Abstract
The nuclear envelope (NE) aids in organizing the interphase genome by tethering chromatin to the nuclear periphery. During mitotic entry, NE–chromatin contacts are broken. Here, we report on the consequences of impaired NE removal from chromatin for cell division of human cells. Using a membrane–chromatin tether that cannot be dissociated when cells enter mitosis, we show that a failure in breaking membrane–chromatin interactions impairs mitotic chromatin organization, chromosome segregation and cytokinesis, and induces an aberrant NE morphology in postmitotic cells. In contrast, chromosome segregation and cell division proceed successfully when membrane attachment to chromatin is induced during metaphase, after chromosomes have been singularized and aligned at the metaphase plate. These results indicate that the separation of membranes and chromatin is critical during prometaphase to allow for proper chromosome compaction and segregation. We propose that one cause of these defects is the multivalency of membrane–chromatin interactions.
INTRODUCTION
The nuclear envelope (NE) is built by a large, specialized membrane sheet of the endoplasmic reticulum (ER) that surrounds and protects chromatin. It consists of two closely juxtaposed membranes that contain large proteinaceous channels termed nuclear pore complexes (NPCs), which serve the selective nucleocytoplasmic exchange of macromolecules. In metazoan cells, an intermediate filament network, the nuclear lamina, is tightly associated with the inner nuclear membrane (INM) and provides mechanical support to the NE. The NE does not only function as protective barrier of the genome, but it also supports the organization of chromatin into spatially separated domains. Whereas actively transcribed gene loci are associated with NPCs, various proteins of the INM and nuclear lamins determine peripheral gene positioning and organization of heterochromatic regions in differentiated cells. NE–chromatin contacts play pivotal roles in the regulation of gene expression, and are important for the maintenance of genome integrity, development and differentiation (Meister and Taddei, 2013; Ptak et al., 2014; Harr et al., 2016; Ungricht and Kutay, 2017; van Steensel and Belmont, 2017).
During open mitosis in metazoans, the NE is dismantled in the process of NE breakdown (NEBD). NEBD is triggered by the activation of the master mitotic protein kinase CDK1/cyclinB (Champion et al., 2017). CDK1-mediated phosphorylation of nuclear lamins and nucleoporins is directly required for the disintegration of the nuclear lamina (Heald and McKeon, 1990; Peter et al., 1990) and NPCs (Laurell et al., 2011; Linder et al., 2017), respectively. Importantly, also the interactions of INM proteins with chromatin are broken in the course of mitotic prophase. As many INM proteins are retained in the nuclear interior by interaction with DNA and chromatin-associated factors in interphase (Ungricht and Kutay, 2015), the dissolution of physical membrane protein–chromatin contacts must precede the progressive partitioning of INM proteins into the ER network (Ellenberg et al., 1997; Yang et al., 1997). It is assumed that changes in posttranslational modifications of both chromatin-associated factors (Fischle et al., 2005; Nichols et al., 2006; Gorjanacz et al., 2007; Tseng and Chen, 2011; Molitor and Traktman, 2014) and INM proteins (Courvalin et al., 1992; Foisner and Gerace, 1993; Dechat et al., 1998; Dreger et al., 1999; Hirano et al., 2005, 2009; Tseng and Chen, 2011; Patel et al., 2014) that occur during mitotic entry contribute to breaking these intricate connections and facilitate membrane dissociation from chromatin. However, direct evidence that these posttranslational modifications are required for membrane removal from chromatin in living cells is lacking. The subsequent spatial separation of membranes from chromatin during prometaphase is aided by microtubule- (MT) dependent forces generated by the minus-end directed motor dynein (Beaudouin et al., 2002; Salina et al., 2002; Muhlhausser and Kutay, 2007; Hebbar et al., 2008; Bolhy et al., 2011; Turgay et al., 2014). During late prometaphase and metaphase, the mitotic ER/NE network is largely excluded from the spindle area, which is further promoted by both an active exclusion mechanism involving the ER membrane proteins REEP3/4 (Schlaitz et al., 2013) and the inhibition of ER-MT linker proteins (Vedrenne et al., 2005; Smyth et al., 2012).
Although INM proteins are known to be phosphorylated in their nucleoplasmic domains during mitosis (Courvalin et al., 1992; Foisner and Gerace, 1993; Dechat et al., 1998; Dreger et al., 1999; Hirano et al., 2005, 2009; Tseng and Chen, 2011; Patel et al., 2014), it is neither established whether their phosphorylation directly contributes to membrane release from chromatin nor what would be the consequences of a failure in membrane removal from chromatin for mitotic progression. To assess the significance of breaking apart the physical contacts between INM proteins and chromatin during early mitosis, we set out to explore how persistent mitotic membrane–chromatin association affects mitosis of mammalian cells. We report that ER/NE membrane–chromatin contacts perturb mitotic chromatin organization, chromosome segregation and cell division as well as nuclear morphology of postmitotic cells.
RESULTS
Tethering of NE/ER membranes to chromatin during mitosis impairs chromosome segregation and cell division
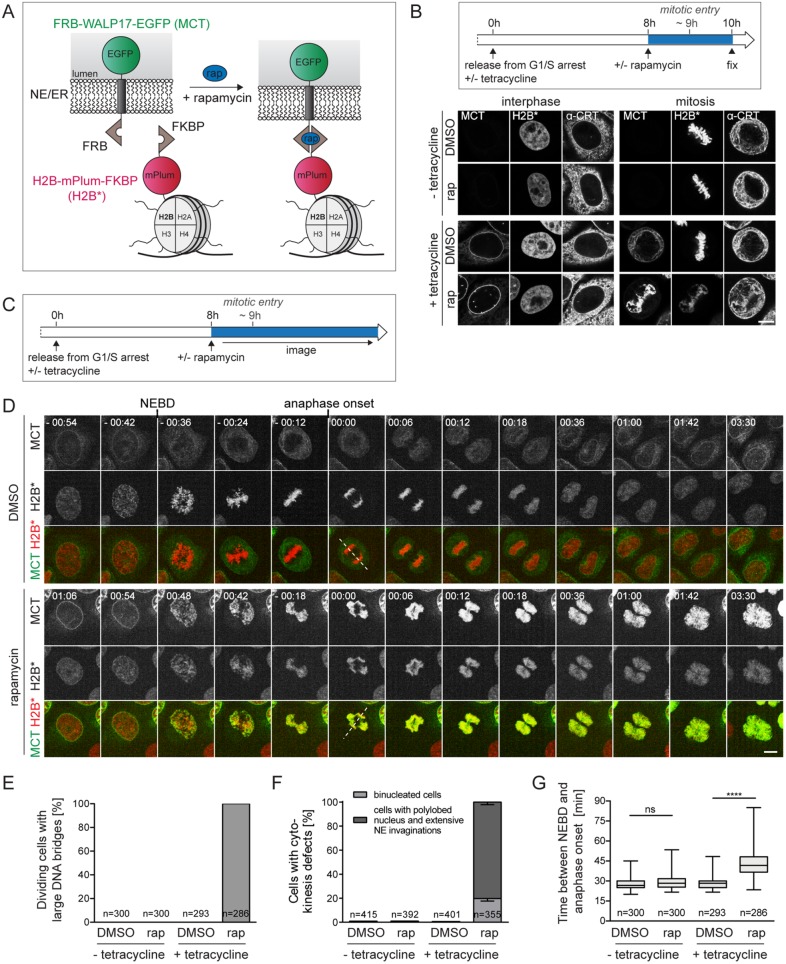
To date, the release of membranes from chromatin during mitotic entry cannot be specifically inhibited. Mutations in INM proteins impairing membrane protein–chromatin dissociation have not been identified. Further, factors like mitotic protein kinases expected to support the dissociation of membrane proteins from chromatin (e.g., CDK1) perform other important mitotic functions. Thus, conventional loss of function experiments would result in pleiotropic defects. To overcome these issues, we exploited a membrane–chromatin tethering approach to prevent the release of membranes from chromatin during mitotic entry. Our membrane–chromatin tethering system (Figure 1A) relies on the rapamycin-dependent interaction between FRB and FKBP attached to a tetracycline-inducible ER membrane protein referred to as the membrane–chromatin tether (MCT) and the core histone H2B, respectively (Ungricht et al., 2015). For visualization, the two modules of the tethering system also contain fluorescent protein tags, that is, an EGFP on the MCT and mPlum on H2B. When the two components were expressed together in HeLa cells (MCT/H2B* cells), the MCT was localized throughout the ER/NE network in the absence of rapamycin. On addition of rapamycin, the MCT accumulated at the NE (Figure 1B, left). NE accumulation of the reporter occurs with very fast kinetics, displaying a half-time of NE enrichment of less than 10 min, as previously determined (Ungricht et al., 2015). Notably, we had used this system before to study protein targeting to the INM and demonstrated that NE accumulation of the EGFP-tagged MCT reflects its INM localization (Ungricht et al., 2015).
