Abstract
The contribution of lineage identity and differentiation state to malignant transformation is controversial. We previously showed that adult neural stem and early progenitor cells give origin to glioblastoma (GBM). Here, we systematically assessed the tumor-initiating potential of adult neural populations at various stages of lineage progression. Cell type-specific tamoxifen inducible Cre recombinase transgenes were used to target GBM relevant tumor suppressors Nf1, Trp53, and Pten in late stage neuronal progenitors, neuroblasts, and differentiated neurons. Mutant mice showed cellular and molecular defects demonstrating the impact of tumor suppressor loss, with mutant neurons being the most resistant to early changes associated with tumor development. However, we observed no evidence of glioma formation. These studies show that increasing lineage restriction is accompanied by decreasing susceptibility to malignant transformation, indicating a GBM cell of origin hierarchy where stem cells sit at the apex and differentiated cell types are least susceptible to tumorigenesis.
Keywords: glioma, glioblastoma, GBM, glioma mouse models, tumor suppressors, cell of origin, mature neurons, neuroblasts, neuronal progenitors, differentiation, tumor susceptibility, malignant transformation
Introduction
Until the 1990’s, the adult mammalian brain was considered a post-mitotic organ. It is primarily composed of mature, specialized neurons with trillions of synaptic connections, along with mature glia. Hence, it was widely accepted that malignant gliomas arose from differentiated CNS cells1. The more recent appreciation of the existence and importance of neural stem cells in the mature brain has prompted a revisiting of the potential source(s) of glioma and the scope of adult cell types along the neuronal lineage that have tumor potential.
In the adult rodent brain, the most abundant neural stem cell niches reside in the subventricular zone (SVZ) of the lateral ventricles and the subgranular layer of the hippocampal dentate gyrus. Stem cells divide symmetrically to self renew and perpetuate the stem cell pool throughout the lifetime of an organism. They also divide assymetrically to give rise to progenitor cells with more restricted self renewal potential. Adult neuronal progenitor cells undergo several rounds of cell division before producing newly born immature neurons, or neuroblasts, that further differentiate into specialized neuronal cell types2–4.
A variety of GBM mouse models have been developed using disease-relevant initiating mutations in tumor suppressors (e.g., Pten, Trp53, Nf1, Ink4a/Arf, Rb) and oncogenes (e.g., EgfrVIII, PDGFRA)5. Our previous studies used a Nestin-creERT2 transgene that specifies expression in adult neural stem and progenitor cells, to drive Nf1, Trp53 and Pten deletion and development of high-grade gliomas with 100% penetrance6. Also, Ascl1-creER™, which targets adult bipotential (neural and oligodendrocyte) progenitors, and the adult oligodendrocyte progenitor cell (OPC)-specific NG2-creER™, can drive GBM7, 8. In vivo ablation of Pten, Trp53, and Rb with a Gfap-creER™ transgene expressed in quiescent neural stem cells as well as mature astrocytes, led to the induction of high-grade gliomas in close proximity to the neurogenic niches, adding experimental support for a stem cell of origin9. Studies that target non-adult cells such as embryonic and early postnatal stem and progenitor cells by viral transduction, transplantation or genetic methods have also produced similar results10. On the other hand, de-differentiation has been proposed as a pathway to gliomagenesis. Cultured early postnatal “astrocytes” overexpressing EGFR together with Ink4a/Arf deletion when transplanted into immunodeficient mice developed tumors11. However, it is unclear whether proliferative postnatal day five neonatal astrocytes faithfully represent in vivo mature adult astrocytes that lack progenitor properties. Injections of a constitutive H-RasV12 or shRNA-expressing lentivirus targeting Nf1, together with Trp53, into brains of adult mice were reported to develop gliomas in mature neurons12. In these studies, oncogenic Ras, or Nf1 and Trp53 knockdowns, were induced broadly, mediated by the H1 and U6 promoters, thus confounding precise analysis of tumor cell of origin.
In this study, we targeted the GBM-associated tumor suppressors Nf1, Trp53 and Pten at progressive stages of adult neural differentiation. Regardless of the cell lineage targeted, we identified molecular and phenotypic evidence of tumor suppressor loss. However, in contrast to the fully penetrant GBM phenotype observed when stem cells or early progenitors were targeted, more differentiated neuronal cells showed reduced defects, demonstrating that increasing lineage restriction is an impediment to glioma formation.
Results
CamKIIα-creER™ Marks Adult, Post-mitotic Neurons
To interrogate the tumor-initiating capacity of adult neuronal lineages, we employed tamoxifen inducible cre transgenes that are expressed in discrete adult neuronal subpopulations to inactivate Nf1, Trp53, and Pten, three of the most commonly mutated genes in human GBM13. The Calcium/Calmodulin Protein Kinase II α (CamKIIα) gene is expressed in mature excitatory neurons in the adult cortex, hippocampus, and striatum14 and its regulatory elements have been used to drive transgene expression with fidelity15, 16. We used CamKIIα-creER™ to target adult, post-mitotic differentiated neurons and confirmed transgene activity and expression by X-gal staining of CamKIIα-creER™;R26-stop-LacZ reporter mice induced with tamoxifen at four weeks of age and examined four weeks later (Figures 1A–1B). Cre-recombined cells were observed in the cortex and hippocampus, with scattered staining in the striatum, thalamus, and cerebellum. We confirmed recombination in specific cell types by double immunoflourescence staining of the β-galactosidase reporter with cell lineage-specific antibodies, including NeuN, to stain mature neurons, but not the astroglial marker, Glial fibrillary acidic protein (Gfap), or the oligodendroglial marker, Adenomatous Polyposis Coli (APC) (Figure 1C). In addition, the CamKIIα-creER™;R26-stop-TdTomato reporter was localized to mature neurons and did not show co-localization with immature markers: Doublecortin, Nestin, Sox2 or Olig2 (Figure 1C). These data confirm that the tamoxifen inducible CamKIIα-creER™ transgene (hereafter iCK-cre) is confined to differentiated neurons in the adult brain.
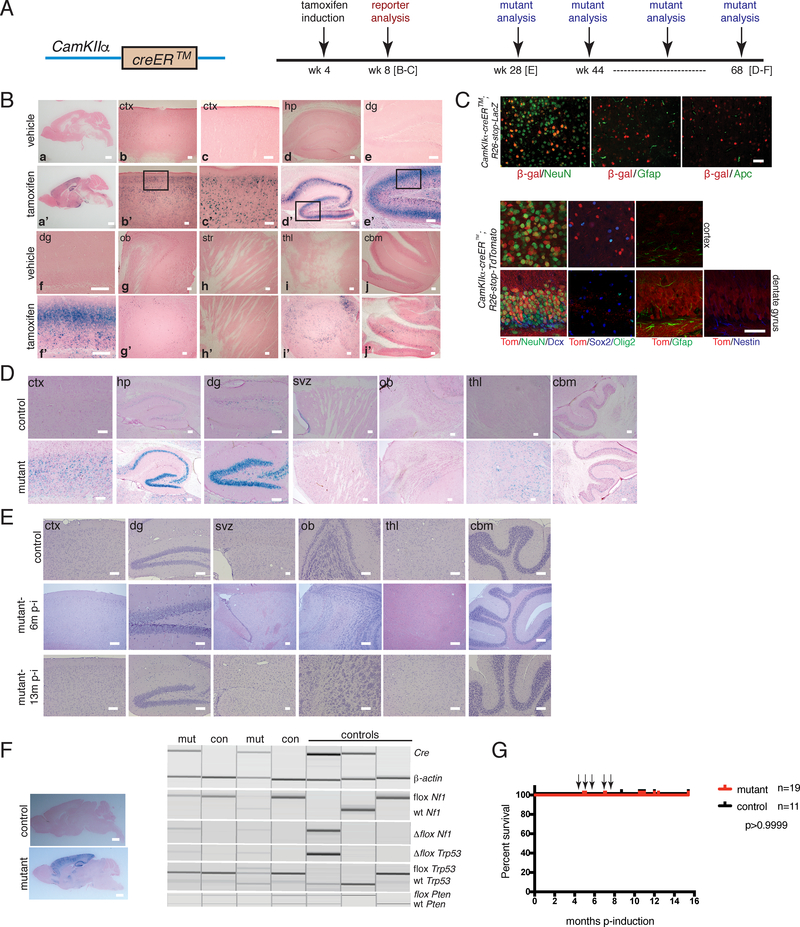
Figure 1. Tumor Suppressor Deletion in Adult Post-mitotic Neurons Does Not Induce Glioblastoma Development.
