Abstract
Soil salinity is a major problem in agriculture. However, crop growth and productivity can be improved by the inoculation of plants with beneficial bacteria that promote plant growth under stress conditions such as high salinity. Here, we evaluated 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity and trehalose accumulation of the plant growth promoting bacterium Pseudomonas sp. UW4. Mutant strains (mutated at acdS, treS, or both) and a trehalose over-expressing strain (OxtreS) were constructed. The acdS mutant was ACC deaminase minus; the treS- strain significantly decreased its accumulation of trehalose, and the double mutant was affected in both characteristics. The OxtreS strain accumulated more trehalose than the wild-type strain UW4. Inoculating tomato plants subjected to salt stress with these strains significantly impacted root and shoot length, total dry weight, and chlorophyll content. The evaluated parameters in the single acdS and treS mutants were impaired. The double acdS/treS mutant was negatively affected to a greater extent than the single-gene mutants, suggesting a synergistic action of these activities in the protection of plants against salt stress. Finally, the OxtreS overproducing strain protected tomato plants to a greater extent under stress conditions than the wild-type strain. Taken together, these results are consistent with the synergistic action of ACC deaminase and trehalose in Pseudomonas sp. UW4 in the protection of tomato plants against salt stress.
Keywords: plant growth-promoting bacteria, saline soils, ACC deaminase, trehalose synthase, ethylene
Introduction
Salinity of agricultural soils is one of the main problems for farmers, since growth and plant production can be adversely affected. Soluble salts decrease the fertility of soils, producing osmotic stress, affecting water balance, and ion homeostasis. This alters the plant’s hormonal status, disturbing transpiration, nutrient acquisition and photosynthesis, among others (Munns and Tester, 2008; Ilangumaran and Smith, 2017). Plants contain various mechanisms to cope with salt stress, including a microbiome associated with the rhizosphere, phyllosphere, and endosphere (Orozco-Mosqueda et al., 2018). A group of organisms that stand out in this plant microbiome are the beneficial bacteria known as plant growth-promoting bacteria (PGPB). These bacteria, such as Azospirillum, Arthrobacter, Azotobacter, Bacillus, Burkholderia, Enterobacter, and Pseudomonas, have been shown to improve salt tolerance in several plant crops (Forni et al., 2017; Bharti and Barnawal, 2019).
Among the arsenal of mechanisms used by PGPB is the presence of the enzyme 1-aminociclopropane-1-carboxylase (ACC) deaminase, which converts ACC into ammonia and α-ketobutyrate. The ACC is a precursor of ethylene in all higher plants, and both ACC and ethylene typically increase in plants when they are under stress. Therefore, the ACC deaminase activity is one of the key traits used by PGPB to decrease ethylene levels under a wide range of stress conditions, including salinity (Mayak et al., 2004; Glick et al., 2007). Direct evidence for this mechanism comes from a study with acdS mutants of Pseudomonas sp. UW4 (ACC deaminase minus), which have lost the ability to promote the elongation of canola roots under gnotobiotic conditions (Li et al., 2000), and in presence of salt, are unable to promote growth of canola plants as the wild-type UW4 strain, which significantly improved plant growth (Cheng et al., 2007). The bacterium Pseudomonas sp. UW4 was isolated from the rhizosphere of common reeds and its genome is completely sequenced. In addition, the UW4 strain has been widely studied for its ability to promote plant growth under different environmental stresses, such as flooding, high concentrations of salt, cold, heavy metals, drought, and biocontrol of phytopathogens (Glick et al., 2007; Duan et al., 2013).
Another mechanism utilized by PGPB to reduce stress and promote growth in plants in the presence of both drought and high levels of salt is the production of trehalose (Suárez et al., 2008). Trehalose is a non-reducing disaccharide in which the two glucose units are linked via an α,α-(1,1)-glycosidic bond. This disaccharide has been isolated from all domains of life including plants, animals, fungi, yeast, archaea, and bacteria (Avonce et al., 2006; Chandra et al., 2011). Trehalose is also industrially produced as it is used in the food, cosmetics, and pharmaceutical industries (Lee et al., 2005; Wang et al., 2014). In addition, this disaccharide can play important and different roles in biological cells, such as, as a signaling molecule, reserve carbohydrate, and protectant against various stresses (e.g., drought, cold, and salt stress; Fernandez et al., 2010). Interestingly, the trehalose content has been strongly associated with the ability of some microorganisms to survive in harsh environments (Kandror et al., 2002; Cheng et al., 2007; Suárez et al., 2008).
Up to five different routes of trehalose biosynthesis have been described, most which include several steps (Paul et al., 2008; Fernandez et al., 2010). The only known route for trehalose synthesis in a single reaction step is the TreS pathway, where trehalose can be synthesized from maltose by trehalose synthase (TreS). Derived from the analysis of the genomic sequence of the strain UW4, it was reported that there may be two routes for the synthesis of trehalose in this bacterium, including treY/treZ, and treS (Duan et al., 2013). TreS interconverts maltose and alpha, alpha-trehalose by transglucosylation. This enzyme has also been found in a wide variety of bacterial species, such as Azospirillum brasilense, Rhizobium etli, Corynebacterium glutamicum, Arthrobacter aurescens, Meiothermus ruber, and Pseudomonas stutzeri (Avonce et al., 2006; Rodríguez-Salazar et al., 2009; Wang et al., 2014), consistent with its importance in survival under abiotic stress. In addition, a higher level of trehalose accumulation has been found in root nodules of Medicago truncatula and Phaseolus vulgaris in response to drought and salt stress, suggesting a key role of trehalose in signaling during plant-bacteria interactions by promoting plant growth, yield, and better adaptation to harsh conditions (López et al., 2008; Suárez et al., 2008). In the study reported herein, we constructed and tested a series of bacterial mutants in order to analyze the synergistic effects of ACC deaminase activity and trehalose accumulation on the ability of the plant growth-promoting bacterium Pseudomonas sp. UW4 to protect tomato (Lycopersicon esculentum cv. Saladette) plants against salt stress during growth.
