Abstract
Viral hepatitis, the leading cause of liver diseases worldwide, is induced upon infection with hepatotropic viruses, including hepatitis A, B, C, D, and E virus. Due to their obligate intracellular lifestyles, culture systems for efficient viral replication are vital. Although basic and translational research on viral hepatitis has been performed for many years, conventional hepatocellular culture systems are not optimal. These studies have greatly benefited from recent efforts on improving cell culture models for virus replication and infection studies. Here we summarize the use of human stem cell-derived hepatocyte-like cells for hepatotropic virus infection studies, including the dissection of virus-host interactions and virus-induced pathogenesis as well as the identification and validation of novel antiviral agents.
1. Introduction
Viral hepatitis manifests itself as continuous liver inflammation and eventually liver injury and hepatic failure. As summarized in Table 1, the major causative agents of viral hepatitis are five hepatotropic viruses, including hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis D virus (HDV), and hepatitis E virus (HEV). HAV and HEV normally spread through contact with contaminated water or food, resulting in an estimated annual incidence of 1.5 million HAV infections and 20 million HEV infections [1, 2]. Both HAV and HEV typically cause acute infections; however, HEV can also cause chronic infections in immunocompromised patients [2]. HBV, HCV, and HDV are transmitted through blood transfusions, organ transplants, sex, and injection behavior [3–5]. Approximately, 10–15% of chronically HBV-infected patients are coinfected with HCV and 5% with HDV [6]. Infection with HBV, HCV, and HDV can cause both self-limited and chronic hepatitis and is the leading cause of liver diseases including fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [3, 4, 7]. In order to prevent disease progression, early diagnosis and treatments are vital. In spite of recent extraordinary advances in the treatment of hepatitis C, based on the success of HCV basic research [8], the need remains to understand the underlying molecular and cellular mechanisms of liver pathogenesis caused by the other hepatotropic viruses. The development of novel specific drugs against hepatotropic virus infection has been a challenging task, partially due to the lack of physiologically relevant cell culture models that can be used for medium/high-throughput drug screening.
Table 1.
Overview of hepatitis viruses.
| HAV | HBV | HCV | HDV | HEV | |
|---|---|---|---|---|---|
| Classification | Picornavirus | Hepadnavirus | Hepacivirus | Deltavirus | Hepevirus |
|
| |||||
| Genome | +ssRNA | dsRNA-RT | +ssRNA | -ssRNA | +ssRNA |
|
| |||||
| Incubation (days) | 20-40 | 45-160 | 15-150 | 30-60 | 15-60 |
|
| |||||
| Transmission | Fecal-oral | Parenteral Perinatal Sexual |
Parenteral Perinatal Sexual |
Parenteral Sexual |
Fecal-oral |
|
| |||||
| Chronicity | Acute | 5-10% chronic1 80% neonates |
70% chronic | Coexistence with HBV | Acute2 (Chronic in immunocompromised patients) |
|
| |||||
| Natural host | Human Chimpanzee Monkey [20] |
Human3 [21] | Human3 [22] | Human3 [23] | Human Animal4 [24] |
|
| |||||
| Carcinogenesis | – | + | + | – | – |
|
| |||||
| Prophylaxis | Vaccine | Vaccine | NA | HBV vaccine | Vaccine5 |
|
| |||||
| Therapy | NA | IFN, NAs | DAAs | IFN | RBV and withdrawal of immunosuppressants |
|
| |||||
| Cure | Self-cure | No | Yes | No | Self-cure (yes) |
15-10% in immunocompetent adults; 2mild in normal patients and severe in pregnant women; 3chimpanzees are susceptible but not naturally infected; 4genotype 3- and 4-specific; 5licensed in China. ss: single-stranded; ds: double-stranded; RT: reverse-transcriptase; IFN: interferon-α; NAs: nucleos(t)ide analogues; DAA: direct-acting antiviral; RBV: ribavirin; NA: not applicable.
Hepatoma cells have been invaluable across the history of hepatotropic virus studies in cell culture. Yet, their aberrant intracellular signaling and metabolic activities limit investigations of the viral and cellular innate immunity interactions as well as effects on cellular proliferation, metabolism, and apoptosis pathways. Furthermore, most hepatoma cell lines lack various functional enzymes, such as CYP450 and other phase I, II, and III drug-metabolizing enzymes, which make them unsuited for the assessment of antiviral drug interactions and metabolism [9, 10]. Therefore, studies using these models are limited in their ability to mimic natural virus-induced pathologies in the liver.
The most authentic cell culture system for hepatotropic virus studies is primary human hepatocytes (PHH). Yet, their use is hindered by limited donor supply, donor-to-donor variability, and rapid dedifferentiation upon plating in cell culture [11]. Given the limitations and challenges of using hepatoma cells and PHHs, hepatocyte-like cells (HLCs) derived from human embryonic stem cells (hESCs) or induced pluripotent stem cell (iPSC) have emerged as a promising cell culture model to study basic and translational liver diseases as well as hepatitis virus infection [12–14]. HLCs have been differentiated from diverse resources, such as hESCs, iPSCs, liver-resident hepatic progenitor cells, and bone marrow-derived mesenchymal stem cells [15]. Differentiated HLCs are functionally characterized by the production of urea, indocyanine green uptake, glycogen storage, and inducible cytochrome P450 (CYP450) activity [16]. In addition, they can rescue liver function after transplantation into animal models [17]. To date, HLC infection models for HBV, HCV, and HEV have been successfully established [17–19]. Here, we summarize the current knowledge on cell culture-based models available for these viruses and highlight the advantages of HLCs derived from stem cell as an improved system for basic and translational viral hepatitis research.
2. Hepatocyte-Like Cells for HBV Infection
Despite the availability of an efficient prophylactic vaccine, HBV infection is still a global public health burden with an estimated 257 million chronically infected people who are at increased risk of developing liver related-fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [25]. HBV contains a partially double-stranded, relaxed circular DNA (rcDNA) genome of approximately 3.2 kb, covalently linked to the HBV polymerase [26]. The rcDNA is delivered into the nucleus after viral entry and converted into fully double-stranded DNA, which is itself converted by ligation into an intracellular HBV replication intermediate called covalently closed circular DNA (cccDNA). cccDNA is responsible for HBV persistence in infected cells [6, 26]. A curative treatment of chronic hepatitis B should therefore target permanent transcriptional silencing or elimination of cccDNA [27].
Currently, treatments for chronic hepatitis B are limited to type 1 interferons (IFN-α) and five approved nucleos(t)ide analogues (NAs) [28]. Due to severe side effects of interferon therapy, only few patients are eligible for treatment, and less than 10% of them show a sustained virological response evidenced as loss of hepatitis B surface antigen (HBsAg) [29]. NAs are the most potent drugs; tenofovir and entecavir can reduce viral DNA, often below the detection limit with low resistance development [30, 31]. However, most patients remain HBsAg-positive even after prolonged treatment and the frequent viral rebound upon therapy withdrawal indicates a need for lifelong treatment [32]. In addition, long-term administration of tenofovir has been associated with Fanconi syndrome, a decrease in bone mineral density and chronic renal tubular damage [33]. Since current antiviral strategies cannot completely eradicate viral infection, an urgent need for the development of novel antiviral therapeutics remains [34].