A. (Left panel) Cartoon of CamKIIa-creER™ (iCK-cre) construct. (Right panel) Timeline of tamoxifen induction and analyses of reporter and mutant strains. B. X-gal staining of tamoxifen-vs. vehicle-induced iCK-cre;R26-stop-LacZ reporter 1 month post-induction (a,a’-whole brain; b,b’-cortex (ctx); c,c’-higher magnification of cortex; d,d’-hippocampus (hp); e,e’-dentate gyrus (dg); f,f’-higher magnification of dentate gyrus; g,g’-olfactory bulb (ob); h,h’-striatum (str); i,i’-thalamus (thl); j,j’-cerebellum (cbm). Scale bars: a,a’=2 mm; b,b’-j,j’=100 μm. C. Immunoflourescence staining of iCK-cre reporters at 1 month post-induction. (Top panel) Immunostaining of iCK-cre;R26-stop-LacZ reporter brain sections using β-galactosidase and cell type-specific markers NeuN (neurons), Gfap (astrocytes), and Apc (oligodendrocytes). (Bottom panel) Immunostaining of iCK-cre;R26-stop-TdTomato reporter brain sections with Tomato and lineage markers NeuN, Dcx (doublecortin), Gfap, Nestin, Sox2 and Olig2. Scale bars=100 μm. D. X-gal staining of iCK-cre mutant vs. control at 12 months post-induction (svz=subventricular zone). Scale bars=100 μm. E. Hematoxylin and eosin (H&E) staining of iCK-cre mutants vs. controls at 6 months and 13 months post-induction. Scale bars=100 μm. F. Genotyping for Cre and recombined, loxP and wild type (wt) alleles of Nf1, Trp53 and Pten (right panel) using micro-dissected mutant (mut) and control (con) brain regions (left panel). Scale bars=2 mm. G. Survival curve of iCK-cre mutants (n=19) vs. controls (n=11). p>0.9999 using Logrank (Mantel-Cox) test. Arrows show n=5 asymptomatic mutant mice that were sacrificed for analysis at 5 to 9 months post-tamoxifen induction for non-health-related reasons. In B-F, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group.
Aged iCK-cre Mutants Do Not Develop Glioma
We bred iCK-cre mice to contain various configurations of Nf1, Trp53 and Pten conditional alleles (iCK-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or iCK-cre;Nf1flox/+;Trp53flox/flox;Ptenflox/+ or iCK-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or iCK-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+ or iCK-cre;Nf1flox/+;Trp53flox/+;Pten+/+). Mice were induced with tamoxifen at four to eight weeks of age along with controls (iCK-cre-negative tumor suppressor conditional mice administered tamoxifen or iCK-cre-positive tumor suppressor conditional mice administered vehicle, sunflower oil), and were aged. iCK-cre mutants appeared indistinguishable from wild type controls and did not exhibit overt developmental or behavioral abnormalities. We confirmed cre-mediated recombination in aged mice by X-gal staining, which showed retention of the β-galactosidase-positive recombined cells, particularly in the cortex and hippocampus in mutants (Figure 1D).
Since the targeted mice did not show adverse symptoms, we performed sequential analysis of mutant brains at different times. Hematoxylin and eosin (H&E) staining of iCK-cre mutants over eighteen months indicated morphologically normal brains with no overt neurological deficits (Figure 1E). We confirmed tumor suppressor loss by analysis of DNA from micro-dissected hippocampi and cortices, and polymerase chain reaction (PCR) genotyping showed the presence of Cre and recombined conditional alleles in Cre-positive mutant vs. Cre-negative control brains (Figure 1F, data not shown). Kaplan-Meier analysis showed similar survival between iCK-cre mutants and controls (p>0.9999; Figure 1G). Hence, deletion of GBM-associated tumor suppressors in adult mature neurons resulted in asymptomatic mice lacking evidence of glioma formation.
We found no evidence of abnormal proliferation in all iCK-cre mutant brain regions examined following short-term bromodeoxyuridine (BrdU) pulses or by Ki67 immunohistochemistry (IHC) (Figure 2A). Apoptosis measured by cleaved caspase 3 IHC also showed no differences (data not shown). However, iCK-cre mutant brains did display abnormal features reflective of signaling pathway alterations. Notably, areas of reactive astrogliosis, evidenced by intense Gfap staining, were present in both young and aged mutant mice (Figures 2A–2B). This phenomenon is reminiscent of the Synapsin1-cre;Nf1flox/flox phenotype which showed global non-cell autonomous astrogliosis17, and likely reflects a paracrine reactive astrocytic response to neuronal loss of tumor suppressors.
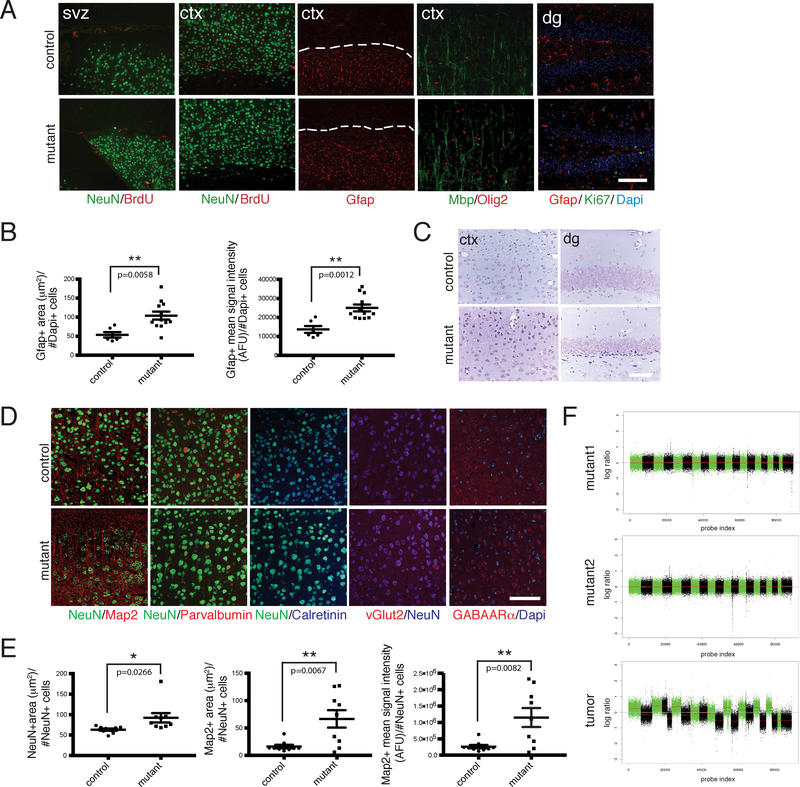
Figure 2. iCK-Cre Mutants Exhibit Astrogliosis and Neuronal Defects.
