Abstract
Purpose
Associations between XRCC1, XRCC3, and ERCC2 gene polymorphism and prognosis have been investigated in several cancers. The aim of this meta-analysis was to assess the prognostic value of XRCC1, XRCC3, and ERCC2 gene polymorphism in hepatocellular carcinoma (HCC).
Methods
A systematic literature search was performed to identify relevant studies in PubMed, Embase, and the Cochrane library up to December 2018. The prognostic values of XRCC1, XRCC3, and ERCC2 polymorphisms in HCC were estimated using crude HRs with 95% CIs.
Results
Ten studies involving 2687 patients were included in the quantitative analysis. There were no statistically significant associations between XRCC1 rs1799782 C>T, XRCC1 rs25487 G>A, and ERCC2 rs1799793 G>A polymorphisms and overall survival (OS). OS was significantly longer for the ERCC2 rs13181 CC genotype than for AA (CC vs. AA: HR = 0.33, 95% CI = 0.15–0.72). A significantly lower OS was observed for patients with the CT genotype compared with the CC genotype at XRCC3 rs861539 (CT vs. CC: HR = 1.64, 95% CI = 1.11–2.42).
Conclusion
The ERCC2 rs13181 A>C polymorphism and XRCC3 rs861539 C>T polymorphism may be predictive markers for prognosis in patients with HCC. Well-designed studies with larger sample sizes are needed to verify our findings.
1. Introduction
Liver cancer is one of the most common malignancies. It was the sixth most commonly diagnosed cancer and the fourth cause of cancer-related death worldwide in 2018 and accounts for approximately 841,000 new cases and 782,000 deaths annually [1]. Hepatocellular carcinoma (HCC) is the most common primary cancer of the liver, accounting for 75–85% of liver cancer cases. According to the stage of HCC, patients receive various treatments, ranging from surgery to radiotherapy, transarterial chemoembolization (TACE), and targeted therapy [2]. Although treatment advances have improved clinical outcomes in the last few decades, the 5-year survival rate has remained at only about 18% from 2005 to 2011 [1].
The development of HCC is a multifactorial process involving an insidious onset, rapid progression, and high mortality rates. The fundamental pathogenic event in carcinogenesis is the accumulation of DNA damage and errors in DNA repair. In response to DNA damage, specific DNA repair mechanisms are activated. The major DNA repair pathways are mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), and double-strand break repair (DSBR) [3]. Polymorphisms in genes involved in DNA repair are likely to play an important role in the prognosis of HCC and are useful factors for determining the risk of cancer progression or recurrence. Various genetic factors are predicted to affect treatment efficiency and prognosis in patients with HCC, such as X-ray cross-complementing group 1 (XRCC1), X-ray repair cross-complementing group 3 (XRCC3), and excision repair cross-complementation group 2 (ERCC2). Many previous studies have investigated the associations between polymorphisms in these genes and prognosis [4–7]. In 2012, Jung et al. [8] found that XRCC1 rs25487, ERCC5 rs2018836, ERCC5 rs3818356, and XRCC4 rs1805377 have significant effects on survival. Another study suggested that ERCC2-312 genotypes but not XRCC1-194 were independent risk factors for poor prognosis in HCC [9]. Han et al. [10] showed that the XRCC1 Gln allele and XRCC3 T allele are related to a poor prognosis in HCC.
Despite a number of recent studies of the relationships between XRCC1, XRCC3, and ERCC2 gene polymorphisms and prognosis in HCC [8–16], the results are inconclusive. We performed a meta-analysis to comprehensively assess the correlation between these polymorphisms and prognosis in HCC.
2. Materials and Methods
2.1. Search Strategy
The present study was conducted according to the PRISMA guidelines for systematic reviews and meta-analyses [17]. A systematic literature search of PubMed, Embase, and the Cochrane library was conducted to identify studies published up to December 2018. The following retrieval strategy was used in accordance with ICPO (liver neoplasms OR hepatocellular cancer OR hepatocellular carcinoma OR liver cancer OR liver carcinoma OR hepatocellular neoplasms) AND (XRCC1 OR X-ray repair cross-complementing group1 OR XRCC3 OR X-ray repair cross-complementing group 3 OR ERCC1 OR excision repair cross-complementation group 1 OR ERCC2 OR excision repair cross-complementing group 2 OR XPD OR xeroderma pigmentosum group D) AND (prognosis OR survival). The reference lists of the identified studies were examined to identify additional studies. When multiple studies evaluated the same population, the most recent and the largest study was included.
