Abstract
The advent of ribosome profiling and other tools to probe mRNA translation has revealed that codon bias — the uneven use of synonymous codons in the transcriptome —serves as a secondary genetic code: a code that guides the efficiency of protein production, the fidelity of translation and the metabolism of mRNAs. Recent advancements in our understanding of mRNA decay have revealed a tight coupling between ribosome dynamics and the stability of mRNA transcripts; this coupling integrates codon bias into the concept of codon optimality, or the effects that specific codons and tRNA concentrations have on the efficiency and fidelity of the translation machinery. In this Review, we first discuss the evidence for codon-dependent effects on translation, beginning with the basic mechanisms through which translation perturbation can affect translation efficiency, protein folding and transcript stability. We then discuss how codon effects are leveraged by the cell to tailor the proteome to maintain homeostasis, execute specific gene expression programmes of growth or differentiation and optimize the efficiency of protein production.
The use of 3-letter codons in mRNA means that 64 possible codons encode a pool of 20 amino acids and translation stop signals. This has led to degeneracy in the genetic code, where multiple codons code for a single amino acid. These synonymous codons are recognized distinctly by the ribosome; in this context, ‘codon optimality’ refers to the non-uniform decoding rate of each of the 61 amino-acid encoding codons by the ribosome. Codon optimality is a function of the stochastic nature of ribosome decoding and the variability of tRNA concentrations. Because of the stochastic recognition of the codon within the ribosome A-site by tRNAs, a codon can be defined (in broad terms) as optimal or non-optimal depending on how efficiently the appropriate cognate tRNA can be selected from the cytoplasmic pool of tRNAs (tRNA pool). Additionally, as codon optimality can be a powerful determinant of translation rates, genomes are seen as having an inherent codon bias. Codon bias, or the propensity for some codons to be disproportionately represented in the transcriptome, is, in part, shaped by codon optimality and is widespread throughout multiple domains of life. Codon bias correlates with tRNA levels in Escherichia coli1, Saccharomyces cerevisiae2, Caenorhabditis elegans3, Drosophila spp.4 and eukaryotes in general5. Codons with strong bias, corresponding to enriched tRNA species, are preferentially found in highly expressed genes6-9. The functional relevance of codon optimality to the speed of ribosome translation remains a topic of intense debate, which has been extensively reviewed elsewhere10-15. Although much of the debate on codon optimality has centred around the influence of codon content on translation efficiency, a substantial body of evidence indicates that codon usage can also influence protein folding and translation fidelity. More recently, codon optimality has been shown to be a powerful determinant of mRNA stability in addition to the translation elongation rate16-18.
Codon usage may also be constrained by factors that are independent of any translational effect, including the need to avoid similarity to functionally important RNA motifs that may interfere with mRNA processing and translation when located in the open reading frame (ORF), like the Shine–Dalgarno sequence19, although these effects probably contribute little to ribosome pausing in general20. The need for proper and unambiguous encoding of splice sites also places a major constraint on codon usage11, as do several evolutionarily neutral factors that influence codon bias in the transcriptome but that are not thought to have any effect on mRNA processing. The GC content of the transcriptome largely matches that of the non-coding genome21. GC-rich areas are more prone to homologous recombination, leading to biased gene conversion, which increases the GC content across transcripts22 and which affects estimates of codon bias because GC-rich codons will tend to be over-represented in ORFs, especially in higher organisms13. However, we are more interested in the extent to which codon usage can have an impact on the dynamic control of gene expression in cells, tissues and developmental stages, and we contrast this with prevailing notions of codon bias as a static entity that operates on evolutionary timescales to fine-tune gene expression.
In this Review, we first discuss the experimental and bioinformatics evidence for codon-dependent effects on translation and on mRNA abundance. We then discuss the functional consequences that arise from the interplay between tRNA pool levels, ribosomes and codon usage, including effects on translation efficiency, protein folding and mRNA stability. Recent advances in our understanding of the molecular machinery that underlies codon effects on translation allow us to shed new light on the long-standing debate over the causes, regulation and consequences of codon effects, with a greater understanding of the relevance of codon biology to cellular homeostasis and disease.
Codons affect translation elongation
For over 20 years, it was assumed that different codons were translated at different speeds. Through the use of a radio-labelled amino acid incorporation assay, it was shown that codon identity could affect the translation elongation rate23. Per-codon elongation rates are crucially dependent on the tRNA pool, as the ribosome will be forced to wait longer for a rare cognate tRNA to enter the ribosome A-site24, and the process of sampling and rejecting near-cognate tRNA species imposes a substantial kinetic cost to elongation in cases where the cognate tRNA species is rare compared with near-cognate tRNAs25. Although this process is thought to represent the major tRNA-dependent cost to elongation, the tRNAs that are efficiently discriminated from near-cognate species tend to be the most abundant; therefore, we will generally mention tRNA abundance without qualification when discussing the link between the tRNA pool and codon-dependent translation effects.
With the advent of ribosome profiling, it became possible to assess ribosome occupancy across the entire transcriptome and to estimate the relative density of ribosomes over specific codons within the ribosome A-site26. If ribosomes do traverse optimal codons more rapidly, then it would be expected that ribosomes would be less abundant where the ribosome A-site is positioned over optimal codons, and this hypothesis was tested in the very first ribosome profiling paper27. Surprisingly, in cells pretreated with the translation elongation inhibitor cycloheximide, no clear correlation was found between tRNA abundance and codon-level ribosome density, and this counter-intuitive finding was replicated in other ribosome profiling experiments28. However, a recent re-analysis of 24 ribosome profiling data sets in yeast showed very elegantly that pretreatment with cycloheximide did not instantly freeze ribosomes in place on an mRNA but instead allowed limited ribosome translocation to continue, thereby distorting ribosome occupancy estimates29,30. In ribosome profiling experiments conducted without cycloheximide pretreatment, there is a clear inverse relationship between tRNA abundance and ribosome occupancy, showing that ribosomes spend less time at optimal codons29,31-33. The exact correlation between tRNA abundance and ribosome occupancy is generally modest and varies considerably between ribosome profiling experiments in yeast, with different profiling experiments showing that tRNA abundance as captured by the tRNA adaptation index (tAI) accounts for between 5% and 25% of the variance in average per-codon ribosome densities. Cycloheximide-free ribosome profiling data sets in metazoans have not yet been analysed for codon effects, making it difficult to extend this analysis beyond yeast. Nevertheless, ribosome profiling in E. coli also failed to show a significant effect of codon bias on ribosome occupancy19.
Additional evidence for the effect of codon content on translation elongation rates can be found in emerging methods for monitoring translation in real time34,35. In a cell-free translation system, a codon-optimized 1.6kb-long mRNA was shown to complete translation 1.5 minutes faster than a non-optimized control36. More recently, a fluorescent-tagged, intracellular antibody co-expressed with an epitope-bearing protein was used to monitor the translation of single mRNAs in real time in mammalian cells. The analysis revealed that the translation rate of codon-optimized mRNAs is 4.9 codons per second, whereas that of non-optimized mRNAs is 3.1 codons per second, an increase of 58% (REF. 37). These results are supported by ribosome runoff experiments, where translation initiation is inhibited and where mRNAs are fractionated by sucrose gradient into ribonucleoprotein (RNP)-, 80S- and polysome-associated pools. Transcripts in the RNP complex fractions are not associated with the translation machinery, whereas transcripts in the 80S and polysome fractions are associated with one or multiple ribosomes, respectively, indicating that they are likely to be actively translated. Following a block of ribosomal initiation, mRNAs with more optimal codons leave the polysome and 80S fractions and appear in the RNP fractions much faster than their non-optimal counterparts, indicating that ribosomes move off these transcripts more rapidly16. Thus, although the exact codon-to-codon effect on translation elongation remains to be quantitatively defined, a growing body of evidence suggests that synonymous codon usage can have a substantial influence on protein synthesis rates.