A. Immunohistochemistry of aged iCK-cre mutant vs. control brain sections using cell type-specific markers Gfap, NeuN, Mbp and Olig2, as well as proliferation markers BrdU (1 hour pulse) and Ki67. Dashed lines show areas with Gfap staining in the cortex. B. Dot plots showing Gfap-positive area (μm2) per number of Dapi-positive cells (left panel; p=0.0058) and Gfap-positive mean signal intensity in AFU (arbitrary fluorescence units) per number of Dapi-positive cells (right panel; p=0.0012) in iCK-cre mutants (n=12) vs. controls (n=6). **p<0.01 using two-tailed unpaired student’s t-test. Data is presented as mean +/− standard error of the mean (SEM). C. Nissl staining of iCK-cre mutant vs. control brain sections. D. Immunoflourescence staining of iCK-cre mutant vs. control brain sections using specialized neuronal markers Map2, Parvalbumin, Calretinin, vGlut2, and GABAARα. E. Quantification of neuronal marker stainings. (Left panel) Dot plot showing the average size of NeuN-positive cells (mean NeuN-positive area per NeuN-positive cell number) in iCK-cre mutants (n=9) vs. controls (n=9). p=0.0266. (Middle panel) Dot plot showing Map2-positive area per NeuN-positive cell number in iCK-cre mutants (n=9) vs. controls (n=9). p=0.0067. (Right panel) Dot plot showing Map2-positive mean signal intensity per NeuN-positive cell number in iCK-cre mutants (n=9) vs. controls (n=9). p=0.0082. *p<0.05, **p<0.01 using two-tailed unpaired Student’s t-test. Data presented as mean +/− SEM. F. Shallow whole genome sequencing of iCK-cre mutants (n=3) compared to controls (n=3). Chromosomal segmentation analysis of mutant recombined brain regions (top 2 panels) and a Synapsin1-cre mutant tumor positive control (bottom panel) showing log ratio of normalized counts between mutant and control on x-axis and probe index on y axis are shown. All scale bars=100 μm. In A and C-D, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group.
Examination of histologic and morphometric features of aged tumor suppressor-deficient neurons showed increased soma size as compared to controls, particularly in the cortex and dentate gyrus (Figures 2C–2E). Nissl staining showed neuronal hypertrophy, with deep staining in mutant brains (Figure 2C) that was confirmed by NeuN IHC (Figures 2D–2E, S1A). We found increased Map2 staining, while other specialized neuronal markers were not significantly altered (Figures 2D–2E, S1B). Some of these neuronal defects are similar to the previously described phenotype of Pten-deficient neurons18. Additional analyses including shallow whole genome sequencing and telomere fluorescent in situ hybridization (FISH) showed evidence of increased chromosomal abnormalities and telomere elongation (Figures 2F, S1C–S1E; Table S1). Overall, these aging experiments showed no evidence of tumor formation or pre-neoplastic lesions in iCK-cre mutants.
Tumor Suppressor Deficiency in Immature Neurons Promotes Abnormal Proliferation
In the adult SVZ, neural stem cells serve as a continuous source of new neurons by forming amplifying progenitor cells that produce neuroblasts, which typically undergo one to two cell divisions4, 19. To determine whether adult, immature neurons can undergo tumorigenesis, we employed the NeuroD1-creERT2 transgene20. Analysis of NeuroD1-creERT2;R26-stop-TdTomato reporter mice verified transgene co-expression with the immature neuronal marker, Doublecortin, especially in the olfactory bulb granular cell layer and striatum, as well as dentate gyrus and cortex (Figures 3A–3C, S2A–S2B). The Tomato reporter also co-expressed with the more differentiated marker NeuN, but not with the early progenitor markers Sox2, Olig2, Gfap or Nestin (Figures 3C, S2B).
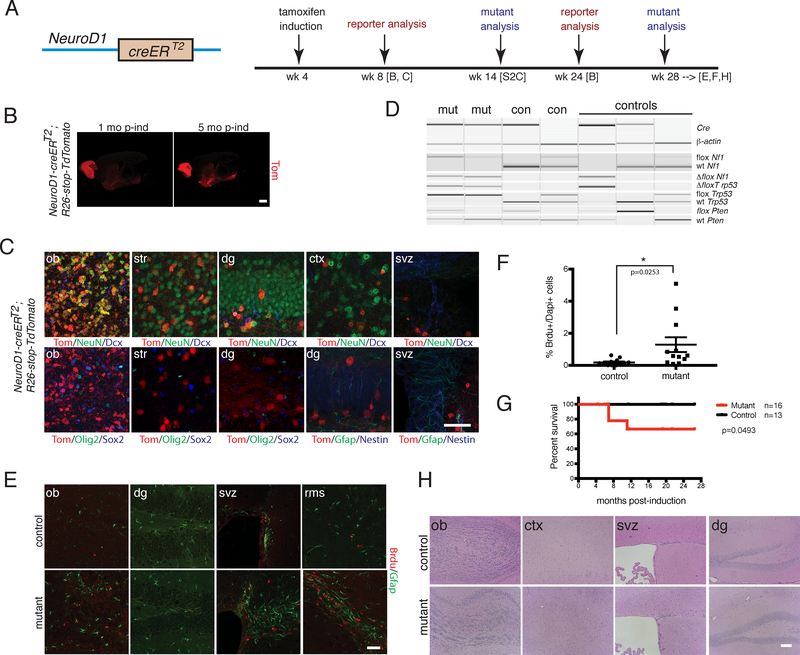
Figure 3. iND-cre Immature Neuron Mutants Show Olfactory Bulb Proliferation Defects.
A. (Left panel) Cartoon of NeuroD1-creERT2 (iND-cre) construct; (Right panel) Timeline of tamoxifen induction and analyses of reporter and mutant strains. B. Immunostaining of iND-cre;R26-stop-TdTomato reporter at 1 month and 5 months post-induction. Scale bar = 2 mm. C. Immunostaining of iND-cre;R26-stop-TdTomato reporter using cell type-specific markers at 1 month post-induction. Scale bar=100 μm. D. PCR genotyping for cre and recombined, wild type and conditional tumor suppressor alleles of iND-cre mutant and control brain regions. E. Immunoflourescence staining of aged iND-cre mutant vs. control after a short term BrdU pulse chase. Scale bar=100 μm. F. Dot plot showing % of BrdU-positive cells per Dapi-positive cells in iND-cre mutants (n=12) vs. controls (n=12). p=0.0253. *p<0.05 using two-tailed unpaired student’s t-test. Data is presented as mean +/− SEM. G. Kaplan-Meier survival curve of iND-cre mutants (n=16) vs. controls (n=13). p=0.0493 using Logrank (Mantel-Cox) test. H. H&E staining of aged iND-cre mutant vs. control mouse brains. Scale bar=100 μm. In B-E and H, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group.
