Abstract
Structure-activity relationship for the inhibition of Schisandra chinensis’s ingredients toward (Uridine-Diphosphate) UDP-glucuronosyltransferases (UGTs) activity was performed in the present study. In vitro incubation system was employed to screen the inhibition capability of S. chinensis’s ingredients, and in silico molecular docking method was carried out to explain possible mechanisms. At 100 μM of compounds, the activity of UGTs was inhibited by less than 90% by schisandrol A, schisandrol B, schisandrin, schisandrin C, schisantherin A, gomisin D, and gomisin G. Schisandrin A exerted strong inhibition toward UGT1A1 and UGT1A3, with the residual activity to be 7.9% and 0% of control activity. Schisanhenol exhibited strong inhibition toward UGT2B7, with the residual activity to be 7.9% of control activity. Gomisin J of 100 μM inhibited 91.8% and 93.1% of activity of UGT1A1 and UGT1A9, respectively. Molecular docking prediction indicated different hydrogen bonds interaction resulted in the different inhibition potential induced by subtle structure alteration among schisandrin A, schisandrin, and schisandrin C toward UGT1A1 and UGT1A3: schisandrin A>schisandrin>schisandrin C. The detailed inhibition kinetic evaluation showed the strong inhibition of gomisin J toward UGT1A9 with the inhibition kinetic parameter (Ki) to be 0.7 μM. Based on the concentrations of gomisin J in the plasma of the rats given with S. chinensis, high herb–drug interaction existed between S. chinensis and drugs mainly undergoing UGT1A9-mediated metabolism. In conclusion, in silico-in vitro method was used to give the inhibition information and possible inhibition mechanism for S. chinensis’s components toward UGTs, which guide the clinical application of S. chinensis.
Keywords: Schisandra chinensis, UDP-glucuronosyltransferases (UGTs), herb–drug interaction, in silico, in vitro
INTRODUCTION
The utilization of herbs becomes more and more popular because of the belief that herbs are natural and safe. However, more and more case reports on the herbal effects strongly increase the hesitation for the application of herbal medicines. For example, the food and drug administration banned the utilization of ephedracontaining herbs due to their adverse effects (Woolf et al., 2005). Herb–drug interaction is another severe adverse effect limiting the clinical application of herbs. For example, the utilization of herbs has been reported to exhibit the influence toward the therapeutic window of many clinical drugs, including warfarin, aspirin, digoxin, and cisplatin (Zhou et al., 2007; Yang et al., 2006).
Schisandra chinensis has long been used as a traditional Chinese medicine in China to treat hepatitis, menstrual dysfunction, and neurasthenia (Xiao et al., 2008). During its clinical application, frequent herb–drug interaction has been reported. For example, the administration of the ethanol extract from Schisandra Wuzhi capsule can significantly affect the pharmacokinetic profile of tacrolimus and paclitaxel, indicated by their increased plasma concentrations (Qin et al., 2010; Jin et al., 2011). The potential reasons might be the inhibition of its ingredients toward drug-metabolizing enzymes or transporters. For example, the known components gomisin B, C, G, and N showed inhibition toward cytochrome P450 3A4-catalyzed N-demethylation of erythromycin (Iwata et al., 2004). The ingredients form S. chinensis can also interact with the transporter P-glycoprotein and alter its activity (Wan et al., 2006).
UDP-glucuronosyltransferases (UGTs), important drug-metabolizing enzymes located in the membrane of endoplasmic reticulum, plays a key role in the phase II metabolic elimination of various endogenous and exogenous substances (Fang et al., 2013; Ma et al., 2014). The metabolism behavior might be significantly affected by the alteration of UGTs’ activity induced by in vivo polymorphism of UGTs or the in vitro influence of xenobiotics toward UGTs’ activity. The disruption of indinavir and sorafenib toward the homeostasis of bilirubin was induced by inhibiting UGT1A1-mediated bilirubin glucuronidation metabolism (Zucker et al., 2001).
In our previous study, the inhibition of S. chinensis’s ingredients deoxyschizandrin and schisantherin A toward UGT isoforms was indicated (Liu et al., 2012). In the present study, structure-activity relationship for the inhibition of S. chinensis’s ingredients toward the activity of UGT isoforms was investigated.
MATERIALS AND METHODS
Chemicals and reagents.