Basic and translational research has been hindered by the absence of in vitro experimental models that feature the physiological condition of hepatocytes and permits efficient HBV and infection. As shown in Table 2, human hepatoma cell lines, such as Huh-7 and HepG2, are widely used as surrogate models for HBV infection, even though they only partially mimic physiological hepatic functions. Stable HBV-integrated hepatoma cell lines have been generated through transfection of human hepatoma cells with an HBV-expressing plasmid [35–38]. Alternative systems were the delivery of the HBV genome by baculoviral or adenoviral vectors, which resulted in sufficient HBV replication and viral particle production [39, 40]. However, these cell lines are not permissive for natural infection as they are unable to mediate early steps of virus infection, including entry, uncoating, and cccDNA formation. Primary human hepatocytes (PHHs) support the full viral replication cycle and serve as the gold standard of HBV infection. However, they have many disadvantages, including high donor variability, short lifespans, and limited availability. Despite many attempts to improve methods for maintaining freshly isolated PHHs, they often rapidly dedifferentiate in culture dishes [19, 41–43]. HepaRG cells are liver progenitor cells that can be differentiated in vitro and then support the whole HBV life cycle, an alternative tool for HBV studies [35, 44]. However, the efficiency of HBV infection in these differentiated HepaRG cells remains lower than in other cell systems. In addition, the differentiated cells contain both hepatocyte and biliary lineages, which affects HBV-host interaction studies in a hepatocyte-specific environment [45]. New HBV infection cell culture models have been developed when human sodium taurocholate cotransporting polypeptide (NTCP) was identified as the HBV entry receptor [46]. NTCP-overexpressing hepatoma cell lines were generated including HepG2-NTCP and Huh-7-NTCP cell lines, which provide an easily accessible platform for HBV-host interaction and antiviral studies [46, 47]. But, as mentioned above, although the entire HBV life cycle is recapitulated, hepatoma cells have altered physiological signaling pathways.
Table 2.
Receptors and infection models of hepatitis viruses.
| HAV | HBV | HCV | HEV | |
|---|---|---|---|---|
| Receptor | TIM-1 [48] | NTCP [46] | CD81, SR-BI, OCLN, CLDN1 [49–52] | Unknown |
| Cell model | HAV | HBV | HCV | HEV |
| PHH | +[53] | +[46] | +[54] | +[55] |
| HepG2 | +[53] | +/– | – | +[56] |
| HepG2NTCP | Unknown | +[46] | – | Unknown |
| HepaRG | Unknown | +[44] | – | +[57] |
| Huh7 | Unknown | +/– | +/– | +[56] |
| Huh7NTCP | Unknown | +[46] | +/– | Unknown |
| Huh7.5.1 | Unknown | – | +[58, 59] | Unknown |
| HLCZ01 | Unknown | +[60] | +[60] | Unknown |
| PLC/PRF5 | Unknown | Unknown | Unknown | +[61] |
| A549 | Unknown | – | – | +[61] |
| iPSC-derived HLC | Unknown | +[19, 62–64] | +[17, 65] | +[18, 66, 67] |
| hESC-derived HLC | Unknown | +[19] | +[17, 65] | +[18, 66, 67] |
TIM-1: T-cell immunoglobulin and mucin domain 1; NTCP: sodium taurocholate cotransporting polypeptide; CD81: cluster of differentiation 81; SR-BI: scavenger receptor class B type I; OCLN: occluding; CLDN1: claudin-1; PHH: primary human hepatocyte; iPSC: induced pluripotent stem cell; hESC: human embryonic stem cell; HLC: hepatocyte-like cell. +: permissive; +/-: barely permissive; -: not permissive.
Recently, iPSC-derived HLCs were reported to support HBV infection [19, 63]. A time-course experiment showed that both a full activation of the transcription machinery and an expression of NTCP on the cell surface are essential to achieving productive HBV infection. This demonstrated the potential of human iPSC-derived HLCs for in vitro studies of HBV biology, yet the infection efficiency remained very low. Although the authors observed temporal induction of interferon-stimulated genes (ISGs) in HBV-infected HLCs, studies from other groups rather support the notion that HBV is a stealth virus both in vitro and in vivo [68–71]. Similarly, Sakurai et al. established human iPSC-derived HLCs that allow about 20% HBV infection efficiency [62]. Xia et al. used an optimized protocol [72] to differentiate the non-colony-type monolayer culture of hESCs and iPSCs to HLCs in 15 days (Figure 1). The HLCs maintained their differentiated state and allowed HBV infection for more than 4 weeks. Importantly, the authors successfully demonstrated that the optimized protocol for HLC differentiation provided an in vitro model capable of supporting HBV spread. Notably, the dedifferentiation process occurred at a slower rate in HLCs than in PHHs, as high expression levels of proviral factors, including NTCP, HNF4A, and RXRA, were maintained for more than 3 weeks, making them a suitable model for long-term HBV infection studies [19]. Knocking down NTCP reduced HBV infection while knocking down antiviral factor APOBEC3A enhanced viral replication, indicating that HLCs constitute an appropriate system for virus-host interaction studies. By using this model, the authors identified two host-targeting agents, genistin and PA452, as novel antivirals. Recently, Nie et al. used iPSCs to generate liver organoids and evaluated their application in studying HBV virus–host interactions [73]. They cultured iPSC-derived endodermal, mesenchymal, and endothelial cells with a chemically defined medium in a three-dimensional (3D) microwell culture system, in which the cells organized themselves to gradually differentiate into a functional liver organoid. They showed that the organoid exhibited stronger hepatic functions than did 2-cultured HLCs with a higher susceptibility to HBV infection [73]. Yuan et al. developed a mouse model to study in vivo HBV infection by engrafting iPSC-derived HLCs into immune-deficient mice [74]. The liver of these mice contains approximately 40% HLCs at week 6 and maintained at this level for at least 14 weeks. After HBV infection, viral replication markers such as HBsAg, HBeAg, RNA, DNA, and cccDNA were detectable in the sera. Furthermore, these mice can be used to test different antivirals [74]. Together, all these studies demonstrated that HLCs fully support HBV infection and virus-host interactions, allowing the identification and validation of novel antiviral agents.