Materials and Methods
Bacterial Strains and Media
Wild-type Pseudomonas sp. strain UW4 (Duan et al., 2013) was grown and maintained aerobically at 30°C in Nutrient Agar (NA) (BD Bioxon). The Escherichia coli strain was grown in Luria-Bertani (LB) medium (BD Bioxon) at 37°C. All strains were routinely maintained at 4°C. Antibiotics were added to the media when needed at the following concentrations (in μg ml-1): Carbenicillin (Cb), 100; Chloramphenicol (Cm), 20; Kanamycin (Km), 50; and Tetracycline (Tc), 10. For selection in cloning experiments, X-Gal (5-bromo-4-chloro-3-indolyl-D-galactoside) (Promega) was added to LB plates at 30 μg/ml.
Construction of Pseudomonas sp. UW4 Mutants
The ACC deaminase minus mutant (acdS-) was previously constructed and characterized (Li et al., 2000). The wild-type Pseudomonas sp. UW4 strain was used to generate a mutation in the treS gene. First, a plasmid with the treS gene, interrupted by a chloramphenicol resistance cassette from pSUP5011 (in the single BamHI site of the treS sequence), was constructed (pJQ200SK/treS::Cmr) and transformed into the genome of the UW4 strain. Subsequently, clones were selected for a homologous double-crossover event that occurred between the wild-type treS gene and the disrupted treS::Cmr gene on the replacement vector. The positive counterselection of clones with subsequent loss of the replacement vector was due to the presence of the sacB gene in the pJQ200SK, and therefore, sucrose sensitivity (Gay et al., 1985). In order to generate an acdS-/treS- double mutant, the aforementioned process was carried out with the exception that, instead of using the wild-type UW4 strain, the acdS- mutant was used. The double mutant was Tcr and Cmr. In all cases, the mutants were checked for gene disruptions by PCR and DNA sequencing. To complement the treS- mutant, a broad-host-range cloning vector (pBBR1MCS-2) was used to carry the functional treS (pBBR1-treS) and express it under a constitutive promoter (derepressed P-lac) of the plasmid. Since the plasmid was a multicopy vector, the respective derivative strain over-expressed the treS gene and was named OxtreS strain. Procedures for genomic DNA isolation, plasmid isolation, gene cloning, and E. coli transformation were carried out using standard protocols (Sambrook and Russel, 2001). Table 1 shows all strains and plasmids used in this work.
Table 1.
Bacterial strains and plasmids used in this work.
| Strain/plasmid | Relevant phenotype | References or source |
|---|---|---|
| Escherichia coli DH5α | SupE44ΔlacU169 (f80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1, Nxr | Life Technologies |
| Escherichia coli TOP10 | F- mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15ΔlacX74 deoR recA1 araD139Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| pBluescriptII SK | ColE1 mcs-lacZ, Apr | Stratagene |
| pBBR1MCS-2 | Kmr | Kovach et al., 1995 |
| pJQ200SK | Suicide vector with lacZα carrying sacB, Gmr | Quandt and Hynes, 1993 |
| pSUP5011 | Cmr | Simon et al., 1983 |
| pBBR1-treS | pBBR1MCS-2 containing treS gene | This work |
| pJQ200SK/treS::Cmr | pJQ200SK containing treS::Cmr | This work |
| Pseudomonas sp. UW4 | Wild-type | Duan et al., 2013 |
| UW4 acdS- | acdS::Tcr | Li et al., 2000 |
| UW4 treS- | treS::Cmr | This work |
| UW4 acdS-/treS- | acdS::Tcr/treS::Cmr | This work |
| UW4 OxtreS | treS overexpressing, cloned in pBBR1MCS-2 | This work |
Survival Experiments of Bacterial Strains on Salt Stress
Survival of Pseudomonas sp. strain UW4, mutants (acdS, treS, and acdS/treS), and the trehalose over-expressing strain (OxtreS) derivative under conditions of salt stress (NaCl) was measured after 48 h of growth on minimal medium (M9) (Sigma), as colony forming units per ml. A pre-inoculum with 1.0 × 106 (± 0.01 × 106) of CFU ml-1 was used for each strain. The minimal medium was supplemented with 0, 0.2, or 0.8 M NaCl, and cultures were incubated with shaking for 48 h at 30°C.
Determination of ACC Deaminase Activity
Plant growth-promoting bacteria containing the ACC deaminase enzyme are able to use ACC as a sole nitrogen source. Therefore, the determination of the ACC deaminase activity in the strains generated in this work was evaluated as previously described by Penrose and Glick (2003). This method quantifies the amount of α-ketobutyrate produced as a result of the cleavage of ACC by ACC deaminase. The final ACC deaminase activity was expressed in μmol α-ketobutyrate mg protein-1 h-1. Wild-type strain Pseudomonas sp. UW4 was used as a positive control in all replicates.
Quantification of IAA Production
The determination of indole-3-acetic acid (IAA) production in bacterial strains used in this work was performed by Gas Chromatography and Mass Spectrometry, as previously reported by Hernández-León et al. (2015). Nevertheless, some modifications for specific estimation in bacteria were applied. The identity of IAA was confirmed by comparison of the retention time in the bacterial extracts with samples of the pure IAA standard (Sigma). For estimation of the IAA amount produced by strains, an individual calibration curve was constructed. The IAA determinations were done in triplicate.