4-Methylumbelliferone (4-MU), 4-methylumbelliferone-β-D-glucuronide (4-MUG), Tris-HCl, 7-hydroxycoumarin, trifluoperazine (TFP, purity≥ 99%), and uridine-5′-diphosphoglucuronic acid trisodium salt (UDPGA) were purchased from Sigma-Aldrich (St Louis, MO). Recombinant human UGT isoforms (UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A10, UGT2B4, and UGT2B7) expressed in baculovirus-infected insect cells were obtained from BD Gentest Corp. (Woburn, MA, USA). Compounds schisandrin A, schisandrol A, schisandrin, schisandrol B, schisandrin C, schisantherin A, schisanhenol, gomisin J, gomisin D, and gomisin G were purchased from Sichuan Weikeqi Company (Chengdu, Sichuan, China). The purity of these compounds was above 95 %. All other reagents were of HPLC grade or of the highest grade commercially available.
Inhibition capability evaluation of S. chinensis’s ingredients toward UGT isoforms.
The inhibition potential of S. chinensis’s ingredients toward various UGT isoforms was investigated using the in vitro recombinant UGT incubation system as previously described (Fang et al., 2013; Ma et al., 2014). For recombinant UGTs (except UGT1A4)-catalyzed 4-MU glucuronidation probe reactions, the incubation system (total volume = 200 μL) contained recombinant human UGT isoforms, 5mM UDPGA, 5mM MgCl2, 50mM Tris–HCl (pH = 7.4), and 4-MU in the absence or presence of different concentrations of various S. chinensis’s components. The used incubation time and protein concentration were previously determined to ensure the reaction rate within the linear range (Fang et al., 2013; Ma et al., 2014). The 4-MU concentration was equal to known Km or S50 values for each UGT isoform. The incubation reaction was initiated through addition of UDPGA to the mixture after a 5 min pre-incubation at 37°C. The reactions were quenched by adding 100 μL acetonitrile with 7-hydroxycoumarin (100 μM) as internal standard. The mixture was centrifuged at 20000 g for 10 min, and an aliquot of supernatant was transferred to an autoinjector vial for HPLC analysis. The HPLC system (Shimadzu, Kyoto, Japan) contained a SCL-10A system controller, two LC-10AT pumps, a SIL-10A autoinjector, and a SPD-10AVP ultraviolet detector. Chromatographic separation was carried out using a C18 column (4.6 × 200 mm, 5 μm, Kromasil) at a flow rate of 1 mL/min and ultraviolet detector at 316nm. The mobile phase consisted of acetonitrile (A) and H2O containing 0.5% (v/v) formic acid (B). The following gradient condition was used: 0–15 min, 95–40% B; 15–20 min, 10% B; 20–30 min, 95% B. The calculation curve was generated by peak area ratio (4-MUG/internal standard) over the concentration range of 4-MUG 0.1–100mM. The curve was linear over this concentration range, with an r2 value > 0.99. The limits of detection and quantification were determined at signal-to-noise ratios of 3 and 10, respectively. The accuracy and precision for each concentration were more than 95%. For UGT1A4-catalyzed TFP glucuronidation reaction, 4 μM (close to its Km value) of TFP was incubated with human recombinant UGT1A4 (0.1mg/mL) at 37°C for 20 min in the absence or presence of S. chinensis’s components.
Inhibition kinetics determination for representative components.
The inhibition kinetic type and parameters were determined through fitting the reaction velocity at different concentrations of S. chinensis’s components and 4-MU or TFP. Lineweaver–Burk and Dixon fitting equations were employed to determine the inhibition type, and the second plot (drawing using the slope of lines from Lineweaver–Burk plot versus the concentrations of inhibitors) was used to calculate the inhibition kinetic parameters (Ki).
Computational modeling.
The two-dimensional structures of the N-terminal domains for UGT1A1 and UGT1A3 were not available in the protein database at this time. Here, we used protein modeling program MODELLER 9v14 to generate models for these two proteins. The complete amino acid sequences for the N-terminal domains of UGT1A1 and UGT1A3 were obtained from (National Center for Biotechnology Information) NCBI database coded as AAG30424 and NP_061966, respectively. Three crystal structures were used as templates for both of the two proteins based on higher identity. The crystal structures of hydroquinone glucosyltransferase (PDB code: 2vce), oleandomycin glycosyltransferase (PDB code: 2iya), and flavonoid glucosyltransferase (PDB code: 2c1x) were selected as templates for UGT1A1, and medicago truncatula UGT85H2 (PDB code: 2pq6), flavonoid glucosyltransferase (PDB code:2c1x), and glucosyltransferase UGT78G1 (PDB code: 3hbf) for UGT1A3.