FIGURE 1:
Tethering of ER membranes to mitotic chromatin impairs chromosome segregation and cell division. (A) Scheme of the chromatin-ER membrane tethering system. FRB-WALP17-EGFP (membrane–chromatin tether, MCT); H2B-mPlum-FKBP (chromatin anchor, H2B*). (B) Flowchart of cell synchronization and drug treatment. Confocal images of interphase and mitotic MCT/H2B* cells. The ER was stained for calreticulin (α-CRT). (C) Flowchart of the experimental setup for D. (D) Time-lapse confocal microscopy of synchronized MCT/H2B* cells progressing through mitosis in the presence of DMSO or 200 nM rapamycin at the indicated times relative to anaphase onset. Quantification of anaphase DNA bridges (E), cytokinesis defects (F), and the time span between NEBD and anaphase onset (G) from time-lapse wide-field microscopy of synchronized MCT/H2B* cells. Note that only 70–80% of all cells express MCT in the presence of tetracycline. Only GFP-positive cells were analyzed. N = 3; mean ± SEM; n, number of cells. Dashed lines represent spindle axes. ****p < 0.0001; ns, not significant. Bars, 10 μm.
To induce the interaction of the MCT with chromatin just before mitotic entry, MCT/H2B* cells were arrested at the G1/S transition by thymidine, released into S phase concomitant with the tetracycline-induced expression of the MCT, and then, approximately one hour before cells started to enter mitosis, 200 nM rapamycin or a solvent control (dimethyl sulfoxide [DMSO]) was added. Cells were allowed to progress into mitosis, fixed, and analyzed by microscopy (Figure 1B, right). In the presence of DMSO, both the MCT and the ER protein calreticulin were distributed throughout the mitotic ER network and excluded from the chromatin/spindle area. In contrast, in the presence of rapamycin, the MCT was strongly enriched on chromatin. Visualization of calreticulin confirmed that tethering of the MCT to H2B* drives the recruitment of the ER network to chromatin, thereby validating that our tethering system enables temporal control over membrane–chromatin connections and mimics conditions of failure in membrane removal from chromatin.
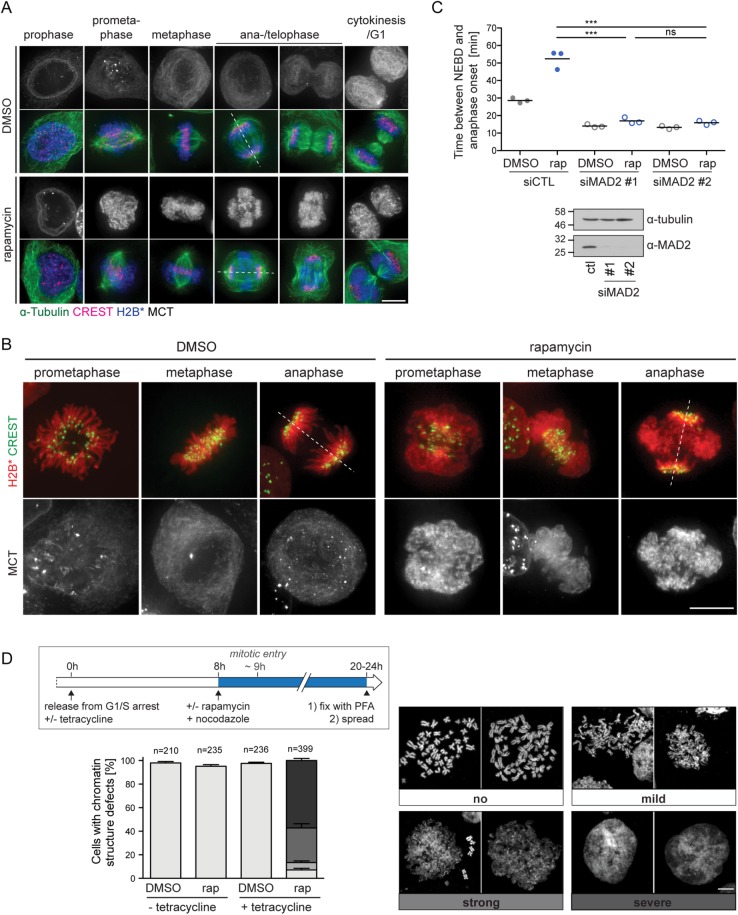
Next, we analyzed the consequences of persistent mitotic MCT-chromatin interactions on mitotic progression and cell division by confocal live-cell microscopy (Figure 1, C and D). Control cells progressed through mitosis properly and divided. In contrast, cells in which the ER/NE membrane network was tethered to chromatin displayed severe chromosome segregation defects, failures in cytokinesis and an aberrant, polylobed nuclear morphology after mitosis. Whereas chromosome congression and alignment at the metaphase plate were not majorly affected, chromatin adopted a characteristic rhomboid-shaped configuration in anaphase, apparently caused by defects in segregating chromosome arms, while the majority of kinetochores was successfully pulled apart (see also Figure 4 later in this article).
FIGURE 4:
Induced membrane-chromatin-contacts perturb the organization of mitotic chromatin. (A) Wide-field fluorescence microscopy of MCT/H2B* cells treated as in Figure 1B. Cells were stained for α-tubulin and kinetochores (CREST). Dashed lines represent spindle axes. (B) Representative wide-field fuorescent images of chromatin organization of DMSO- or rapamycin-treated MCT/H2B* cells progressing through mitosis. Kinetochores were immunostained using the CREST antibody. (C) Quantification of the time span between NEBD and anaphase onset of MCT/H2B* cells (as in Figure 1G) treated with either control or MAD2 siRNAs for 48 h. N = 3; mean ± SEM; n = 150 per condition. Bottom, immunoblot analysis of MAD2 depletion. (D) Analysis of MCT/H2B* cells with respect to mitotic chromatin structure. Flowchart of the cell synchronization protocol combined with drug treatment used for the generation of mitotic chromatin spreads (top). Representative confocal images of MeOH-fixed metaphase spreads from nocodazole-arrested MCT/H2B* cells. Spreads were counterstained with Hoechst. The chromosome structure defects were classified into four categories, ranging from a classical thread-like shape (“no”) to a disorganized amorphous cloud (“severe”). The quantification of each phenotype observed after the indicated treatment is shown in the bottom panel (N = 4; mean ± SEM; n, number of analyzed cells). Bars, 10 μm.
The observed defects were quantified by monitoring dividing MCT/H2B* cells every 5 min using wide-field microscopy. Of MCT/H2B*-expressing cells treated with rapamycin, 96% showed chromosome segregation defects (Figure 1E), the large majority of which failed to undergo cytokinesis (Figure 1F). Moreover, rapamycin-exposed MCT/H2B* cells entered anaphase with a slight delay of ∼15 min compared with control conditions (Figure 1G). Importantly, when the MCT was not expressed, cells progressed through mitosis properly and divided without obvious perturbations in the presence of 200 nM rapamycin, excluding that our regime of rapamycin exposure causes defects in chromatin segregation (Figure 1, E–G). Taken together, these results demonstrate that a failure in breaking physical NE–chromatin interactions at mitotic entry induces strong defects in chromosome segregation and cytokinesis.