NeuroD1-creERT2 (hereafter iND-cre) mutants were bred to contain the tumor suppressor conditional alleles (iND-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or iND-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or iND-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+ or iND-cre;Nf1flox/+;Trp53flox/+;Pten+/+) and induced at 4 to 8 weeks of age. We confirmed cre-mediated recombination by PCR genotyping (Figure 3D). As early as 10 weeks post-tamoxifen induction, we found increased BrdU incorporation in iND-cre mutants (Figure S2C), which persisted in older mice (Figures 3E–3F, S2D). A proportion of the iND-cre mutants developed masses outside the brain, likely due to transgene expression in the peripheral nervous system, including cranial and dorsal root ganglia20, and other tissues such as muscle, and were sacrificed (Figure 3G; data not shown). However, the cohort of mutant mice that survived 11 months or more showed no evidence of glioma formation (Figures 3E, 3G–3H). We did observe astrogliosis as well as increased BrdU-positive cells in older iND-cre mutants that appeared to be transient in nature, as no appreciable intracranial masses, histologic abnormalities, or differences in expression of other lineage markers developed (Figures 3E–3F, 3H, S2D–S2E). Thus, tumor suppressor-deficient immature neurons exhibit extended cell division but no obvious histologic abnormalities, and remain incapable of undergoing frank transformation and tumorigenesis.
Tumor Suppressor Targeting of Advanced Neuronal Progenitors Causes Abnormal Proliferation
Having established the inability of adult mature and immature neurons to initiate glioma in response to tumor suppressor ablation, we next moved upstream on the progenitor pathway to late stage neuronal progenitors. We used a Dlx1-creERT2 knock-in mouse (hereafter iDlx-cre) to specifically target tumor suppressors in adult neuronal progenitors21. Neural stem cell cultures derived from SVZ of iDlx-cre mice were previously shown to produce neurospheres when grown in serum-free conditions22, and the transgene has been used to study developing progenitors and GABAergic interneurons21. We confirmed cre activity in adult neuronal progenitors using the iDlx-cre;R26-stop-TdTomato reporter by tamoxifen induction at 4 to 8 weeks of age. Examination at 1 month and 5 months post-tamoxifen induction showed expression in the dentate gyrus, particularly in the granule cell layer, the glomerular layer in the olfactory bulb, cortex and cerebellum (Figures 4A–4C, S3A). Tomato reporter-positive cells co-express the progenitor markers Sox2 and Nestin and their downstream lineage markers Doublecortin and NeuN, but not markers of upstream or other progenitor lineage phases such as Gfap and Olig2 (Figures 4C, S3B). Thus, iDlx-cre activity is confined to advanced stage neuronal progenitors.
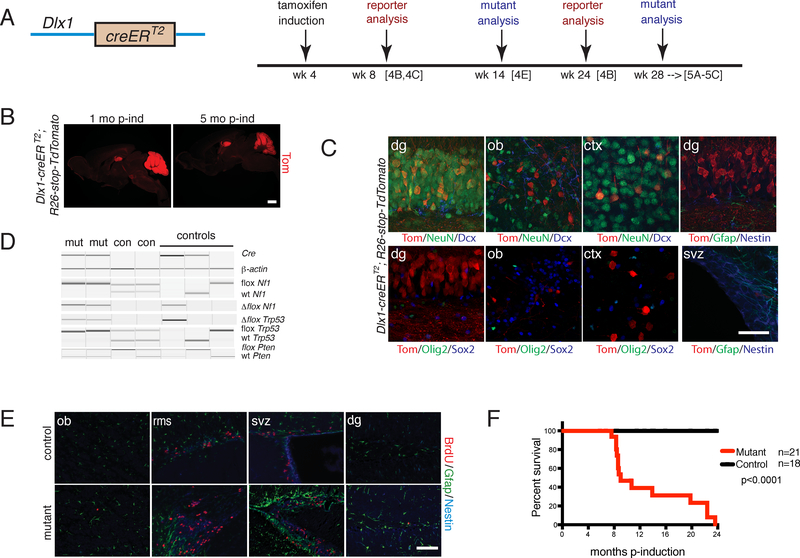
Figure 4. Adult Neuronal Progenitor iDlx-cre Targeted Mutants Exhibit Early Defects.
A. (Left panel) Cartoon of iDlx-cre (Dlx1-creERT2) construct; (Right panel) Timeline of tamoxifen induction and analyses of reporter and mutant strains. B. Immunostaining of iDlx-cre;R26-stop-TdTomato reporter at 1 month and 5 months post-induction. Scale bar=2 mm. C. Immunohistochemistry of iDlx-cre reporter at 1 month post-induction using cell type-specific markers. Scale bar=100 μm. D. PCR genotyping for cre and tumor suppressor wild type, conditional and recombined alleles of mutant vs. control tissue. E. Immunoflourescence staining of iDlx-cre mutant vs. control given a short BrdU pulse at 10 weeks post-induction. Scale bar=100 μm. F. Kaplan-Meier survival curve of iDlx-cre mutants (n=21) vs. controls (n=18) p<0.0001 using Logrank (Mantel-Cox) test. In B-E, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group.
We bred iDlx-cre to contain the tumor suppressor conditional alleles (iDlx-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or iDlx-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or iDlx-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+ or iDlx-cre;Nf1flox/+;Trp53flox/+;Pten+/+), performed tamoxifen induction at 4 to 8 weeks of age, and confirmed cre-mediated recombination by PCR genotyping (Figure 4D). BrdU pulse chase experiments indicated abnormal SVZ and RMS proliferation beginning at 10 weeks post-induction (Figure 4E). We also observed increased Gfap staining in these regions and shortened life spans (Figure 4F). Despite the evident abnormal brain proliferation and hyperplasia observed from 6 months to more than one year post tamoxifen induction (Figures 5A–5C, S4A), careful brain pathology surveys did not identify evidence of intracranial tumors (Figure 5C). The Dlx1 gene is expressed outside the brain, including in muscle and endocrine glands (www.proteinatlas.org). Likely related to this non-CNS expression, a small subset of mutant mice exhibited limb paralysis and tumors in the head and extremities. Histologic analysis of the extracranial tumors from both the iDlx-cre and iND-cre mice show high expression of smooth muscle actin and desmin, consistent with a mesenchymal origin23 (data not shown). However, the dominant cohort of mutant mice that were aged more than one year post-tamoxifen induction showed no evidence of intracranial tumor formation.
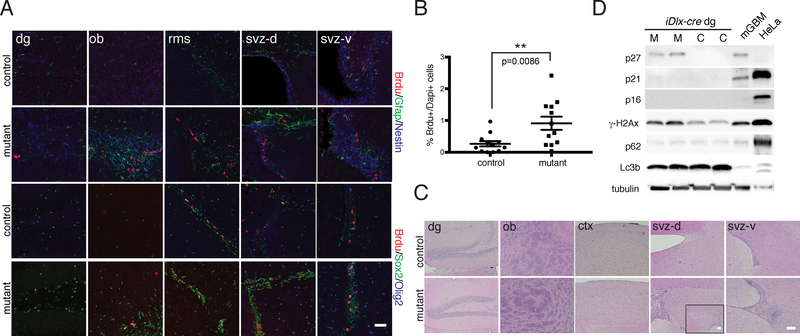
Figure 5. iDlx-cre Mutants exhibit Hyperplastic Lesions and Molecular Phenotypes.