2.2. Inclusion and Exclusion Criteria
The following inclusion criteria were applied. (1) Patients in the study were diagnosed with HCC. (2) Genotype frequencies could be extracted for at least one of the five polymorphisms. (3) Studies provided sufficient data for the prognostic effects in patients with HCC. The following exclusion criteria were applied: (1) studies with repeated data, (2) studies lacking sufficient data, and (3) studies of patients with carcinomas other than the liver.
2.3. Data Extraction and Quality Assessment
Two investigators extracted data independently, including information on authors, year, country, ethnicity, number of patients, SNP sites, quality assessment scores, and HRs with 95% confidence intervals (CIs). If the HR and 95% CI could not be obtained directly, they were extracted using the methods of Parmar [18], Tierney [19], and Williamson [20]. Discrepancies were resolved by consensus. The Newcastle-Ottawa scale (NOS) was used to evaluate the quality of the identified studies [21]. The NOS includes 3 categories, selection (0–4 points), comparability (0–2 points), and outcome (0–3 points) [22]. The total scores ranged from 0 to 9. NOS scores of 0–3, 4–6, and 7–9 were considered low, moderate, and high.
2.4. Statistical Analysis
The strength of the associations between five gene polymorphisms and overall survival (OS) in HCC was estimated using crude HRs with 95% CIs. The Chi squared-based Q-test and I2 statistic were utilized to evaluate heterogeneity. When P < 0.10 or I2 > 25% for the Q-test, a random effects model (DerSimonian and Laird method) was used to evaluate the pooled HR. Otherwise, a fixed-effects model (Mantel-Haenszel method) was used [23]. In addition, a sensitivity analysis was used to estimate the stability of our results by omitting one study at a time and recalculating the pooled HR. Funnel plots and Egger's test were used to assess publication bias [24]. All statistical analyses were implemented using STATA (version 12.0; STATA Corporation, College Station, TX, USA) and two-sided P-values were obtained.
3. Results
3.1. Literature Search
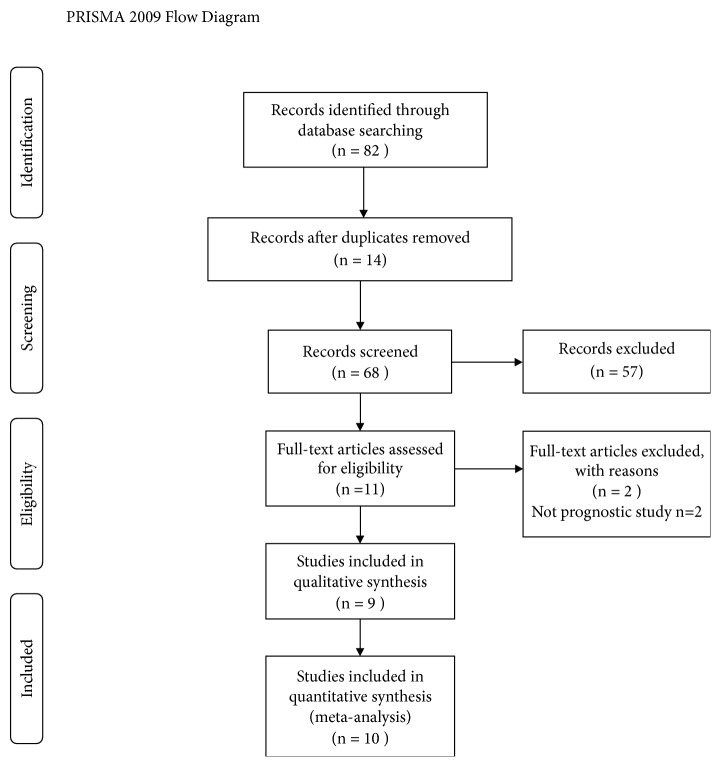
The study search and selection strategies are presented in Figure 1. In total, 82 potentially relevant studies were retrieved from PubMed, Embase, and the Cochrane library. Ten duplicate studies were excluded and 57 studies were excluded after reading the titles and abstracts. After further review of the full-length texts, 2 studies did not examine prognosis and were excluded. Finally, we identified 9 eligible articles including 10 studies for inclusion in the meta-analysis.