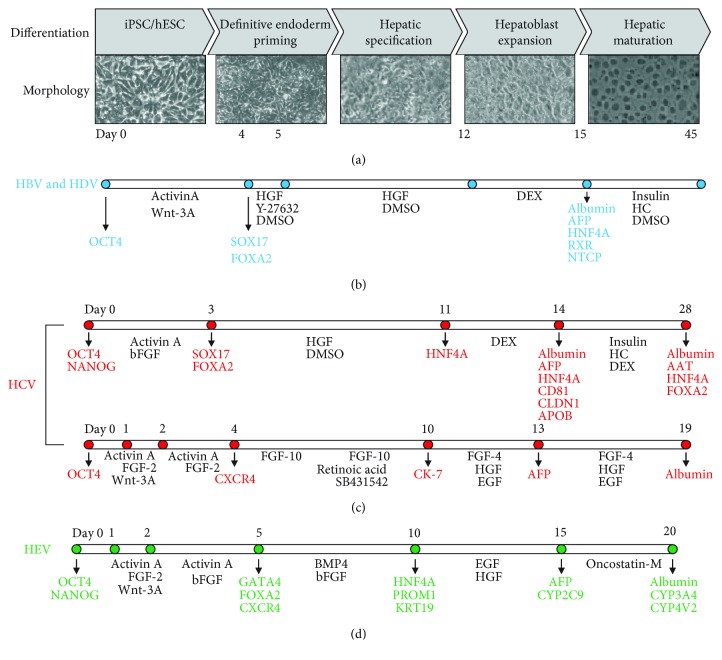
Figure 1.
Stem cell-derived hepatocyte-like cells support virus research of HBV, HCV, and HEV. (a) During differentiation, iPSC or hESC undergo definitive endoderm induction, hepatic specification, hepatoblast expansion, and hepatic maturation to become HLCs that are permissive for infection with hepatitis viruses. (b) For HBV infection, cells are treated with activin A and Wnt-3A enhancer for 4 days and HGF and Rock inhibitor Y-27632 for one day, followed by HGF for 1 week. Hepatoblast cells are administered with DEX for 3 days and can be used for HBV infection at this point. The infected cells can be maintained in the presence of insulin, HC, and DMSO for another 1 month [19]. (c) For HCV application, iPSC/hESC is differentiated using two methods. Upper: this arm is similar to (b) (a, [17]). Lower: cells are treated with activin A and FGF-2 until day 4 when FGF-10 is added for 2 days and then coadded with retinoic acid and TGF-β inhibitor SB431542 until day 10. Cells are further treated with FGF-4, HGF, and EGF for about 10 days [65]. (d) On day 5, endoderm is formed by activin A, FGF-2, and bFGF and further differentiated by BMP4 and bFGF until day 10. Cells are terminally differentiated by EGF and HGF to hepatocyte-like cells. Cells can be infected with HEV or maintained in the medium containing oncostatin-M [18, 75]. Abbreviations: AAT: α-1-antitrypsin; AFP: alpha fetoprotein; APOB: apolipoprotein B; BMP4: bone morphogenetic protein 4; Cd81: cluster of differentiation 81; CK-7: cytokeratin 7; CLDN1: claudin-1; CXCR4: C-X-C chemokine receptor type 4; CYP: cytochrome 450 enzyme; DEX: dexamethasone; DMSO: dimethyl sulfoxide; EGF: epidermal growth factor; FGF: fibroblast growth factor; FOXA2: forkhead box protein A2 (also known as hepatocyte nuclear factor 3-beta); GATA4: GATA-binding protein 4; HC: hydrocortisone; hESC: human embryonic stem cells; HGF: hepatocyte growth factor; HNF: hepatocyte nuclear factor; iPSC: induced pluripotent stem cells; KRT19: cytokeratin 19; NANOG: Nanog homeobox; NTCP: sodium taurocholate cotransporting polypeptide; OCT4: octamer-binding transcription factor 4; PROM1: prominin 1; RXR: retinoid X receptor; SOX17: sex determining region Y-box 17; TGF: tumor growth factor.
3. Hepatocyte-Like Cells for HCV Infection
Around 71 million people worldwide are chronically infected with HCV, which increases their risk of progressive liver disease [76]. Although the standard of care for chronic HCV infection has been dramatically improved through direct-acting antiviral agents (DAAs), it still poses significant problems, including treatment failure in some patient groups and limited access to therapy due to high cost of treatment. Further, a protective vaccine is still in need. Notably, 15–45% of HCV-infected individuals are able to clear the virus within six months without intervention, but the underlying mechanisms remain unknown [77, 78].
To address these scientific questions and develop new anti-HCV drugs, various in vitro HCV-infection models have been developed. JFH-1 is a unique cell culture- (cc-) adapted strain of HCV to study the complete viral life cycle in the hepatoma system [79]; however, it does not reflect the variability and diversity of HCV infection in patients [80]. The Huh-7.5.1 cell line derived from Huh-7 cells that carries a defect retinoic-inducible gene I (RIG-I), a critical player in viral genome recognition and host immune response, is frequently used for JFH-1 HCVcc infection [81].
Several studies have independently validated that HLCs are competent to support HCV infection (Table 2). In preliminary research, HLCs were generated from iPSCs differentiated with growth factors and by adenovirus delivery of SOX17, HEX, and HNF4A. All HCV entry receptors were expressed on these HLCs, including CD81, SR-B1, claudin-1, and occludin, which allowed the entry and replication of HCV pseudoparticles and subgenomic replicons, respectively [82]. Subsequently, other groups demonstrated that HLCs are not only permissive to different forms of cell culture-adapted HCV (viral pseudoparticles and JFH-1; HCVpp and HCVcc) but also showed detectable infection with different serum-derived HCV genotypes 1a, 1b, 2, 3, and 4; this is not possible with hepatoma cell lines since they are not permissive for infection with patient isolates [17, 83].
HLCs support the complete life cycle of HCV genotype 2a for up to 21 days (Figure 1) [65, 68]. Another feature of HLCs is that HCV can spread from infected cells to adjacent cells, suggesting possible direct cell-to-cell transmission of HCV, as has been described previously in HuH-7 cells [17]. A recent study used human iPSCs derived from human mesenchymal stem cells that were subsequently differentiated into HLCs with polycistronic OSKM-reprogramming factors. These HLCs supported the entire life cycle of wild-type HCV (genotype 1a, 1b, 3a, 3b, 6f, and 6n) isolated from patients and achieved increasing infection rates by incubating cells with α-tocopherol. The released HCV viral particles could infect both naïve HLCs and HuH-7 cells and were susceptible to treatment with IFN-α, ribavirin, or sofosbuvir [85].
Although HLCs represent a unique and highly relevant model to study HCV infection in vitro and in vivo, particularly in the context of a patient-specific genetic background, some limitations of this model remain to be addressed. First, the production of viral particles remains very low compared with reported levels from HuH-7 cell lines [86]. It has been shown that the permissive and persistent infection of HCV in hepatic progenitor cells is affected by liver-specific microRNA-122 and cellular cytokines [87, 88]. By inhibiting the JAK/STAT pathway to block IFN responses, viral infection and replication were improved in HLCs [89]. Additionally, higher HCV replication levels were observed in STAT2- but not STAT1-deficient HLCs [89]. Even after JAK/STAT pathway inhibitor treatment, HLCs demonstrated intact type III interferon and ISG responses. This suggests that HLCs may be a suitable model to study the HCV-host interaction [86]. Multiple mutations in different regions of the viral genome of JFH-1 HCVcc enhanced the titers in HuH-7 cells, and infections were maintained in an animal model, but the appearance of mutations has not been investigated in the HLCs. A further concern in the HLC model is the observed variability between cell lines with timing or cytokine concentrations necessary for hepatocyte differentiation. Thus, iPSC differentiation protocols and HLC culture conditions need to be optimized. For example, humanized liver chimeric mice based on human hepatocyte (such as HLCs) engraftment were reported to support HCV infection [90]. Engrafting HLCs in vivo to produce human liver chimeric mouse models has been fraught with low efficiencies [91]. By using an optimized hepatocyte differentiation protocol on transgenic mice, which carry the uPA (urokinase-type plasminogen activator) gene driven by the major urinary protein promoter onto a SCID (severe combined immunodeficiency)/beige background, HLCs differentiated from both hESCs and patient-specific iPSCs were able to engraft and undergo further maturation in vivo. Productive and chronic HCV infection in these repopulated liver injury models can be launched with high-dose inoculations (1,000 CID50 per mouse) [17]. Although challenges remain, robust cell culture and animal models for serum-derived HCV using HLCs provide remarkable systems for investigating HCV life cycle and HCV-associated hepatocellular carcinoma development.