A. Immunoflourescence staining of iDlx-cre mutant vs. control with BrdU (after a short term pulse) and lineage markers (svz-d=dorsal SVZ region, svz-v=ventral SVZ region). B. Quantification of % BrdU-positive cells in iDlx-cre mutants (n=12) vs. controls (n=12). p=0.0086. **p<0.01 using two-tailed unpaired Student’s t-test. Data is presented as mean +/− SEM. C. H&E staining of aged iDlx-cre mutant vs. control mouse brains reveal regions of hypercellularity (>28 weeks post-induction). Inset in mutant svz-d showing magnified lateral ventricle. D. Western blot analysis of iDlx-cre mutants and controls using DNA damage (gH2Ax), senescence (p16, p21, p27) and autophagy (Lc3b, p62) markers. Mouse GBM (mGBM) and HeLa cell lysates were used as controls. All scale bars=100 μm. In A and C-D, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group
In addition, iDlx-cre mutants showed increased CNS expression of the progenitor markers Sox2, Nestin and Doublecortin (Figures 5A, S4B). The evidence for persistent progenitor proliferation in the absence of tumor development can be explained by either constant apoptosis, sustained differentiation, or some combination thereof. We did not observe increased apoptosis as measured by cleaved caspase 3 staining, whereas long term BrdU pulse/chases showed BrdU+ cell migration into the olfactory bulb (data not shown). Thus, tumor suppressor targeting of adult neuronal progenitors elicits histologic and proliferative abnormalities reminiscent of the pre-tumorigenic state observed in previous glioma models6, 7, 24.
The molecular pathways linked to p53, Nf1 and Pten are not confined to cell proliferation or survival. The Ras-Nf1 and Phosphatidyl-inositol-3-kinase (PI3K)-Pten pathways impinge on many effectors of cellular function including migration, cytoskeletal remodeling, metabolism, endocytosis and autophagy. P53 serves not only as a cell cycle check point but also mediates many cellular stress responses including DNA damage and senescence25. To verify that tumor suppressor loss elicited downstream effectors, we examined specific molecular events in mutant brains.
Western blotting for γ-H2AX showed increased DNA damage in the dentate gyrus of iDlx-cre mutant neurons (Figure 5D) and confirmed elevated p-Erk and p-Akt (Figure S4C). Both iDlx-cre and iCK-cre mutants showed increased senescence marker p27 expression (Figure 5D; data not shown). We did not observe changes in p16 or p21, nor autophagy (Lc3b, p62) or SASP (senescence-associated secretory phenotype) markers (Figure 5D; data not shown), while telomere FISH showed shorter telomeres in iDlx-cre mutants (Figures S4D–S4F). Thus, cells of the neuronal lineage targeted by iDlx-cre and iCK-cre exhibit multiple pathway abnormalities consequent to tumor suppressor loss without eliciting tumor development.
Ectopic Synapsin1-cre expression in Oligodendrocyte and Neural Progenitors drives GBM formation
In this study, we also examined tumor suppressor recombination mediated by a Synapsin1-cre transgene (hereafter Syn-cre). Syn-cre becomes active in the developing CNS at embryonic day 13.5 (E13.5) and was previously employed to study the function of the Nf1 gene in neurons, under conditions that required predominant but not absolute or exclusive expression in neurons17. In the present scenario where, as for iCK-cre, exclusive mature neuronal recombination was intended, a detailed analysis of Syn-cre expression was performed using the TdTomato reporter. While, as previously documented17, a majority of Tomato-positive cells co-label with the neuronal marker NeuN, we found that a substantial subset of Tomato positive cells also marked both oligodendrocyte and neural progenitors, and astrocytes in the adult brain. Tomato-positive cells co-labelled with the oligodendrocyte progenitor marker Olig2 and the neural progenitor marker Sox2, particularly in the cortex near the corpus callosum border and with Gfap-positive elsewhere in the brain, including the dentate gyrus (Figure 6A; data not shown). Thus, the Syn-cre transgene has unexpected non-neuronal expression in adult CNS progenitors. When Syn-cre was bred to contain various configurations of the Nf1, Trp53 and Pten conditional alleles (Syn-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or Syn-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or Syn-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+ or Syn-cre;Nf1flox/+; Trp53flox/+;Pten+/+), mutant mice that were homozygous for both Nf1 and Trp53 exhibited shortened lifespan compared to mutants that were heterozygous for both or all three alleles (Figure 6B). Double knockout mice (Syn-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or Syn-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+) did not develop tumors nor did they exhibit proliferative defects (Figures 6C–6D). However, we observed reactive astrogliosis similar to iCK-cre mutants and as previously reported for Syn-cre;Nf1flox/flox mutants17. Double or triple heterozygous mice (Syn-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or Syn-cre;Nf1flox/+;Trp53flox/+;Pten+/+), on the other hand, developed tumors starting at 5 months of age (Figure 6B). These tumors showed histologic features of glioblastoma, with expression of the GBM markers Gfap, Ki67, Olig2, Sox2 and Pdgfra, with some cells positive for NeuN (Figure 6E). We confirmed that these tumors arose from cre-mediated recombination by X-gal staining, PCR genotyping, and immunostaining (Figure 6F; data not shown).
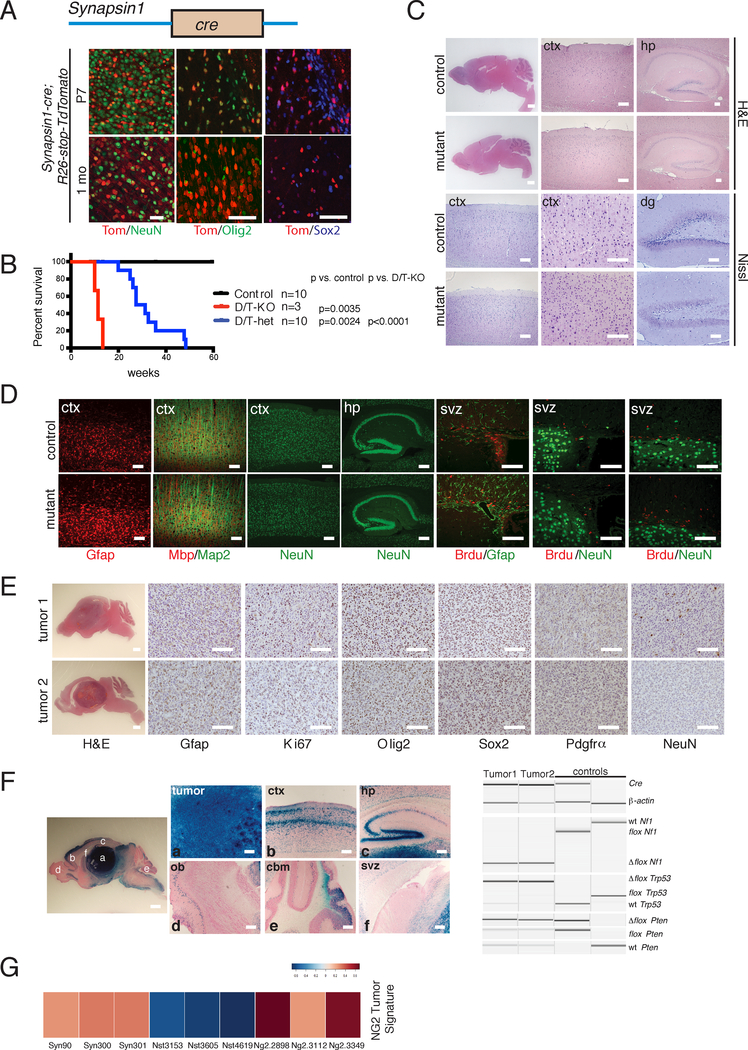
Figure 6. Syn-cre Mutants Exhibit Ectopic Recombination in Neural Progenitors and OPCs.