Persistent ER/NE–chromatin connections cause postmitotic aberrations of nuclear morphology
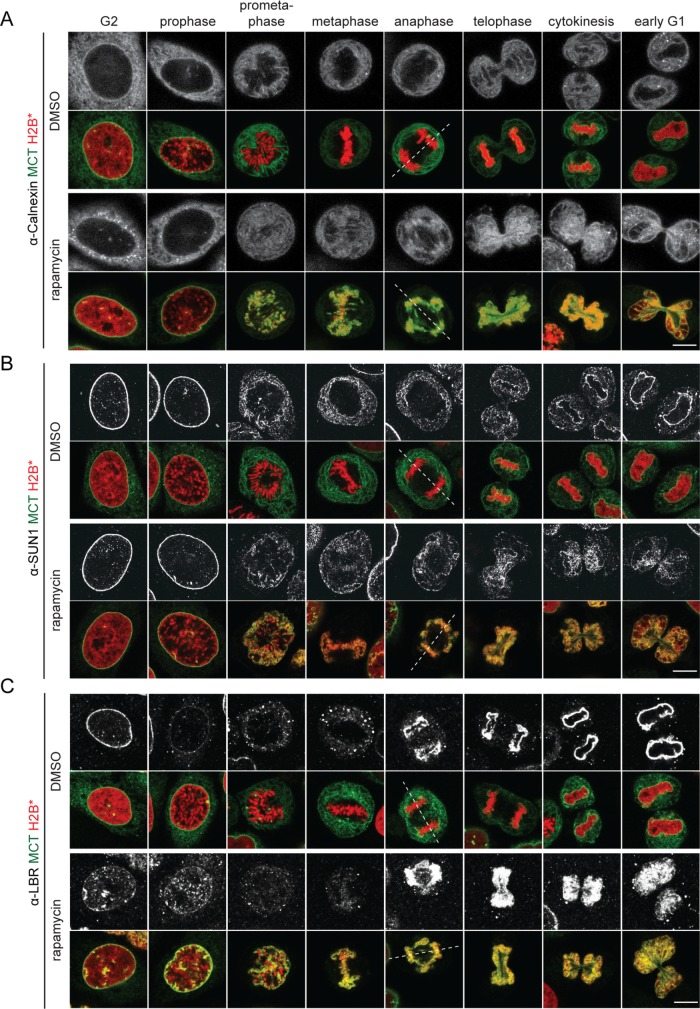
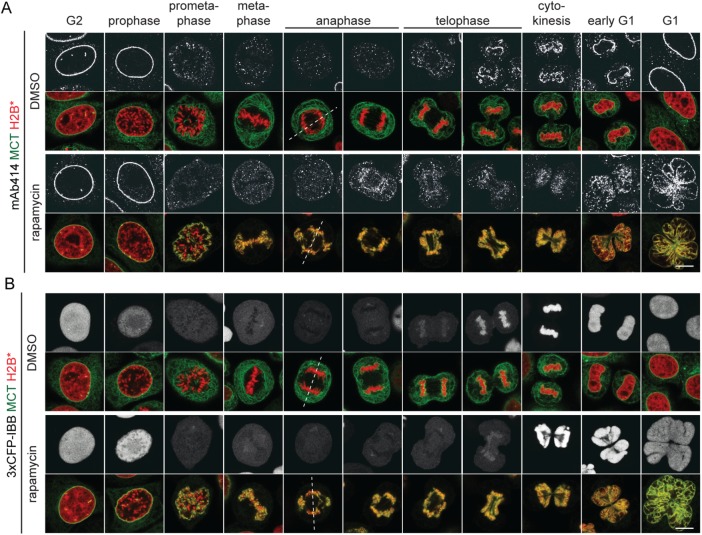
To analyze how persistent membrane-chromatin contacts affect the mitotic localization of endogenous ER and NE proteins, MCT/H2B* cells were released into mitosis, fixed and immunostained for the ER membrane protein calnexin (CNX), and the INM proteins SUN1 and LBR. CNX localized evenly throughout the mitotic ER network in both control and rapamycin-treated MCT/H2B* cells, marking the chromatin-attached ER network in the latter (Figure 2A). The partitioning of the INM proteins SUN1 and LBR from the INM into the mitotic ER network seemed unaffected, and like for CNX, a fraction of both factors localized to the chromatin-associated ER in rapamycin-treated MCT/H2B* cells (Figure 2, B and C). Strikingly, however, inspection of postmitotic cells revealed that nuclear morphology was severely perturbed (Figures 2, 3, and 1D). The NE of cells that had progressed through mitosis with persistent membrane-chromatin contacts appeared strongly polylobed, reflecting the irregular shape of the underlying chromatin mass (Figures 1D and 2, B and C). We further noted that the staining of nucleoporins by the mAb414 antibody at the chromatin contours during telophase and early G1 was less accentuated than in control cells (Figure 3A). Despite these shortcomings, nuclei were competent for nuclear import of a 3xCFP-IBB fusion protein, indicating that NE sealing and NPC assembly were not severely affected (Figure 3B). Taken together, these data demonstrate that endogenous INM proteins are successfully released into the ER network in early mitosis despite the presence of the MCT. Yet cells with persistent membrane–chromatin contacts display chromosome segregation defects and an aberrant postmitotic nuclear morphology, indicative of defects in the relaxation of chromatin into its normal, ball-shaped interphase configuration.
FIGURE 2:
ER and NE membrane proteins are distributed throughout the mitotic ER in the presence of MCT-induced chromatin–membrane contacts. MCT/H2B* cells were treated as outlined in Figure 1B, immunostained using anti-calnexin (A), anti-SUN1 (B), and anti-LBR (C) antibodies, and analyzed by confocal microscopy. Chromatin configuration was used to assign cells to the indicated cell cycle stages. Dashed lines represent spindle axes. Bars, 10 μm.
FIGURE 3:
Persistent, MCT-induced chromatin–membrane contacts induce an aberrant postmitotic nuclear morphology but do not affect postmitotic NPC assembly. MCT/H2B* cells were treated as in Figure 1B. (A) Immunostaining of FG repeat Nups using the mAb414 antibody. (B) Analysis of MCT/H2B*-3xCFP-IBB expressing cells. Dashed lines represent spindle axes. Bars, 10 μm.
Membrane–chromatin tethering impacts on mitotic chromatin organization but does not impair activation of the spindle assembly checkpoint
The observed defects in chromosome segregation (Figures 1–3) prompted us to investigate the integrity of the mitotic spindle in cells with membrane-tethered chromatin. The localization of microtubules and kinetochores was analyzed by immunofluorescence using α-tubulin and CREST antibodies, respectively. Remarkably, metaphase spindle microtubule morphology seemed normal, and kinetochores were aligned at the metaphase plate in both rapamycin- and DMSO-treated MCT/H2B* cells (Figure 4A). However, the morphology of membrane-associated chromatin was aberrant compared with control cells (Figure 4B). Specifically, puffs of chromatin, likely emerging from chromosome arms, often dangled outside of the spindle in metaphase cells (Figure 4, A and B). During sister chromatid segregation in anaphase, an entangled mass of chromatin decorated the circumference of the spindle midzone and failed to move poleward.
As a defective spindle assembly checkpoint (SAC) might induce chromosome segregation defects (Musacchio and Salmon, 2007), we assessed its activity. In both rapamycin- and DMSO-treated MCT/H2B* cells, down-regulation of the SAC factor MAD2 caused a precocious onset of anaphase as expected (Figure 4C), demonstrating that the SAC is active in cells with persistent membrane–chromatin interaction. Thus, access of SAC factors to kinetochores seems not perturbed by persistent MCT contacts. Immunofluorescence analysis further confirmed that MAD2 localized to unattached kinetochores during (pro)metaphase and was released when cells had progressed into anaphase, demonstrating a normal SAC response (Supplemental Figure S1B). Inspection of membrane–chromatin contacts by confocal fluorescence microscopy indeed revealed that kinetochores were not fully covered by membranes (Supplemental Figure S1A). Consistent with the delay in anaphase onset of rapamycin-treated MCT/H2B* cells, the number of MAD-positive dots in metaphase was slightly increased (Supplemental Figure S1C), indicative of some delay in proper microtubule attachment to kinetochores. We also observed a higher frequency of lagging kinetochores during anaphase, although the majority of kinetochores remained unaffected (Supplemental Figure S1D).