Figure 1.
The flow chart of included studies in this meta-analysis.
3.2. Characteristics of Eligible Studies
The characteristics of studies included in the meta-analysis are shown in Table 2. There were 8 studies of Asian populations and 2 studies of Caucasians. The sample sizes of eligible studies ranged from 50 to 708. Five studies reported an association between the XRCC1 rs25487 G>A polymorphism and prognosis in HCC, 3 studies reported an association for XRCC1 rs1799782 C>T, 2 studies reported an association for ERCC2 rs13181 A>C, 3 studies reported an association for ERCC2 rs1799793 G>A, and 2 studies reported an association for XRCC3 rs861539 C>T (Table 1). The NOS scores ranged from 5 and 9, demonstrating that the quality of the eligible studies was acceptable.
Table 2.
Characteristics of studies included in this meta-analysis.
| Study | Year | Country | Ethnicity | Number of patients | Age | HBV | SNP loci | scores of quality evaluation |
|---|---|---|---|---|---|---|---|---|
| Jung1 [8] | 2012 | Korea | Asian | 708 | 53.3 ± 8.3 | + | xrcc1 rs25487 | 8 |
| Jung2 [8] | 2012 | Korea | Asian | 314 | 53.3 ± 8.3 | + | xrcc1 rs25487 | 8 |
| Han [10] | 2012 | China | Asian | 112 | 50.8±8.5 | mixed | xrcc1 rs25487, xrcc3 rs861539 | 7 |
| Yue [11] | 2013 | China | Asian | 231 | 50.9±9.6 | mixed | xrcc1 rs25487, xrcc1 rs1799782, ercc2 rs13181, ercc2 rs1799793 | 7 |
| Wu [12] | 2014 | China | Asian | 218 | 52.2 ± 8.5 | + | xrcc1 rs25487, xrcc1 rs1799782, ercc2 rs13181, ercc2 rs1799793 | 6 |
| Wang [13] | 2016 | China | Asian | 308 | 53( 25–80) | Mixed | xrcc1 rs25487,ercc2 rs13181 | 7 |
| Yu [14] | 2016 | China | Asian | 485 | ⩽60,418; >60,67 | + | xrcc1 rs25487 | 7 |
| Santonocito [15] | 2017 | Italy | Caucasian | 89 | 66.3±10.5 | mixed | xrcc1 rs25487 | 6 |
| Guan [9] | 2017 | China | Asian | 172 | 50.4±4.8 | Not reported | xrcc1 rs1799782, ercc2 rs1799793 | 6 |
| Avadanei [16] | 2018 | romania | Caucasian | 50 | ⩽65,28; >65,22 | mixed | xrcc3 rs861539 | 5 |
Table 1.
Polymorphism involved in this study.
| Genes | Polymorphisms | NCBI SNP ID | Allele | genotypes | References |
|---|---|---|---|---|---|
| XRCC1 | G28152A (Arg399Gln) |
rs25487 | Ga | GG | [8, 10–15] |
| GA | |||||
| Ab | AA | ||||
| C26304T (Arg194Trp) | rs1799782 | C | CC | [9, 11, 12] | |
| CT | |||||
| T | TT | ||||
| ERCC2 | G934A (Asp312Asn) | rs1799793 | G | GG | [9, 11–13] |
| GA | |||||
| A | AA | ||||
| A2251C (Lys751Gln) | rs13181 | A | AA | [9, 11, 12] | |
| AC | |||||
| C | CC | ||||
| XRCC3 | C722T (Thr241Met) |
rs861539 | C | CC | [10, 16] |
| CT | |||||
| T | TT |
awild allele; bmutant allele.