A. (Top panel) Cartoon of Synapsin1-cre (Syn-cre) construct. (Bottom panel) Immunoflourescence staining of Syn-cre;R26-stop-TdTomato reporter mouse brain using cell lineage markers at P7 and 1 month of age. Scale bars=100 μm. B. Kaplan-Meier survival curve of double/triple knockout (D/T-KO: Syn-cre;Nf1flox/flox;Trp53flox/flox;Ptenflox/+ or Syn-cre;Nf1flox/flox;Trp53flox/flox;Pten+/+; n=3), double/triple heterozygotes (D/T-het: Syn-cre;Nf1flox/+;Trp53flox/+;Ptenflox/+ or Syn-cre;Nf1flox/+; Trp53flox/+;Pten+/+; n=10), and controls (n=10). D/T-KO vs. control: p=0.0035; D/T-het vs. control: p=0.0024; D/T-KO vs. D/T-het: p<0.0001 using Logrank (Mantel-Cox) test. C. H&E and Nissl staining of double/triple knockout mutant brains vs. controls. Scale bars=100 μm except for whole brain=2 mm. D. Immunoflourescence staining of double/triple knockout mutant brains vs. controls using lineage and proliferation markers. Scale bars=100 μm. E. H&E and immunohistochemistry of representative brain tumors using cell lineage and proliferation markers. Scale bars=100 μm except for whole brain=2 mm. F. Recombination of tumor suppressors in Syn-cre Mutants. (Left panel) X-gal staining of a Syn-cre mutant brain section showing different brain regions. (Right panel) PCR genotyping of Syn-cre mutants and controls. Scale bars=100 μm except for whole brain=2 mm. G. Heat map of NG2-creER™ GBM gene expression signature in Syn-cre tumors (n=3, leftmost), as compared with Nestin-creERT2 (n=3, middle) and NG2-creER™ (n=3, rightmost) tumors. In A and C-F, experiments were independently repeated with similar results at least n=3 times using at least n=3 different mouse tissue samples for each group.
We have previously shown that adult OPCs can give origin to GBM with characteristic cellular, histologic, and molecular features (Type 2) that are distinct from those derived from the adult neural stem cell-driven Nestin-creERT2 mutants (Type 1)7, 8. Syn-cre tumors exhibit striking similarities to Type 2 GBM, including well-defined borders indicative of low infiltration, high Olig2 and Pdgfrα and reduced Gfap expression with preferential location in the ventral brain. These data, taken together together with the iCK-cre studies, raise the possibility that Syn-cre-induced GBM does not originate in neurons. When we compared the transcriptional profiles of Syn-cre tumors with that of Type 1 (Nestin-creERT2) and Type 2 (NG2-creER™) tumors by RNA sequencing, Syn-cre tumor profiles preferentially clustered with Type 2 tumors and showed enrichment for the NG2-creER™ GBM signature, including expression of genes associated with the oligodendrocytic lineage (Figures 6G, S5; Table S2). The present data, taken as a whole, provide support for the contention that the tumor cell of origin for Syn-cre-derived GBM is the OPC lineage and not mature neurons.
Discussion
The rodent brain is composed of over three hundred million cells with approximately 60% neurons and 40% glia, in addition to the many essential, non-CNS specific resident cells26. In contrast, the pool of adult CNS stem and progenitor cells comprises a minimal fraction. Glioblastoma is the most common adult primary brain malignancy and historical thinking based on anecdotal evidence and histologic associations conferred the origin of these tumors to astrocytes1. The advent of genetically engineered mouse models prompted revisiting of this issue, and the data has increasingly implicated CNS stem cells and their early progeny as the most likely source of GBM6, 7, 24, 27.
Our previous studies provided indirect evidence for the reduced tumorigenic capacity of non-stem/progenitor cells. Here, we directly address the tumorigenic potential of increasingly differentiated CNS neuronal lineage cells, by employing well characterized conditional Cre recombinase driver transgenes. The data show that deletion of three of the most commonly mutated tumor suppressors in human GBM can be effectively accomplished in each of the targeted cell types. In contrast to the 100% GBM incidence elicited in neural stem cells, the phenotypic consequences of tumor suppressor deletion in more differentiated cells were insufficient to drive GBM formation. Taken together with studies associating stem cells and early progenitors with GBM, these data indicate an inverse relationship between GBM initiating potential and differentiation state of neuronal cell types, where stem cells are most susceptible and mature neurons the most resistant to malignant transformation (Figure 7). The GBM hierarchy shares similarities with that of leukemias, where mutations may accumulate in long-lived hematopoietic stem cells or confer self-renewal capacity to specific progenitor populations28. Our studies demonstrate that tumor suppressor-deficient cells beyond the early progenitor state are phenotypically altered despite not undergoing malignant transformation. Better insight into the mechanisms that prevent more differentiated cells from re-entering the cell cycle could advance our understanding of intrinsic cellular anti-cancer barriers.
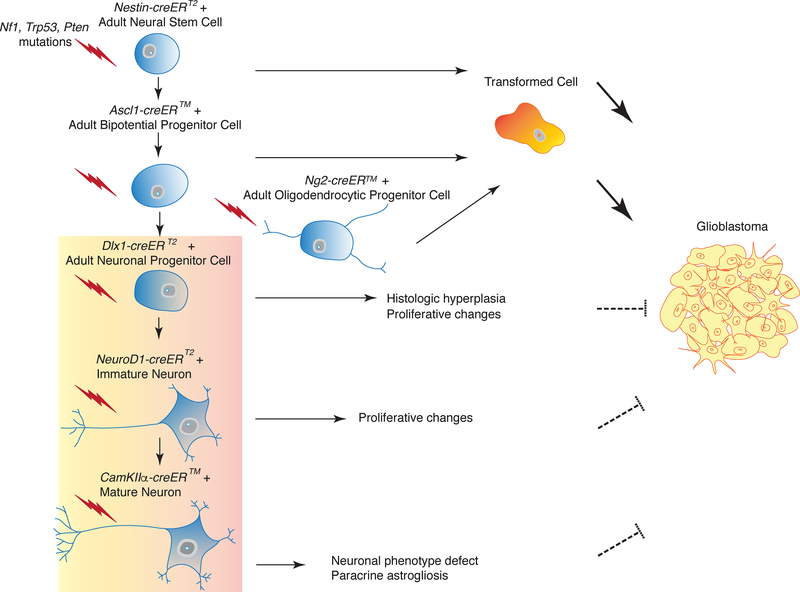
Figure 7. Model for Tumor Suppressor-Mediated Transformation of Adult Neural Lineage Cells.
Targeting of tumor suppressor mutations (Nf1, Trp53, Pten, symbolized by lightning) in neural stem cells (Nestin-creERT2), bi-potential progenitors (Ascl1-creER™) and oligodendrocyte progenitors (NG2-creER™) leads to transformation and glioma development, whereas targeting of late stage neuronal progenitors (Dlx1-creERT2), newly born neurons (NeuroD1-creERT2) and post-mitotic, differentiated neurons (CamKIIα-creER™) does not. The latter three mouse models show decreasing phenotypic defects with increasing lineage differentiation.