Trehalose Quantification
The accumulation of the disaccharide trehalose in wild-type UW4, the isogenic mutants, and the trehalose overexpressing strain was measured by high performance liquid chromatography (HPLC). Briefly, cell cultures of 250 ml were prepared in minimal medium (M9) with 4% glycerol and the culture was left for 48 h under agitation at 30°C. The medium supernatant was filtered (MF-Millipore membrane filters 0.22 μm) and 1.5 ml of sterile deionized water was added. Trehalose concentration was determined by HPLC with a carbohydrate analysis column Aminex HPX-87C (Bio-Rad Labs, Richmond, CA, United States). A standard of pure trehalose (Sigma) was employed as a reference to identify the peak of the disaccharide.
Evaluation of Plant Growth-Promotion in the Presence of Salt Stress
Greenhouse experiments with tomato plants (L. esculentum cv. Saladette) were performed according to the methods described by Rojas-Solís et al. (2018). The experiments were carried out in pots (6 cm tall × 5 cm wide) with sterile peat moss (Sphaigne, Canada), with or without irrigation with a salt solution (0.2 M NaCl). Previous screening experiments with different salt stresses (0, 0.1, 0.2, 0.4, and 0.8 M NaCl) demonstrated that a concentration of 0.2 M of NaCl was considered a saline stress for tomato plants after comparing the growth and chlorophyll parameters of these plants with control plants devoid of salt stress. Subsequently, tomato seeds were germinated in vitro, and after 1 week, seedlings of the same size were selected and transplanted into pots (one plant per pot). Bacterial inoculants dissolved in sterile deionized water were applied every week after pot transplantation according to the experimental design, which also included treatments without bacterial inoculations. The concentration of bacterial inoculants was adjusted such that their optical density at 600 nm was 1 (∼0.75–1 × 108). The plants were irrigated every 3 days with deionized water throughout the whole experiment.
Each of the experimental treatments (Control, Control+NaCl, Control+each of the 5 strains, and Control+NaCl+each of the 5 strains) included 12 plants. Since the experiment was repeated twice, the total experimental units were 288. The effect of each of the bacterial inoculants on the root length, aerial parts, total dry weight, and chlorophyll concentration was evaluated after 5 weeks of plant growth. The chlorophyll concentration was measured in three leaves from each plant, as previously reported by Rojas-Solís et al. (2018).
Statistical Analysis
As mentioned above, plant growth promoting experiments were performed twice. The results were analyzed using Statistica 6.0 software (StatSoft Inc., 2001). The Student’s t-test was used to compare the means of two groups, and the ANOVA and Duncan’s means separation test were used for multiple comparisons (p < 0.05).
Results
Survival of Wild-Type and Mutants on Salt Stress
Survival of the wild-type strain Pseudomonas sp. UW4, the isogenic mutants (acdS, treS, and acdS/treS), and the trehalose over-expressing strain (OxtreS) derivative were evaluated in two salt concentrations: 0.2 and 0.8 M NaCl (Table 2). The results show that, in all strains, very little significant (p < 0.05) decrease in the survival was observed at 0.2 M NaCl. However, at 0.8 M, a decrease in the survival of all strains was noted. It should be noted that in mutants treS and acdS/treS, the decrease in survival was more marked, and a four- to fivefold decrease in survival relative to the wild-type was observed.
Table 2.
Survival of Pseudomonas sp. strain UW4, mutants, and overexpressing strain derivatives.
| UW4 derivative strains | Survival without saline stress (0 M NaCl) | Survival on saline stress (0.2 M NaCl) | Survival on saline stress (0.8 M NaCl) |
|---|---|---|---|
| Wild-type | 2.2 × 108 ( ± 0.23 × 108) a | 1.2 × 108 ( ± 0.16 × 108) a | 2.8 × 106 ( ± 0.39 × 106) b |
| acdS- | 1.1 × 108 ( ± 0.18 × 108) a | 0.8 × 108 ( ± 0.68 × 108) a | 4.5 × 104 ( ± 0.24 × 104) c |
| treS- | 0.9 × 107 ( ± 0.77 × 107) b | 0.5 × 107 ( ± 0.22 × 107) b | 1.8 × 103 ( ± 0.54 × 103) c |
| acdS-/treS- | 0.2 × 107 ( ± 0.48 × 107) b | 0.1 × 107 ( ± 0.21 × 107) b | 3.8 × 102 ( ± 0.39 × 102) d |
| OxtreS | 3.2 × 108 ( ± 0.13 × 108) a | 1.0 × 108 ( ± 0.13 × 108) a | 2.1 × 107 ( ± 0.23 × 107) b |
Survival was measured after 48 h of growth in minimal media as colony forming units per ml; pre-inoculum of each strain contained 1.0 × 106 ( ± 0.01 × 106) of CFU ml-1. The standard deviation is shown in parentheses (±). Different letters indicate significant differences at α = 0.05.