For better understanding the molecular interactions between ligand and protein, the docking between flexible small molecule and rigid protein was performed using Autodock Version 4.2. Three ligands (schisandrin A, schisandrin, and schisandrin C) were docked into proteins UGT1A1 and UGT1A3, respectively. In the preparatory phase of docking process, the non-polar hydrogen atoms of UGT1A1 and UGT1A3 were merged, and the Kollman and Gasteiger charges were assigned to protein by Autodock Tools. The Kollman and Gasteiger partial charges were also added to schisandrin A, schisandrin, and schisandrin C. The Autogrid Version 4.2 was used to calculate the grid maps for the proteins in the docking process. The grid dimensions covering the entire protein-binding site were 90 × 90 × 90 grid points with a spacing of 0.375 Å in each dimension. Lamarckian genetic algorithm with default parameter settings was used to perform docking simulations. The best docking conformation for protein–ligand was selected for post-docking analysis, and the interactions between protein and ligand were analyzed as well.
RESULTS
Inhibition profiles of S. chinensis’s ingredients toward various UGT isoforms
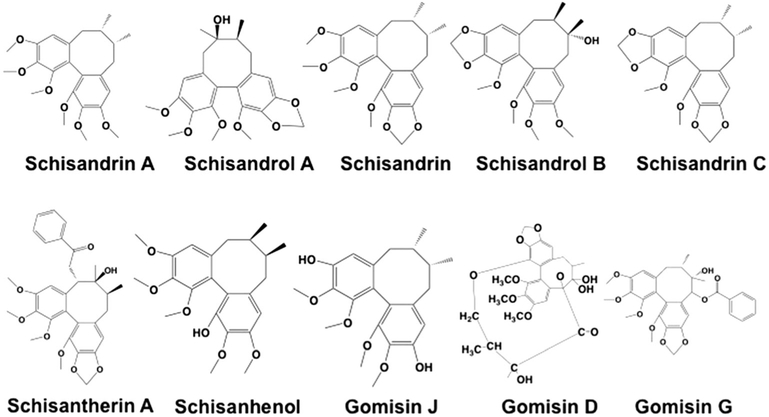
The structures of S. chinensis’s ingredients tested in this study were given in Fig. 1. Used to screen the inhibition potential of S. chinensis’s ingredients toward the activity of UGTs were 100 μM of compounds. The results (Table 1) were given as the residual activity (% residual activity = (the activity at 100 μM of compounds/the control activity at 0 μM compounds)*100%). At 100 μM of compounds, the activity of UGTs was inhibited by less than 90% by schisandrol A, schisandrol B, schisandrin, schisandrin C, schisantherin A, gomisin D, and gomisin G. Schisandrin A exerted strong inhibition toward UGT1A1 and UGT1A3, with the residual activity to be 7.9% and 0% of control activity. Schisanhenol exhibited strong inhibition toward UGT2B7, with the residual activity to be 7.9% of control activity. Gomisin J of 100μM inhibited 91.8% and 93.1% of activity of UGT1A1 and UGT1A9, respectively.
Figure 1.
The molecular structures of schisandrin A, schisandrol A, schisandrin, schisandrol B, schisandrin C, schisantherin A, schisanhenol, gomisin J, gomisin D, and gomisin G.
Table 1.
Initial screening of Schisandra chinensis’s ingredients (100 μM) toward various important UGT isoforms
| 1A1 | 1A3 | 1A4 | 1A6 | 1A7 | 1A8 | 1A9 | 1A10 | 2B4 | 2B7 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Schisandrin A | 7.9 | 0 | 20.4 | 35.3 | 59.5 | 253.2 | 81.8 | 36.5 | 30.8 | 32.4 |
| Schisandrol A | 51.2 | 88.1 | 72.3 | 121.6 | 83.6 | 215.8 | 105.2 | 89.4 | 88.5 | 97.4 |
| Schisandrin | 26.3 | 37.0 | 18.9 | 35.5 | 31.5 | 96.4 | 93.2 | 50.4 | 23.9 | 31.6 |
| Schisandrol B | 66.1 | 57.5 | 59.4 | 81.1 | 40.1 | 155.4 | 63.7 | 65.9 | 54.2 | 62.0 |
| Schisandrin C | 61.5 | 50.4 | 57.3 | 111.2 | 71.2 | 81.4 | 54.8 | 78.5 | 33.9 | 38.1 |
| Schisantherin A | 18.6 | 12.9 | 47.7 | 108.4 | 72.9 | 137.7 | 106.8 | 55.4 | 70.4 | 56.7 |
| Schisanhenol | 17.6 | 17.7 | 27.8 | 12.8 | 36.4 | 88.8 | 46.2 | 60.8 | 20.8 | 7.9 |
| Gomisin J | 8.2 | 81.2 | 34.6 | 36.0 | 26.0 | 140.2 | 6.9 | 35.8 | 18.1 | 36.7 |
| Gomisin D | 10.1 | 14.9 | 50.3 | 93.9 | 45.7 | 169.1 | 74.1 | 57.0 | 77.7 | 80.7 |
| Gomisin G | 17.9 | 25.9 | 13.1 | 97.7 | 72.0 | 119.0 | 101.3 | 96.9 | 76.9 | 51.2 |
The values shown in this table are the residual activity, which is calculated using the following equation: % residual activity = the activity at 100 μM of S. chinensis’s ingredients/the control activity at 0 μM of S. chinensis’s ingredients*100%.