3.3. Quantitative Synthesis
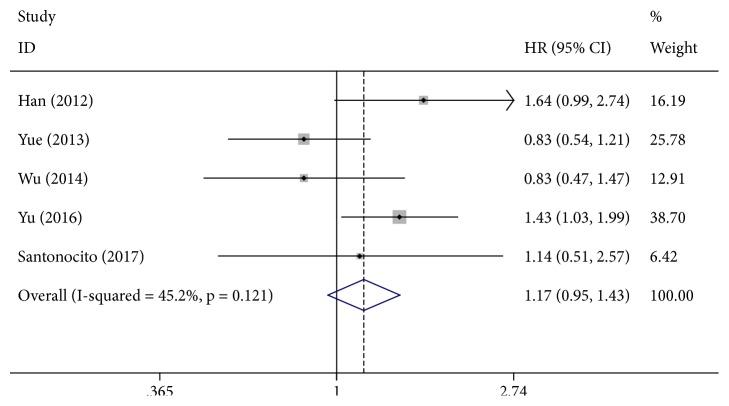
Eight studies involving 2465 patients were included in the final analysis of the relationship between the XRCC1 rs25487 G>A polymorphism and OS in HCC (Table 3). Our meta-analysis showed that there were no statistically significant associations between the XRCC1 rs25487 G>A polymorphism and the OS (AA vs. GG: HR = 0.97, 95% CI = 0.51–1.87; GA vs. GG: HR = 1.17, 95% CI = 0.95–1.43, Figure 2; AA+GA vs. GG: HR = 1.25, 95% CI = 0.83–1.88).
Table 3.
Meta-analysis of the association between XRCC1, ERCC2, and XRCC3 and overall survival for HCC patients.
| Genetic comparisons | No. of studies |
Test of association | Model | Test of heterogeneity | |
|---|---|---|---|---|---|
| HR(95% CI) | P | I2(%) | |||
| xrcc1 rs25487 | |||||
| AA vs. GG | 5 | 0.97(0.51-1.87) | R | 0.001 | 82.2 |
| GA vs. GG | 5 | 1.17(0.95-1.43) | F | 0.121 | 45.2 |
| AA+GA vs. GG | 5 | 1.25(0.83-1.88) | R | 0.002 | 76.3 |
| xrcc1 rs1799782 | |||||
| TT vs. CC | 3 | 0.72(0.48-1.08) | F | 0.849 | 0 |
| CT vs. CC | 3 | 0.88(0.63-1.22) | F | 0.853 | 0 |
| ERCC2 rs13181 | |||||
| CC vs. AA | 2 | 0.33(0.15-0.72) | F | 0.884 | 0 |
| CA vs. AA | 2 | 0.83(0.62-1.12) | F | 0.971 | 0 |
| ERCC2 rs1799793 | |||||
| AA vs. GG | 3 | 0.74(0.49-1.11) | F | 0.840 | 0 |
| GA vs. GG | 3 | 0.93(0.67-1.28) | F | 0.780 | 0 |
| XRCC3 rs861539 | |||||
| CT vs. CC | 2 | 1.64(1.11-2.42) | F | 46.4 | 12 |
HR: hazard ratio; CI: confidence interval; vs: versus; F: fixed effect model; R: random effect model.
Figure 2.
Forest plot for the association between XRCC1 rs25487 and overall survival for HCC patients (GA VS. GG).
Three studies with a total of 621 patients with advanced HCC were eligible for analyses of the association between the XRCC1 rs1799782 C>T polymorphism and the OS. No significant association was detected between XRCC1 rs1799782 C>T and prognosis in HCC (TT vs. CC: HR = 0.72, 95% CI = 0.48–1.08; CT vs. CC: HR = 0.88, 95% CI = 0.63–1.22).
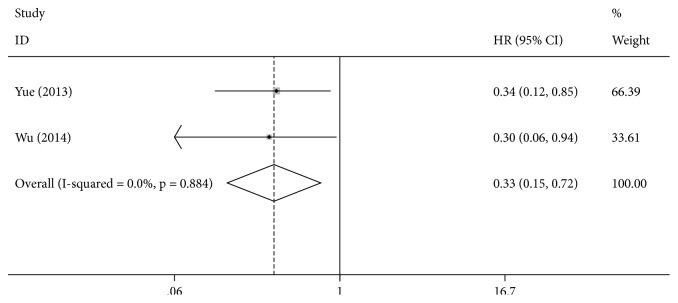
Two eligible studies were identified for the analysis of the correlation between the ERCC2 rs13181 A>C polymorphism and the OS. ERCC2 rs13181 CC genotype carriers had a significantly longer OS than that of AA genotype carriers (CC vs. AA: HR = 0.33, 95% CI = 0.15–0.72, Figure 3), but there was no significant difference between the CC genotype and the AA genotype (CA vs. AA: HR =0.83, 95% CI = 0.62–1.12).