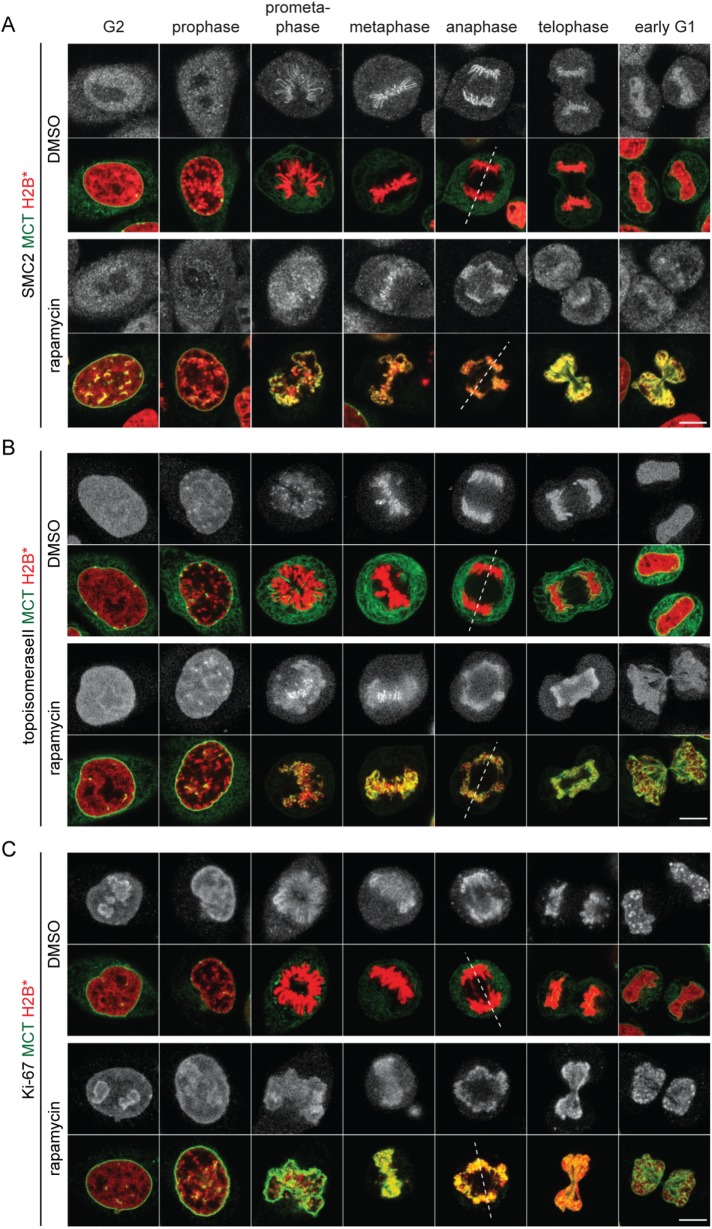
In light of the aberrant mitotic chromatin morphology, we analyzed mitotic chromosome spreads prepared from nocodazole-arrested MCT/H2B* cells. These confirmed the structural disorganization of chromosomes from mitotic cells with persistent chromatin-membrane contacts, whereas chromosomes from control cells displayed the expected condensed, X-shaped morphology (Figure 4D and Supplemental Figure S2). Note that chromatin association of key factors implicated in the condensation and singularization of mitotic chromosomes such as condensin I, condensin II, topoisomerase II, Ki-67, and chromokinesins (Samejima et al., 2012; Kschonsak and Haering, 2015; Booth et al., 2016; Booth and Earnshaw, 2017; Cuylen et al., 2016; Piskadlo and Oliveira, 2016; Gibcus et al., 2018; Walther et al., 2018) seemed unperturbed (Figure 5 and Supplemental Figure S3B), indicating that access of these factors to chromatin is not obviously inhibited by the MCT. Also the phosphorylation of histone H3 at Ser10 was normal (Supplemental Figure S3C). Thus, abundant membrane–chromatin contacts may interfere with the functionality of chromatin condensation factors or otherwise impair the maintenance of the condensed state. Notably, there was also no obvious sign of DNA damage (Supplemental Figure S3D). Taken together, membrane tethering to mitotic chromatin causes defects in mitotic chromatin organization and massive chromatin bridges during anaphase, whereas kinetochore functionality seems largely unaffected, as indicated by the efficient SAC response and the poleward movement of the bulk of kinetochores.
FIGURE 5:
Chromatin condensation factors can be efficiently loaded on mitotic chromatin in the presence of the MCT. Synchronized MCT/H2B* cells were released into mitosis as outlined in Figure 1B. Cells were treated with DMSO or rapamycin 8 h after release, fixed 2 h after the treatment, and immunostained using anti-SMC2 (A), anti-topoisomeraseII (B) and anti–Ki-67 (C) antibodies and analyzed by confocal microscopy. Chromatin configuration was used to assign cells to the indicated cell cycle stages. Dashed lines represent spindle axes. Bars, 10 μm.
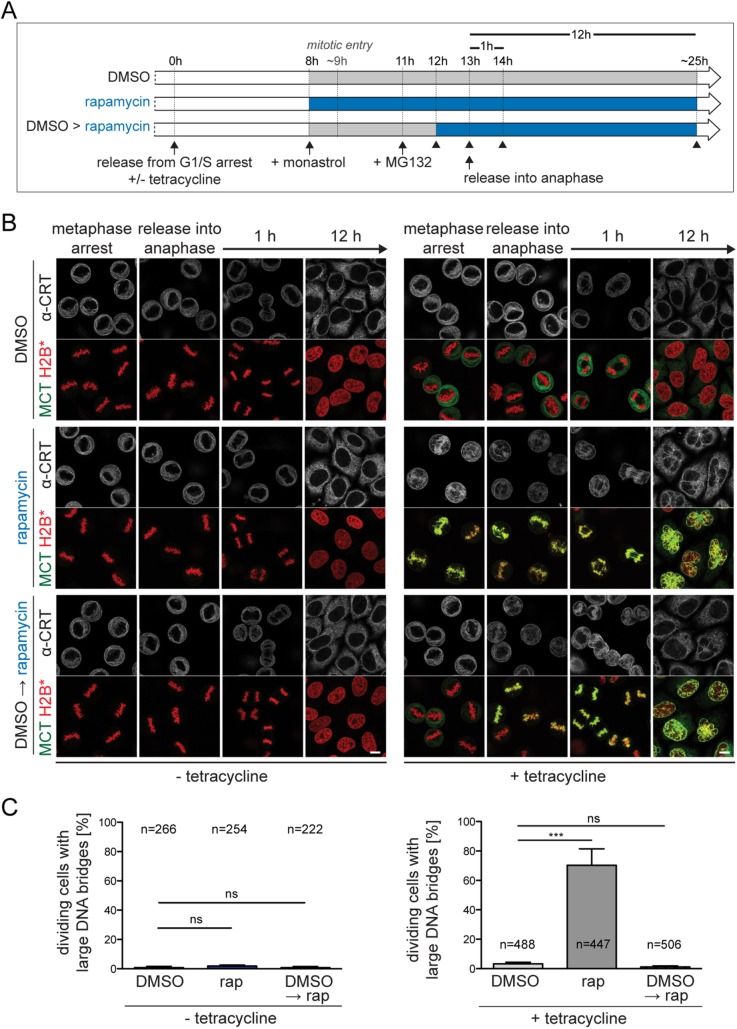
Tethering ER/NE membranes to metaphase chromosomes does not impair their segregation
To investigate whether the observed defects in chromosome segregation and nuclear morphology arise from membrane contacts in early or late mitosis, we compared cells in which membrane tethering was induced either before mitotic entry or only later, that is, during a metaphase arrest imposed by sequential treatment with monastrol and MG132 (Figure 6). As expected, chromosomes in cells without MCT expression (solvent control DMSO) segregated successfully and cells divided with fully resolved chromosome arms, whereas membrane–chromatin tethering prior to mitotic entry (rapamycin) caused severe segregation defects. Strikingly, when MCT-mediated membrane tethering to chromatin was induced in metaphase-arrested cells (DMSO→rapamycin), only 1% of the cells displayed unresolved chromosome arms and failed to complete cytokinesis. Yet nuclei showed morphological defects after mitosis, that is, membrane invaginations into chromatin or a polylobed shape.
FIGURE 6:
Chromosome segregation and cell division are not affected when membrane–chromatin tethering is induced in metaphase. (A) Flowchart of experimental approach in B. Cells were fixed at the indicated time points (see arrowheads). (B) Confocal images of synchronized MCT/H2B* cells fixed at the indicated time points. (C) Quantification of large DNA bridges in cells 1 h after release into anaphase. Note that all dividing cells regardless of MCT expression level were analyzed (without tetracycline: N = 2; mean ± SD; n, number of cells; with tetracycline: N = 4; mean ± SEM). Bars, 10 μm.
To exclude that the strong defects observed on tethering of membranes to chromatin during early mitosis are merely a byproduct of excessive expression of the MCT, we reduced its expression to a level similar to that of LAP2β (Supplemental Figure S4A), a well-characterized human INM protein present at ∼1.3 × 106 molecules/HeLa cell (Itzhak et al., 2016). Even when expressed at low level, similar to that of a single INM protein, and at a 50-fold reduced rapamycin concentration, thereby weakening the retention of the MCT on chromatin (Ungricht et al., 2015), we still observed striking defects in mitotic chromatin organization, binucleation and nuclei displaying an aberrant morphology (Supplemental Figure S4, B and C).