Contrary to the present results, it has been reported that mature neurons can undergo dedifferentiation to give rise to glioblastoma12. Mouse models of brain tumors were generated using stereotactic injection of lentiviruses expressing either oncogenic Ras or shRNAs to Nf1 together with Trp53 under non-specific promoters and utilizing the Syn-cre transgene as an independent reporter for neurons. Our present results demonstrating Syn-cre transgene expression in OPCs and neural progenitors that are known cells of origin for GBM7 broaden the scope of interpretation of the preceding studies, raising the possibility that these tumors did not arise from de-differentiation of mature neurons12.
We selected mutations in TP53, NF1 and PTEN by virtue of their inclusion in a large proportion of human GBMs13, which are also the same mutations used in previous studies described12, 29. Our data do not address whether other classes and combinations of mutations would render differentiated neurons tumorigenic. Compelling studies have demonstrated that a Ras oncogene can render astrocytic cells tumorigenic in the experimental setting. However, Ras mutations are exceedingly rare in the repertoire of GBM mutations, making it unlikely that those conditions would occur in the physiologic setting30. Reports of reprogramming of neurons into stem cells have used early postnatal neuronal cultures with the Yamanaka factors (Oct3/4, Klf4, Sox2, Oct4) and involved concurrent p53 suppression and overexpression of repressor element-1 repressing transcription (REST), illustrating the difficulty in forcing these cells to re-enter the cell cycle, even in this extremely artificial in vitro setting31. However, it is possible that other oncogenic mutations and other tumor suppressor combinations not tested here, may have capacity to transform differentiated cell types. In leukemia, different cells along the hematopoietic lineage have been shown to harbor differential sensitivities to specific oncogenic mutations. Depending on the model system employed, MLL-AF9 mutations transform committed progenitors, whereas others such as BCR-ABL are oncogenic only when expressed in stem cells32. Hence, the identity of the cell of origin, which is also related to the developmental stage and intrinsic properties of the cell lineage, are equally important as the initiating event.
The present data implicate a small fraction of cells in the brain with GBM potential. The relative paucity of transformable cells in the brain as compared to epithelial organs such as lung or colon may contribute to the much lower incidence of CNS tumors compared to other cancer types. These theories have been supported by studies that suggest that cancer incidence is proportionally related to the size of the stem cell pool33. Indeed, many reports have recorded the presence of neural stem cells in the adult brain34. However, recent controversy has arisen by contrasting reports regarding the relative abundance of adult human neural stem cells. One report claimed complete decline in progenitor populations in the adult human CNS35, while another showed persistent neurogenesis, albeit with decline, throughout life36. Examples of robust adult neurogenesis in other species including the seasonal maturation of high vocal center CNS nuclei in songbirds, as well as SVZ and hippocampal neurogenesis in rodents3, 4, primates37 and humans38, challenge the notion that neurogenesis would somehow disappear in the most advanced stages of CNS evolution. Along this line, a recent report using whole exome and single cell sequencing of matched GBM patient brain tissue, showed a lineage relationship in tumor evolution arising from SVZ cells with low-level driver mutations39. These recent findings, together with our current and previous data6, 7, 24, 27, are most consistent with a stem/progenitor cell of origin for GBM while dedifferentiation from post-mitotic neurons or neuroblasts is a less likely event.
The incidence of cancer increases proportionally with age. For a solid tumor to arise, a single tumor cell of origin must accumulate a cohort of oncogenic and tumor suppressive driver mutations. These requirements are consonant with the notion that self-renewing stem cells or closely related progeny are a more plausible source of tumors compared to post-mitotic cells. These are the cells that retain cell cycle activity over prolonged periods, albeit with reduced frequency, allowing for sequential accumulation of mutations over time. Moreover, the mechanisms involved in causing functional “transformation” of a stem cell to become a tumor seem less constraining than those envisioned to force a terminally differentiated cell to overcome the transcriptional and epigenetic barriers that govern specialized function, as differentiated cells possess innate programs to prevent cell cycle re-entry. Accordingly, numerous studies have explored the possibility of differentiation therapy to inhibit tumor cell proliferation, and a few examples already exist in the clinical setting40, 41. Understanding these anti-tumorigenic mechanisms may continue to reveal effective anti-cancer therapeutic targets, especially for these fatal cancers.
Methods
Mouse Experiments
All mouse experiments were approved and performed according to the guidelines of the Institutional Animal Care and Use Committee of UT Southwestern Medical Center and Memorial Sloan Kettering Cancer Center. Mice with cre transgenes and conditional Nf1, p53 and/or Pten alleles with the R26-stop-lacZ and R26-stop-TdTomato reporters were maintained on a mixed 129Svj/C57Bl6/B6CBA background24, 27 Male and female mice with inducible transgenes were administered tamoxifen or vehicle (sunflower oil) at 4 to 8 weeks of age, as previously described6. Prior to sacrifice, mice were injected with BrdU intraperitoneally at 100 mg/kg 1 hour prior to perfusion or harvest.
Histology and Tumor Analysis
Mice were perfused and fixed with 4% paraformaldehyde. For paraffin blocks, 5 μm sections were cut and every fifth slide was stained with H&E. Brain sections were independently examined by S.A.L. and D.K.B., a board-certified neuropathologist, and tumor diagnosis was determined based on the World Health Organization criteria42. Brains used for X-gal staining were post-fixed in 2% PFA overnight. Fifty-μm vibratome sections were stained in X-gal solution, and sections were counterstained by nuclear fast red, as previously described16. Brains used for frozen sections were incubated in sucrose, embedded in OCT and sectioned at 12 um. For PCR genotyping, we micro-dissected tissue from mutants and controls using either vibratome or paraffin sections, or frozen tissue. DNA was extracted using the Qiagen DNeasy extraction kit or the Qiagen FFPE DNA extraction kit, following manufacturer’s instructions. PCR was performed using primers for recombined Nf1 and Pten alleles24 and Trp5343 as previously described.
Immunohistochemistry and Western Blot Analysis
Paraffin sections were deparaffinized, rehydrated, and subjected to citrate-based antigen retrieval. In other cases, 50-μm vibratome sections were used for staining. Primary antibodies against the following were used as follows: GFAP (DAKO; BD Biosciences), Ki67 (Novocastra), nestin (BD Biosciences), Olig2 (Chemicon), β-galactosidase (ICN), Doublecortin (Santa Cruz), NeuN (Chemicon), MBP (Sternberger), GFP (Chemicon), PDGFRα (Santa Cruz), NG2 (Millipore), CNPase (Millipore), APC (Calbiochem), BrdU (Abcam), Map2 (Sigma), Parvalbumin (Swant), Calretinin (Millipore), vGlut2 (Millipore), GABAARα (Millipore), Synapsin (Millipore), Sox2 (Abcam), γ-H2Ax (Cell Signaling), p21 (BD), p27 (Cell Signaling), p16 (Abcam), Lc3b (Novus Biologicals), p62 (MBL), pAKT (Cell Signaling), pErk (Cell Signaling) and pS6 (Cell Signaling). We used both immunofluorescence staining using Cy2, Cy3, or Cy5 (Jackson Labs) and biotin-streptavidin-Alexa Fluor-conjugated secondary antibodies (Molecular Probes), as well as horseradish peroxidase-based Vectastain ABC Kit (Vector Lab). The same antibodies were used for immunoblotting for γ-H2Ax, p21, p27, p16, Lc3b and p62. All antibodies were verified using appropriate positive and negative controls. Detailed information on all antibodies used are in the Supplementary Information section.