4. Hepatocyte-Like Cells for HEV Infection
HEV is recognized as an important global health problem [92]. HEV is a nonenveloped positive-strand RNA virus of the Hepeviridae family, which is divided into two genera: Orthohepevirus and Piscihepevirus [93]. The Orthohepevirus genus is further divided into four species A, B, C, and D. Human-infecting HEV strains belong to the Orthohepevirus A, which include human-restricted genotypes (gt) 1 and 2 as well as zoonotic genotypes 3, 4, and 7. The human-restricted genotypes are transmitted fecal-orally and sporadically lead to large waterborne outbreaks in developing countries with poor sanitation (reviewed in [94]). These infections are mostly acute and self-resolving but can cause an increased virulence in pregnant women, leading to a 25% maternal mortality in the third trimester [95]. For the zoonotic viruses, infected animals serve as reservoirs and can transmit HEV through the consumption of infected meat [94]. These zoonotic species of HEV cause acute and chronic diseases in immunocompromised patients [92]. Reducing immunosuppression in combination with using off-label ribavirin is the only available treatment, but treatment resistances have been reported [96–98]. A high-efficacy vaccine has been developed and licensed in China but is not available elsewhere [24].
The 7.2 kb polyadenylated HEV genome contains three partially overlapping open reading frames (ORF1-3) (reviewed in [2]). ORF1 encodes the viral replicase, ORF2 for the capsid, and ORF3 for a small protein involved in virus assembly and secretion [2]. A range of different expression systems have been used to study HEV without resulting in authentic virus replication [67]. In this regard, HEV behaves like other hepatotropic viruses, in that they grow poorly in cell culture, which has severely hampered molecular studies, leaving many fundamental aspects of its life cycle poorly understood [67].
Breakthroughs in developing robust HEV cell culture systems have been made through the isolation of specific viral strains with improved replication efficiency and the identification of compatible cell lines [99]. After serial passaging in these cell lines, the isolated strains accumulated mutations and/or insertions, which increased their ability to replicate. For example, a gt3 HEV virus, the Kernow-C1 strain, was isolated from a chronic HEV patient [100] and serially passaged six times (passage 6) in the hepatoma cell line HepG2 [101]. A virus with an insertion derived from the human 40S ribosomal protein S17 in the ORF1 region became the dominant species with greater in vitro replication ability and broadened host range [101]. Similarly, other strains with insertions into ORF1 have been reported with enhanced viral fitness in vitro [102–104]. These adapted clones enabled molecular HEV studies and yielded valuable insights into HEV biology. Yet, this approach is limited to gt3 and 4 viruses and, to our knowledge, has not been successful for other genotypes.
Studies have proposed utilizing HLCs differentiated from iPSC/hESC as an alternative HEV cell culture model to hepatoma cells and PHHs (Figure 1) [18, 66]. HLCs can be infected with the adapted p6 strain [18, 66, 75]. Transition studies showed that germ layer cells support intracellular HEV replication but not infection [66]. Only when endodermal cells were differentiated to immature hepatocytes did they become susceptible for HEV infection [18]. This strongly suggested that virus entry, governed by the expression of a yet unknown cellular protein, was the limiting factor which could also be the key determinant of HEV tissue tropism [66]. In addition, HLCs are readily permissive for HEV isolates from animals infected with gt1-4 without prior adaptation [18]. Surprisingly, an early, nonadapted passage of the Kernow-C1 strain replicated better than the adapted p6 strain in HLCs. This suggests that acquired mutations in cell culture attenuate viral replication in more physiologically relevant systems. HLCs therefore enable studies of not only authentic HEV replication but also pan-genotype HEV biology. The high degree of heterogeneity among HEV genotypes has not been fully explored to date, but HLCs may now provide a reproducible platform to study such differences. As such, viral or cellular determinants that may define a host range and infections across species barriers have not been defined yet and are one of the many poorly understood topics in the field.
Employing precise editing technologies, such as CRISPR (cluster regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9), allows for rapid and efficient genome-editing of relevant host factors in stem cells to explore their importance in HEV replication pathways. Using CRISPR-Cas9, we identified at least one striking difference between nonadapted and adapted HEV replication [18]. The host factor cyclophilin A, which was previously reported to restrict HEV replication [18], only inhibited the cell culture-adapted p6 clone but not the original Kernow-C1 isolate replication in HLCs [18]. Corroborating evidence has shown discrepancies in drug responsiveness between in vitro and in vivo conditions; molecules (i.e., mycophenolic acid and rapamycin) that affected adapted HEV replication in cell culture [105, 106] failed to show any effect in patients [107]. If this discrepancy is due to alterations of the viral genome or a reflection of in vivo complexity as opposed to viral replication studies in a single cell type, it can be now explored using HLCs. With recent efforts in identifying novel compounds that inhibit HEV replication [75, 108–110], validation in the HLCs system will become more and more relevant.
Several studies suggest that host genetics determine susceptibility to HEV infection [111–113]. The ability to study replication of nonadapted HEV isolates in tandem with autologous, patient-derived iPSCs enables personalized models of HEV infection [67]. This may provide patient-tailored platforms to test potential treatments in vitro, especially for chronic HEV patients who have already developed resistance against RBV. An alternative treatment approach that we are currently exploring is based on the use of nonpathogenic adeno-associated viruses (AAV) combined with CRISPR-Cas9 to deliver short hairpin RNA (shRNA) to downregulate HEV replication. In this scenario, genetic vaccination would be achieved by transducing patient-derived iPSCs prior to HLC differentiation and transplantation into the liver of chronic HEV patients to establish a genetically protected hepatocyte population. Alternatively, hepatotropic AAVs will allow direct delivery of target shRNAs in vivo. These approaches are not restricted to HEV, as HLCs are permissive for HCV and HBV isolates [63, 65] (Table 1). With that, HLCs provide a uniquely reproducible and genetically tractable cellular system in which to perform coinfection studies as widely used hepatoma cell lines vary in their virus permissiveness (Table 2).