Molecular docking to understand the interaction between S. chinensis’s compounds and UGT isoforms
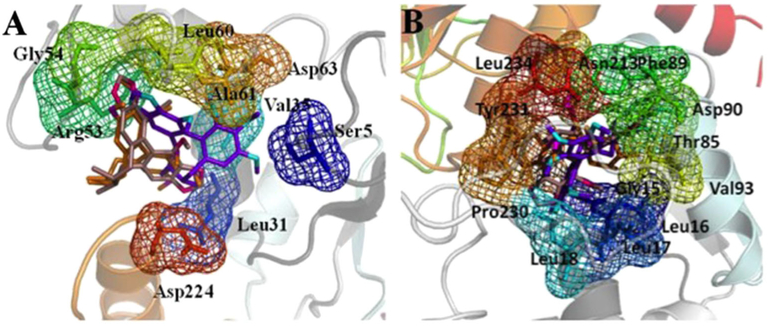
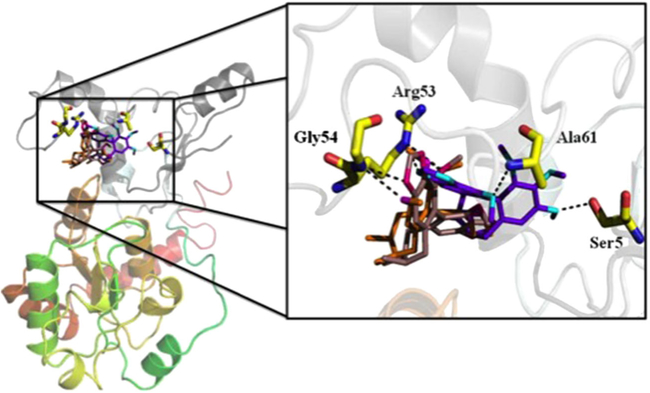
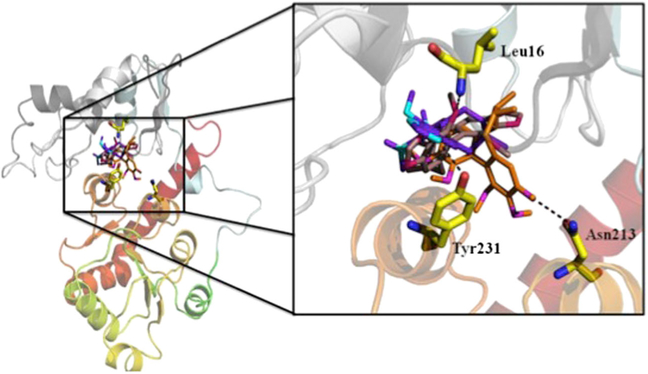
The homology modeling for protein UGT1A1 and UGT1A3 was performed by molecular docking, which explored the interactions between the two proteins and three kinds of S. chinensis’s ingredients schisandrin, schisandrin C, and schisantherin A. At present, it is a great task to recognize the ligand binding domains of UGT1A1 and UGT1A3. In the automatic mode of molecular docking, the active pockets of UGT1A1 and UGT1A3 for ligands were generated. The active site of protein UGT1A1 is comprised of residue Ser5, Leu31, Val35, Arg53, Gly54, Leu60, Ala61, Asp63, and Asp224, which were shown in Fig. 2A. The ligands schisandrin A, schisandrin, and schisandrin C were docked into the active pocket of UGT1A1, respectively. The conformations with analogical bind to the proteins for the three inhibitors and top rank in the ten conformations obtaining from molecular docking were selected. The interactions between inhibiters and UGT1A1 in the cavity of active pocket were shown in Fig. 3. Schisandrin A and schisandrin C generated hydrogen bonds with the side chain of residue Arg53 and Gly54 of protein UGT1A1, and schisandrin formed hydrogen bonds with residue Ser5, Ala62, and the side chain of Arg53. Inhibitor schisandrin also tend to interact with residue Leu60 and Asp63. The active pocket of UGT1A3 was shown in Fig. 2B. Inhibiters schisandrin A, schisandrin, and schisandrin C were docked into protein UGT1A3, respectively, and the active pocket of UGT1A3 was composed of residue Gly15, Leu16, Leu17, Leu18, Thr85, Phe89, Asp90, Val93, Asn213, Pro230, Tyr231, and Leu234. In active pocket, the hydrogen bonds between the three inhibiters and UGT1A3 were shown (Fig. 4). When inhibitor schisandrin A bound into protein UGT1A3, the hydrogen bond was formed between schisandrin A and the side chain of residue Asn213 of UGT1A3, schisandrin A tends to form a hydrogen bond with residue Tyr231 as well. Schisandrin adopts the conformation that formed hydrogen bond with the side chain of residue Tyr231, which also tend to form interactions with residue Gly15, Leu16, and Leu18. The hydrogen bond was formed between schisandrin C and residue Leu16 of UGT1A3. Schisandrin C also tends to form hydrogen bonds with residue Leu16, Leu17, The85, Asn90, and Tyr231. We also computed the binding free energy between