Abstract
Objective
Turnover of cartilage endplate extracellular matrix (ECM) may play an important role in disc degeneration and low back pain (LBP). However, the expression pattern of pro‐inflammatory factors, matrix metalloproteinases (MMP), and tissue inhibitors of metalloproteinases (TIMP) in the cartilage endplates (CEP) of intervertebral discs (IVD) is not understood. We aimed to examine the transcriptional levels of MMP, TIMP, and interleukins (IL), and the correlations between them.
Methods
Thirty degenerated cartilage endplate samples from patients with LBP who underwent lumbar fusion surgery were included in the degenerated group. Ten patients without LBP history who underwent lumbar surgery because of vertebral burst fractures were included in the control group. The degenerative severity of the samples was evaluated by MRI, and hematoxylin–eosin and safranin O‐fast green (SO‐FG) staining. Real‐time polymerase chain reaction (RT‐PCR) was used to detect the mRNA levels of MMP‐1, MMP‐3, MMP‐9, MMP‐13, TIMP‐1, TIMP‐2, TIMP‐3, IL‐1α, IL‐1β, and IL‐6. The correlations between the levels of these genes were tested using Spearman’s rho test.
Results
Hematoxylin–eosin and SO‐FG staining confirmed a decrease in cell number and proteoglycans in the degenerated cartilage endplate. MRI showed significant signal changes in degenerated cartilage endplates. Patients in the degenerated group showed a higher rate of endplate Modic changes when compared with the control group. MMP‐3, MMP‐9, TIMP‐3, IL‐1α, and IL‐1β were elevated with statistical significance, while MMP‐1, MMP‐13, TIMP‐1, TIMP‐2, and IL‐6 were changed without statistical significance or remained unchanged. Expression of MMP‐3 was positively correlated with IL‐1α (Spearman coefficient, 0.486; P < 0.05); expression of TIMP‐3 was positively correlated with MMP‐9, IL‐1α, and IL‐1β (Spearman coefficient, 0.577, 0.407, and 0.571, respectively; P < 0.05).
Conclusion
MMP‐3, MMP‐9, TIMP‐3, IL‐1α, and IL‐1β may play a role in the process of cartilage endplate degeneration. MMP‐3 may be regulated by IL‐1α, and TIMP‐3 might be associated with MMP‐9 and regulated by IL‐1α and IL‐1β.
Keywords: Cartilage endplate, Degenerative disc disease, Interleukins, MMP, TIMP
Introduction
Low back pain (LBP) is one of the most common complaints in outpatients, leading to chronic pain and lost working hours, and high costs for treatment1. Intervertebral disc degeneration has been proven to be one of the most important causes of LBP2, 3. The cartilage endplate comprises the superior and inferior boundaries of the intervertebral disc and is the major nutrient supply channel for the intervertebral disc4. As decreased nutrient supply is associated with intervertebral disc degeneration, it is reasonable to assume that changes in the cartilage endplate may exhibit a marked effect on disc degeneration. Besides, the degenerated cartilage endplate is a potential source of inflammatory mediators, such as tumor necrosis factor (TNF)‐α, interleukin‐1β (IL‐1β), IL‐6, and macrophage inhibition factor (MIF), and stimulates adjacent disc tissue like nucleus pulposus5.
Cartilage endplate extracellular matrix (ECM) degradation is among the most important processes of degeneration. The main component of cartilage endplates is type II collagen and aggrecan. As major constituents of the cartilage endplates (CEP), degradation of collagen II and aggrecan are regarded as a central feature in cartilage endplate degeneration, wherein matrix metalloproteinase (MMP)‐13 is the principal protease responsible for type II collagen degradation, and a disintegrin and metalloproteinase with thrombospond in motifs‐5 (ADAMTS‐5) is the main aggrecan‐degrading proteinase; both are thought to play critical roles in CEP degeneration6.