Translation efficiency
The striking impact of codon optimality on the expression levels of genes in heterologous systems led to the hypothesis that codon bias could influence translation efficiency, or the total number of protein molecules produced per transcript in a given time1. This hypothesis is bolstered by the observation that codon bias is often more pronounced in highly expressed genes and that evolutionary conservation of codon bias is more marked in these genes7,38-41. Although it is clear that codon optimization has a profound effect on gene expression in a host of different heterologous gene expression systems42,43, the evidence for a substantial relationship between codon content and translation efficiency in endogenous genes is often contradictory.
With the advent of high-throughput proteomics, it became clear that the relationship between mRNA and protein expression levels was far from perfect, suggesting that post-transcriptional regulation is important for tuning protein output. In early proteomic experiments, codon bias was implicated as a major factor determining protein levels in E. coli44 and yeast45, but it has been unclear to what extent translation efficiency linked codon bias to protein levels, as neither of the early studies looked at protein:mRNA ratios as a measure of translation efficiency. Early shotgun proteomics experiments coupled with microarray-based mRNA expression analyses allowed us to assess this link and revealed that codon bias correlated very poorly with translation efficiency in E. coli46, S. cerevisiae47 and a human cell line48. These findings were confirmed by a study of large sets of GFP constructs with varying codon content, which found that codon optimality did not contribute to variation in protein levels49,50. However, re-analysis correcting for RNA folding energy revealed a modest correlation between the codon bias of a GFP gene and its translation efficiency. Furthermore, a much weaker though significant relationship between codon bias and translation efficiency was uncovered in S. cerevisiae51.
Much of the difficulty in parsing the effect of codon bias on translation efficiency comes from the plethora of other factors that may influence codon usage. The folding energy of mRNAs has a marked impact on translation efficiency, especially near the start codon, as more stable RNA secondary structures require more energy to unfold before translation initiation49,52,53. In E. coli, non-optimal codons are associated with decreased folding energy because they tend to be AT-rich compared with their optimal counterparts and are therefore associated with increased translation efficiency when located near the 5′end of the coding DNA sequence (CDS)50. This suggests that codon optimality must be balanced against the need for lax secondary structure at the 5′ end of the CDS in order to maximize translation efficiency, and this may confound the impact of codons on translation efficiency51, although it is important to note that the folding energy of secondary structures is insufficient to explain the link between codon bias and ribosome elongation rates36.
Direct in vivo measurements of translation rates37,54,55 and computer simulations of ribosome initiation and elongation56 demonstrated that translation initiation is generally the rate-limiting step in protein synthesis. If initiation is rate-limiting, then any change in elongation rates will simply lead to faster ribosome runoff, and not to any change in the rate of protein production per molecule of mRNA. Slow initiation is necessary to ensure that mRNAs do not become crowded with ribosomes, which can result in ribosome collisions and other events that decrease the efficiency of translation57,58. If initiation is rate-limiting, then the apparent link between codon content and translation efficiency indicates a coupling of elongation and initiation. Any slowing of ribosomes over non-optimal codons may propagate back to other ribosomes on the same transcript, affecting initiation by decreasing the rate at which ribosomes can move off from the initiation site of a message58,59. In keeping with this hypothesis, codon bias can influence translation efficiency in yeast, and the inclusion of as few as eight non-optimal codons near the 5′ end of the CDS is sufficient to produce a significant decrease in protein expression, and the inclusion of a large non-optimal stretch in the 3′ part of a gene can also produce a marked decrease in protein production, consistent with a model of ribosomes crowding behind non-optimal codon sequences and interfering with initiation60 (FIG. 1a). These findings are consistent with previous work in E. coli showing that the capacity of a non-optimal codon to decrease protein production was dependent on its position near the 5′ end of a CDS61. There is also evidence from simulations of translation dynamics that transcripts that contain stretches of non-optimal codons have a codon-imposed upper limit on their achievable translation efficiency, whereas translation efficiency is solely a function of initiation rate for transcripts that are not constrained by non-optimal codon content62.
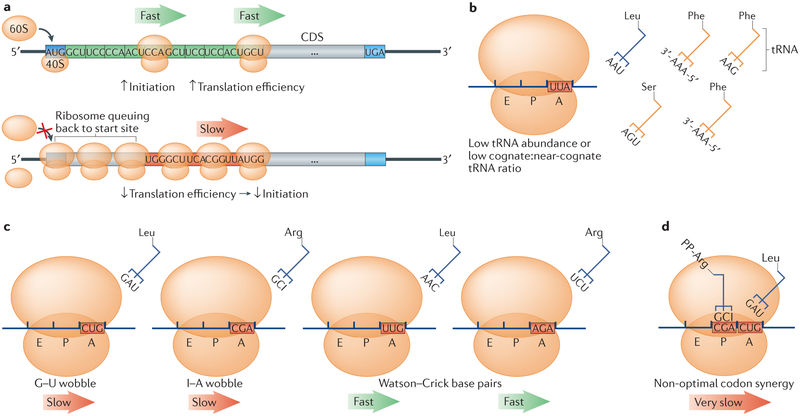
Figure 1 ∣. The codon content of an mRNA can influence translation via tRNA-dependent mechanisms.
a ∣ Optimal codons (green; top) allow rapid ribosome translocation, thereby clearing the 5′ end of the coding DNA sequence (CDS) to allow the assembly of the 80S ribosome from its 40S and 60S subunits over the start codon. Stretches of non-optimal codons (red; bottom) can slow ribosome translocation as the ribosomes wait for a rare cognate tRNA. The non-optimal codon stretch can create ribosome crowding that can eventually result in the formation of a queue that, should it stretch back far enough, could inhibit translation initiation, thereby modulating the overall rate of protein production per transcript, or translation efficiency61. Thus, although translation initiation may be the rate-limiting step for the majority of transcripts56, non-optimal codons can still modulate translation efficiency by directly influencing initiation60. b–d ∣ Representation of the modes through which ribosome elongation can be slowed in a codon-dependent manner. When the codon–cognate tRNA species (blue) is rare in the tRNA pool or in low abundance relative to near-cognate tRNA species (yellow), slow translation can result owing to the increased time required until the correct codon is successfully bound by the ribosome and accommodated, as shown in part b. Part c shows that codons that must be decoded by tRNA species that do not form canonical Watson–Crick base pairs at the third position of the codon will often cause a slowing of the ribosome, due to the increased time required to incorporate wobble tRNAs. Two common wobble interactions in yeast are shown, with a G–U wobble interaction in the decoding of the non-optimal Leu codon CUG and a hypoxanthine (I) modification at the wobble position in the tRNA that decodes the non-optimal Arg codon CGA. We contrast these with traditional Watson–Crick base pairs (on the right-hand side), which are associated with more efficient translation. Part d shows that non-optimal codon doublets are associated with the capacity to induce ribosome slowing to a degree greater than the sum of their parts66. PP, polypeptide.