the three inhibitors and protein UGT1A1 and UGT1A3 (Table 2). The binding free energy values of schisandrin A, schisandrin, and schisandrin C with protein UGT1A1 were −7.48, −7.15, and −7.00, respectively, which is in accordance with the experimental result that the rank of the inhibition activity for the three inhibitors is schisandrin A>schisandrin>schisandrin C. We compared the binding affinities of the three inhibitors with UGT1A3 as well, and the rank of the binding free energy for schisandrin A, schisandrin, and schisandrin C is equivalent to the in vitro assays results.
Figure 2.
The active pockets of protein UGT1A1 (A) and UGT1A3 (B) for binding with schisandrin A, schisandrin, and schisandrin C. The C atoms of schisandrin A are colored with oranges, and N atoms colored with magenta. The C atoms of schisandrin are colored with purple, and N atoms colored with cyans. The C and N atoms of schisandrin C show violet and hot pink, respectively. All the three inhibitors are shown in stick, and the residues in the active pockets of proteins are shown in stick and mesh. This figure is available in color online at wileyonlinelibrary.com/journal/ptr.
Figure 3.
The UGT1A1 structure in complex with schisandrin A, schisandrin, and schisandrin C. An enlarged representation of the interactions between UGT1A1 and the three inhibitors are demonstrated, and hydrogen bond is shown by a black line. This figure is available in color online at wileyonlinelibrary.com/journal/ptr.
Figure 4.
The UGT1A3 structure in complex with schisandrin A, schisandrin, and schisandrin C. The hydrogen bonds are shown in the enlarged presentation. This figure is available in color online at wileyonlinelibrary.com/journal/ptr.
Table 2.
Docking results of schisandrin A, schisandrin, and schisandrin C with protein UGT1A1 and UGT1A3
| Inhibitor | Intermolecular energy (kcal/mol) |
Internal energy (kcal/mol) |
Torsional free energy (kcal/mol) |
Unbound system’s energy (kcal/mol) |
Binding free energy (kcal/mol) |
|---|---|---|---|---|---|
| UGT1A1 | |||||
| Schisandrin A | −9.27 | −1.49 | 1.79 | −1.49 | −7.48 |
| Schisandrin | −8.34 | −1.08 | 1.19 | −1.08 | −7.15 |
| Schisandrin C | −7.59 | −0.61 | 0.60 | −0.61 | −7.00 |
| UGT1A3 | |||||
| Schisandrin A | −10.25 | −1.50 | 1.79 | −1.50 | −8.46 |
| Schisandrin | −9.48 | −0.99 | 1.19 | −0.99 | −8.29 |
| Schisandrin C | −8.63 | −0.46 | 0.60 | −0.46 | −8.04 |
Binding free energy = intermolecular energy + internal energy + torsional free energy + unbound system’s energy.