Our previous work confirmed that ADAMTS‐5 is upregulated in degenerated cartilage endplates with Modic changes7. It is well recognized in articular cartilage that activation of MMP leads to degradation and loss of ECM8. Increasing evidence has also shown that pro‐inflammatory cytokines, such as interleukin‐1 (IL‐1) and tumor necrosis factor‐α (TNF‐α), could stimulate expression of MMP9, while tissue inhibitor of metalloproteinase (TIMP) could act as specific inhibitor of the proteolytic activity of MMP by binding to active forms of MMP10. However, few studies focus on the expression of pro‐inflammatory factors, MMP, and TIMP in cartilage endplate of intervertebral discs (IVD).
In this study, we investigated the expression pattern of pro‐inflammatory factors, MMP and TIMP in the cartilage endplates of intervertebral discs and compared the difference between normal and degenerated cartilage endplates. In addition, we examined the transcriptional levels of MMP, TIMP, and IL, and the correlations between them. Thus, the aim of the present study was to explore the potential pathogenesis of cartilage endplates and IVD degeneration.
Subjects and Methods
Cartilage End Plate Collection
The present study was approved by the Ethics Committee of Sir Run Run Shaw Hospital, affiliated to the Zhejiang University School of Medicine. Informed consent was obtained from all participants included in the study. The study included 30 degenerated cartilage endplate samples from patients with severe lumbocrural pain and diagnosed as having lumbar disc herniation, spinal stenosis or lumbar spondylolisthesis. They underwent lumbar fusion surgery and were included in the degenerated group. Ten patients without LBP history who underwent lumbar surgery because of vertebral burst fractures were included in the control group. All samples were collected in our hospital from January 2012 to May 2013. Preoperative lumbar MRI of patients from both groups were collected and used for Modic changes classification. Both the superior endplate and inferior endplate of the surgical lumbar disc were collected. The collected samples weighted approximately 5 g; once collected, nucleus pulposus and annulus fibrosus were dissected away immediately, under microscopic examination, and refrigerated in liquid nitrogen or fixed in 4% buffered p‐formaldehyde solution for 24 h.
There were no differences in gender and body mass index (BMI) between degenerated and control groups. The patients in the control group were younger than in the degenerated group, although the difference showed no significance (41.70 vs 55.75 years old; U = 106, P = 0.113).
RNA Extraction
Cartilage endplate tissue was frozen in liquid nitrogen and ground into a fine powder using a mortar and pestle. Total RNA was extracted according to the manufacturer’s protocols using RNeasy Mini Kits (Qiagen, Valencia, CA). Total RNA quantity was measured by spectrophotometer. Complementary DNA (cDNA) was synthesized using 0.5 μg RNA, 4 μL dNTP Mix, 4 μL 5 × RT Buffer, 2 μL Prime Mix, 2 μL DTT and 1 μL HiFiScript. RNase free dH2O was added up to a volume of 20 μL. RT‐PCR was performed on an ABI Prism 7500 96‐well plate (Applied Biosystems, Foster City, CA, USA) using UltraSYBR Mixture (KWBIO, China). The total volume (20 μL) of each PCR reaction consisted of 10 μL UltraSYBR Mixture, 7 μL ddH2O, 2 μL complementary DNA, and 0.5 μL each of forward and reverse primers (Table 1).
Table 1.