The abundance of tRNAs is not the only factor that influences ribosome elongation dynamics. The tRNA pool is complex, and the concentration of a tRNA species relative to its near-cognate tRNAs can also influence ribosome dynamics, as the ribosome slowly samples, proofreads and rejects more of the tRNA pool before incorporating the correct tRNA species25 (FIG. 1b). Another key influence on ribosome elongation is wobble (non-Watson–Crick) base pairing between the first (5′) base of the tRNA anticodon and the third (3′) base of the mRNA codon. Wobble base pairing arises when a codon lacks a cognate tRNA and must be decoded by a synonymous tRNA species or when a tRNA undergoes a modification at the first position of the anticodon (FIG. 1c). These wobble interactions are relatively unfavourable compared with Watson–Crick base pairing63, are consistently found to be under-represented in coding sequences64,65 and show high levels of ribosome A-site occupancy in ribosome profiling experiments33, indicating that ribosomes tend to move very slowly off these codons.
It is also noteworthy that a narrow focus on individual codons is misleading, as the sequence and amino acid context of a codon in the A-site of a ribosome will modify the effect of that codon on elongation dynamics. The ribosome does not interact with one codon at a time, and there is strong evidence to suggest that codon pairs act synergistically to slow ribosome movement66 (FIG. 1d). The codon pairs that influence translation generally involve at least one codon that interacts with its cognate tRNA via a wobble interaction, and when suboptimal codons occur together, they can destabilize protein production66,67. The emerging emphasis on non-optimal codon doublets underscores the importance of considering codons in the context of their surroundings. It is also important to consider the amino acid identity of a codon and that of the preceding codons, as this can have a profound effect on translation dynamics not only at the A-site68,69 but also at the exit tunnel of the ribosome70-73, giving rise to the concept of optimal and non-optimal amino acids9.
Protein folding
Several lines of evidence suggest that suboptimal codons can be used to slow translation at key structural motifs in order to facilitate proper protein folding (FIG. 2). The idea that co-translational polypeptide folding is dictated in part by the rate of protein synthesis is long-standing74, and early analysis of codon usage around protein structural elements showed that codon usage was likely to be an important contributor to slowing protein synthesis to allow for proper folding75. Experiments in E. coli showed that replacing rare codons with optimal codons increased overall protein production; however, these substitutions also increased protein misfolding, as assessed by different assays76,77, including protein solubility assays, where effects were seen with as few as two synonymous codon substitutions78. More recently, nuclear magnetic resonance was used to directly probe the structures of proteins produced by altered codon usage, revealing clear changes in protein conformational states of proteins translated from mRNAs with differing codon content, in addition to changes in cysteine oxidation and in vitro protein stability79.
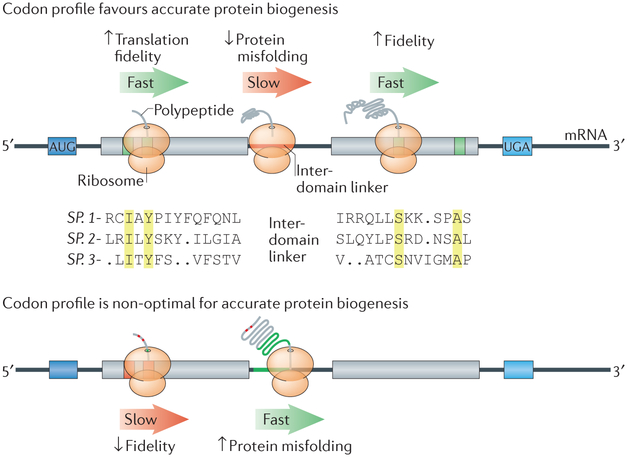
Figure 2 ∣. Codon optimality in a transcript can be used to optimize protein folding.
A stretch of non-optimal codons (red line; top) in an inter-domain linker region can slow translation elongation to allow proper folding of the emerging polypeptide into a functional protein domain36,76,85. When codon optimality is high in this region (green line; bottom), protein misfolding can occur. Conversely, ribosomes quickly translate optimal codons (green boxes; top)81,86,87, which often encode highly conserved residues (marked in yellow). Optimal codons are less prone to reading errors because they correspond to tRNA species with a high cognate:near-cognate tRNA ratio94, thereby ensuring high translation fidelity of the most functionally important residues. When important amino acids are encoded by non-optimal codons (red boxes; bottom), missense errors are more likely to occur during translation.
Increasing the concentration of rare tRNAs resulted in increased protein misfolding, underlining the importance of cognate tRNA concentrations to protein folding77. Moreover, a Neurospora crassa cell-free translation system was used to show that although a codon-optimized luciferase mRNA completed translation elongation more rapidly than the wild-type mRNA and produced more protein, a large proportion of the protein was non-functional, and thus the accumulation of functional protein was much slower using the optimized transcript36. Optimization of 200 codons encoding a key luciferase structural domain was sufficient to produce the non-functional protein, indicating that the use of slow codons in this key region is necessary for the proper folding of the protein. Applying this principle in the opposite direction, it was also shown that transgene protein misfolding can be minimized by replacing some optimal codons with less optimal codons at key structural positions80.
Bioinformatics analysis of codon usage near protein secondary and tertiary structures has supported these results33,81,82, suggesting that non-optimal codons are widely used to promote co-translational folding and that we can generalize the principle from the handful of genes that have been systemically studied. These analyses also show us that optimal and non-optimal codons cluster at different locations in mRNAs to maximize translation fidelity and efficiency. In E. coli, regions of non-optimal codons were located immediately downstream of structural domains in the majority of genes surveyed83, and non-optimal codons may be more associated with secondary structures84,85. Ribosomes tend to move less rapidly over mRNA regions that encode linkers of structural domains, further underlining the connection between codon usage, ribosome translation rates and successful folding of newly synthesized domains without interference from downstream amino acid sequences33.
Other studies have fitted these findings into a more nuanced picture of conserved codon usage that is associated with a host of structural features81. Although non-optimal codons are clustered in protein domain linker regions, highly conserved regions of mRNAs associated with key residues of structural domains show a trend towards increased codon optimality82,86-88 (FIG. 2), suggesting that codons with minimal mistranslation rates were selected for at sites where a mistranslation event would be most likely to result in a dysfunctional protein87. This is consistent with evidence that codons that are considered optimal are much less prone to mistranslation than their non-optimal counterparts89-93, although codon effects on missense translation errors are more sensitive to the cognate:near-cognate tRNA ratio than to tRNA abundance per se94, as near-cognate codons of high abundance will necessarily compete with the cognate codon for incorporation95. Underscoring the importance of codon usage in preventing errors in protein production, it was shown that proteins that are most prone to misfolding but that cannot rely on a chaperone system to ensure their proper folding were those with the greatest degree of optimal codon enrichment at structurally conserved sites96, suggesting that codon bias functions in parallel with chaperones to minimize protein misfolding. The tendency for optimal codon usage is also more pronounced in genes for which the consequences of a translation error would be more energetically costly to the cell, for example, long ORFs, which require more energy for protein synthesis86.
The research linking codon optimality and protein folding discussed above shows that both non-optimal and optimal codons seem to be involved in ensuring that protein products behave appropriately. These mechanisms need not be mutually exclusive, with optimal codons selected for at sites where mistranslation is most costly and non-optimal codons selected for downstream of domains where failure of co-translational folding is most costly. Non-optimal codons may also be implicated in maintaining other aspects of protein biogenesis, such as discouraging the formation of insoluble polypeptide intermediates and protein aggregates97 or modulating the ability of the polypeptide to form helical structures within the ribosomal exit tunnel, as reviewed elsewhere98-101.