Beyond HLCs, stem cell technology may also provide a platform to study other aspects of HEV biology. HEV mainly infects the liver but likely additional tissues [114] as some patients experience extrahepatic manifestations including neurological disorders, thrombocytopenia, renal injury, and other conditions [115]. This is further corroborated by the observation that HEV can replicate in vitro in nonhepatic cell types, such as lung [116], neuronal [117], and placental [118] cell lines. Stem cells, with their intrinsic ability to give rise to cells of various lineages, may help to define the determinants of HEV tissue tropism [66]. In conclusion, knowledge on the viral life cycle of HEV and virus-host interactions, i.e., on systemic and cellular levels, remains scarce. Studies of pan-genotype HEV biology in a physiologically relevant cell system such as HLCs, which support authentic HEV infection and replication, shall significantly advance our understanding of HEV biology. This will facilitate and promote the development of specific anti-HEV therapies.
5. Conclusions and Future Directions
HLCs derived from hiPSC or hESC provide a promising tool to study the biology of hepatotropic viruses and to screen novel antiviral treatments in the future. We summarized current progress in developing HLCs that support the entire life cycles of HBV, HCV, and HEV (Figure 1). HLCs constitute a novel cell culture model that is more physiologically relevant than immortalized hepatoma cell lines. Beyond that, the use of HLCs may help overcome two major limitations of PHHs: donor-to-donor variability and long-term culture to study chronic infection. Specifically, HLCs support high-efficiency and long-term HBV replication and, remarkably, virus spread [19]. In terms of the nature of diverse genotypes and high replicative mutations of RNA viruses, such as HCV and HEV, HLCs allow pan-genotype permissiveness and even support direct infection with patient-derived isolates that have not been adapted in cell culture. Engrafting HLCs into immunosuppressive liver injury mouse models, like the uPA/SCID mice, may facilitate studies of antiviral evaluation and virus-host interaction in vivo.
Coinfections of hepatitis viruses (e.g., HBV, HCV, and HEV) occur in patients. However, the exact modes of coinfection are poorly described due to the single permissiveness of available culture models (Table 2). HLCs therefore constitute a universal tool, in which to study how two or more hepatitis viruses modulate host factor(s) such as MAVS and cyclophilin. This may provide information on how and in which order coinfected patients could be treated.
HLCs derived from iPSCs are of less societal and ethical concern than PHHs or fetal tissue-derived hepatocytes are. In addition, HLCs serve as a powerful tool to assess the influence of genetic factors on virus infection, as HLCs can be generated from iPSCs with a diverse genetic background.
Despite these advantages and optimization of available protocols, HLC differentiation remains time-consuming and complicated. Although HLCs are more physiologically relevant than many hepatoma cell lines, they retain an immature phenotype and cannot fully recapitulate hepatocyte functions. Perhaps, differentiation under 3D-culture conditions may improve this and yield HLCs that resemble PHHs more closely. Supporting this, Gieseck et al. have demonstrated that hepatocyte-specific genes are higher expressed in HLCs, when cultured in 3D conditions [119].
Ultimately, HLCs provide a personalized platform for viral hepatitis studies. For patients who do not respond to available treatments, personalized iPSC-derived HLCs are the best model to study the host determinants and validate second-line antivirals. Taken together, HLCs provide an important tool for studying the life cycle of hepatitis viruses, in spite of the distinct replicative nature of HBV, HCV, and HEV. The development of HLCs derived from stem cells has opened a new era and provides a physiologically relevant system to advance our understanding of the viral life cycles. This will ultimately contribute to the development of novel therapeutic strategies towards the elimination of viral hepatitis.
Acknowledgments
Y Xia was funded by the State Key Laboratory of Virology of China. This program has been supported with an educational grant via the Gilead Sciences Research Scholars Program in Liver Disease Asia. B Qu was financed by the SFB/TRR179 program—project number 272983813—funded by the German Research Foundation. C Zhang was supported by a fellowship from Deutsches Zentrum für Infektionsforschung. VL Dao Thi was funded by CHS Stiftung and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer 240245660-SFB1129. We thank Andrew Freistaedter and Xianfang Wu for the critical reading of the manuscript.
Contributor Information
Viet Loan Dao Thi, Email: VietLoan.DaoThi@med.uni-heidelberg.de.
Yuchen Xia, Email: yuchen.xia@nih.gov.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
- 1.Lemon S. M., Ott J. J., Van Damme P., Shouval D. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. Journal of Hepatology. 2018;68(1):167–184. doi: 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 2.Nimgaonkar I., Ding Q., Schwartz R. E., Ploss A. Hepatitis E virus: advances and challenges. Nature Reviews Gastroenterology & Hepatology. 2017;15(2):96–110. doi: 10.1038/nrgastro.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginzberg D., Wong R. J., Gish R. Global HBV burden: guesstimates and facts. Hepatology International. 2018;12(4):315–329. doi: 10.1007/s12072-018-9884-8. [DOI] [PubMed] [Google Scholar]
- 4.Chevaliez S., Pawlotsky J. M. New virological tools for screening, diagnosis and monitoring of hepatitis B and C in resource-limited settings. Journal of Hepatology. 2018;69(4):916–926. doi: 10.1016/j.jhep.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Rizzetto M. Hepatitis D virus: introduction and epidemiology. Cold Spring Harbor Perspectives in Medicine. 2015;5(7, article a021576) doi: 10.1101/cshperspect.a021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto W. K., Lo Y. R., Pawlotsky J. M., Yuen M. F. Chronic hepatitis B virus infection. Lancet. 2018;392(10161):2313–2324. doi: 10.1016/S0140-6736(18)31865-8. [DOI] [PubMed] [Google Scholar]
- 7.Liver (Hepatocellular) Cancer Prevention (PDQ(R)): Health Professional Version, PDQ Cancer Information Summaries. Bethesda, MD, USA: National Cancer Institute (US); 2002. [PubMed] [Google Scholar]
- 8.Marshall A. D., Pawlotsky J. M., Lazarus J. V., Aghemo A., Dore G. J., Grebely J. The removal of DAA restrictions in Europe - one step closer to eliminating HCV as a major public health threat. Journal of Hepatology. 2018;69(5):1188–1196. doi: 10.1016/j.jhep.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 9.Blight K. J., McKeating J. A., Marcotrigiano J., Rice C. M. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. Journal of Virology. 2003;77(5):3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sa-ngiamsuntorn K., Wongkajornsilp A., Kasetsinsombat K., et al. Upregulation of CYP 450s expression of immortalized hepatocyte-like cells derived from mesenchymal stem cells by enzyme inducers. BMC Biotechnology. 2011;11(1):p. 89. doi: 10.1186/1472-6750-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y., Wang X., Nyberg S. Potential and challenges of induced pluripotent stem cells in liver diseases treatment. Journal of Clinical Medicine. 2014;3(3):997–1017. doi: 10.3390/jcm3030997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Si-Tayeb K., Noto F. K., Nagaoka M., et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song Z., Cai J., Liu Y., et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Research. 2009;19(11):1233–1242. doi: 10.1038/cr.2009.107. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y. F., Tseng C. Y., Wang H. W., Kuo H. C., Yang V. W., Lee O. K. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55(4):1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zakikhan K., Pournasr B., Vosough M., Nassiri-Asl M. In vitro generated hepatocyte-like cells: a novel tool in regenerative medicine and drug discovery. Cell Journal. 2017;19(2):204–217. doi: 10.22074/cellj.2016.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vosough M., Omidinia E., Kadivar M., et al. Generation of functional hepatocyte-like cells from human pluripotent stem cells in a scalable suspension culture. Stem Cells and Development. 2013;22(20):2693–2705. doi: 10.1089/scd.2013.0088. [DOI] [PubMed] [Google Scholar]