Together, these findings indicate that detachment of membranes from chromatin during early mitosis is necessary to ensure accurate chromosome segregation and division. Further, the persistent presence or a precocious recruitment of membranes to chromatin affects nuclear morphology, suggesting that reconfiguration of chromatin into its interphase state and NE reformation during mitotic exit are usually well coordinated. Interestingly, similar, but less severe mitotic phenotypes have been previously observed on depletion of the ER proteins REEP3/4, which causes an invasion of ER membranes into the spindle and onto chromosomes, a slight increase in chromosome segregation defects, a reduced spatial separation of daughter nuclei and slight NE aberrations in postmitotic cells (Schlaitz et al., 2013).
Defects in chromosome segregation are mimicked by soluble, bivalent chromatin binding proteins
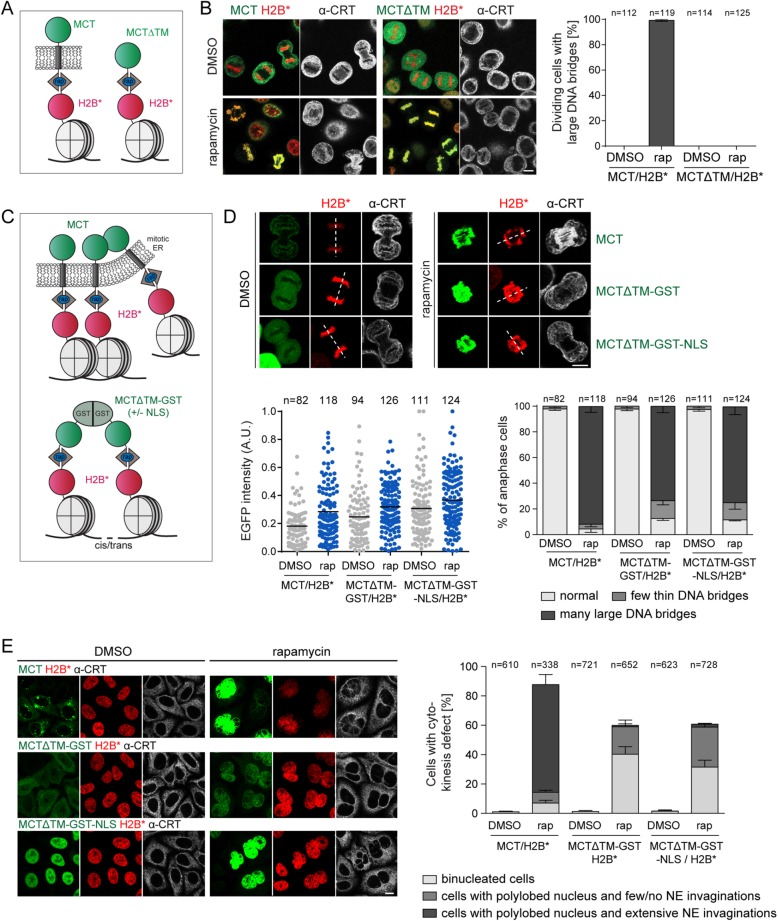
To unambiguously demonstrate that the persistent association of membranes with chromatin causes the observed chromosome segregation defects, we generated a derivative of the MCT lacking the transmembrane domain (MCTΔTM) (Figure 7A). Although the soluble MCTΔTM protein was efficiently recruited to mitotic chromatin in MCTΔTM/H2B* cells in the presence of rapamycin, anaphase chromosome segregation proceeded normally, without DNA bridges (Figure 7B). These findings confirm that the tethering of ER/NE membranes to mitotic chromatin leads to the observed chromatin segregation errors.
FIGURE 7:
Membrane-induced chromatin segregation defects can be mimicked by soluble, bivalent chromatin-binding proteins. (A) Schematic comparison of the MCT and a soluble derivative lacking the transmembrane domain (MCT∆TM). (B) Synchronized MCT/H2B* and MCT∆TM/H2B* cells were released into mitosis as in Figure 1B, fixed, stained for calreticulin, and analyzed by confocal microscopy. The number of dividing cells with large DNA bridges was quantified (N = 3; mean ± SEM; n, number of analyzed GFP-positive cells). (C) Depiction of chromatin cross-linking by the MCT and MCT∆TM-GST(-NLS). (D, E) MCT/H2B*, MCTΔTM-GST/H2B*, and MCTΔTM-GST-NLS/H2B* cells were synchronized, treated with DMSO or 200 nM rapamycin before mitotic entry, and fixed in anaphase or after mitosis. (D) Confocal images of MCT/H2B*, MCTΔTM-GST/H2B* or MCTΔTM-GST-NLS/H2B* anaphase cells. Relative GFP intensities of all analyzed cells (N = 3; mean ± SEM; n, total number of cells) and quantification of chromosome segregation defects. (E) Confocal images of MCT/H2B*, MCTΔTM-GST/H2B*, or MCTΔTM-GST-NLS/H2B* after mitosis. The indicated postmitotic aberrations were quantified (N = 3; mean ± SEM; n, number of cells). CRT: calreticulin; rap: rapamycin. Dashed lines represent spindle axes. Bars, 10 μm.
There exist several, nonexclusive possibilities to explain why a failure in the removal of membranes from chromatin during mitotic entry perturbs mitotic chromatin organization and segregation. For example, membranes could exert a negative effect by their “biochemical” environment by exposing mitotic chromatin to membrane-associated enzymes like protein phosphatases. However, we did not detect changes in the mitotic phosphorylation status of histone H3 (Supplemental Figure S3C), although we cannot exclude that modification of other factors on chromatin is affected. However, membranes could also present a physical impediment. For instance, the tethered ER/NE membrane network might interfere with the accessibility of chromatin. Arguing against a general loss of accessibility, MAD2 bound to kinetochores on SAC activation (Supplemental Figure S1B), and the loading of chromatin condensation factors like condensin I, which binds chromatin during prometaphase (Hirota et al., 2004; Gerlich et al., 2006), seemed unperturbed (Figure 5 and Supplemental Figure S3B). Alternatively, multisite attachment of chromosomes to membranes could cross-link chromatin to the surface of the ER/NE network such that the singularization and segregation of chromosomes is hampered.
To test whether cross-linking of chromatin can in principle contribute to the observed phenotypes, we generated a soluble variant of MCTΔTM forced into a dimeric configuration by fusion to GST (MCTΔTM-GST), potentially competent of cross-linking chromatin in cis or trans (Figure 7C). Note that the GST fusion protein is too large to efficiently pass NPCs and to reach chromatin until NPCs are disassembled during NEBD (Supplemental Figure S5). Therefore, we also tested a derivative that can efficiently enter nuclei even before NEBD (MCTΔTM-GST-NLS). Strikingly, similar to MCT/H2B*-expressing cells, both MCTΔTM-GST/H2B* and MCTΔTM-GST-NLS/H2B* cells displayed chromatin bridges during anaphase in the vast majority of cells (Figure 7D), indicating that exposure of chromatin to a bivalent chromatin-binding protein during prometaphase is sufficient to perturb proper chromosome segregation. Interestingly, the analysis of mitotic chromatin spreads revealed that chromosome architecture differed between cells with the MCT membrane tether and the soluble, GST-based constructs. Chromatin of MCTΔTM-GST/H2B* and MCTΔTM-GST-NLS/H2B* cells in the presence of rapamycin appeared condensed, and chromosomes were clustered together compared with control conditions (Supplemental Figure S6). Thus, cross-linking of chromatin can cause chromatin bridges in the absence of obvious condensation defects.
Inspection of postmitotic MCTΔTM-GST/H2B* and MCTΔTM-GST-NLS/H2B* cells showed that many cells failed to undergo cytokinesis (Figure 7E). However, compared with cells expressing the membrane tether, NE morphology was less severely affected and most nuclei adopted a normal interphase shape without obvious NE invaginations. Thus, cross-linking of chromatin by abundant bivalent tethers is sufficient to induce chromatin bridges in anaphase and cell division defects, reminiscent of the MCT. In contrast though, multivalent tethering of membranes to chromatin can additionally cause defects in chromatin compaction and induces stronger postmitotic NE aberrations.
DISCUSSION
Cells that enter mitosis in the presence of persistent NE/ER membrane–chromatin contacts display drastic chromosome segregation defects, characterized by failures in separating chromosome arms during spindle elongation in anaphase. Whereas kinetochore capture during (pro)metaphase proceeds by-and-large normally and cells enter anaphase only with a slight delay, chromatin cannot be fully segregated, and puffs of chromatin stay associated with membranes, mostly at the circumference of the spindle midzone. In late anaphase and telophase, there are excessive DNA bridges, and cells display cytokinesis problems. Postmitotic nuclei display striking morphological alterations and appear polylobed. In contrast, when membranes are only tethered to chromatin later, during metaphase, chromosome segregation during anaphase is not perturbed. Also in this case, postmitotic nuclear morphology remains affected. Collectively, our results indicate that the removal of NE membranes from chromatin in early mitosis is required for faithful chromosome segregation.