Sequences of primers for real‐time polymerase chain reaction
| Genes | Sense | Sequence 5′→3′ | Accession number |
|---|---|---|---|
| MMP‐1 | F | GAAGAATGATGGGAGGCAAGT | NM_001145938.1 |
| R | GAGGACAAACTGAGCCACATC | ||
| MMP‐3 | F | AACATCCAAAAACGCCAGAC | NM_002422.4 |
| R | GGAAGTTCTGGCCAAAATGA | ||
| MMP‐9 | F | GATGCGTGGAGAGTCGAAAT | NM_004994.2 |
| R | CTATCCAGCTCACCGGTCTC | ||
| MMP‐13 | F | TCTTCGGCTTAGAGGTGACTG | NM_002427.3 |
| R | CAGAGGAGTTACATCGGACCA | ||
| TIMP‐1 | F | CTTCTGGCATCCTGTTGTTG | NM_003254.2 |
| R | AGAAGGCCGTCTGTGGGT | ||
| TIMP‐2 | F | AAGCGGTCAGTGAGAAGGAA | NM_003255.4 |
| R | TCTCAGGCCCTTTGAACATC | ||
| TIMP‐3 | F | ACCTGCCTTGCTTTGTGACT | NM_000362.4 |
| R | GGCGTAGTGTTTGGACTGGT | ||
| IL‐1α | F | GTATGTGACTGCCCAAGATGAA | NM_000575.4 |
| R | CACACCCAGTAGTCTTGCTTTG | ||
| IL‐1β | F | ACAGATGAAGTGCTCCTTCCA | NM_000576.2 |
| R | GTCGGAGATTCGTAGCTGGAT | ||
| IL‐6 | F | GACAGCCACTCACCTCTTCAG | NM_000600.4 |
| R | CATCCATCTTTTTCAGCCATC | ||
| IL‐8 | F | TTGCCAAGGAGTGCTAAAGAA | NM_000584.3 |
| R | GCCCTCTTCAAAAACTTCTCC | ||
| SDHA | F | AGACCTAAAGCACCTGAAGACG | NM_001294332.1 |
| R | ATCAATCCGCACCTTGTAGTCT |
IL, interleukin; MMP, matrix metalloproteinases; TIMP, tissue inhibitors of metalloproteinases
Real‐time Polymerase Chain Reaction
Real‐time polymerase chain reaction was performed at 95°C for 10 min (activation), 95°C for 10 s, 60°C for 20 s, 72°C for 20 s for 40 cycles (amplification), and 72°C for 1 min (final extension). As our previous experiments showed that succinate dehydrogenase complex, subunit A is a comparably far more stable internal control gene than the other genes that were tested, our data were normalized to succinate dehydrogenase complex, subunit A11.
Histology
Cartilage endplates, after being fixed in 4% buffered p‐formaldehyde for 24 h, decalcified in 10% ethylene diamine tetraacetic acid for 1 month, and embedded in paraffin, were cut into three serial sections, so that each specimen was 4‐μm thick. These sections were then stained with hematoxylin–eosin (H&E) and safranin O‐fast green (SO‐FG) for observing cell density, morphology, matrix degeneration, and proteoglycans content.
Statistical Analysis
The Mann–Whitney U‐test was used to evaluate the patients’ age, BMI, and changes in mRNA levels between the degenerated group and the control group. Spearman’s rho test was used to analyze the correlation between mRNA expression of IL, MMP, and TIMP. Data analysis was performed using SPSS 16.0 (SPSS, Chicago, IL). All values were expressed as mean ± standard deviation. P < 0.05 indicated a significant difference between groups.
Results
Radiological and Morphological Changes in Degenerated Cartilage Endplate
Modic changes are MRI signal intensity changes in the vertebral bone marrow and reflect lesions in endplates. Nine patients in the degenerated group (n = 30) showed no Modic change in preoperative lumbar MRI, 2 Modic I changes, 17 Modic II changes, and 2 Modic III changes. All patients in the control group (n = 10) showed no Modic change, indicating that endplate Modic change may be related to cartilage endplate degeneration.
The results of MRI as well as H&E and SO‐FG staining demonstrated apparent changes in patients with degenerated cartilage endplates (Fig. 1A–H). MRI of the degenerated group (Fig. 1C,D) showed uneven and broken cartilage endplates with significant signal changes in subchondral bone and bone marrow, which we referred to as Modic changes. Decrease in T2WI signal intensity of nucleus pulposus (NP), disappearance of the demarcation between NP and annulus fibrosus, and narrowing of the intervertebral space compared with the trauma control group were also observed (Fig. 1A,B). H&E staining (Fig. 1E,F) showed a large decrease in cell number in the degenerated cartilage endplates compared with the control group, with the shape of the chondrocytes also changed from round small cells to spindle‐like cells in the degenerated disc group. SO‐FG staining demonstrated a sharp reduction in the content of proteoglycans, as shown in Fig. 1G,H.