- 17.Carpentier A., Tesfaye A., Chu V., et al. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. Journal of Clinical Investigation. 2014;124(11):4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X., Dao Thi V. L., Liu P., et al. Pan-genotype hepatitis E virus replication in stem cell-derived hepatocellular systems. Gastroenterology. 2018;154(3):663–674.e7. doi: 10.1053/j.gastro.2017.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y., Carpentier A., Cheng X., et al. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. Journal of Hepatology. 2017;66(3):494–503. doi: 10.1016/j.jhep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balayan M. S. Natural hosts of hepatitis A virus. Vaccine. 1992;10(Suppl 1):S27–S31. doi: 10.1016/0264-410X(92)90537-T. [DOI] [PubMed] [Google Scholar]
- 21.Dandri M., Petersen J. Mechanism of hepatitis B virus persistence in hepatocytes and its carcinogenic potential. Clinical Infectious Diseases. 2016;62(Supplement 4):S281–S288. doi: 10.1093/cid/ciw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaender S., Brown R. J., Pietschmann T., Steinmann E. Natural reservoirs for homologs of hepatitis C virus. Emerging Microbes & Infections. 2014;3(3, article e21) doi: 10.1038/emi.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldabe R., Suarez-Amaran L., Usai C., Gonzalez-Aseguinolaza G. Animal models of chronic hepatitis delta virus infection host-virus immunologic interactions. Pathogens. 2015;4(1):46–65. doi: 10.3390/pathogens4010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T., Li S. W., Zhang J., Ng M. H., Xia N. S., Zhao Q. Hepatitis E vaccine development: a 14 year odyssey. Human Vaccines & Immunotherapeutics. 2012;8(6):823–827. doi: 10.4161/hv.20042. [DOI] [PubMed] [Google Scholar]
- 25.Hutin Y., Nasrullah M., Easterbrook P., et al. Access to treatment for hepatitis B virus infection-Worldwide, 2016. American Journal of Transplantation. 2018;18(10):2595–2598. doi: 10.1111/ajt.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valaydon Z. S., Locarnini S. A. The virological aspects of hepatitis B. Best Practice & Research Clinical Gastroenterology. 2017;31(3):257–264. doi: 10.1016/j.bpg.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Xia Y., Liang T. J. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology. 2019;156(2):311–324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghany M. G. Current treatment guidelines of chronic hepatitis B: the role of nucleos(t)ide analogues and peginterferon. Best Practice & Research Clinical Gastroenterology. 2017;31(3):299–309. doi: 10.1016/j.bpg.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Trépo C., Chan H. L. Y., Lok A. Hepatitis B virus infection. The Lancet. 2014;384(9959):2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 30.Lai C. L., Wong D., Ip P., et al. Reduction of covalently closed circular DNA with long-term nucleos(t)ide analogue treatment in chronic hepatitis B. Journal of Hepatology. 2017;66(2):275–281. doi: 10.1016/j.jhep.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Zoulim F., Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137(5):1593–1608.e2. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 32.Chevaliez S., Hezode C., Bahrami S., Grare M., Pawlotsky J. M. Long-term hepatitis B surface antigen (HBsAg) kinetics during nucleoside/nucleotide analogue therapy: finite treatment duration unlikely. Journal of Hepatology. 2013;58(4):676–683. doi: 10.1016/j.jhep.2012.11.039. [DOI] [PubMed] [Google Scholar]
- 33.Han Y., Zeng A., Liao H., Liu Y., Chen Y., Ding H. The efficacy and safety comparison between tenofovir and entecavir in treatment of chronic hepatitis B and HBV related cirrhosis: a systematic review and meta-analysis. International Immunopharmacology. 2017;42:168–175. doi: 10.1016/j.intimp.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Schinazi R. F., Ehteshami M., Bassit L., Asselah T. Towards HBV curative therapies. Liver International. 2018;38(Suppl 1):102–114. doi: 10.1111/liv.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia Y., Stadler D., Lucifora J., et al. Interferon-gamma and tumor necrosis factor-alpha produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology. 2016;150(1):194–205. doi: 10.1053/j.gastro.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 36.Sureau C., Romet-Lemonne J. L., Mullins J. I., Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47(1):37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 37.Sells M. A., Chen M. L., Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proceedings of the National Academy of Sciences. 1987;84(4):1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ladner S. K., Otto M. J., Barker C. S., et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrobial Agents and Chemotherapy. 1997;41(8):1715–1720. doi: 10.1128/AAC.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaney W. E.t., Isom H. C. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28(4):1134–1146. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- 40.Sprinzl M. F., Oberwinkler H., Schaller H., Protzer U. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. Journal of Virology. 2001;75(11):5108–5118. doi: 10.1128/JVI.75.11.5108-5118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heslop J. A., Rowe C., Walsh J., et al. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Archives of Toxicology. 2017;91(1):439–452. doi: 10.1007/s00204-016-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fraczek J., Bolleyn J., Vanhaecke T., Rogiers V., Vinken M. Primary hepatocyte cultures for pharmaco-toxicological studies: at the busy crossroad of various anti-dedifferentiation strategies. Archives of Toxicology. 2013;87(4):577–610. doi: 10.1007/s00204-012-0983-3. [DOI] [PubMed] [Google Scholar]
- 43.Zeisberg M., Kramer K., Sindhi N., Sarkar P., Upton M., Kalluri R. De-differentiation of primary human hepatocytes depends on the composition of specialized liver basement membrane. Molecular and Cellular Biochemistry. 2006;283(1-2):181–189. doi: 10.1007/s11010-006-2677-8. [DOI] [PubMed] [Google Scholar]
- 44.Gripon P., Rumin S., Urban S., et al. Infection of a human hepatoma cell line by hepatitis B virus. Proceedings of the National Academy of Sciences. 2002;99(24):15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hantz O., Parent R., Durantel D., Gripon P., Guguen-Guillouzo C., Zoulim F. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. The Journal of General Virology. 2009;90(1) Part 1:127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- 46.Yan H., Zhong G., Xu G., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1, article e00049 doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwamoto M., Watashi K., Tsukuda S., et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochemical and Biophysical Research Communications. 2014;443(3):808–813. doi: 10.1016/j.bbrc.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 48.Feigelstock D., Thompson P., Mattoo P., Zhang Y., Kaplan G. G. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. Journal of Virology. 1998;72(8):6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pileri P., Uematsu Y., Campagnoli S., et al. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 50.Scarselli E., Ansuini H., Cerino R., et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. The EMBO Journal. 2002;21(19):5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans M. J., von Hahn T., Tscherne D. M., et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 52.Ploss A., Evans M. J., Gaysinskaya V. A., et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung P. S., Hong S.-H., Lee J., et al. CXCL10 is produced in hepatitis A virus-infected cells in an IRF3-dependent but IFN-independent manner. Scientific Reports. 2017;7(1):p. 6387. doi: 10.1038/s41598-017-06784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Molina S., Castet V., Pichard-Garcia L., et al. Serum-derived hepatitis C virus infection of primary human hepatocytes is tetraspanin CD81 dependent. Journal of Virology. 2008;82(1):569–574. doi: 10.1128/JVI.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oshiro Y., Yasue H., Takahashi K., et al. Mode of swine hepatitis E virus infection and replication in primary human hepatocytes. Journal of General Virology. 2014;95(Part 12):2677–2682. doi: 10.1099/vir.0.068452-0. [DOI] [PubMed] [Google Scholar]