Codon usage controls mRNA stability
In yeast, transcript half-lives vary considerably, from under a minute to over an hour16,102. Although factors that contribute to mRNA stability or decay have been identified for many transcripts, including microRNAs103,104 and structural elements in the 3′ or 5′ untranslated regions105-107, much of the observed variation in transcript stability remains unexplained. The introduction of rare codons into a small subset of genes leads to a marked decrease in mRNA stability, suggesting a link between translation dynamics and mRNA decay108-111. However, early transcriptome-wide analyses of mRNA decay in yeast failed to yield a clear link between the codon adaptation index (CAI) of a transcript and its half-life102,112.
To re-evaluate the role of codon usage in dictating mRNA stability, a heat-sensitive allele of RNA Pol II was used to drive transcription shut-off followed by measurements of transcript abundance using high-throughput sequencing at specific time points after the cessation of transcription16. Rather than relying on existing measures of codon bias, the contribution of each codon to mRNA stability was independently assessed. The analysis revealed that many codons are preferentially enriched in stable transcripts, whereas others are associated with unstable transcripts. The link between codon frequency and transcript stability is expressed as the codon stability coefficient (CSC), defined as the Pearson correlation coefficient between codon frequency in a transcript and the half-life of the transcript. The per-codon CSC values calculated in yeast correlated well with the tAI, which is a measure of how efficiently a codon will be translated given the tRNA pool of a cell2. Furthermore, altering the codon optimality of a transcript could increase or decrease its stability by as much as tenfold, and decreasing the codon optimality of a gene construct could decrease ribosome elongation rates, as well as overall translation efficiency16.
A relationship between codon content and mRNA stability has since been uncovered in E. coli113, Schizosaccharomyces pombe17 and zebrafish9,114, as well as in human, mouse and rat cell lines (S. Martin, G.H., N. Al-Husaini and J.C., unpublished observations), suggesting that codon usage represents a conserved mechanism for directly encoding mRNA stability.
Similarly to translation efficiency and protein folding accuracy, the ability of synonymous codon usage to affect the dynamics of translating ribosomes ultimately underpins the effect of codon optimality on mRNA stability. In budding yeast, the DEAD-box helicase Dhh1 is necessary for the influence of codon optimality on mRNA stability, and Dhh1 preferentially associates with mRNAs enriched in non-optimal codons (FIG. 3). This association is mediated by the number of slow-moving ribosomes on a transcript, and it appears that Dhh1 may preferentially bind to these slow-moving ribosomes, thereby triggering mRNA decay18.
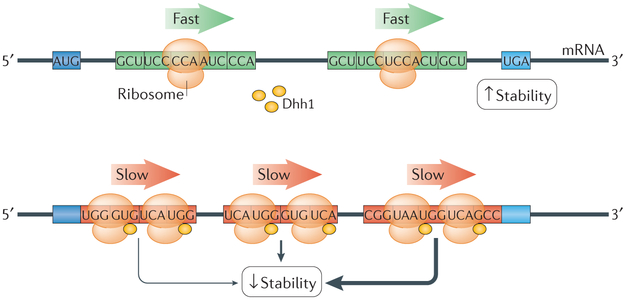
Figure 3 ∣. Non-optimal codons decrease mRNA stability in a Dhh1-dependent mechanism.
In budding yeast, fast translation over optimal codons (green) is coupled with high mRNA stability. The slowing of ribosomes over non-optimal codons (red) recruits the DEAD-box helicase and decapping activator Dhh1 to the ribonucleoprotein, thereby initiating mRNA decay18. The polarity of the effect of the association of Dhh1 with the ribosome is emphasized, as stretches of non-optimal codons at the 3′ region of transcripts appear to induce mRNA decay more robustly.
The polarity of codon effects
Codon optimality can influence a variety of cellular processes, including protein output and mRNA stability. Highly expressed genes may leverage optimal codon content to maximize protein output, thereby minimizing the waste of energy and the cytotoxicity associated with mistranslated proteins, or to decrease transcript turnover. The extent to which a particular transcript can leverage synonymous codons to gain some but not all of these advantages is currently unknown, but the data presented thus far suggest that codon effects are dependent on the location of optimal or non-optimal codon stretches within the ORF. We have discussed above how optimal codons can have a beneficial effect on protein folding if they are located at highly conserved, structurally and functionally important domains87, whereas they are deleterious to co-translational folding when they enable the rapid synthesis of polypeptides located downstream of such domains83. The effect of synonymous codon choice on translation efficiency involves a highly context-dependent and complex interdependence between translation elongation and initiation; the experimental evidence shows that codon effects on translation efficiency are most potent near the 5′ end of a transcript60,61, whereas non-optimal codons have the greatest capacity to decrease mRNA stability near the 3′ end of a transcript18 (FIG. 3). This apparent polarity of codon effects suggests that codon optimization can be used to independently fine-tune unrelated aspects of gene expression, depending on where in a transcript the optimal or non-optimal codons occur62, thus increasing the functional utility of the synonymous codon spectrum to a cell.
Functional relevance
Codon optimality helps to coordinate the expression patterns of functionally related genes. The expression levels of interacting proteins are much more closely matched than their encoding mRNAs115, but functionally related mRNAs often have very similar patterns of codon usage116, and they decay at similar rates16,102. In yeast, enzymes involved in glycolysis are uniformly enriched in high-optimality codons. This pattern of codon usage may reflect the need of the cell to maintain high levels of metabolically important enzymes, such as those involved in glycolysis, by stabilizing transcripts and/or maximizing protein yield per mRNA. However, proteins involved in transient responses to stimuli, such as the pheromone response, are enriched in non-optimal codons (FIG. 4). This may reflect selective pressure to keep the levels of such regulatory gene products low, and the instability conveyed by high levels of non-optimal codon content helps to ensure that responses to transient stimuli can be quickly curtailed when the stimulus is withdrawn. One well-established example of this is the clock genes, which control circadian rhythms in various organisms, including in cyanobacteria117 and Neurospora crassa118. In both organisms, the clock genes are markedly deficient in optimal codons, and their non-optimality is crucial for their ability to drive circadian rhythms.
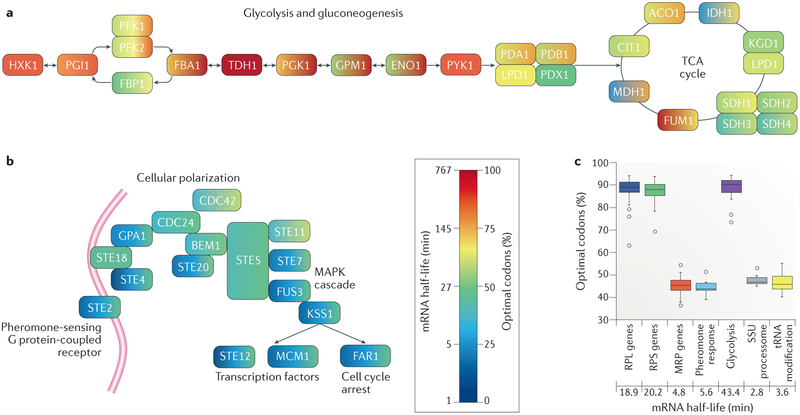
Figure 4 ∣. Codon content and mRNA stability are matched in transcripts encoding functionally-related proteins.
a,b ∣ Genes that encode interconnected components of biochemical pathways tend to have a similar codon content and similar mRNA half-lives. Each enzyme in the two biochemical pathways is represented by two colours. The colour on the left corresponds to the mRNA half-life of each transcript in yeast, based on previously published mRNA half-life data16, and the colour on the right corresponds to the codon content, which is defined as the percentage of optimal codons in the transcript. Codon optimality was determined by tRNA adaptation index (tAI) values calculated in yeast2, and codons with corresponding tAI values greater than the median were considered optimal. Part a shows a simplified representation of the glycolysis and gluconeogenesis enzymes, as well as enzymes of the tricarboxylic acid (TCA) cycle; metabolic intermediates were omitted for clarity. Most enzymes are encoded by highly stable transcripts with a high proportion of optimal codons, underscoring the importance of this fundamental pathway in cell growth and survival. By contrast, part b shows that transcripts encoding proteins involved in the mating pheromone response pathway are remarkably unstable and depleted of optimal codons, which is consistent with the transient nature of this response. c ∣ Codon optimality and median mRNA half-life for a number of gene ontology terms, showing that genes with similar functions (for example, in pheromone response, glycolysis and tRNA modification) or that encode components of a large cellular apparatus (including the ribosome (RPL and RPS genes), the mitochondrial ribosome (MRP genes) or the small subunit processome (SSU genes)), tend to share similar codon optimality profiles. RPL, large subunit ribosomal proteins; RPS, small subunit ribosomal proteins. Part c is reproduced with permission from REF. 16, Elsevier.