But why would persistence of membrane–chromatin contacts during prometaphase obstruct proper chromosome segregation? Most likely, membranes physically impede the condensation, singularization, and segregation of chromosomes by multivalent cross-linking of chromatin. Although the loading of factors known to promote singularization and condensation of chromosomes such as condensins, topoisomerase II and Ki-67 onto chromatin seemed unaffected, we noticed chromosome condensation defects. In light of recent theoretical and experimental support for condensin-mediated chromatin loop extrusion as the basis for chromatin compaction in prometaphase (Goloborodko et al., 2016; Gibcus et al., 2018; Walther et al., 2018), we assume that multivalent cross-linking of chromatin to membranes may impair condensin I-mediated chromatin compaction during prometaphase, perhaps by preventing the threading of chromatin fibers through condensin I rings. Failure in condensin I function is in turn expected to diminish chromatin decatenation by topoisomerase II (Baxter et al., 2011; Baxter and Aragon, 2012; Piskadlo and Oliveira, 2016), which would lead to persistent chromatin entanglements. Chromatin organization defects induced by persistent membrane–chromatin contacts were clearly apparent by the dumbbell-shaped organization of the metaphase plate, and cells progressed into anaphase in the presence of extensive chromatin bridges between the separating masses of chromosomes, explaining the cell division defect. In contrast, induction of membrane contacts to chromatin in cells during metaphase did not lead to chromosome segregation problems, likely because chromosomes had already been properly compacted, singularized and spatially positioned.
Interestingly, chromatin condensation defects have been noted in an early study investigating mitotic defects induced by expression of a lamin A/C mutant that prevents lamina disassembly (Heald and McKeon, 1990). In light of our results obtained with the membrane-chromatin tether, it is possible that these similarities arise from the multivalent attachment of large structures incompatible with the action of chromatin condensation factors in both cases. Notably, soluble, GST-based bivalent chromatin cross-linking proteins induced anaphase chromatin bridges and cytokinesis failure in the absence of obvious chromosome compaction defects, highlighting that mere cross-linking of chromatin is sufficient to induce chromosome segregation errors and binucleation in the absence of large-scale chromosome compaction defects. The multivalency of membrane-chromatin contacts may exaggerate these cross-linking-based defects by additionally impairing both chromatin reorganization during mitosis and the pulling of chromosome arms, attached at multiple sites to the membranous cocoon formed by the mitotic ER, to the spindle poles.
So far, the mechanisms that promote the dissociation of INM proteins from chromatin during mitotic entry remain poorly characterized. In interphase, INM proteins may contact chromatin in different ways, that is, by direct binding of DNA (Ulbert et al., 2006), by binding of core chromatin factors like histones (Hirano et al., 2012), or by association with peripheral chromatin components like the barrier-to-autointegration factor BAF (Brachner and Foisner, 2011). Of these, the mitotic modulation of the DNA-BAF-INM protein network is the best understood. The protein kinase VRK1 phosphorylates BAF in early mitosis, impairing its interaction with DNA and reducing its affinity to INM proteins of the LEM (Lap2β, emerin, Man1) domain family (Molitor and Traktman, 2014). Depletion of VRK1 from human somatic cells indeed inhibits BAF release from DNA during mitotic entry (Molitor and Traktman, 2014). However, metaphase chromosomes are devoid of membranes in VRK-depleted cells (Samwer et al., 2017), highlighting that blocking BAF phosphorylation by VRK1 is insufficient to prevent the dissociation of LEM domain proteins. Mitotic modifications of LEM domain proteins may contribute in addition (Hirano et al., 2005, 2009). The example of the three-layered DNA-BAF-LEM protein network illustrates that changes on two interfaces (BAF-LEM and BAF-DNA) contribute to breaking NE-chromatin contacts in this case. Interestingly, loss of VRK1 causes severe chromosome segregation errors (Molitor and Traktman, 2014; Samwer et al., 2017). In light of our findings, this could be explained by a cross-linking of DNA by BAF homodimers in a scenario similar to our soluble GST-based bivalent chromatin cross-linkers, or by a somewhat retarded release of LEM domain proteins and thus membranes from chromatin. Clearly, our work motivates a detailed future analysis of how the various interactions between INM proteins with DNA and histones are modulated during mitotic entry.
NEBD during open mitosis has been mainly considered a mechanism to disrupt the barrier between chromatin and microtubules to allow for spindle assembly. In this study, we demonstrated that the breakdown of the NE, and especially the disintegration of NE membrane protein–chromatin connections, has far more direct implications on the process of cell division than anticipated. This is not only relevant for open mitosis, as chromatin also dissociates from the NE in organisms with closed mitosis such as yeasts (Kanoh, 2013). Interestingly, it has been reported that NE attachment of chromatin constrains the topoisomerase II-dependent resolution of DNA intertwines in budding yeast (Titos et al., 2014). The dissociation of mitotic NE–chromatin contacts may thus be a universal requirement in eukaryotes to ensure proper chromosome disentanglement for cell division. If so, then the resolution of NE–chromatin contacts must be ensured to prevent binucleation, which can eventually promote genomic instability and tumorigenesis in mammals (Fujiwara et al., 2005; Davoli and de Lange, 2011).
MATERIALS AND METHODS
Molecular cloning and cell lines
The DNA plasmids encoding the MCT (HA-RRSR-TEV-FRB-WALP17-EGFP in pcDNA5-FRT/TO; Invitrogen) and H2B-FKBP-mPlum (in pIRES-puro2; Clontech) (H2B*) have been described previously (Ungricht et al., 2015). A DNA fragment encoding 3xCFP-IBB (Ungricht et al., 2015) was subcloned into the pIRES-neo3 vector (Clontech) and used for random genome integration into the MCT/H2B* HeLa cell line. To generate an MCTΔTM, a DNA fragment encoding HA-TEV-RRSR-FRB-EGFP was cloned into the EcoRV and NotI sites of the vector pcDNA5-FRT/TO. pcDNA5-FRT/TO-based plasmids encoding FRB-EGFP-GST and FRB-EGFP-GST-NLS(PKKKRKV) were generated by PCR amplification of the respective DNA fragments using MCT-pcDNA5-FRT/TO and pGEX-2T (GE Healthcare Life Sciences) as templates, followed by insertion in the AflII/AflII, EcoRV/NotI and NotI/ApaI sites of pcDNA5-FRT/TO, respectively. To generate tetracycline-inducible cell lines, pcDNA5-based constructs were stably integrated into HeLa FRT/TetR cells.
Cell culture and inhibitor treatments
HeLa cell lines were grown in DMEM containing 10% fetal calf serum and 100 μg/ml penicillin/streptomycin at 37°C with 5% CO2 in a humidified incubator. Transgenic cell lines were supplemented with 0.5 mg/ml G418, 1 μg/ml puromycin, and/or 0.3 mg/ml hygromycin. To synchronize HeLa cells in mitosis, cells were treated for 16–18 h with 3 mM thymidine (Sigma-Aldrich) and released from G1/S arrest for 10 h. To arrest cells in prometaphase, 100 ng/ml nocodazole (Sigma-Aldrich) was added 8 h after thymidine release for the indicated periods of time. To arrest cells in metaphase, cells were treated 8 h after thymidine release with 100 μM of the Eg5 inhibitor monastrol (Sigma-Aldrich) for 3 h, released, and treated with 10 μM MG132 (Sigma-Aldrich) for 2 h. Expression of the MCT and MCTΔTM constructs was generally induced with 200 ng/ml tetracycline (Invitrogen), unless indicated otherwise, after the release from G1/S arrest for 8–10 h. To avoid strong MCT overexpression during long experiments (i.e., live-cell imaging over 6–12 h, immunofluorescence analysis of postmitotic cells and chromatin spreads of cells treated with nocodazole for 20–24 h), tetracycline concentration was reduced to 5 ng/ml after the initial induction with 200 ng/ml tetracycline. To obtain comparable expression levels, MCT and MCTΔTM-GST(-NLS) expression was induced by addition of 5 and 10 ng/ml tetracycline, respectively, 16 h before the release from G1/S arrest. To induce chromatin-membrane tethering before mitotic entry, 200 nM rapamycin (Sigma-Aldrich) (or other concentrations as indicated in Supplemental Figure S4) was added to synchronized cells 8 h after thymidine release. To induce chromatin-membrane tethering during metaphase, 200 nM rapamycin was added to MG132-arrested cells. Live-cell imaging experiments were performed at 37°C with 5% CO2 in Lab-TekII chambers (Thermo Fisher Scientific), Ibidi chambers (Ibidi) or 96-well imaging plates (Greiner). All cell lines were tested negative for mycoplasma using PCR-based testing.
siRNA and plasmid transfections
Plasmid and small interfering RNA (siRNA) transfections were performed using X-tremeGENE 9 (Roche) and INTERFERin (Polyplus) transfection reagents, respectively, according to the manufacturer’s protocol. The following siRNA oligonucleotides were purchased from QIAGEN: Allstar negative control, siMAD2#1 (CAGAAAGCTATCCAGGATGAA) and siMAD2#2 (ATGGATATTTGTACTGTTTAA). Cells were treated with 20 nM siRNA oligonucleotides and imaged 48 h after siRNA transfection.