Analysis of ACC Deaminase Activity and IAA Synthesis
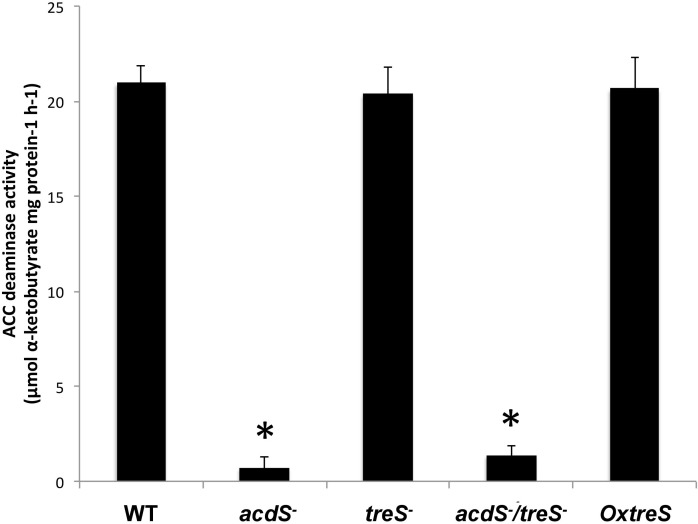
The ACC deaminase activity was analyzed either in strain UW4 or the ACC deaminase minus mutant. The wild-type strain showed the ability to use ACC as a sole source of nitrogen, while the adcS- strain showed no ACC deaminase activity (Figure 1). The mutant strains treS and OxtreS did not show any change in ACC deaminase activity, while the double mutant acdS/treS did not show any ACC deaminase activity.
FIGURE 1.
Evaluation of ACC (1-aminocyclopropane-1-carboxylate) deaminase activity in wild-type (WT) bacterium Pseudomonas sp. UW4; its derivative mutant strains in the acdS, treS, and acdS/treS genes; and the overexpressing OxtreS (Trehalose synthase) strain. The figure represents the results of at least three independent experiments. Asterisks indicate that the means of the samples are different from the WT at P < 0.05.
When monitoring the production of indoleacetic acid in the analyzed mutants, no significant changes were observed compared to the wild-type strain of Pseudomonas sp. UW4. The results of this analysis are shown in Table 3.
Table 3.
Indole acetic acid (IAA) production of Pseudomonas sp. UW4 and derivative strains.
| UW4 derivative strains | IAA production (μg/ml) |
|---|---|
| Wild-type | 3.13 ± 0.6 |
| acdS- | 2.92 ± 0.4 |
| treS- | 3.08 ± 0.5 |
| acdS-/treS- | 2.55 ± 0.9 |
| OxtreS | 2.89 ± 0.7 |
The standard deviation is shown in parentheses (±). Statistical analysis indicated no significant differences at α = 0.05.
Production of Trehalose
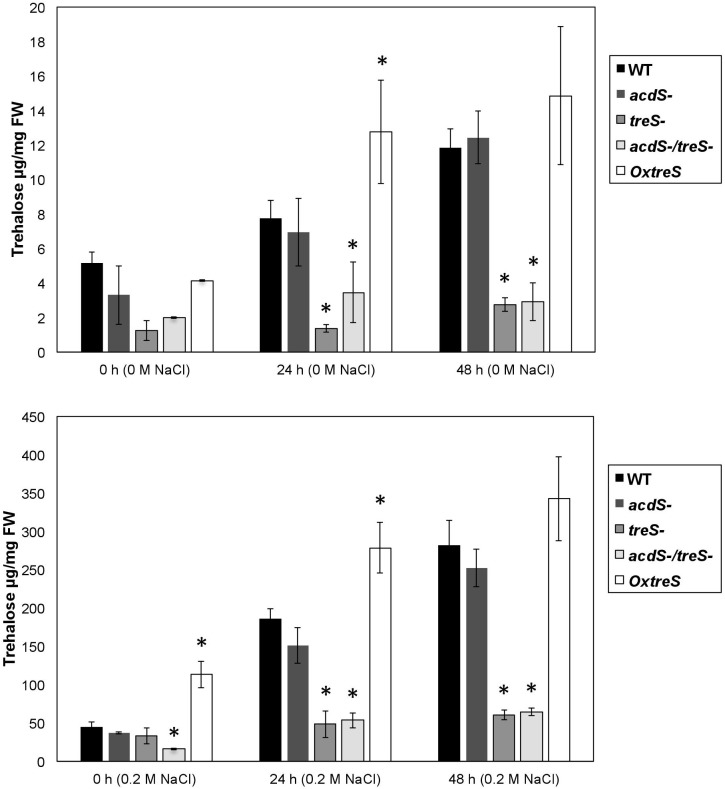
The accumulation of trehalose after 0, 24, and 48 h without salt conditions can be observed in Figure 2A. Specifically, the wild-type and the ACC deaminase minus mutant strains increased trehalose production at 24 and 48 h of growth, while OxtreS showed an even greater accumulation than the other strains, even in unsalted media. On the other hand, the treS and acdS/treS mutants significantly decreased their accumulation of trehalose compared with the strains not affected in the treS gene.
FIGURE 2.
Analysis of trehalose accumulation in wild-type (WT) Pseudomonas sp. UW4; its derivative mutants acdS, treS, and acdS/treS; and the overexpressing OxtreS (OxtreS) strain without (Upper panel) and with salt (Lower panel). The figures exhibit the results of at least three independent experiments. Asterisks indicate that the means of the samples are different from the WT at P < 0.05.
In Figure 2B, the quantification of trehalose is observed in growth media supplemented with 0.2 M NaCl. It should be noted that the accumulation of trehalose in the overexpressing strain was greater than that in the other strains, including the wild-type strain, which increased its accumulation after 24 and 48 h. The treS and acdS/treS mutants also exhibited a slight increase in the accumulation of trehalose in the presence of salt; however, this increase was less marked than in the wild-type strain. This suggests that there are other routes for the production of trehalose in Pseudomonas sp. UW4; nevertheless, the mutation in the treS gene significantly reduced the production of trehalose under high-salinity conditions, which correlates with a lower survival of bacteria that contained this mutation in media with NaCl (Table 2).