The evidence thus indicates that codon bias has the capacity to ensure that genes requiring consistently high levels of expression give rise to highly stable mRNAs that produce proteins efficiently and accurately, whereas genes that generally require low expression levels give rise to unstable mRNAs that produce proteins less efficiently. However, gene expression requirements change as tissues develop and differentiate. If the set of optimal codons is static as the cell develops and responds to stimuli, then codon optimization is necessarily limited to a baseline optimization of gene expression that would not interfere with the changing demands of the cell. This would necessarily limit the utility of differential codon usage, and it is therefore unsurprising that genes with similar expression patterns across tissues and environmental conditions tend to have similar codon biases119,120. That is, genes that are expressed together tend to have similar codon compositions. We have discussed above how tRNA concentrations mediate the relationship between codon usage and ribosome dynamics; this has led to the hypothesis that tRNA concentrations can be dynamically regulated to alter gene expression in a tissue-specific manner121-123.
Changes in tRNA pools help to maintain cellular homeostasis.
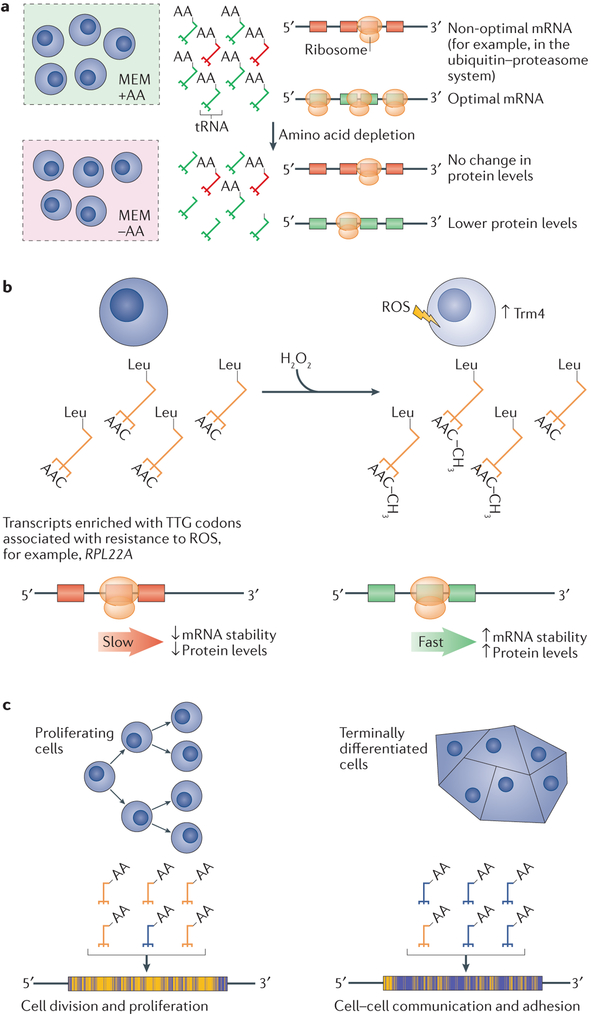
Perhaps the best characterized example of the influence of tRNA pools on gene expression is the cellular response to amino acid scarcity. In E. coli, the amino acid leucine is decoded by six different isoacceptor tRNAs, and the cognate codons for leucine show marked codon bias. The charged population of optimal tRNA species decoding leucine markedly declines in conditions of leucine starvation, whereas the charged pool of non-optimal leucine tRNAs remains relatively constant124; these findings were extended to several other sets of amino acid isoacceptor tRNAs125. This change in tRNA expression levels effectively switches codon optimality and favours the expression of previously non-optimal transcripts. Among the transcripts favoured in amino acid-depleted conditions are those responsible for amino acid biosynthesis and protein recycling. Thus, although numerous changes in transcription and translation do occur to maintain homeostasis in the face of amino acid deprivation, the tRNA pool can change dynamically in response to environmental stresses to adapt to the translational needs of the mRNAs required to restore homeostasis126 (FIG. 5a). In human embryonic kidney cells, charged tRNA pools are altered in response to amino acid starvation to favour the translation of mRNAs involved in protein recycling by the ubiquitin–proteasome pathway, suggesting that adaptation of tRNA pools to the translational demands of proteins involved in homeostasis is conserved across multiple domains of life127. tRNAs are subject to a wide array of post-transcriptional modifications, many of which are required for accurate and efficient translation128,129, but perhaps most intriguingly, there is growing evidence that some of these post-transcriptional modifications at the wobble position of certain tRNA species are sensitive to the cellular milieu130,131. These modifications can be dynamically regulated to bias the codon preferences of the tRNA pool in favour of mRNAs required to mount a robust response to cellular stresses such as oxidative stress132-135 (FIG. 5b).
Figure 5 ∣. Various conditions can alter tRNA pools in different ways to support the translation of mRNAs necessary to maintain homeostasis or to favour the current gene expression programme.
a ∣ Amino acid (AA) depletion can alter the relative charging of synonymous tRNA species125 that favour the production of proteins that help to restore the availability of amino acids, for example, components of the ubiquitin–proteasome system127, b ∣ In conditions of oxidative stress in budding yeast, such as an increase in the levels of reactive oxygen species (ROS), Trm4-dependent methylation of the wobble cytidine in tRNALeu(CAA) is upregulated to favour the translation of mRNAs enriched with TTG Leu codons, such as 60S ribosomal protein L22-A (RPL22A), which is known to confer resistance to ROS132. c ∣ Proliferating cells such as induced pluripotent stem cells and neoplasm-derived cell lines have a tRNA pool that is distinct from cells that have undergone terminal differentiation and that have entered a postmitotic state. This tRNA pool favours the production of proteins involved in translation, DNA replication and the cell cycle over proteins involved in cell–cell adhesion, tissue patterning and multicellular functions139. MEM, minimum essential medium.
Codon usage and cell proliferation.