Figure 3.
Forest plot for the association between ERCC2 rs13181 and overall survival for HCC patients (CC VS.AA).
Three studies were applicable for analyzing the association between the ERCC2 rs1799793 G>A polymorphism and the OS. The pooled results showed no significant association for any genetic model (AA vs. GG: HR = 0.74, 95% CI = 0.49–1.11; GA vs. GG: HR = 0.93, 95% CI = 0.67–1.28).
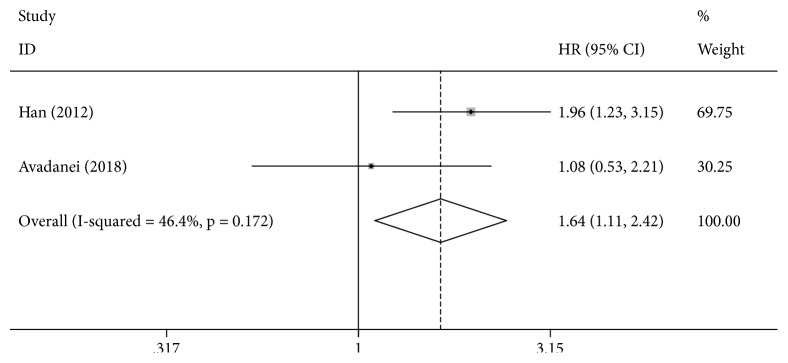
Data from 2 studies were eligible for the analysis of the association between the XRCC3 rs861539 C>T polymorphism and the OS. A significantly lower OS was observed for the CT genotype compared with the CC genotype (CT vs. CC: HR = 1.64, 95% CI = 1.11–2.42, Figure 4).
Figure 4.
Forest plot for the association between XRCC3 rs861539 and overall survival for HCC patients (CT VS.CC).
3.4. Publication Bias and Sensitivity Analysis
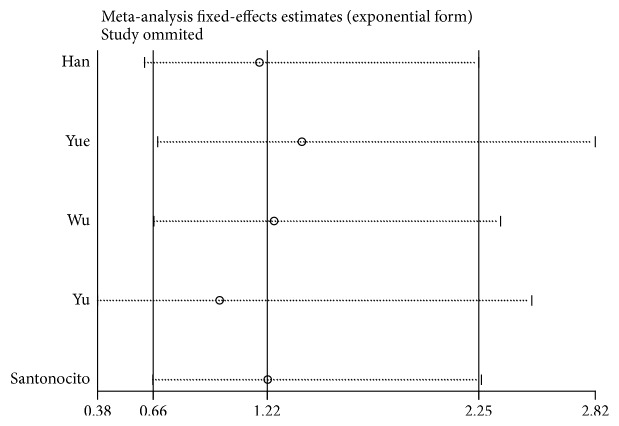
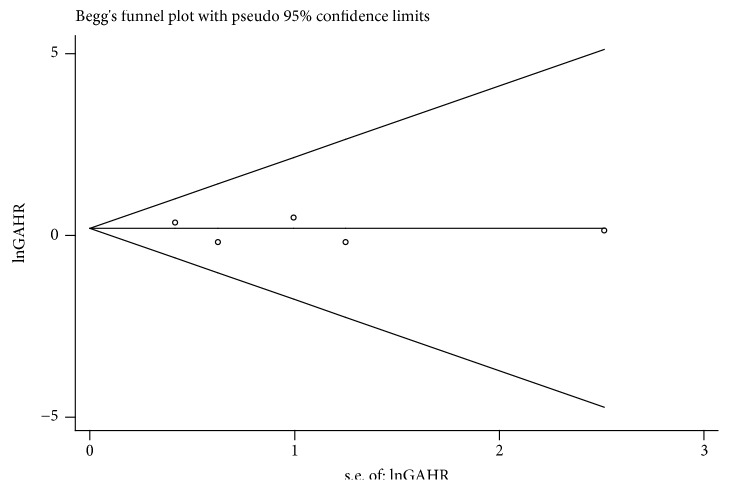
Funnel plots and Egger's tests were used to assess publication bias. The funnel plot did not show apparent asymmetry in the overall population. Egger's test demonstrated funnel plot symmetry for the XRCC1 rs25487 G>A polymorphism (AA vs. GG, P = 0.585; GA vs. GG, P = 0.653, Figure 5; AA+GA vs. GG, P = 0.802). In addition, we performed a sensitivity analysis to estimate the stability of our results by omitting one study at a time and recalculating the pooled HR. No single study changed the corresponding pooled HR and 95% CI (Figure 5), indicating that the results of our meta-analysis were statistically robust (Figure 6).