Immuno-FISH
For analysis of telomere length, we used a modified Telomere FISH protocol as was previously described44. We performed immunofluorescence staining on paraffin sections and proceeded with the FISH protocol starting at the methanol fixation step. Images were acquired using the Leica Confocal system and processed for quantification as previously described44.
RNA-Sequencing Analysis
Primary tumor and control brain tissue were harvested from symptomatic mutant and age-matched control mice and flash frozen in liquid nitrogen. RNA was extracted using Qiagen Lipid Tissue RNeasy Mini Kit and performed according to manufacturer’s instructions. Total RNA was submitted to the Weill Cornell Genomics Core for RNA Sequencing Analysis. RNA sequencing data was analyzed using Bioconductor version 3.0 under R3.13. Differentially expressed transcripts between Type 1 and Type 2 tumors were analyzed by exactTest function in edgeR package version 3.8.6 and determined by false discovery rate (FDR) <0.01 and two-fold change. Type 1 signatures were highly expressed genes in Nestin-creERT2 tumors and Type 2 signatures were highly expressed genes in NG2-creER™ tumors, as previously described7. Enrichment scores for the NG2 Type 2 signature were calculated by GSVA package version 1.14.1. Dimension reduction analysis was done using Locally Linear Embedding (LLE) algorithm from RDRToolbox package version 1.16.0 and heatmaps were generated by gplots package version 3.0.1.
Shallow Whole Genome Sequencing Analysis
Tissue samples were micro-dissected from X-gal-stained vibratome sections to show regions of recombination within the brain. Extracted DNA was subsequently submitted to the MSKCC Genomics Core for QC and Shallow Whole Genome Sequencing, which was performed at 125 base pair end reads and 10 million reads/sample. FASTQ files were mapped to the target genome using the BWA mapper and the BAM files were then processed using the seqDNAcopy library to first get pairwise counts for the target sample and control samples. Adapted bin sizes were chosen so that 75% of the bins have at least 100 counts in the control sample. The data was then segmented using seqDNAcopy’s seqsegment method (https://github.com/veseshan/seqDNAcopy). Gene ontology analysis of gene events was performed using PANTHER (www.pantherdb.org )45 version 13.1.
Statistical Analysis
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications7, 24 Blinding and randomization were performed in all experiments. Data distribution was assumed to be normal but this was not formally tested. No animals or data points were excluded from the analyses for any reason. Statistical analysis between groups was performed using two-tailed unpaired Student’s t-test. Kaplan-Meier survival curves were analyzed using Logrank (Mantel-Cox) test. For dot plot figures presented, the center line represents the mean +/− SEM. Additional details are found in the Life Science Reporting Summary and Supplementary Information.
Data Availability
Sequence data that supports the findings in this paper is publicly accessible in Gene Expression Omnibus as GSE117258. The data that supports the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
The authors would like to thank D. Laks for processing of the RNA Sequencing Data and members of the Parada laboratory for helpful suggestions and discussion. We thank N. Socci, E. Feng, and V. Boyko for their help in various analyses, and the MSKCC Genomics and Bioinformatics Cores and the Weill Cornell Genomics and Epigenomics Cores for their assistance. This work was supported in part by the Children’s Tumor Foundation Young Investigator Award and NIH T32 Postdoctoral Training Grant (2T32CA124334–06; PI: Jerry Shay) to S.R.A.L. L.F.P. is a recipient of NIH R01 grant CA131313–01A1 and NCI R35 grant CA210100.
Footnotes
Competing Interests Statement
The authors declare the following financial competing interests: L.F.P. has advisory/consulting relationships with Bio-Thera Solutions (2013–2018), Howard Hughes Medical Institute Scientific Advisory Board (2006–2023), and the National Cancer Institute Board of Scientific Advisors (2013–2018).
Accession Codes
Sequencing data is publicly available in Gene Expression Omnibus (GEO) as GSE117258.
References
- 1.Sanai N, Alvarez-Buylla A & Berger MS Neural stem cells and the origin of gliomas. N Engl J Med 353, 811–822 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Bond AM, Ming GL & Song H Adult Mammalian Neural Stem Cells and Neurogenesis: Five Decades Later. Cell Stem Cell 17, 385–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gage FH & Temple S Neural stem cells: generating and regenerating the brain. Neuron 80, 588–601 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Lim DA & Alvarez-Buylla A The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb Perspect Biol 8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huse JT & Holland EC Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol 19, 132–143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15, 45–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alcantara Llaguno SR, et al. Adult Lineage-Restricted CNS Progenitors Specify Distinct Glioblastoma Subtypes. Cancer Cell 28, 429–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galvao RP, et al. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proc Natl Acad Sci U S A 111, E4214–4223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19, 305–316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holland EC, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25, 55–57 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1, 269–277 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338, 1080–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith LC Calcium/calmodulin-dependent protein kinase II: an unforgettable kinase. J Neurosci 24, 8391–8393 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luikart BW, et al. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J Neurosci 25, 3774–3786 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, et al. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev 15, 859–876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron 50, 377–388 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM & Alvarez-Buylla A Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Aprea J, Nonaka-Kinoshita M & Calegari F Generation and characterization of Neurod1-CreER(T2) mouse lines for the study of embryonic and adult neurogenesis. Genesis 52, 870–878 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mich JK, et al. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. Elife 3, e02669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oien KA & Dennis JL Diagnostic work-up of carcinoma of unknown primary: from immunohistochemistry to molecular profiling. Ann Oncol 23 Suppl 10, x271–277 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Kwon CH, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res 68, 3286–3294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herculano-Houzel S & Lent R Isotropic fractionator: a simple, rapid method for the quantification of total cell and neuron numbers in the brain. J Neurosci 25, 2518–2521 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 8, 119–130 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horton LB, et al. Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site. Antimicrob Agents Chemother 56, 5667–5677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcantara Llaguno SR & Parada LF Cell of origin of glioma: biological and clinical implications. Br J Cancer 115, 1445–1450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knobbe CB, Reifenberger J & Reifenberger G Mutation analysis of the Ras pathway genes NRAS, HRAS, KRAS and BRAF in glioblastomas. Acta Neuropathol 108, 467–470 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. Reprogramming of postnatal neurons into induced pluripotent stem cells by defined factors. Stem Cells 29, 992–1000 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6, 587–596 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Tomasetti C & Vogelstein B Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat Med 4, 1313–1317 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Sorrells SF, et al. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boldrini M, et al. Human Hippocampal Neurogenesis Persists throughout Aging. Cell Stem Cell 22, 589–599 e585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gould E, Reeves AJ, Graziano MS & Gross CG Neurogenesis in the neocortex of adult primates. Science 286, 548–552 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Curtis MA, et al. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science 315, 1243–1249 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Lee JH, et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 560, 243–247 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Warrell RP Jr., de The H, Wang ZY & Degos L Acute promyelocytic leukemia. N Engl J Med 329, 177–189 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Sell S Stem cell origin of cancer and differentiation therapy. Crit Rev Oncol Hematol 51, 1–28 (2004). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 42.Louis DN, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 131, 803–820 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Lin SC, et al. Somatic mutation of p53 leads to estrogen receptor alpha-positive and - negative mouse mammary tumors with high frequency of metastasis. Cancer Res 64, 3525–3532 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Flores I, et al. The longest telomeres: a general signature of adult stem cell compartments. Genes Dev 22, 654–667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45, D183–D189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that supports the findings in this paper is publicly accessible in Gene Expression Omnibus as GSE117258. The data that supports the findings of this study are available from the corresponding authors upon request.