The deregulation of tRNA pools was found in some cancers136,137, and the advent of ribosome profiling has revealed that translation dynamics are altered in neoplastic cells138. Given the wide-ranging influences of tRNA pools on translation dynamics, there may be a causal relationship between altered tRNA pools and the proliferative potential of cancer cells. A remarkable coherence between the proliferative status of a cell and the composition of its tRNA pool was shown to exist by using microarrays to measure tRNA levels and by using data from histone-targeted chromatin immunoprecipitation followed by sequencing (ChIP–seq) or microarray analysis (ChIP–chip) from the ENCODE projects, as well as genomic Pol III occupancy maps, to identify open chromatin and Pol III activity around tRNA genes in order to infer their transcription status139. The tRNA species that were upregulated in proliferating cells, including in cancer cells, were matched to the codon content of cell-autonomous genes involved in processes such as the cell cycle and cell growth, whereas tRNA species enriched in terminally differentiated cells were matched to the codon content of genes involved in coordinating the arrangement of cell ensembles, such as cell-adhesion genes (FIG. 5c). This finding suggests that cells have the ability to alter their tRNA content by controlling the chromatin structure near tRNA genes126, thereby enabling widespread regulation of entire classes of genes. This mechanism appears to be co-opted in cancer to favour cell type-independent functions of growth and proliferation, underscoring the clinical relevance of codon optimality and tRNA regulation140.
Much of the work demonstrating that tRNA pools are dynamically regulated to support the gene expression needs of the cell was performed before the link between codon content and mRNA stability was fully appreciated. It is therefore unknown to what extent changing tRNA pools affects translation efficiency or transcript stability, although it is likely that both are affected. During the maternal-to-zygotic transition (MZT) in early embryonic development, maternal mRNAs must be cleared rapidly after the onset of zygotic transcription, and this is mediated by the relative codon content of the expressed maternal and zygotic genes9,114. Consistent with findings in yeast, mRNA decay in zebrafish embryos and in Xenopus laevis embryos is strongly positively correlated with relative enrichment in non-optimal codons, where optimal codons are those with enriched cognate tRNA species, underlying the importance of codon bias in development9,114. These findings extend the emerging body of evidence linking lower rates of ribosome translocation to the mRNA decay machinery141.
Conclusions and future perspectives
In this Review, we have discussed the diverse functional consequences of degeneracy in the genetic code. The existence of multiple codons for the same amino acid allows the cell to fine-tune the progress of the ribosome as it translates a message by manipulating the relative concentrations of isoacceptor tRNAs. The tRNA species that are scarce in the cell will take longer to arrive at the A-site of the ribosome and necessarily cause delay in ribosome translocation over the cognate codon, thereby influencing the fidelity of translation and protein folding, the amount of protein produced and the stability of the mRNA. Exactly how the cognate:near-cognate tRNA ratio influences elongation dynamics and the fidelity of the translation apparatus requires more research, especially to understand the more nuanced features of the tRNA pool that influence some of the codon-dependent phenotypes we have discussed. The extent to which codon content will influence any of these processes is crucially dependent on the location of high- or low-codon-optimality stretches within the transcript16,36 and the sequence context of these codons66, along with other factors that have not yet been identified. The heterogeneity and context-dependence of codon effects has undoubtedly contributed to much of the ambiguity in the literature on codon optimality, and there is no reason to think that codons need be the primary determinant of mRNA stability and translation efficiency in all mRNAs, given the plethora of other factors at play in the control of ribosomal dynamics. However, it seems clear from the latest bioinformatics and experimental results that codons can and do markedly affect gene expression in different ways.
As we have emphasized throughout this Review, an emerging consensus on mRNA decay implicates translation dynamics as a key mediator of different mRNA decay processes, including decay related to non-optimal codon content18 and microRNA-mediated decay104,142. The discovery of a specific protein, Dhh1, that constantly monitors ribosome elongation rates in yeast allows us to reframe codon optimality as something more than an epiphenomenal optimization mechanism of the translation of highly expressed genes: it is a mechanism to control mRNA decay rates and, by extension, mRNA levels in the cell.
We have discussed above how dynamic tRNA pools cause codon-mediated changes in translation to drive large-scale changes in gene expression. More work is required to understand how trans-acting mediators of codon-driven mRNA decay function in the context of the ribosome to read out changes in elongation dynamics and how these mediators might be regulated to fine-tune codon effects on decay in response to developmental stages, proliferative states or cellular stress. It is also unknown to what extent trans-acting factors such as Dhh1 influence other codon-mediated effects, including translation efficiency. Nevertheless, the recent string of findings linking codon content to a diverse array of cellular processes, from development and differentiation through proliferation and neoplastic transformation to the response to amino acid starvation and oxidative stress, suggests that synonymous codon usage truly represents an influential secondary genetic code.
Ribosome A-site.
The part of the ribosome where the amino acid-charged tRNA complex initially binds and is recognized by the mRNA codon triplet.
Open reading frame (ORF).
The portion of a transcript with the potential to be translated and yield a protein; it is flanked by a start codon (AUG) and a stop codon (UAG, UAA or UGA).
tRNA adaptation index (tAI).
A normalized measure of how well a codon or a set of codons is adapted to the cellular tRNA pool.
Ribonucleoprotein (RNP).
RNA in association with its binding proteins. In the context of polysome analysis, this excludes the ribosome.
80S.
The fully assembled eukaryotic ribosome, named after its sedimentation coefficient. In the context of polysome analysis, this is taken to include the associated mRNA.
Polysome.
A complex of mRNA and two or more ribosomes. In polysome analysis, the polysome fraction is often further divided into sub-fractions based on sedimentation rate, with faster sedimentation rates corresponding to mRNAs with more ribosomes.
Shotgun proteomics.
A method for assaying the identities and quantities of proteins within a complex protein mixture using high-performance liquid chromatography and mass spectrometry (HPLC–MS).
Coding DNA sequence (CDS).
The portion of a gene or transcript that is translated in the cell to yield a functional protein.
Missense translation errors.
The incorporation of an incorrect amino acid into a protein due to the accommodation of a non-cognate tRNA species during translation.
Codon adaptation index (CAI).
A measure of the extent to which a transcript is biased towards the use of codons enriched in the transcriptome as a whole, over less commonly used synonymous codons.
Maternal-to-zygotic transition (MZT).
The activation of zygotic gene expression, which is accompanied by the degradation of the maternally supplied transcripts.
Acknowledgements
The authors thank the members of the laboratories of K. E. Baker and J.C. for their helpful discussions. Funding was provided by NIGMS to J.C. (GM118018 and GM125086) and to G.H. (GM007250).
Footnotes
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ikemura T Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151, 389–409 (1981). [DOI] [PubMed] [Google Scholar]
- 2.dos Reis M, Savva R & Wernisch L Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 32, 5036–5044 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duret L tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16, 287–289 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Moriyama EN & Powell JR Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 45, 514–523 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Sabi R & Tuller T Modelling the efficiency of codon–tRNA interactions based on codon usage bias. DNA Res. 21, 511–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp PM & Li W-H An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24, 28–38 (1986). [DOI] [PubMed] [Google Scholar]
- 7.Dong H, Nilsson L & Kurland CG Co-variation of trna abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Roth AC Decoding properties of tRNA leave a detectable signal in codon usage bias. Bioinformatics 28, i340–i348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazzini AA et al. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J. 35, 2087–2103 (2016).Codon usage varies between maternal and zygotic mRNAs in X. laevis and zebrafish and contributes to the MZT by targeting maternally loaded transcripts for degradation.
- 10.Rocha EPC Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 14, 2279–2286 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamary JV, Parmley JL & Hurst LD Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7, 98–108 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Gingold H & Pilpel Y Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, 481 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plotkin JB & Kudla G Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novoa EM & Ribas de Pouplana L Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 28, 574–581 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Quax Tessa, E. F., Claassens Nico, J., Söll D& van der Oost J Codon bias as a means to fine-tune gene expression. Mol. Cell 59, 149–161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Presnyak V et al. Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124 (2015).Codon usage within a transcript determines mRNA stability. Codons corresponding to abundant tRNA species are enriched in more stable mRNAs, whereas rare codons are enriched in unstable mRNAs.