Tomato Growth-Promotion on Salt Stress
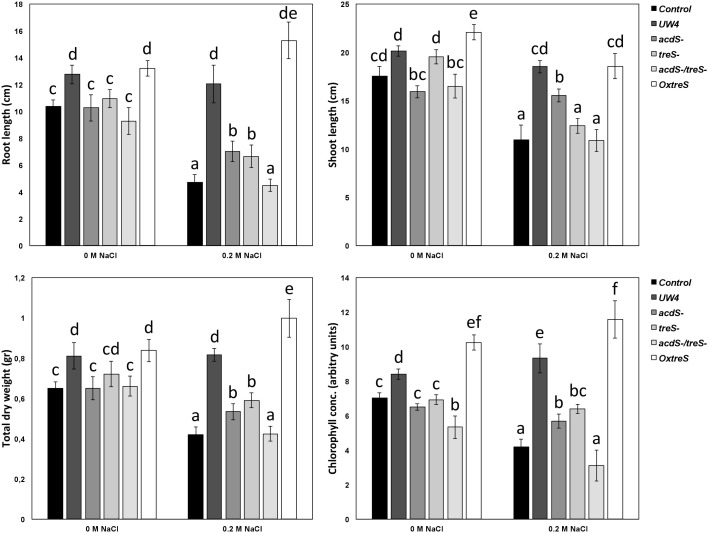
Bacterial ACC deaminase activity and trehalose production has been evaluated (separately) in several studies (Li et al., 2000; Cheng et al., 2007; Suárez et al., 2008). However, in this study, the synergy of both processes in promoting tomato plant growth was evaluated using plant the growth-promoting strain Pseudomonas sp. UW4 and its different mutants in pot experiments. The wild-type strain UW4 maintained its growth-promoting capacities under conditions of salt stress in tomato plants by increasing plant root and shoot length, total dry weight, and chlorophyll content to levels similar to those in plants that were not subjected to NaCl stress (Figure 3). The acdS and treS mutants showed a similar trend (to one another) when used to inoculate plants under salt stress, failing to improve the parameters evaluated in tomato plants. Surprisingly, plants inoculated with the double mutant showed a beneficial effect in three parameters, namely root length, total dry weight, and chlorophyll content, showing similar values to control plants subjected to uninoculated saline stress. In addition, inoculation of plants with the double mutant resulted in lower values than with plants inoculated with each of the single acdS and treS mutants. Finally, the inoculation of the OxtreS strain showed a significant improvement in root length (greater than the wild-type UW4 strain), whereas for the other evaluated parameters, it showed beneficial effects that were statistically similar to those observed for the wildtype UW4 strain (Figure 4).
FIGURE 3.
Evaluation of tomato (Lycopersicon esculentum cv Saladette) growth promoting effects by the inoculation of Pseudomonas sp. UW4 WT, single acdS and treS mutants, double mutant acdS/treS, and the overexpressing OxtreS (TreS) strain. Bars represent the mean ± SE values (n = 12). Different letters indicate significant differences (p < 0.05; Duncan’s multiple range test).
FIGURE 4.
Composite picture of tomato (L. esculentum cv Saladette) plants inoculated with the indicated bacterial strains. Control plants contained no salt or bacteria. C+NaCl (0.2 M) indicates plants irrigated with salt. See the Section “Materials and Methods” for details of the experiment. The rest of the panels show plants + salt (0.2 M) + each of the strains (WT Pseudomonas sp. UW4, acdS-, treS-, acdS- /treS-, and OxtreS).
Discussion
Organisms have evolved different mechanisms to counteract the damaging effects of different adverse environmental conditions, such as soil salinity, which is an enormous problem in agriculture. During salt stress, plants increase their ethylene synthesis, thereby inhibiting their growth and productivity (Glick et al., 2007). One strategy to reduce ethylene levels in plants has been the inoculation of plants with bacteria that contain ACC deaminase activity. Thus, it has been observed that bacteria of different genera, such as Achromobacter, Azospirillum, Brevibacterium, Bacillus, or Pseudomonas, can mitigate the toxic effects of high-salinity, increasing biomass, chlorophyll content, and production (Bacilio et al., 2004; Mayak et al., 2004; Cheng et al., 2007; Siddikee et al., 2011; Ali et al., 2014). However, not all bacteria that help mitigate the toxic effects of salt on plants contain the genetic machinery essential for ACC deaminase activity (Blaha et al., 2006; Duan et al., 2009; Ramadoss et al., 2013); other beneficial bacterial mechanisms can help to mitigate the inhibitory effects of high salt. For example, PGPB that produce IAA under saline conditions may supply an additional amount of this phytohormone to plants. The supplementary IAA may help to stimulate root growth and partially reverse the growth inhibiting effects of salt stress in both shoot and root growth, as well as improving other physiological parameters like chlorophyll content (Duca et al., 2018). In this regard, the mutants constructed here did not show any impairment in IAA synthesis, since all of them showed similar production when compared to the wild-type strain (Table 3). Therefore, the beneficial role of IAA (or other compounds) still produced by the mutants cannot be excluded from the low level of beneficial effects observed in tomato plants during inoculation.
Another relevant and widely studied mechanism of how bacteria facilitate plant growth during salt and drought stress is the production of metabolites such as trehalose (Avonce et al., 2006). Trehalose is a non-reducing disaccharide that is widely present in different groups of organisms and plays diverse roles such as an energy source molecule, in addition to being important during osmotic, heat, and desiccation stress tolerance (Iordachescu and Imai, 2008). Moreover, trehalose synthesis is also triggered by salinity stress in many rhizobacteria (Suárez et al., 2008) and the importance of trehalose in the osmostress response has been well established in several rhizobia (Domínguez-Ferreras et al., 2009; Sugawara et al., 2010; Reina-Bueno et al., 2012).