Figure 5.
Sensitivity analysis between xrcc1 rs25487 and overall survival for HCC patients (GA VS.GG).
Figure 6.
Funnel plot between xrcc1 rs25487 and overall survival for HCC patients (GA VS.GG).
4. Discussion
DNA repair systems play a fundamental role in maintaining the integrity of genomic DNA. Polymorphisms in genes related to DNA repair mechanisms, including BER (e.g., XRCC1), NER (e.g., ERCC2), and DSBR (e.g., XRCC3), may be instrumental in carcinogenesis, drug responses, treatment efficiency, and survival in HCC, thus affecting prognosis.
XRCC1 is located on chromosome 19q13.2 and belongs to the BER pathway. It constantly repairs DNA base lesions and single-strand breaks caused by endogenous and exogenous mutagens; it has a central role in the BER pathway by interacting with other DNA repair proteins [25]. More than 300 single nucleotide polymorphisms have been identified in XRCC1. Among them, the XRCC1 rs25487 G>A and rs1799782 C>T polymorphisms are the most well studied. Variation in XRCC1 expression may modulate cancer sensitivity, clinical treatment efficiency, and prognosis. However, in our study, we found that neither XRCC1 rs25487 G>A nor rs1799782 C>T influenced prognosis in HCC. A sensitivity analysis indicated that the results were statistically robust. Heterogeneity was detected in the AA vs. GG and AA+GA vs. GG model for the XRCC1 rs25487 G>A polymorphism, indicating variability. Heterogeneity may have been caused by different study characteristics, such as ethnicity, HBV status, sample size, or cure method. The random effects model was used to determine the pooled HR. Because only 1 in 5 studies focused on Caucasians and data for African populations were not available, studies of different ethnicities are needed to further evaluate this locus.
NER is a powerful and complicated DNA damage removal pathway. The ERCC2 enzyme, a critical element in NER, contributes to DNA repair by participating in DNA unwinding and structurally identifying DNA lesions, including large adducts and thymidine dimmers. rs13181 A>C and rs1799793 G>A are important polymorphisms in the ERCC2 gene. Previous research has shown that the wild-type genotype AA is associated with a higher DNA repair capacity as compared to that of the CC genotype in the rs13181 polymorphism [26]. Qian et al. suggested that the ERCC2 rs13181 A>C polymorphism has prognostic value in patients with colorectal cancer undergoing oxaliplatin-based chemotherapy [27]. A meta-analysis demonstrated that the rs13181 A>C and rs1799793 G>A polymorphisms are significantly correlated with the response to chemotherapy for patients with osteosarcoma [28]. Considering the importance of the NER pathway in tumors, polymorphisms in ERCC2 were expected to have an influence on prognosis, but this was only assessed in two studies. Although we did not find that the ERCC2 rs1799793 G>A polymorphism is associated with OS, our meta-analysis demonstrated that patients with the ERCC2 rs13181 CC genotype have significantly longer OS than that of AA genotype carriers. Further studies of the association between ERCC2 polymorphisms and response to chemotherapy for patients with HCC are needed.
The XRCC3 protein, a member of the DSBR pathway, plays an important role in homologous recombination and is therefore critical for chromosomal integrity and the stability of the genome. In 2012, Han et al. [10] found that individuals with XRCC3 CT (HR = 1.96, 95% CI = 1.23–3.15) and TT genotypes (HR = 2.98, 95% CI = 1.77–7.54) have a significantly higher risk of HCC than that of individuals with the XRCC3 CC genotype. In addition, Avadanei and collaborators [16] evaluated the rs861539 C>T polymorphism and observed a better survival only for the homozygote genotype (TT) compared to the heterozygote genotype (CT) but no differences between heterozygote TC and wild-type CC genotypes. We also performed a comprehensive evaluation of the XRCC3 rs861539 C>T polymorphism and OS. We found a significantly lower OS for the CT genotype than the CC genotype. We suspected that the XRCC3 rs861539 C>T polymorphism is a promising marker for prognosis in HCC. However, we did not perform a systematic literature review to comprehensively evaluate the effects of XRCC3 TT and TC genotypes on prognosis in HCC owing to the lack of data. The relation between XRCC3 and prognosis in HCC should be evaluated in future work. Because only two eligible studies were included in the meta-analysis, our results should be interpreted with caution. Studies with larger sample sizes are needed to obtain more definitive conclusions.