- 17.Harigaya Y & Parker R Analysis of the association between codon optimality and mRNA stability in Schizosaccharomyces pombe. BMC Genomics 17, 895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radhakrishnan A et al. The DEAD-box protein Dhh1p couples mRNA decay and translation by monitoring codon optimality. Cell 167, 122.e9–132.e9 (2016).The DEAD-box helicase Dhh1 is a necessary link between non-optimal codon content and RNA decay.
- 19.Li G-W, Oh E & Weissman JS The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad F, Woolstenhulme CJ, Green R & Buskirk AR Clarifying the translational pausing landscape in bacteria by ribosome profiling. Cell Rep. 14, 686–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SL, Lee W, Hottes AK, Shapiro L & McAdams HH Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl Acad. Sci. USA 101, 3480–3485 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galtier N, Piganeau G, Mouchiroud D & Duret L GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159, 907–911 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sørensen MA & Pedersen S Absolute in vivo translation rates of individual codons in Escherichia coli. J. Mol. Biol. 222, 265–280 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Koutmou KS, Radhakrishnan A & Green R Synthesis at the speed of codons. Trends Biochem. Sci. 40, 717–718 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Chu D, Barnes DJ & von der Haar T The role of tRNA and ribosome competition in coupling the expression of different mRNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 39, 6705–6714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15, 205–213 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Ingolia NT, Ghaemmaghami S, Newman JRS& Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian W, Yang J-R, Pearson NM, Maclean C& Zhang J Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 8, e1002603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussmann JA, Patchett S, Johnson A, Sawyer S & Press WH Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLOS Genet. 11, e1005732 (2015).Cycloheximide pretreatment, which is common in ribosome profiling, is shown to markedly distort the true distribution of ribosomes over transcripts, thereby explaining earlier findings of a lack of a relationship between rare codons and ribosome density.
- 30.Gerashchenko MV & Gladyshev VN Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 42, e134 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardin J et al. Measurement of average decoding rates of the 61 sense codons in vivo. ELife 3, e03735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lareau LF, Hite DH, Hogan GJ & Brown PO Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. ELife 3, e01257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg DE et al. Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14, 1787–1799 (2016).This paper shows how ribosome sequencing data can be used to gain a quantitative understanding of translation, revealing that ribosomes do in fact spend more time on average over non-optimal codons.
- 34.Chekulaeva M & Landthaler M Eyes on translation. Mol. Cell 63, 918–925 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Iwasaki S & Ingolia NT Seeing translation. Science 352, 1391–1392 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Yu C-H et al. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59, 744–754 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan X, Hoek Tim, A., Vale Ronald, D. & Tanenbaum Marvin, H. Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016).Leveraging a novel system for probing translation rates of single molecules in real time, this study reveals that codon composition has a marked impact on elongation rates in vivo.
- 38.Akashi H Synonymous codon usage in Drosophila melanogaster. natural selection and translational accuracy. Genetics 136, 927–935 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell JR & Moriyama EN Evolution of codon usage bias in Drosophila. Proc. Natl Acad. Sci. USA 94, 7784–7790 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urrutia AO & Hurst LD Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics 159, 1191–1199 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z & Nielsen R Mutation-selection models of codon substitution and their use to estimate selective strengths on codon usage. Mol. Biol. Evol. 25, 568–579 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Gustafsson C, Govindarajan S & Minshull J Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Burgess-Brown NA et al. Codon optimization can improve expression of human genes in Escherichia coli: a multi-gene study. Protein Expr. Purif. 59, 94–102 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Lithwick G & Margalit H Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 13, 2665–2673 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghaemmaghami S et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Lu P, Vogel C, Wang R, Yao X & Marcotte EM Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Li G-W, Burkhardt D, Gross C& Weissman Jonathan, S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel C et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudla G, Murray AW, Tollervey D & Plotkin JB Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman DB, Church GM & Kosuri S Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Tuller T et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141, 344–354 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Gu W, Zhou T & Wilke COA Universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput.Biol. 6, e1000664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsao D, Shabalina SA, Gauthier J, Dokholyan NV & Diatchenko L Disruptive mRNA folding increases translational efficiency of catechol-O-methyltransferase variant. Nucleic Acids Res. 39, 6201–6212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morisaki T et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Wu B, Eliscovich C, Yoon YJ & Singer RH Translation dynamics of single mRNAs in live cells an neurons. Science 352, 1430–1435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah P, Ding Y, Niemczyk M, Kudla G& Plotkin Joshua, B. Rate-limiting steps in yeast protein translation. Cell 153, 1589–1601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarai Y, Margaliot M & Tuller T On the ribosomal density that maximizes protein translation rate. PLoS ONE 11, e0166481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitarai N, Sneppen K & Pedersen S Ribosome collisions and translation efficiency: optimization by codon usage and mrna destabilization. J. Mol. Biol. 382, 236–245 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Potapov I, Mäkelä J, Yli-Harja O & Ribeiro AS Effects of codon sequence on the dynamics of genetic networks. J. Theor. Biol. 315, 17–25 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Chu D et al. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34 (2014).This paper shows that non-optimal codon usage can create a backlog of ribosomes that effectively feeds back on translation initiation by preventing new ribosome assembly near the start codon.
- 61.Rosenberg AH, Goldman E, Dunn JJ, Studier FW & Zubay G Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol 175, 716–722 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciandrini L, Stansfield I & Romano MC Ribosome traffic on mRNAs maps to gene ontology: genome-wide quantification of translation initiation rates and polysome size regulation. PLoS Comput. Biol. 9, e1002866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Curran JF Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 23, 683–688 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulmer M The selection-mutation-drift theory of synonymous codon usage. Genetics 129, 897–907 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartl DL, Moriyama EN & Sawyer SA Selection intensity for codon bias. Genetics 138, 227–234 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamble CE, Brule CE, Dean KM, Fields S& Grayhack EJ Adjacent codons act in concert to modulate translation efficiency in yeast. Cell 166, 679–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coleman JR et al. Virus attenuation by genome-scale changes in codon pair bias. Science 320, 1784–1787 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charneski CA & Hurst LD Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 11, e1001508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabi R & Tuller T A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics 16, S5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu J & Deutsch C Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pavlov MY et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl Acad. Sci. 106, 50–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson DN & Beckmann R The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 21, 274–282 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Peil L et al. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl Acad. Sci. 110, 15265–15270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purvis IJ et al. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. J. Mol. Biol. 193, 413–417(1987). [DOI] [PubMed] [Google Scholar]
- 75.Thanaraj T & Argos P Protein secondary structural types are differentially coded on messenger RNA. Protein Sci. 5, 1973–1983 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Komar AA, Lesnik T & Reiss C Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 462, 387–391 (1999). [DOI] [PubMed] [Google Scholar]
- 77.Zhang G, Hubalewska M & Ignatova Z Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol. 16, 274–280 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Cortazzo P et al. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 293, 537–541 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Buhr F et al. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell 61, 341–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spencer PS, Siller E, Anderson JF & Barral JM Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 422, 328–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pechmann S & Frydman J Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 20, 237–243 (2013).The interdependence of tRNA supply and demand helps to shape codon optimality, and optimal and non-optimal codons are distributed in stereotyped patterns throughout elements encoding protein secondary structures.