In this work, using tomato as a model plant, we tested the hypothesis that the ACC deaminase activity of Pseudomonas sp. UW4 and the production of trehalose by this bacterium play a synergistic role in protecting plants from the growth inhibitory effects of high salinity. The results of this study first confirmed that the mutant of strain UW4 that does not contain ACC deaminase activity does not exhibit growth-promoting effects on tomato, consistent with the results observed in other studies, where ACC deaminase minus mutants did not confer resistance to salt stress in canola (Li et al., 2000; Cheng et al., 2007) or tomato (Ali et al., 2014).
Some bacteria contain up to five disaccharide biosynthesis pathways (Avonce et al., 2006; Paul et al., 2008). Therefore, a first mutation was generated in the treS gene of Pseudomonas sp. UW4. When analyzing the accumulation of trehalose in this mutant, it was observed that a large percentage of the total accumulated trehalose was decreased (Figure 3). As mentioned earlier, there appear to be two routes of synthesis of trehalose in the bacterium Pseudomonas sp. UW4, so the treS pathway would be the most important for the production of trehalose in this strain (Duan et al., 2013), although it would be necessary to generate other mutants to determine the validity of such a hypothesis. This result also agrees with previous work showing that there are multiple routes of trehalose synthesis and its synthesis is not completely abolished by abolishing a single pathway (Suárez et al., 2008). Nevertheless, the mutation in treS affected the beneficial effect of the strain UW4 when it was used to inoculate tomato plants, since, compared to the control plants inoculated with the wild strain, significant decreases in the length of the tomato plant roots and shoots, chlorophyll concentration, and total dry weight were observed. This suggests that the TreS pathway in strain UW4 plays a key role in the production of trehalose and in subsequently mitigating the toxic effects of salinity in tomato plants. Other studies have shown the importance of this route in bacteria such as Mycobacterium smegmatis (Pan et al., 2004). Also, in a recent work, a new treS gene (TreS) derived from a metagenomic analysis of a saline-alkaline soil was identified (Jiang et al., 2013). The protein (TreS) was characterized as follows: it showed good production of trehalose from maltose, was stable, and possessed different biochemical characteristics compared to other TreSs (Jiang et al., 2013). The treS gene from Pseudomonas putida KT2440 has also exhibited good potential for trehalose production (Wang et al., 2014). Streeter and Gomez (2006) analyzed three trehalose biosynthetic pathways in Bradyrhizobium, either in a free-living state or during symbiosis with plants. Interestingly, these authors observed that TreS was the dominant enzyme in bacteroids, although the substrate for TreS, maltose, was present in very low concentration in nodules. The precise role(s) of the alternative routes of trehalose synthesis in bacteria and how they affect plant-bacteria interactions remains to be elaborated.
To test the hypothesis that bacterial ACC deaminase activity and trehalose accumulation act synergistically in the protection of plants against toxic saline effects, a double mutant, acdS/treS Pseudomonas sp. UW4, was constructed. The results show that the double mutant was significantly impaired in the beneficial effects of the wild-type bacterium on the length of tomato plant roots, total plant dry weight, and chlorophyll content, in the presence of salt, and in general, these plants behaved similarly to control (uninoculated) plants subjected to saline stress. It is worth mentioning that inoculation with each of the single acdS or treS mutants showed better protection against saline conditions than plants inoculated with the double mutant acdS/treS, suggesting a synergistic role of these traits.
Stearns et al. (2012) analyzed the genetic response of Arabidopsis to the inoculation of ACC deaminase positive and negative strains of Pseudomonas sp. UW4. The microarray results from those experiments demonstrated that the transcription of genes involved in plant hormone regulation, stress response, and secondary metabolism was modified in plants by the presence of the bacterial strains, whereas the upregulation of genes for auxin response factors and the downregulation of stress response genes was observed only in the presence of the ACC deaminase positive strain. In those experiments, the trehalose biosynthetic genes (TPS/TPP) in plants were weakly downregulated (-1.7 to -1.8-fold change) by the inoculation of both ACC deaminase positive and negative strains, suggesting a link between the bacterium and the plant’s trehalose metabolism, but not a direct link to the ACC deaminase activity. It would be interesting to further evaluate the mutants of Pseudomonas sp. UW4 generated in the present study (treS, acdS/treS, and OxtreS) in relation to the gene expression and metabolism of plants in order to explore possible novel signaling pathways in plant-bacteria interactions.
In the present work, a second strain with the treS gene was constructed to overproduce RNA copies of the treS gene (OxtreS strain). The OxtreS strain significantly overproduced trehalose compared to the wild-type stain, growing either in media supplemented or not with NaCl. Trehalose overaccumulation was most evident after 24 h of growth. After 48 h of growth, trehalose accumulation in the OxtreS strain was only slightly (not significantly) higher than in the wild-type strain (UW4). It is worth mentioning here that wild-type UW4 showed good accumulation of trehalose during growth after 24 and 48 h in media with 0.2 M of NaCl. When plants were inoculated with the OxtreS trehalose overexpressing strain, it was notable that the resultant plants displayed improved root length, dry weight, and chlorophyll content when compared with the wild-type strain. These results agree with other studies where the previously mentioned strategies protect plants of agronomic interest under conditions of stress (Orozco-Mosqueda et al., 2018). In fact, it has been proposed that the physiological responses of plants to salinity are similar to other environmental stresses such as drought, and that therefore, they may share common stress-tolerance pathways (Munns and Tester, 2008; Forni et al., 2017). For example, the overexpression of trehalose-6-phosphate synthase gene (otsA) has been analyzed in Rhizobium etli, where the modified strain improved drought tolerance and grain yield in P. vulgaris (Suárez et al., 2008). Similarly, a genetically engineered strain of a plant growth-promoting strain of Azospirillum with an improved level of trehalose biosynthesis increased both drought tolerance and plant biomass in maize plants (Rodríguez-Salazar et al., 2009).