- 56.Okamoto H. Culture systems for hepatitis E virus. Journal of Gastroenterology. 2013;48(2):147–158. doi: 10.1007/s00535-012-0682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogee S., Talbot N., Caperna T., Bouquet J., Barnaud E., Pavio N. New models of hepatitis E virus replication in human and porcine hepatocyte cell lines. Journal of General Virology. 2013;94, Part 3:549–558. doi: 10.1099/vir.0.049858-0. [DOI] [PubMed] [Google Scholar]
- 58.Wakita T., Pietschmann T., Kato T., et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Medicine. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindenbach B. D., Evans M. J., Syder A. J., et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 60.Yang D., Zuo C., Wang X., et al. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proceedings of the National Academy of Sciences. 2014;111(13):E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka T., Takahashi M., Kusano E., Okamoto H. Development and evaluation of an efficient cell-culture system for hepatitis E virus. Journal of General Virology. 2007;88(3) Part 3:903–911. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- 62.Sakurai F., Mitani S., Yamamoto T., et al. Human induced-pluripotent stem cell-derived hepatocyte-like cells as an in vitro model of human hepatitis B virus infection. Scientific Reports. 2017;7(1) doi: 10.1038/srep45698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shlomai A., Schwartz R. E., Ramanan V., et al. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proceedings of the National Academy of Sciences. 2014;111(33):12193–12198. doi: 10.1073/pnas.1412631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaneko S., Kakinuma S., Asahina Y., et al. Human induced pluripotent stem cell-derived hepatic cell lines as a new model for host interaction with hepatitis B virus. Scientific Reports. 2016;6(1, article 29358) doi: 10.1038/srep29358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X., Robotham J. M., Lee E., et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathogens. 2012;8(4, article e1002617) doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helsen N., Debing Y., Paeshuyse J., et al. Stem cell-derived hepatocytes: a novel model for hepatitis E virus replication. Journal of Hepatology. 2016;64(3):565–573. doi: 10.1016/j.jhep.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 67.Dao Thi V. L., Wu X., Rice C. M. Stem cell-derived culture models of hepatitis E virus infection. Cold Spring Harbor Perspectives in Medicine. 2019;9(3, article a031799) doi: 10.1101/cshperspect.a031799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng X., Xia Y., Serti E., et al. Hepatitis B virus evades innate immunity of hepatocytes but activates cytokine production by macrophages. Hepatology. 2017;66(6):1779–1793. doi: 10.1002/hep.29348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mutz P., Metz P., Lempp F. A., et al. HBV bypasses the innate immune response and does not protect HCV from antiviral activity of interferon. Gastroenterology. 2018;154(6):1791–1804.e22. doi: 10.1053/j.gastro.2018.01.044. [DOI] [PubMed] [Google Scholar]
- 70.Suslov A., Boldanova T., Wang X., Wieland S., Heim M. H. Hepatitis B virus does not interfere with innate immune responses in the human liver. Gastroenterology. 2018;154(6):1778–1790. doi: 10.1053/j.gastro.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 71.Wieland S., Thimme R., Purcell R. H., Chisari F. V. Genomic analysis of the host response to hepatitis B virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(17):6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carpentier A., Nimgaonkar I., Chu V., Xia Y., Hu Z., Liang T. J. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Research. 2016;16(3):640–650. doi: 10.1016/j.scr.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nie Y. Z., Zheng Y. W., Miyakawa K., et al. Recapitulation of hepatitis B virus-host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine. 2018;35:114–123. doi: 10.1016/j.ebiom.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan L., Liu X., Zhang L., et al. A chimeric humanized mouse model by engrafting the human induced pluripotent stem cell-derived hepatocyte-like cell for the chronic hepatitis B virus infection. Frontiers in Microbiology. 2018;9:p. 908. doi: 10.3389/fmicb.2018.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dao Thi V. L., Debing Y., Wu X., et al. Sofosbuvir inhibits hepatitis E virus replication in vitro and results in an additive effect when combined with ribavirin. Gastroenterology. 2016;150(1):82–85.e4. doi: 10.1053/j.gastro.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 76.Modin L., Arshad A., Wilkes B., et al. Epidemiology and natural history of hepatitis C virus infection among children and young people. Journal of Hepatology. 2019;70(3):371–378. doi: 10.1016/j.jhep.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 77.Gower E., Estes C., Blach S., Razavi-Shearer K., Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. Journal of Hepatology. 2014;61(1) Supplement:S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 78.Chevaliez S., Pawlotsky J. M. HCV genome and life cycle. In: Tan S. L., editor. Hepatitis C viruses: Genomes and Molecular Biology. Norfolk, UK: Horizon Bioscience; 2006. [PubMed] [Google Scholar]
- 79.Kato T., Date T., Miyamoto M., et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125(6):1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 80.Date T., Kato T., Miyamoto M., et al. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. Journal of Biological Chemistry. 2004;279(21):22371–22376. doi: 10.1074/jbc.M311120200. [DOI] [PubMed] [Google Scholar]
- 81.Breiman A., Grandvaux N., Lin R., et al. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. Journal of Virology. 2005;79(7):3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshida T., Takayama K., Kondoh M., et al. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochemical and Biophysical Research Communications. 2011;416(1-2):119–124. doi: 10.1016/j.bbrc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Ge D., Fellay J., Thompson A. J., et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 84.Schwartz R. E., Trehan K., Andrus L., et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proceedings of the National Academy of Sciences. 2012;109(7):2544–2548. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sa-Ngiamsuntorn K., Wongkajornsilp A., Phanthong P., et al. A robust model of natural hepatitis C infection using hepatocyte-like cells derived from human induced pluripotent stem cells as a long-term host. Virology Journal. 2016;13(1):p. 59. doi: 10.1186/s12985-016-0519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou X., Sun P., Lucendo-Villarin B., et al. Modulating innate immunity improves hepatitis C virus infection and replication in stem cell-derived hepatocytes. Stem Cell Reports. 2014;3(1):204–214. doi: 10.1016/j.stemcr.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y. P., Gottwein J. M., Scheel T. K., Jensen T. B., Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5' UTR. Proceedings of the National Academy of Sciences. 2011;108(12):4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lanford R. E., Hildebrandt-Eriksen E. S., Petri A., et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327(5962):198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schobel A., Rosch K., Herker E. Functional innate immunity restricts Hepatitis C Virus infection in induced pluripotent stem cell-derived hepatocytes. Scientific Reports. 2018;8(1):p. 3893. doi: 10.1038/s41598-018-22243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kawahara T., Toso C., Douglas D. N., et al. Factors affecting hepatocyte isolation, engraftment, and replication in an in vivo model. Liver Transplantation. 2010;16(8):974–982. doi: 10.1002/lt.22099. [DOI] [PubMed] [Google Scholar]