The inhibition kinetics for the inhibition of representative component of S. chinensis toward UGTs’ isoforms
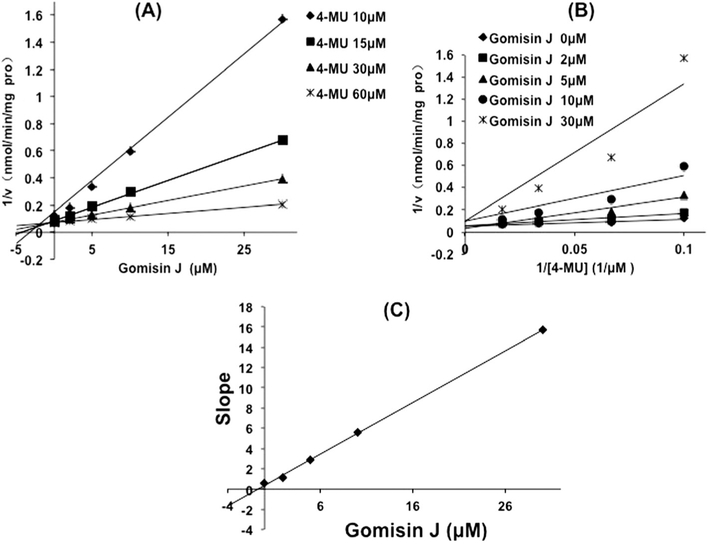
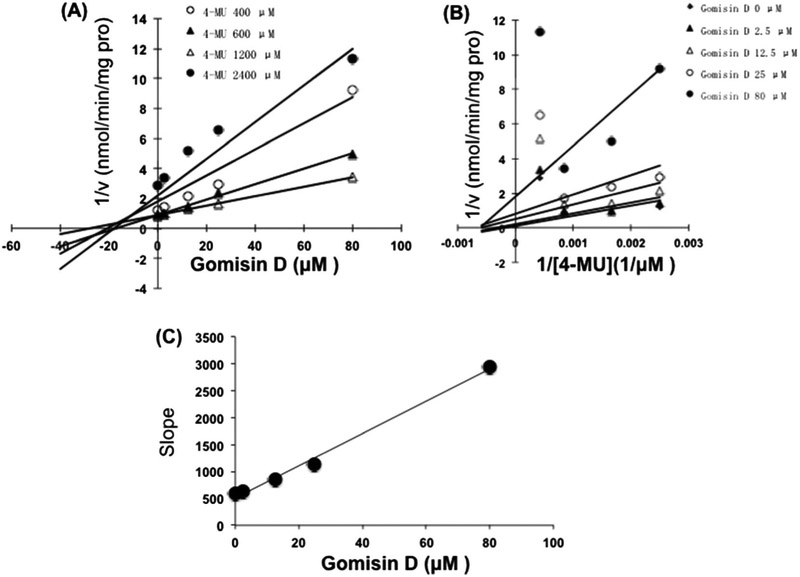
The inhibition of gomisin D toward UGT1A3 and gomisin J toward UGT1A9 was determined as the representative samples. As shown in Fig. 5, Dixon plot (Fig. 5A) and Lineweaver–Burk plot (Fig. 5B) showed that gomisin J competitively inhibited the activity of UGT1A9, and the inhibition kinetic parameter (Ki) was calculated to be 0.7 μM (Fig. 5C). Gomisin D noncompetitively inhibited the activity of UGT1A3, and the inhibition kinetic parameter (Ki) was determined to be 16.9 μM (Fig. 6C).
Figure 5.
Inhibition kinetic evaluation of gomisin J toward UGT1A9-catalyzed glucuronidation of 4-methylumbelliferone (4-MU). (A) Dixon plot for the inhibition of gomisin J toward UGT1A9-catalyzed glucuronidation of 4-MU. (B) Lineweaver–Burk plot for the inhibition of gomisin J toward UGT1A9-catalyzed glucuronidation of 4-MU. (C) The second plot for the inhibition of gomisin J toward UGT1A9-catalyzed glucuronidation of 4-MU.
Figure 6.
Inhibition kinetic evaluation of gomisin D toward UGT1A3-catalyzed glucuronidation of 4-methylumbelliferone (4-MU). (A) Dixon plot for the inhibition of gomisin D toward UGT1A3-catalyzed glucuronidation of 4-MU. (B) Lineweaver–Burk plot for the inhibition of gomisin D toward UGT1A3-catalyzed glucuronidation of 4-MU. (C) The second plot for the inhibition of gomisin D toward UGT1A3-catalyzed glucuronidation of 4-MU.