In conclusion, this work demonstrated that ACC deaminase and trehalose synergistically protect tomato plants against salt stress during the interaction with the PGPB Pseudomonas sp. UW4. It remains to be elucidated whether the inoculation of other plants with these mutants affects plants other than tomato in a similar manner and in particular whether appropriately engineered bacteria might facilitate grain or fruit production of plant growing under different stresses.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
MO-M conducted the experiments, analyzed the data, and prepared the figures and/or tables. JD, MD, and EZ conducted the experiments and prepared the figures and/or tables. JC-G analyzed the data and reviewed drafts of the manuscript. BG conceived and designed the experiments, analyzed the data, and reviewed drafts of the manuscript, and approved the final draft. GS conceived and designed the experiments, wrote the manuscript, and approved the final draft. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Aurora Flores and Daniel Rojas for their technical help.
Footnotes
Funding. GS thanks the CIC-UMSNH for the supporting research projects (2018–2019) and the CONACYT-Mexico (Propuesta A1-S-15956). MO-M thanks the CONACYT-Mexico for a postdoctoral fellowship (Propuesta No. 232234) at the University of Waterloo.
References
- Ali S., Charles T. C., Glick B. R. (2014). Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 80 160–167. 10.1016/j.plaphy.2014.04.003 [DOI] [PubMed] [Google Scholar]
- Avonce N., Mendoza-Vargas A., Morett E., Iturriaga G. (2006). Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109 10.1186/1471-2148-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacilio M., Rodriguez H., Moreno M., Hernandez J. P., Bashan Y. (2004). Mitigation of salt stress in wheat seedlings by a gfp-tagged Azospirillum lipoferum. Biol. Fertil. Soils 40 188–193. 10.1007/s00374-004-0757-z [DOI] [Google Scholar]
- Bharti N., Barnawal D. (2019). “Amelioration of salinity stress by PGPR: ACC deaminase and ROS scavenging enzymes activity,” in PGPR Amelioration in Sustainable Agriculture, eds Singh A. K., Kumar A., Singh P. K. (Cambridge: Woodhead Publishing; ), 85–106. 10.1016/b978-0-12-815879-1.00005-7 [DOI] [Google Scholar]
- Blaha D., Prigent-Combaret C., Mirza M. S., Moënne-Loccoz Y. (2006). Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol. Ecol. 56 455–470. 10.1111/j.1574-6941.2006.00082.x [DOI] [PubMed] [Google Scholar]
- Chandra G., Chater K. F., Bornemann S. (2011). Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 157 1565–1572. 10.1099/mic.0.044263-0 [DOI] [PubMed] [Google Scholar]
- Cheng Z., Park E., Glick B. R. (2007). 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can. J. Microbiol. 53 912–918. 10.1139/W07-050 [DOI] [PubMed] [Google Scholar]
- Domínguez-Ferreras A., Soto M. J., Pérez-Arnedo R., Olivares J., Sanjuán J. (2009). Importance of trehalose biosynthesis for Sinorhizobium meliloti osmotolerance and nodulation of Alfalfa roots. J. Bacteriol. 191 7490–7499. 10.1128/JB.00725-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Jiang W., Cheng Z., Heikkila J. J., Glick B. R. (2013). The complete genome sequence of the plant growth-promoting bacterium Pseudomonas putida UW4. PLoS One 8:e58640 10.1371/journal.pone.0058640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Müller K. M., Charles T. C., Vesely S., Glick B. R. (2009). 1-aminocyclopropane-1-carboxylate (ACC) deaminase genes in rhizobia from southern Saskatchewan. Microb. Ecol. 57 423–436. 10.1007/s00248-008-9407-6 [DOI] [PubMed] [Google Scholar]
- Duca D. R., Rose D. R., Glick B. R. (2018). Indole acetic acid overproduction transformants of the rhizobacterium Pseudomonas sp. UW4. Antonie van Leeuwenhoek 111 1645–1660. 10.1007/s10482-018-1051-7 [DOI] [PubMed] [Google Scholar]
- Fernandez O., Béthencourt L., Quero A., Sangwan R. S., Clément C. (2010). Trehalose and plant stress responses: friend or foe? Trends Plant Sci. 15 409–417. 10.1016/j.tplants.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Forni C., Duca D., Glick B. R. (2017). Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 410 335–356. 10.1007/s11104-016-3007-x [DOI] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. (1985). Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164 918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R., Cheng Z., Czarny J., Duan J. (2007). Promotion of plant growth by ACC deaminase-containing soil bacteria. Eur. J. Plant Pathol. 119 329–339. 10.1007/978-1-4020-6776-1_8 [DOI] [Google Scholar]
- Hernández-León R., Rojas-Solís D., Contreras-Pérez M., del Carmen Orozco-Mosqueda M., Macías-Rodríguez L. I., Reyes-de la Cruz H., et al. (2015). Characterization of the antifungal and plant growth-promoting effects of diffusible and volatile organic compounds produced by Pseudomonas fluorescens strains. Biol. Control 81 83–92. 10.1016/j.biocontrol.2014.11.011 [DOI] [Google Scholar]
- Ilangumaran G., Smith D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8:1768 10.3389/fpls.2017.01768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordachescu M., Imai R. (2008). Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 50 1223–1229. 10.1111/j.1744-7909.2008.00736.x [DOI] [PubMed] [Google Scholar]