Several limitations of our study should be addressed. First, the analyses of ERCC2 and XRCC3 polymorphisms were based on only a few eligible studies. Second, the results of our comprehensive analyses were based on unadjusted estimates owing to the lack of adjusted data. Third, ethnicity, therapeutic regimen, and HBV infection status are very important factors for stratified analyses. However, few studies provided sufficient data for subgroups, making such stratified analyses impossible.
5. Conclusions
In conclusion, the ERCC2 rs13181 A>C polymorphism and XRCC3 rs861539 C>T polymorphisms are potential prognostic markers in HCC. The ERCC2 rs13181 CC genotype was associated with a significantly longer OS than that of the AA genotype. The carriers of the CT genotype had a poor prognosis compared with that of the CC genotype carriers. Well-designed studies with larger sample sizes are needed to verify our findings.
Disclosure
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Hailiang Li conceived and designed the study. Yan Zhao, Erjiang Zhao, Junhui Zhang, and Junli Ma performed study selection and data extraction. Yan Zhao, Erjiang Zhao, Yuanyuan Chen, and Junhui Zhang analyzed the data. Yan Zhao and Hailiang Li were involved in manuscript drafting and revision. Yan Zhao and Erjiang Zhao contributed equally to the paper.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ulahannan S. V., Duffy A. G., Mcneel T. S., et al. Earlier presentation and application of curative treatments in hepatocellular carcinoma. Hepatology. 2014;60(5):1637–1644. doi: 10.1002/hep.27288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtin N. J. DNA repair dysregulation from cancer driver to therapeutic target. Nature Reviews Cancer. 2012;12(12):801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z., Xiang Q., Mu G., et al. XRCC1 polymorphism and overall survival in ovarian cancer patients treated with platinum-based chemotherapy. Medicine. 2018;97(45):p. e12996. doi: 10.1097/MD.0000000000012996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D.-J., Xiao D. Association between the XRCC1 polymorphisms and clinical outcomes of advanced NSCLC treated with platinum-based chemotherapy: A meta-analysis based on the PRISMA statement. BMC Cancer. 2017;17(1):p. 501. doi: 10.1186/s12885-017-3487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu M., Xu L., Yang X., et al. XRCC3 Thr241Met is associated with response to platinum-based chemotherapy but not survival in advanced non-small cell lung cancer. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0077005.e77005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M., Zhao Y., Zhao E., Wang K., Lu W., Yuan L. Predictive value of two polymorphisms of ERCC2, rs13181 and rs1799793, in clinical outcomes of chemotherapy in gastric cancer patients: a meta-analysis. Disease Markers. 2018;2018:12. doi: 10.1155/2018/3947626.3947626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung S. W., Park N. H., Shin J. W., et al. Polymorphisms of DNA repair genes in Korean hepatocellular carcinoma patients with chronic hepatitis B: possible implications on survival. Journal of Hepatology. 2012;57(3):621–627. doi: 10.1016/j.jhep.2012.04.039. [DOI] [PubMed] [Google Scholar]
- 9.Guan Q., Chen Z., Chen Q., Zhi X. XRCC1 and XPD polymorphisms and their relation to the clinical course in hepatocarcinoma patients. Oncology Letters. 2017;14(3):2783–2788. doi: 10.3892/ol.2017.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han X., Xing Q., Li Y., et al. Study on the DNA repair gene XRCC1 and XRCC3 polymorphism in prediction and prognosis of hepatocellular carcinoma risk. Hepato-Gastroenterology. 2012;59(119):2285–2289. doi: 10.5754/hge12096. [DOI] [PubMed] [Google Scholar]
- 11.Yue A.-M., Xie Z.-B., Guo S.-P., Wei Q.-D., Yang X.-W. Implication of polymorphisms in DNA repair genes in prognosis of hepatocellular carcinoma. Asian Pacific Journal of Cancer Prevention. 2013;14(1):355–358. doi: 10.7314/APJCP.2013.14.1.355. [DOI] [PubMed] [Google Scholar]