- 91.Schwartz R. E., Fleming H. E., Khetani S. R., Bhatia S. N. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnology Advances. 2014;32(2):504–513. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.von Wulffen M., Westholter D., Lutgehetmann M., Pischke S. Hepatitis E: still waters run deep. Journal of Clinical and Translational Hepatology. 2018;6(1):40–47. doi: 10.14218/JCTH.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forni D., Cagliani R., Clerici M., Sironi M. Origin and dispersal of hepatitis E virus. Emerging Microbes & Infections. 2018;7(1):p. 11. doi: 10.1038/s41426-017-0009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kamar N., Izopet J., Pavio N., et al. Hepatitis E virus infection. Nature Reviews Disease Primers. 2017;3, article 17086 doi: 10.1038/nrdp.2017.86. [DOI] [PubMed] [Google Scholar]
- 95.Navaneethan U., Al Mohajer M., Shata M. T. Hepatitis E and pregnancy: understanding the pathogenesis. Liver International. 2008;28(9):1190–1199. doi: 10.1111/j.1478-3231.2008.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Todt D., Gisa A., Radonic A., et al. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut. 2016;65(10):1733–1743. doi: 10.1136/gutjnl-2015-311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Debing Y., Gisa A., Dallmeier K., et al. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology. 2014;147(5):1008–1011.e7. doi: 10.1053/j.gastro.2014.08.040. [DOI] [PubMed] [Google Scholar]
- 98.Lhomme S., Kamar N., Nicot F., et al. Mutation in the hepatitis E virus polymerase and outcome of ribavirin therapy. Antimicrobial Agents and Chemotherapy. 2016;60(3):1608–1614. doi: 10.1128/AAC.02496-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meister T. L., Bruening J., Todt D., Steinmann E. Cell culture systems for the study of hepatitis E virus. Antiviral Research. 2019;163:34–49. doi: 10.1016/j.antiviral.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 100.Shukla P., Nguyen H. T., Torian U., et al. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proceedings of the National Academy of Sciences. 2011;108(6):2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shukla P., Nguyen H. T., Faulk K., et al. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. Journal of Virology. 2012;86(10):5697–5707. doi: 10.1128/JVI.00146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen H. T., Torian U., Faulk K., et al. A naturally occurring human/hepatitis E recombinant virus predominates in serum but not in faeces of a chronic hepatitis E patient and has a growth advantage in cell culture. Journal of General Virology. 2012;93(3) Part 3:526–530. doi: 10.1099/vir.0.037259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Johne R., Reetz J., Ulrich R. G., et al. An ORF1-rearranged hepatitis E virus derived from a chronically infected patient efficiently replicates in cell culture. Journal of Viral Hepatitis. 2014;21(6):447–456. doi: 10.1111/jvh.12157. [DOI] [PubMed] [Google Scholar]
- 104.Debing Y., Ramiere C., Dallmeier K., et al. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. Journal of Hepatology. 2016;65(3):499–508. doi: 10.1016/j.jhep.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 105.Zhou X., Wang Y., Metselaar H. J., Janssen H. L. A., Peppelenbosch M. P., Pan Q. Rapamycin and everolimus facilitate hepatitis E virus replication: revealing a basal defense mechanism of PI3K-PKB-mTOR pathway. Journal of Hepatology. 2014;61(4):746–754. doi: 10.1016/j.jhep.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 106.Wang Y., Zhou X., Debing Y., et al. Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology. 2014;146(7):1775–1783. doi: 10.1053/j.gastro.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 107.Kamar N., Lhomme S., Abravanel F., et al. An early viral response predicts the virological response to ribavirin in hepatitis E virus organ transplant patients. Transplantation. 2015;99(10):2124–2131. doi: 10.1097/TP.0000000000000850. [DOI] [PubMed] [Google Scholar]
- 108.Todt D., Moeller N., Praditya D., et al. The natural compound silvestrol inhibits hepatitis E virus (HEV) replication in vitro and in vivo. Antiviral Research. 2018;157:151–158. doi: 10.1016/j.antiviral.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaushik N., Subramani C., Anang S., et al. Zinc salts block hepatitis E virus replication by inhibiting the activity of viral RNA-dependent RNA polymerase. Journal of Virology. 2017;91(21) doi: 10.1128/JVI.00754-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Glitscher M., Himmelsbach K., Woytinek K., et al. Inhibition of hepatitis E virus spread by the natural compound Silvestrol. Viruses. 2018;10(6):p. 301. doi: 10.3390/v10060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang L., Yesupriya A., Chang M. H., Teshale E., Teo C. G. Apolipoprotein E and protection against hepatitis E viral infection in American non-Hispanic blacks. Hepatology. 2015;62(5):1346–1352. doi: 10.1002/hep.27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hazam R. K., Deka M., Kar P. Role of nitric oxide synthase genes in hepatitis E virus infection. Journal of Viral Hepatitis. 2014;21(9):671–679. doi: 10.1111/jvh.12186. [DOI] [PubMed] [Google Scholar]
- 113.Mishra N., Arankalle V. A. Association of polymorphisms in the promoter regions of TNF-alpha (-308) with susceptibility to hepatitis E virus and TNF-alpha (-1031) and IFN-gamma (+874) genes with clinical outcome of hepatitis E infection in India. Journal of Hepatology. 2011;55(6):1227–1234. doi: 10.1016/j.jhep.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 114.Pischke S., Hartl J., Pas S. D., Lohse A. W., Jacobs B. C., Van der Eijk A. A. Hepatitis E virus: infection beyond the liver? Journal of Hepatology. 2017;66(5):1082–1095. doi: 10.1016/j.jhep.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 115.Kamar N., Marion O., Abravanel F., Izopet J., Dalton H. R. Extrahepatic manifestations of hepatitis E virus. Liver International. 2016;36(4):467–472. doi: 10.1111/liv.13037. [DOI] [PubMed] [Google Scholar]
- 116.Takahashi M., Tanaka T., Takahashi H., et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. Journal of Clinical Microbiology. 2010;48(4):1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Drave S. A., Debing Y., Walter S., et al. Extra-hepatic replication and infection of hepatitis E virus in neuronal-derived cells. Journal of Viral Hepatitis. 2016;23(7):512–521. doi: 10.1111/jvh.12515. [DOI] [PubMed] [Google Scholar]
- 118.Knegendorf L., Drave S. A., Dao Thi V. L., et al. Hepatitis E virus replication and interferon responses in human placental cells. Hepatology Communications. 2018;2(2):173–187. doi: 10.1002/hep4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gieseck R. L., III, Hannan N. R. F., Bort R., et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9(1, article e86372) doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]