Immunofluorescence staining
Cells were grown on glass coverslips, washed with phosphate-buffered saline (PBS), fixed with 1 or 4% paraformaldehyde (in PBS) for 10 min, and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After blocking with 2% bovine serum albumin (BSA) in PBS for at least 1 h, cells were incubated with primary antibodies, diluted in 2% BSA in PBS for 1 h at room temperature (RT) or overnight at 4°C, washed three times with PBS, and incubated with secondary antibodies in 2% BSA in PBS for 1 h at RT. After immunostaining, cells were washed with PBS and mounted on microscopic slides with VectaShield (VectorLabs) for microscopic analysis. The following primary antibodies were used in this study: anti-calnexin (Abcam, ab22595), anti-calreticulin (ThermoScientific, PA3-900), CREST (ImmunoVision, HCT0100), anti–Ki-67 (Abcam, ab16667), anti-KIF4 (Abcam, ab3815), anti-KIF22 (Abcam, ab222187), anti-LBR (abcam, ab32535), mAb414 (Abcam, ab24609), anti-MAD2 (Bethyl Laboratories, A300-301A), anti-topoisomeraseII (Abcam, ab109524), anti–α-tubulin (Sigma, T5168), and anti–γ-H2AX (Millipore, 05-636-l). The antibody against human SUN1 has been previously described (Sosa et al., 2013). For secondary antibodies, Alexa Fluor–conjugated antibodies (goat, 1:300; Life Technologies) were used.
Mitotic spreads
Nocodazole-arrested cells were harvested by mitotic shake-off and mitotic chromatin spreads were performed as previously described (Hanisch et al., 2006). Spreads were counterstained with Hoechst, washed three times with PBS, and mounted on microscopic slides with VectaShield for microscopic analysis.
Image acquisition, live-cell imaging, and image analysis
Confocal fluorescence images were acquired with either an upright LSM880 microscope (Zeiss) or an inverted SP2 AOBS (Leica). The LSM880 microscope is operated by the ZEN Black 2012 software and equipped with two standard fluorescent photomultiplier tube (PMT) detectors and a GaAsP detector. Experiments were performed with a 63 × 1.4NA, oil, DIC Plan-Apochromat objective (Figures 1B, 2, 3, 4D, 5, and 7 and Supplemental Figures S1A and S2–S6). The SP2 AOBS microscope is operated by the LCS software and equipped with three standard PMT detectors. Experiments were performed with a 63 × 1.4NA, oil, HCX Plan-Apochromat objective (Figure 6). Wide-field fluorescence images were acquired with a DeltaVision microscope (Olympus IX71) connected to a Roper CoolSnap HQ camera (Photometrics). Image stacks (0.2-μm plane interval) were acquired with a 100 × 1.4NA DIC Oil PlanApo objective (Figure 4, A and B, and Supplemental Figure S1B). The Linux-based DeltaVision Sofware SoftWorx (version 4.1.0) was used to operate the microscope and to perform deconvolution (three iterations) and maximum intensity projection of the image stacks.
We used two fully automated systems equipped with environmental control (37°C, 5% CO2) to perform live-cell imaging for 5–8 h; an ImageXpress microscope (Molecular Devices) for low-resolution widefield imaging (Figure 4C) and an Eclipse T1 (Nikon) for low-resolution widefield imaging (Figure 1, E–G) as well as high-resolution confocal imaging (Figure 1D and Supplemental Movie S1). Low resolution, wide-field time-lapse imaging was performed with either an Eclipse T1 inverted microscope using a 20 × 0.75 CFI Plan Apo VC objective (Zeiss) or with the ImageXpress microscope (equipped with a Photometrics CoolSNAP HQ digital charge-coupled device camera) using a Plan Fluor 10×, NA 0.3 air objective (Nikon). Fluorescent dyes were illuminated with LED lamps at low light intensity and single-plane images were acquired every 5 min. The ImageXpress system is operated by the MetaXpress 5 software and metamorph macros (PlateScan software package) developed in the laboratory of Daniel Gerlich (IMP, Vienna). High resolution, confocal live-cell imaging was performed with a spinning disk confocal microscope (UltraVIEW ERS; PerkinElmer) using a 60 × 1.4 CFI Plan Apo λ Oil objective (Zeiss) and a spinning disk with 50-µm pinhole size. This Nipkow spinning disk setup is equipped with the Yokogawa Confocal Scanner Unit CSU-W1-T2, two sCMOS cameras (Orca Flash 4.0 V2) and is operated by the VisiVIEW (“Metamorph”) software. Image stacks (5 z slices, 2.5-μm plane interval) were acquired every 2 min.
Movie S1.
Time lapse microscopy of rapamycin-treated MCT/H2B* cells. Confocal time lapse microscopy of synchronized MCT/H2B* cells treated with 200 nM rapamycin before mitotic entry as in Figure 1D. Note that in this polyclonal MCT/H2B* cell line only about 70-80% of cells express the MCT. Bar: 10 μm.
Image analysis
Images were analyzed and processed using the FIJI software (ImageJ; National Institutes of Health). Mitotic duration was scored based on time-lapse microscopy by analyzing the time span between NEBD (defined by the onset of chromosome movements at the end of prophase) and anaphase onset (defined by the onset of sister chromatid separation). Movies were generated after performing maximal intensity projection of all z slices. MCT expression levels were determined based on the GFP intensity in confocal images of mitotic cells. Anti-calreticulin immunofluorescence signal was used to estimate cell size. Quantitative immunoblots were analyzed with ImageJ.
Statistical methods
Unpaired two-tailed t tests were run on PRISM (6.0; GraphPad Software), and p values less than 0.001 are indicated by four asterisks (****p ≤ 0.0001), three asterisks (***p ≤ 0.001); p values less than 0.01 by two asterisks (**p ≤ 0.01); p values less than 0.05 by one asterisk (*p ≤ 0.05); and p values higher than 0.05 are marked as “not significant” (ns). Box plots show median, lower, and upper quartiles (line and box), 10th and 90th percentiles (whiskers). Scatter plots show the single data points together with the mean (line). Graphs were generated in PRISM.
Subcellular fractionation and immunoblotting
Nocodazole-arrested, monoclonal MCT/H2B* cells (with 90–100% MCT-expressing cells after tetracycline induction) were harvested by mitotic shake off and washed once with PBS. The cells were resuspended in 500 μl of lysis buffer (10 mM HEPES, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, 0.25% Triton X-100, protease inhibitor mix) and homogenized 15 times through a 27G needle, yielding a crude cell lysate (total cell lysate – input). The chromatin fraction was collected by low-speed centrifugation at 1300 × g for 5 min. The chromatin pellet was washed twice with lysis buffer and resuspended in SDS–PAGE sample buffer (chromatin fraction). The supernatant was centrifuged at 16,000 × g for 20 min, yielding a cleared cytoplasmic fraction. Total cell lysate, cytoplasmic and chromatin fractions were analyzed by Western blotting (Supplemental Figure S3).
For Western blot analysis, whole-cell lysates (or subcellular fractions) were resuspended in SDS–PAGE sample buffer, separated on 8–15% SDS–PAGE gels and transferred onto a nitrocellulose membrane by semidry blotting, followed by Ponceau staining. For immunostaining, the following primary antibodies were used: anti–CAP-D2 and anti–CAP-D3 (Bethyl Laboratories, A300-601A and A300-604A), anti-H3S10p (Cell Signaling, 9701S), anti-MAD2 (Bethyl Laboratories, A300-301A) and anti–α-tubulin (Sigma, T5168), anti-SMC2 (abcam, ab10412) and anti–topoisomerase IIα (Millipore, MAB4197). The antibodies against GFP (Turgay et al., 2014) and Nup53 (Linder et al., 2017) have been previously described. As secondary antibodies, rabbit anti-mouse or goat anti-rabbit horseradish peroxidase–conjugated antibodies (Sigma) were used. For quantitative analysis of immunoblots, a Fusion FX system (Vilber Lourmat) was used.