- 12.Wu J. S., Chen Y. P., Wang L. C., et al. Implication of polymorphisms in DNA repair genes with an increased risk of hepatocellular carcinoma. Genetics and Molecular Research. 2014;13(2):3812–3818. doi: 10.4238/2014.May.16.5. [DOI] [PubMed] [Google Scholar]
- 13.Wang X.-C., Wang F., Quan Q.-Q. Roles of XRCC1/XPD/ERCC1 polymorphisms in predicting prognosis of hepatocellular carcinoma in patients receiving transcatheter arterial chemoembolization. Genetic Testing and Molecular Biomarkers. 2016;20(4):176–184. doi: 10.1089/gtmb.2015.0267. [DOI] [PubMed] [Google Scholar]
- 14.Yu L., Liu X., Han C., et al. XRCC1 rs25487 genetic variant and TP53 mutation at codon 249 predict clinical outcomes of hepatitis B virus-related hepatocellular carcinoma after hepatectomy: a cohort study for 10 years’ follow up. Hepatology Research. 2016;46(8):765–774. doi: 10.1111/hepr.12611. [DOI] [PubMed] [Google Scholar]
- 15.Santonocito C., Scapaticci M., Nedovic B., et al. XRCC1 Arg399Gln gene polymorphism and hepatocellular carcinoma risk in the Italian population. The International Journal of Biological Markers. 2017;32(2):e190–e194. doi: 10.5301/jbm.5000241. [DOI] [PubMed] [Google Scholar]
- 16.Avadanei E.-R., Giusca S.-E., Negura L., Caruntu I.-D. Single nucleotide polymorphisms of XRCC3 gene in hepatocellular carcinoma - relationship with clinicopathological features. Polish Journal of Pathology. 2018;69(1):73–81. doi: 10.5114/pjp.2018.75340. [DOI] [PubMed] [Google Scholar]
- 17.Moher D., Liberati A., Tetzlaff J., Altman D. G., The PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Annals of Internal Medicine. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 18.Parmar M. K. B., Torri V., Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in Medicine. 1998;17(24):2815–2834. doi: 10.1002/(SICI)1097-0258(19981230)17:24<2815::AID-SIM110>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Tierney J. F., Stewart L. A., Ghersi D., Burdett S., Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8, article no. 16 doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson P. R., Smith C. T., Hutton J. L., Marson A. G. Aggregate data meta-analysis with time-to-event outcomes. Statistics in Medicine. 2002;21(22):3337–3351. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 21.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. European Journal of Epidemiology. 2010;25(9):603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 22.Bi H., Gan Y., Yang C., Chen Y., Tong X., Lu Z. Breakfast skipping and the risk of type 2 diabetes: a meta-analysis of observational studies. Public Health Nutrition. 2015;18(16):3013–3019. doi: 10.1017/s1368980015000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22(4):719–748. doi: 10.1093/jnci/22.4.719. [DOI] [PubMed] [Google Scholar]
- 24.Egger M., Smith G. D., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehouse C. J., Taylor R. M., Thistlethwaite A., et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104(1):107–117. doi: 10.1016/S0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 26.Xiao S., Cui S., Lu X., et al. The ERCC2/XPD Lys751Gln polymorphism affects DNA repair of benzo[a]pyrene induced damage, tested in an in vitro model. Toxicology in Vitro. 2016;34:300–308. doi: 10.1016/j.tiv.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Qian Y.-Y., Liu X.-Y., Pei D., et al. The XPD Lys751Gln polymorphism has predictive value in colorectal cancer patients receiving oxaliplatin-based chemotherapy: a systemic review and meta-analysis. Asian Pacific Journal of Cancer Prevention. 2014;15(22):9699–9706. doi: 10.7314/APJCP.2014.15.22.9699. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H., Ge J., Hong H., Bi L., Sun Z. Genetic polymorphisms in ERCC1 and ERCC2 genes are associated with response to chemotherapy in osteosarcoma patients among Chinese population: a meta-analysis. World Journal of Surgical Oncology. 2017;15(1, article no. 75) doi: 10.1186/s12957-017-1142-3. [DOI] [PMC free article] [PubMed] [Google Scholar]