DISCUSSION
Our previous study showed that S. chinensis’s ingredients deoxyschizandrin and schisantherin A exerted inhibitory potential toward UGT1A3 (Liu et al, 2012). Therefore, the deeper understanding of structure–activity relationship for the inhibition of S. chinensis’s ingredients was carried out in the present study. The results showed that the subtle alteration of structures could significantly change the inhibition potential toward UGT isoforms. We used schisandrin A, schisandrin, and schisandrin C as the examples and found that the alteration of methoxy group into methylenedioxy group significantly weakened the inhibition potential toward UGT1A1 and UGT1A3. In silico, the alteration for the structures of these three ligands induced the alteration of binding capability according to the binding free energy between ligands and proteins, which may be associated with the changes of hydrogen bonds between inhibitors and proteins. When schisandrin A, schisandrin, and schisandrin C bind into UGT1A1, all the three inhibitors formed hydrogen bonds with residue Arg53 of UGT1A1. For schisandrin, the methylenedioxy group formed hydrogen bond with Arg53; however, other groups formed hydrogen bonds with Arg53 for schisandrin A and schisandrin C. The methylenedioxy groups of schisandrin A and schisandrin C formed hydrogen bonds with the residue Gly54; however, the hydrogen bond was not generated with residue Gly54. These factors might together result in the inhibition orders for schisandrin A, schisandrin, and schisandrin C. For the interaction between these ligands with UGT1A3, the different hydrogen bonds were formed. Schisandrin A formed hydrogen bond with residue Asn213, the orientation of which toward residue Asn213, whereas schisandrin and schisandrin C were different from schisandrin A, which oriented to residues Tyr231 and Leu16, and made the formation of hydrogen bonds different from schiandrin A, thus affecting the final inhibition potential of these three compounds toward UGT1A3.
The representative kinetic behavior was determined for the inhibition of gomisin J toward UGT1A9, and gomisin D toward UGT1A3. Competitive inhibition of gomisin J toward UGT1A9 was demonstrated, and the inhibition parameter was calculated to be 0.7 μM. When the rats were given with S. chinensis at a dose of 30 mg/kg through intravenous administration, the maximum concentration of gomisin J was approximately 2 μg/mL (5.1 μM) (Kim et al., 2014). Based on the drug–drug interaction threshold ([I]/Ki > 1, highly possible), the inhibition of gomisin J toward UGT1A9 might result in the high drug-drug interaction potential between S. chinensis and UGT1A9 substrates. UGT1A9 is a key UGT isoform involved in the metabolism of clinical drugs, including propofol (Liang et al., 2011) and arbidol (Song et al, 2013). Therefore, the potential herb–drug interaction between S. chinensis and drugs mainly undergoing UGT1A9-mediated metabolism should be paid much attention. It should be noted that in vitro-in vivo extrapolation +B was not performed for gomisin D due to no report for the in vivo concentration of gomisin D.
The conclusion of inhibition intensity of S. chinensis’s ingredients toward different UGT isoforms was shown in Fig. 7, and the strongest inhibitors for UGT1A1 were schisandrin A and gomisin J (the inhibition potential above 90%), for UGT1A3 was schisandrin A, for UGT1A9 was gomisin J, and for UGT2B7 was schisanhenol. This information figure can be employed to guide the clinical utilization of drugs when in combination with S. chinensis. Together with the previous studies (Ma et al., 2014; Gao et al., 2014), the in silico-in vitro method for elucidating the inhibition potential of compounds toward UGT isoforms was demonstrated to be a good method to elucidate the inhibition mechanism of a series of compounds with the similar structure toward UGT isoforms.
Figure 7.
A brief conclusion of the inhibition potential of various Schisandra chinensis’s ingredients toward UGT isoforms. The red color represents the inhibition ability above 90% at 100 μM of S. chinensis’s ingredients, and the black color represents the inhibition ability below 90% at 100 μM of S. chinensis’s ingredients. This figure is available in color online at wileyonlinelibrary.com/journal/ptr.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 81202586, 81202587, 81202588, 81303146).
Footnotes
Conflict of Interest
The authors have declared that there are no conflicts of interest.