Figure 1.
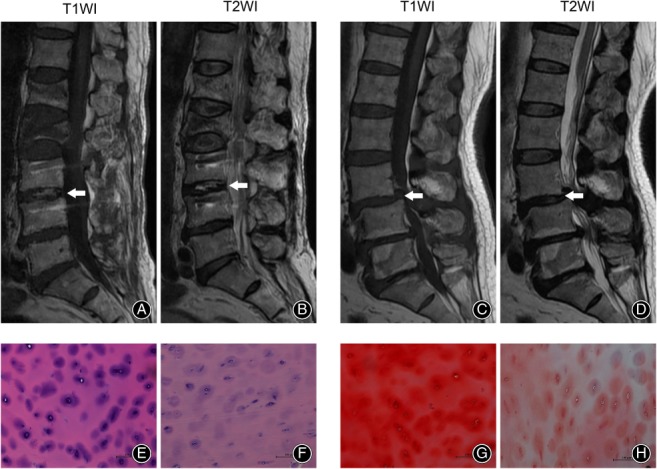
MRI and histological changes in cartilage endplates of control and degenerated group. (A, B) A 76‐year‐old male patient with L2 fracture. A sample of cartilage endplate of L2–L3 was collected for our study (white arrow). (C, D) A 65‐year‐old male patient with degenerative disc disease. A sample of cartilage of L3–L4 was collected for our study (white arrow). (E, F) Hematoxylin–eosin‐stained cartilage endplate samples of the control group (E) and the degenerated group (F). (G, H) Safranin O stained cartilage endplate samples of the control group (G) and degenerated group (H). Scale bar: 100 μm.
mRNA Levels of Matrix Metalloproteinases, Tissue Inhibitors of Metalloproteinases, and Interleukins in Control and Degenerated Cartilage Endplate
Among the MMP, expression of MMP‐3 and MMP‐9 were elevated with statistical significance, while MMP‐1 was upregulated without statistical significance and expression of MMP‐13 was largely unchanged between control and degenerated cartilage endplates.
Transcriptional levels of TIMP‐1 and TIMP‐2 were slightly downregulated, but this finding was not statistically significant, while TIMP‐3 was significantly upregulated in degenerated cartilage endplate.
Interleukin‐1α and IL‐1β showed a trend toward an increase in degenerated cartilage endplate with significant difference, while the expression of IL‐6 remained unchanged (Table 2).
Table 2.
mRNA levels of MMP, TIMP, and IL in control and degenerated cartilage endplate (mean ± SD)
| Genes | Control group (n = 10) | Degenerated group (n = 30) | P‐value* |
|---|---|---|---|
| MMP1 | 3.48 ± 4.00 | 13.23 ± 37.87 | 0.49 |
| MMP3 | 24.56 ± 18.68 | 2695.21 ± 5681.59 | 0.02 |
| MMP9 | 0.54 ± 0.46 | 2.29 ± 1.70 | 0.04 |
| MMP13 | 2.15 ± 4.24 | 1.93 ± 2.89 | 0.52 |
| TIMP1 | 2.48 ± 2.92 | 1.43 ± 2.02 | 0.35 |
| TIMP2 | 2.28 ± 2.34 | 1.34 ± 0.93 | 0.57 |
| TIMP3 | 2.61 ± 1.99 | 24.14 ± 41.88 | 0.03 |
| IL‐1α | 0.97 ± 0.60 | 28.00 ± 79.76 | 0.03 |
| IL‐1β | 4.53 ± 5.60 | 586.65 ± 1740.10 | 0.04 |
| IL‐6 | 142.99 ± 151.10 | 66.90 ± 124.57 | 0.34 |
Mann–Whitney U‐test. IL, interleukin; MMP, matrix metalloproteinases; TIMP, tissue inhibitors of metalloproteinases
Spearman’s rho test also showed that expression of MMP‐3 was positively correlated with that of IL‐1α (Spearman coefficient, 0.486; P < 0.05) and expression of TIMP‐3 was positively correlated with that of IL‐1α, IL‐1β, and MMP‐9 (Spearman coefficient, 0.407, 0.571, and 0.577, respectively; P < 0.05).