Supplementary Material
Acknowledgments
We thank I. Zemp for critical reading; C. Ashiono for excellent assistance; D. Gerlich, M. Gotta, C. Lehner, M. Linder, and P. Meraldi for stimulating discussions; and the ETH Zurich Microscopy platform ScopeM for continuous support. This work was funded by a European Research Council Advanced Grant (322582, NucEnv) to U.K.
Abbreviations used:
- CNX
calnexin
- CRT
calreticulin
- FKBP
FK506 binding protein
- FRB
FKBP-rapamycin-binding
- IBB
importin-β–binding (domain of importin-α)
- IF
immunofluorescence
- INM
inner nuclear membrane
- MCT
membrane–chromatin tether
- MT
microtubule
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- noco
nocodazole
- NPC
nuclear pore complex
- NUP
nucleoporin
- rap
rapamycin
- SAC
spindle assembly checkpoint
- tet
tetracycline
- TM
transmembrane domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0609) on December 26, 2018.
REFERENCES
- Baxter J, Aragon L. (2012). A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet , 110–117. [DOI] [PubMed] [Google Scholar]
- Baxter J, Sen N, Martinez VL, De Carandini ME, Schvartzman JB, Diffley JF, Aragon L. (2011). Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science , 1328–1332. [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell , 83–96. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. (2011). A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol , 855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Beckett AJ, Molina O, Samejima I, Masumoto H, Kouprina N, Larionov V, Prior IA, Earnshaw WC. (2016). 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol Cell , 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth DG, Earnshaw WC. (2017). Ki-67 and the chromosome periphery compartment in mitosis. Trends Cell Biol , 906–916. [DOI] [PubMed] [Google Scholar]
- Brachner A, Foisner R. (2011). Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem Soc Trans , 1735–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion L, Linder MI, Kutay U. (2017). Cellular reorganization during mitotic entry. Trends Cell Biol , 26–41. [DOI] [PubMed] [Google Scholar]
- Courvalin JC, Segil N, Blobel G, Worman HJ. (1992). The lamin B receptor of the inner nuclear membrane undergoes mitosis-specific phosphorylation and is a substrate for p34cdc2-type protein kinase. J Biol Chem , 19035–19038. [PubMed] [Google Scholar]
- Cuylen S, Blaukopf C, Politi AZ, Muller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA, Gerlich DW. (2016). Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature , 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. (2011). The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol , 585–610. [DOI] [PubMed] [Google Scholar]
- Dechat T, Gotzmann J, Stockinger A, Harris CA, Talle MA, Siekierka JJ, Foisner R. (1998). Detergent-salt resistance of LAP2alpha in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J , 4887–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger M, Otto H, Neubauer G, Mann M, Hucho F. (1999). Identification of phosphorylation sites in native lamina-associated polypeptide 2 beta. Biochemistry , 9426–9434. [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol , 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. (2005). Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature , 1116–1122. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. (1993). Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell , 1267–1279. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. (2005). Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature , 1043–1047. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. (2006). Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr Biol , 333–344. [DOI] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, et al. (2018). A pathway for mitotic chromosome formation. Science , eaao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloborodko A, Imakaev MV, Marko JF, Mirny L. (2016). Compaction and segregation of sister chromatids via active loop extrusion. Elife , e14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanacz M, Klerkx EP, Galy V, Santarella R, Lopez-Iglesias C, Askjaer P, Mattaj IW. (2007). Caenorhabditis elegans BAF-1 and its kinase VRK-1 participate directly in post-mitotic nuclear envelope assembly. EMBO J , 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Wehner A, Nigg EA, Sillje HH. (2006). Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell , 448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Gonzalez-Sandoval A, Gasser SM. (2016). Histones and histone modifications in perinuclear chromatin anchoring: from yeast to man. EMBO Rep , 139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McKeon F. (1990). Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell , 579–589. [DOI] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. (2008). Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol , 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. (2012). Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J Biol Chem , 42654–42663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Iwase Y, Ishii K, Kumeta M, Horigome T, Takeyasu K. (2009). Cell cycle-dependent phosphorylation of MAN1. Biochemistry , 1636–1643. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Segawa M, Ouchi FS, Yamakawa Y, Furukawa K, Takeyasu K, Horigome T. (2005). Dissociation of emerin from barrier-to-autointegration factor is regulated through mitotic phosphorylation of emerin in a xenopus egg cell-free system. J Biol Chem , 39925–39933. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. (2004). Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci , 6435–6445. [DOI] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, Borner GH. (2016). Global, quantitative and dynamic mapping of protein subcellular localization. Elife , e16950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J. (2013). Release of chromosomes from the nuclear envelope: a universal mechanism for eukaryotic mitosis? Nucleus , 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak M, Haering CH. (2015). Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. BioEssays , 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell , 539–550. [DOI] [PubMed] [Google Scholar]
- Linder MI, Kohler M, Boersema P, Weberruss M, Wandke C, Marino J, Ashiono C, Picotti P, Antonin W, Kutay U. (2017). Mitotic disassembly of nuclear pore complexes involves CDK1- and PLK1-mediated phosphorylation of key interconnecting nucleoporins. Dev Cell , 141–156 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A. (2013). Building silent compartments at the nuclear periphery: a recurrent theme. Curr Opin Genet Dev , 96–103. [DOI] [PubMed] [Google Scholar]
- Molitor TP, Traktman P. (2014). Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol Biol Cell , 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhausser P, Kutay U. (2007). An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown. J Cell Biol , 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. (2007). The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol , 379–393. [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Wiebe MS, Traktman P. (2006). The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Mol Biol Cell , 2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JT, Bottrill A, Prosser SL, Jayaraman S, Straatman K, Fry AM, Shackleton S. (2014). Mitotic phosphorylation of SUN1 loosens its connection with the nuclear lamina while the LINC complex remains intact. Nucleus , 462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. (1990). In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell , 591–602. [DOI] [PubMed] [Google Scholar]
- Piskadlo E, Oliveira RA. (2016). Novel insights into mitotic chromosome condensation. F1000Res , 1807, doi: 10.12688/f1000research.8727.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak C, Aitchison JD, Wozniak RW. (2014). The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol , 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell , 97–107. [DOI] [PubMed] [Google Scholar]
- Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF, et al. (2012). Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol , 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwer M, Schneider MWG, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, Gerlich DW. (2017). DNA cross-bridging shapes a single nucleus from a set of mitotic chromosomes. Cell , 956–972 e923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaitz AL, Thompson J, Wong CC, Yates JR, 3rd, Heald R. (2013). REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell , 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth JT, Beg AM, Wu S, Putney JW, Jr, Rusan NM. (2012). Phosphoregulation of STIM1 leads to exclusion of the endoplasmic reticulum from the mitotic spindle. Curr Biol , 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Kutay U, Schwartz TU. (2013). Structural insights into LINC complexes. Curr Opin Struct Biol , 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titos I, Ivanova T, Mendoza M. (2014). Chromosome length and perinuclear attachment constrain resolution of DNA intertwines. J Cell Biol , 719–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng LC, Chen RH. (2011). Temporal control of nuclear envelope assembly by phosphorylation of lamin B receptor. Mol Biol Cell , 3306–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Champion L, Balazs C, Held M, Toso A, Gerlich DW, Meraldi P, Kutay U. (2014). SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J Cell Biol , 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW. (2006). Direct membrane protein–DNA interactions required early in nuclear envelope assembly. J Cell Biol , 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. (2015). Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol , 687–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. (2015). Establishment of NE asymmetry-targeting of membrane proteins to the inner nuclear membrane. Curr Opin Cell Biol , 135–141. [DOI] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. (2017). Mechanisms and functions of nuclear envelope remodelling. Nat Rev Mol Cell Biol , 229–245. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Belmont AS. (2017). Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell , 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne C, Klopfenstein DR, Hauri HP. (2005). Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell , 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther N, Hossain MJ, Politi AZ, Koch B, Kueblbeck M, Odegard-Fougner O, Lampe M, Ellenberg J. (2018). A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J Cell Biol , 2309–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. (1997). Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol , 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.