REFERENCES
- Fang ZZ, Cao YF, Hu CM, et al. 2013. Structure–inhibition relationship of ginsenosides towards UDP-glucuronosyltransferases (UGTs). Toxicol Appl Pharmacol 267(2): 149–154. [DOI] [PubMed] [Google Scholar]
- Gao X, Qu H, Ai CZ, et al. 2014. Regulation profile of phosphatidylcholines (PCs) and lysophosphatidylcholines (LPCs) components towards UDP-glucuronosyltransferases (UGTs) isoforms. Xenobiotica 26: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H, Tezuka Y, Kadota S, Hiratsuka A, Watabe T. 2004. Identification and characterization of potent CYP3A4 inhibitors in Schisandra fruit extract. Drug Metab Dispos 32(12): 1351–1358. [DOI] [PubMed] [Google Scholar]
- Jin J, Bi HC, Hu JQ, Zeng, et al. 2011. Effect of Wuzhi tablet (Schisandra sphenanthera extract) on the pharmacokinetics of paclitaxel in rats. Phytother Res 25: 1250–1253. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lee HJ, Kim CY, Han CY, Chin YW, Choi YH. 2014. Simultaneous determination of nine lignans from Schisandra chinensis extract using ultra-performance liquid chromatography with tandem mass spectrometry in rat plasma, urine and gastrointestinal tract samples: application to the pharmacokinetic study of Schisandra chinensis. J Sep Sci 37: 2851–2863. [DOI] [PubMed] [Google Scholar]
- Liang SC, Ge GB, Liu HX, et al. 2011. Determination of propofol UDP-glucuronosyltransferase (UGT) activities in hepatic microsomes from different species by UFLC-ESI-MS. J Pharm Biomed Anal 54(1): 236–241. [DOI] [PubMed] [Google Scholar]
- Liu C, Cao YF, Fang ZZ, et al. 2012. Strong inhibition of deoxyschizandrin and schisantherin A toward UDP-glucuronosyltransferase (UGT) 1A3 indicating UGT inhibition-based herb–drug interaction. Fitoterapia 83(8): 1415–1419. [DOI] [PubMed] [Google Scholar]
- Ma HY, Sun DX, Cao YF, et al. 2014. Herb–drug interaction prediction based on the high specific inhibition of andrographolide derivatives towards UDP-glucuronosyltransferase (UGT) 2B7. Toxicol Appl Pharmacol 277(1): 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XL, Bi HC, Wang XD, et al. 2010. Mechanistic understanding of the different effects of Wuzhi Tablet (Schisandra sphenanthera extract) on the absorption and first-pass intestinal and hepatic metabolism of Tacrolimus (FK506). Int J Pharm 389: 114–121. [DOI] [PubMed] [Google Scholar]
- Song JH, Fang ZZ, Zhu LL, et al. 2013. Glucuronidation of the broad-spectrum antiviral drug arbidol by UGT isoforms. J Pharm Pharmacol 65(4): 521–527. [DOI] [PubMed] [Google Scholar]
- Wan CK, Zhu GY, Shen XL, Chattopadhyay A, Dey S, Fong WF. 2006. Gomisin A alters substrate interaction and reverses P-glycoprotein-mediated multidrug resistance in HepG2-DR cells. Biochem Pharmacol 72(7): 824–837. [DOI] [PubMed] [Google Scholar]
- Woolf AD, Watson WA, Smolinske S, Litovitz T. 2005. The severity of toxic reactions to ephedra: comparisons to other botanical products and national trends from 1993–2002. Clin Toxicol 43(5): 347–355. [DOI] [PubMed] [Google Scholar]
- Xiao WL, Huang SX, Wang RR, et al. 2008. Nortriterpenoids and lignans from Schisandra sphenanthera. Phytochemistry 69(16): 2862–2866. [DOI] [PubMed] [Google Scholar]
- Yang XX, Hu ZP, Duan W, Zhu YZ, Zhou SF. 2006. Drug–herb interactions: eliminating toxicity with hard drug design. Curr Pharm Des 12(35): 4849–4864. [DOI] [PubMed] [Google Scholar]
- Zhou SF, Zhou ZW, Li CG, et al. 2007. Identification of drugs that interact with herbs in drug development. Drug Discov Today 12(15–16): 664–673. [DOI] [PubMed] [Google Scholar]
- Zucker SD, Qin X, Rouster SD, et al. 2001. Mechanism of indinavirinduced hyperbilirubinemia. Proc Natl Acad Sci U S A 98: 12671–12676. [DOI] [PMC free article] [PubMed] [Google Scholar]