We also compared the gene expression levels among patients in the control group and in the degenerated group with different types of Modic changes. However, as shown in Table 3, no difference was found in MMP, TIMP, and IL expressions among the patients in the degenerated group with different types of Modic changes and the control group, except for IL‐6 (P < 0.01). As further analysis shows in Table 4, IL‐6 was found to be significantly upregulated in patients of the degenerated group with Modic I changes compared with other Modic types and the control group (P < 0.01).
Table 3.
Gene expression levels of MMP, TIMP, and IL in control and degenerated cartilage endplate (mean ± SD)
| Genes | Control group (n = 10) | Degenerated samples without Modic changes (n = 8) | Degenerated samples with Modic I (n = 2) | Degenerated samples with Modic II (n = 18) | Degenerated samples with Modic III (n = 2) | F‐value (one‐way ANOVA) |
|---|---|---|---|---|---|---|
| MMP1 | 3.48 ± 4.00 | 33.34 ± 73.01 | 10.42 ± 5.10 | 3.84 ± 3.85 | 8.00 ± 5.80 | 0.66 |
| MMP3 | 24.56 ± 18.68 | 420.10 ± 901.29 | 8474.95 ± 11894.12 | 3476.32 ± 6323.95 | 225.82 ± 149.35 | 0.26 |
| MMP9 | 0.54 ± 0.46 | 1.67 ± 1.41 | 3.23 ± 2.95 | 2.60 ± 1.75 | 1.33 ± 1.20 | 0.36 |
| MMP13 | 2.15 ± 4.24 | 0.16 ± 0.23 | 1.61 ± 0.75 | 2.41 ± 4.27 | 2.81 ± 1.06 | 0.95 |
| TIMP1 | 2.48 ± 2.92 | 0.54 ± 0.61 | 5.36 ± 4.51 | 1.04 ± 0.74 | 1.11 ± 0.37 | 0.10 |
| TIMP2 | 2.28 ± 2.34 | 1.05 ± 1.20 | 1.95 ± 0.89 | 1.31 ± 1.00 | 1.40 ± 0.25 | 0.81 |
| TIMP3 | 2.61 ± 1.99 | 12.02 ± 13.45 | 15.40 ± 17.90 | 31.16 ± 53.41 | 24.18 ± 0.72 | 0.73 |
| IL‐1α | 0.97 ± 0.60 | 2.70 ± 1.19 | 11.38 ± 10.86 | 42.18 ± 100.92 | 8.65 ± 10.70 | 0.75 |
| IL‐1β | 4.53 ± 5.60 | 61.59 ± 55.27 | 385.22 ± 533.40 | 798.02 ± 2294.12 | 1251.88 ± 1645.36 | 0.73 |
| IL‐6 | 142.99 ± 151.10 | 16.89 ± 11.24 | 325.11 ± 91.73 | 17.85 ± 21.35 | 6.36 ± 2.27 | 0.00 |
Table 4.
Comparison of interleukin‐6 expression among the patients in the degenerated group with different types of Modic changes and the control group (P‐value)
| Groups | Control group | Degenerated samples without Modic change | Degenerated samples with Modic I | Degenerated samples with Modic II | Degenerated samples with Modic III |
|---|---|---|---|---|---|
| Control group | ‐ | 0.017 | 0.000 | 0.791 | 0.865 |
| Degenerated samples without Modic change | 0.017 | ‐ | 0.003 | 0.001 | 0.023 |
| Degenerated samples with Modic I | 0.000 | 0.003 | ‐ | 0.000 | 0.000 |
| Degenerated samples with Modic II | 0.791 | 0.001 | 0.000 | ‐ | 0.988 |
| Degenerated samples with Modic III | 0.865 | 0.023 | 0.000 | 0.988 | ‐ |
Discussion
In this study, we collected both trauma control and degenerated human intervertebral cartilage samples and investigated the expression of MMP, TIMP, and IL. Our results showed that MMP‐3, MMP‐9, TIMP‐3, IL‐1α, and IL‐1β were expressed at different levels in degenerated cartilage endplates, while the expression of MMP‐13, TIMP‐1, TIMP‐2, and IL‐6 remained unchanged. Furthermore, the mRNA level of MMP‐3 was positively correlated with that of IL‐1α, and expression of TIMP‐3 was positively correlated with that of MMP‐9, IL‐1α, and IL‐1β.