- 82.Zhou M Wang T, Fu J, Xiao G & Liu Y Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol. Microbiol. 97, 974–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang G & Ignatova Z Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS ONE 4, e5036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saunders R & Deane CM Synonymous codon usage influences the local protein structure observed. Nucleic Acids Res. 38, 6719–6728 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chaney JL et al. Widespread position-specific conservation of synonymous rare codons within coding sequences. PLOS Comput. Biol. 13, e1005531 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stoletzki N & Eyre-Walker A Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 24, 374–381 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Drummond DA & Wilke CO Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134, 341–352 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou T, Weems M & Wilke CO Translationally optimal codons associate with structurally sensitive sites in proteins. Mol. Biol. Evol. 26, 1571–1580 (2009).Optimal codons are found disproportionately in structurally important sites in proteins, often within the solvent-isolated core. This tendency is conserved across multiple domains of life.
- 89.Dix DB & Thompson RC Codon choice and gene expression: synonymous codons differ in translational accuracy. Proc. Natl Acad. Sci. USA 86, 6888–6892 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thomas LK, Dix DB & Thompson RC Codon choice and gene expression: synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc. Natl Acad. Sci. USA 85, 4242–4246 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kramer EB & Farabaugh PJ The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang Y, Koonin EV, Lipman DJ& Przytycka TM Selection for minimization of translational frameshifting errors as a factor in the evolution of codon usage. Nucleic Acids Res. 37, 6799–6810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kramer EB, Vallabhaneni H, Mayer LM& Farabaugh PJ A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 16, 1797–1808 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fluitt A, Pienaar E & Viljoen H Ribosome kinetics and aa-tRNA competition determine rate and fidelity of peptide synthesis. Comput. Biol. Chem. 31, 335–346 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah P & Gilchrist MA Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLOS Genet. 6, e1001128 (2010).The ratio of cognate:near-cognate tRNA species is much more influential in dictating translation fidelity than the abundance of cognate tRNAs, as high levels of near-cognate tRNAs will compete for incorporation even if the cognate tRNA is also highly abundant.
- 96.Warnecke T & Hurst LD GroEL dependency affects codon usage — support for a critical role of misfolding in gene evolution. Mol. Syst. Biol. 6, 340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jacobson GN & Clark PL Quality over quantity: optimizing co-translational protein folding with non-‘optimal’ synonymous codons. Curr. Opin. Struct. Biol. 38, 102–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Komar AA A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 34, 16–24 (2009).This paper provides a comprehensive appraisal of the ways in which codon usage can influence protein folding dynamics, with an emphasis on the interplay between codon usage, tRNA pools and co-translational folding.
- 99.Angov E Codon usage: nature’s roadmap to expression and folding of proteins. Biotechnol. J. 6, 650–659 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rodnina MV The ribosome in action: tuning of translational efficiency and protein folding. Protein Sci. 25, 1390–1406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chaney JL & Clark PL Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 44, 143–166 (2015). [DOI] [PubMed] [Google Scholar]
- 102.Wang Y et al. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA 99, 5860–5865 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fabian MR, Sonenberg N & Filipowicz W Regulation of mRNA translation and stability by microRNAs. Annu. Rev Biochem. 79, 351–379 (2010). [DOI] [PubMed] [Google Scholar]
- 104.Djuranovic S, Nahvi A & Green R miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grosset C et al. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103, 29–40 (2000). [DOI] [PubMed] [Google Scholar]
- 106.Geisberg Joseph, V., Moqtaderi Z, Fan X, Ozsolak F & Struhl K Global analysis of mRNA isoform half-lives reveals stabilizing and destabilizing elements in yeast. Cell 156, 812–824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen C-YA & Shyu A-B Emerging themes in regulation of global mRNA turnover in cis. Trends Biochem. Sci. 42, 16–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoekema A, Kastelein RA, Vasser M & de Boer HA Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol. Cell. Biol. 7, 2914–2924 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caponigro G, Muhlrad D & Parker R A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13, 5141–5148 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu W, Sweet TJ, Chamnongpol S, Baker KE& Coller J Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461, 225–229 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sweet T, Kovalak C & Coller J The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 10, e1001342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carlini DB Context-dependent codon bias and messenger RNA longevity in the yeast transcriptome. Mol. Biol. Evol. 22, 1403–1411 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Boël G et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 529, 358–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mishima Y & Tomari Y Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61, 874–885 (2016). [DOI] [PubMed] [Google Scholar]
- 115.Tuller T, Kupiec M & Ruppin E Determinants of protein abundance and translation efficiency in S. cerevisiae. PLoS Comput. Biol. 3, e248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Akashi H Translational selection and yeast proteome evolution. Genetics 164, 1291–1303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu Y et al. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495, 116–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou M et al. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495, 111–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Najafabadi HS, Goodarzi H & Salavati R Universal function-specificity of codon usage. Nucleic Acids Res. 37,7014–7023 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Olivares-Hernández R, Bordel S & Nielsen J Codon usage variability determines the correlation between proteome and transcriptome fold changes. BMC Syst. Biol. 5, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Plotkin JB, Robins H & Levine AJ Tissue-specific codon usage and the expression of human genes. Proc. Natl Acad. Sci. USA 101, 12588–12591 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dittmar KA, Goodenbour JM & Pan T Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Camiolo S, Farina L & Porceddu A The relation of codon bias to tissue-specific gene expression in Arabidopsis thaliana. Genetics 192, 641–649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Elf J, Nilsson D, Tenson T & Ehrenberg M Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300, 1718–1722 (2003). [DOI] [PubMed] [Google Scholar]
- 125.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M & Pan T Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6, 151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wohlgemuth SE, Gorochowski TE & Roubos JA Translational sensitivity of the Escherichia coli genome to fluctuating tRNA availability. Nucleic Acids Res. 41, 8021–8033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saikia M et al. Codon optimality controls differential mRNA translation during amino acid starvation. RNA 22, 1719–1727 (2016).The translation of gene products involved in protein recycling and amino acid mobilization in conditions of amino acid deprivation is maintained through the use of rare codons in ubiquitin–proteasome mRNAs and the uncharacteristic stability of the levels of charged tRNAs complementary to these rare codons.
- 128.Nedialkova Danny, D. & Leidel Sebastian, A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zinshteyn B & Gilbert WV Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9, e1003675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deng W et al. Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet 11, e1005706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ranjan N & Rodnina MV tRNA wobble modifications and protein homeostasis. Translation 4, e1143076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chan CT et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012).Under conditions of oxidative stress, Trm4-dependent methylation of the wobble cytidine in tRNALeu(CAA) is upregulated, thereby enhancing the translation of the TTG Leu codon, which is enriched in genes required for effective resistance to oxidative conditions. A defect in this tRNA modification pathway increases susceptibility to oxidative damage.
- 133.Chan CTY et al. a quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fernández-Vázquez J et al. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9, e1003647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Endres L, Dedon PC & Begley TJ Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 12, 603–614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pavon-Eternod M et al. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37, 7268–7280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shin S-H et al. Implication of leucyl-tRNA synthetase 1 (LARS1) over-expression in growth and migration of lung cancer cells detected by siRNA targeted knock-down analysis. Exp. Mol. Med. 40, 229–236 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hsieh AC et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gingold H et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).Proliferating cells express a tRNA pool that favours the translation of pro-growth mRNAs, whereas differentiated cells express a different tRNA pool, which favours the translation of mRNAs that support cell–cell adhesion and communication.
- 140.Grewal SS Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim. Biophys. Acta 1849, 898–907 (2015). [DOI] [PubMed] [Google Scholar]
- 141.Richter Joel, D. & Coller J Pausing on polyribosomes: make way for elongation in translational control. Cell 163, 292–300 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bazzini AA, Lee MT & Giraldez AJ Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]