOI] [PubMed] [Google Scholar]
- 35.Franks PW, Jablonski KA, Delahanty LM, McAteer JB, Kahn SE, Knowler WC, Florez JC: Assessing gene-treatment interactions at the FTO and INSIG2 loci on obesity-related traits in the Diabetes Prevention Program. Diabeto/ogia 2008, 51:2214–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O’Connell JR, Ducharme JL, Hines S, Sack P, Naglieri R et al. : Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med 2008, 168:1791–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, Winkler TW, Chu AY, Mahajan A, Hadley D et al. : Genome-wide physical activity interactions in adiposity — a meta-analysis of 200,452 adults. PLoS Genet 2017, 13:e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young AI, Wauthier F, Donnelly P: Multiple novel gene-by-environment interactions modify the effect of FTO variants on body mass index. Nat Commun 2016, 7:12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmad S, Varga TV, Franks PW: Gene × environment interactions in obesity: the state of the evidence. Hum Hered 2013, 75:106–115. [DOI] [PubMed] [Google Scholar]
- 40.Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK, Liang L, Curhan GC, Pasquale LR, Wiggs JL et al. : Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ 2014, 348: g1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR,Ridker PM, Hunter DJ, Willett WC, Rimm EB et al. : Sugar-sweetened beverages and genetic risk of obesity. N Eng/J Med 2012, 367:1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Study suggesting that sugar-sweetened beverages are associated with higher BMI in those at higher obesity genetic risk.
- 42.Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G et al. : Gene-obesogenic environment interactions in the UK Biobank study. Int J Epidemiol 2017. 10.1093/ije/dyw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Franks PW: Commentary: Mining gene-lifestyle interactions in UK Biobank: all that glitters isn’t gold. Int J Epidemiol 2017, 46:576–577. [DOI] [PubMed] [Google Scholar]
- 44.Livingstone KM, Celis-Morales C, Papandonatos GD, Erar B, Florez JC, Jablonski KA, Razquin C, Marti A, Heianza Y, Huang T et al. : FTO genotype and weight loss: systematic review and meta-analysis of 9563 individual participant data from eight randomised controlled trials. BMJ 2016, 354:i4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papandonatos GD, Pan Q, Pajewski NM, Delahanty LM, Peter I, Erar B, Ahmad S, Harden M, Chen L, Fontanillas P et al. : Genetic predisposition to weight loss and regain with lifestyle intervention: Analyses from the Diabetes Prevention Program and the Look AHEAD randomized controlled trials. Diabetes 2015, 64:4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franks PW, Poveda A: Lifestyle and precision diabetes medicine: will genomics help optimise the prediction, prevention and treatment of type 2 diabetes through lifestyle therapy? Diabetologia 2017, 60:784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D, Ericson U, Koivula RW, Chu AY, Rose LM et al. : Gene × physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS Genet 2013, 9: e1003607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, Dohnalova L, Braverman S, Rozin S, Malitsky S et al. : Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 2016, 540:544–551. [DOI] [PubMed] [Google Scholar]
- 49.Ashley EA: Towards precision medicine. Nat Rev Genet 2016, 17:507–522. [DOI] [PubMed] [Google Scholar]; • Review showing the steps towards the implementation of precision medicine.
- 50.Thompson FE, Subar AF, Loria CM, Reedy JL, Baranowski T: Need for technological innovation in dietary assessment. JAm Diet Assoc 2010, 110:48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Perez I, Posma JM, Gibson R, Chambers ES, Hansen TH,Vestergaard H, Hansen T, Beckmann M, Pedersen O, Elliott P et al. : Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol 2017, 5:184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Clinical trial demonstrating that urine metabolomics profile can identify different dietary patterns.
- 52.Franks PW, McCarthy MI: Exposing the exposures responsible for type 2 diabetes and obesity. Science 2016, 354:69–73. [DOI] [PubMed] [Google Scholar]; •• Review about the interplay between genetic and lifestyle factors in type 2 diabetes.
- 53.Atabaki-Pasdar N, Ohlsson M, Shungin D, Kurbasic A, Ingelsson E, Pearson ER, Ali A, Franks PW: Statistical power considerations in genotype-based recall randomized controlled trials. Sci Rep 2016, 6:37307. [DOI] [PMC free article] [PubMed] [Google Scholar]