- Jiang L., Lin M., Zhang Y., Li Y., Xu X., Li S., et al. (2013). Identification and characterization of a novel trehalose synthase gene derived from saline-alkali soil metagenomes. PLoS One 8:e77437 10.1371/journal.pone.0077437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandror O., DeLeon A., Goldberg A. L. (2002). Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. U.S.A. 99 9727–9732. 10.1073/pnas.142314099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., II, et al. (1995). Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166 175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Lee K.-H., Kim C.-G., Lee S.-Y., Kim G.-J., Park Y.-H., et al. (2005). Cloning and expression of a trehalose synthase from Pseudomonas stutzeri CJ38 in Escherichia coli for the production of trehalose. Appl. Microbiol. Biotechnol. 68 213–219. 10.1007/s00253-004-1862-1865 [DOI] [PubMed] [Google Scholar]
- Li J., Ovakim D. H., Charles T. C., Glick B. R. (2000). An ACC deaminase minus mutant of Enterobacter cloacae UW4No longer promotes root elongation. Curr. Microbiol. 41 101–105. 10.1007/s002840010101 [DOI] [PubMed] [Google Scholar]
- López M., Tejera N. A., Iribarne C., Lluch C., Herrera-Cervera J. A. (2008). Trehalose and trehalase in root nodules of Medicago truncatula and Phaseolus vulgaris in response to salt stress. Physiol. Plant 134 575–582. 10.1111/j.1399-3054.2008.01162.x [DOI] [PubMed] [Google Scholar]
- Mayak S., Tirosh T., Glick B. R. (2004). Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 42 565–572. 10.1016/j.plaphy.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59 651–681. 10.1146/annurev.arplant.59.032607.092911 [DOI] [PubMed] [Google Scholar]
- Orozco-Mosqueda M., del C., del Carmen Rocha-Granados M., Glick B. R., Santoyo G. (2018). Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208 25–31. 10.1016/j.micres.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Pan Y. T., Koroth Edavana V., Jourdian W. J., Edmondson R., Carroll J. D., Pastuszak I., et al. (2004). Trehalose synthase of Mycobacterium smegmatis: purification, cloning, expression, and properties of the enzyme. Eur. J. Biochem. 271 4259–4269. 10.1111/j.1432-1033.2004.04365.x [DOI] [PubMed] [Google Scholar]
- Paul M. J., Primavesi L. F., Jhurreea D., Zhang Y. (2008). Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59 417–441. 10.1146/annurev.arplant.59.032607.092945 [DOI] [PubMed] [Google Scholar]
- Penrose D. M., Glick B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 118 10–15. 10.1034/j.1399-3054.2003.00086.x [DOI] [PubMed] [Google Scholar]
- Quandt J., Hynes M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127 15–21. 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
- Ramadoss D., Lakkineni V. K., Bose P., Ali S., Annapurna K. (2013). Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. SpringerPlus 2:6 10.1186/2193-1801-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Bueno M., Argandoña M., Nieto J. J., Hidalgo-García A., Iglesias-Guerra F., Delgado M. J., et al. (2012). Role of trehalose in heat and desiccation tolerance in the soil bacterium Rhizobium etli. BMC Microbiol. 12:207 10.1186/1471-2180-12-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Salazar J., Suárez R., Caballero-Mellado J., Iturriaga G. (2009). Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 296 52–59. 10.1111/j.1574-6968.2009.01614.x [DOI] [PubMed] [Google Scholar]
- Rojas-Solís D., Zetter-Salmón E., Contreras-Pérez M., del Carmen Rocha-Granados M., Macías-Rodríguez L., Santoyo G. (2018). Pseudomonas stutzeri E25 and Stenotrophomonas maltophilia CR71 endophytes produce antifungal volatile organic compounds and exhibit additive plant growth-promoting effects. Biocat. Agri. Biotechnol. 13 46–52. 10.1016/j.bcab.2017.11.007 [DOI] [Google Scholar]
- Sambrook J., Russel D. W. (2001). Molecular Cloning: a Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Siddikee M. A., Glick B. R., Chauhan P. S., jong Yim W., Sa T. (2011). Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol. Biochem. 49 427–434. 10.1016/j.plaphy.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Simon R., Prisfer U., Puhler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- StatSoft, Inc. (2001). Statistica. Data Analysis Software System, Version, 8. Available at: http://www.statsoft.com [Google Scholar]
- Stearns J. C., Woody O. Z., McConkey B. J., Glick B. R. (2012). Effects of bacterial ACC deaminase on Brassica napus gene expression. Mol. Plant Microbe Interact. 25 668–676. 10.1094/MPMI-08-11-0213 [DOI] [PubMed] [Google Scholar]
- Streeter J. G., Gomez M. L. (2006). Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 72 4250–4255. 10.1128/aem.00256-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez R., Wong A., Ramírez M., Barraza A., Orozco M. D. C., Cevallos M. A., et al. (2008). Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol. Plant Microbe Interact. 21 958–966. 10.1094/MPMI-21-7-0958 [DOI] [PubMed] [Google Scholar]
- Sugawara M., Cytryn E. J., Sadowsky M. J. (2010). Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl. Environ. Microbiol. 2010 1071–1081. 10.1128/AEM.02483-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Jia S., Dai K., Liu H., Wang R. (2014). Cloning and expression of a trehalose synthase from Pseudomonas putida KT2440 for the scale-up production of trehalose from maltose. Can. J. Microbiol. 60 599–604. 10.1139/cjm-2014-2330 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.