Matrix Metalloproteinase‐3 and Matrix Metalloproteinase‐9 Are the Major Matrix Metalloproteinases Involved in the Degeneration of Cartilage Endplate
To the best of our knowledge, MMP are critical proteolytic enzymes in cartilage turnover, and they are responsible for the degrading of almost all the components of the cartilage ECM12, 13. MMP‐1, MMP‐3, MMP‐9, and MMP‐13 are the major MMP involved in the degeneration of articular cartilage and can be upregulated by inflammatory factors13, 14, 15. MMP‐1 and MMP‐13 are collagenases that account for most collagen turnover in cartilage, and they are the major rate‐limiting enzymes, especially MMP‐13, as the proteolytic enzyme of type II collagen13. MMP‐3 is an important proteinase in cartilage degradation as a collagenase, aggrecanase, and proteinase for other non‐collagen cartilage ECM components, such as fibronectin, elastin, and laminin16. As a collagenase, MMP‐3 has 5–10 times the type II collagen degrading activity compared with MMP‐1. MMP‐9 is a gelatinase that further degrades denatured collagen or gelatin after the cleaving work of collagenase. These MMP are reported to be significantly increased with pathological conditions of articular cartilage, such as rheumatoid arthritis (RA) and osteoarthritis (OA)13. However, in the present study of cartilage endplates, expression of MMP‐1 and MMP‐13 were not upregulated in degenerated samples. As MMP‐1 is synthesized in synovial cells in articular and synovial tissue, which is absent in intervertebral discs, it is not surprising that MMP‐1 was not upregulated. The results of MMP‐13 expression may suggest that the role of MMP‐13 in cartilage endplate degeneration is not as important as that in articular cartilage. As MMP‐1 and MMP‐13 are major rate‐limiting collagenases, this implies that the decline in anabolism may exceed the elevation in catabolism in collagen metabolism when the disc degenerates.
Tissue Inhibitors of Metalloproteinase‐3 Was Upregulated in Degenerated Cartilage Endplate
Tissue inhibitors of metalloproteinases are physiologic inhibitors of MMP, which are produced by the same connective tissue that produce MMP17. A cysteine residue of TIMP may chelate the zinc ion of the MMP active site to inhibit the activity of MMP18. As TIMP can block all the MMP and are synthesized in chondrocytes, they are considered potential therapeutic targets for cartilage degeneration. In normal articular cartilage, TIMP levels are marginally higher than proteinases levels, while in OA cartilage, TIMP are upregulated but the elevated proteinases exceed TIMP. This is proposed as a possible contributing cause of cartilage break down19. In OA and RA cartilage, TIMP‐3 is regarded as the leading candidate13, 20. It can block the activity of both ADAMTS and MMP. Our results in cartilage endplates showed that TIMP‐3, but not TIMP‐1 and TIMP‐2, was upregulated in degenerated cartilage endplates and positively correlated with MMP‐9, IL‐1α, and IL‐1β, implying that TIMP‐3 is the most important TIMP in the degenerative process of cartilage endplates and it is associated with the expression of MMP‐9 and regulation of the inflammatory factors IL‐1α and IL‐1β.
Interleukin‐1α interleukin‐1β Were Upregulated in Degenerated Cartilage Endplate
The role of inflammatory factors in cartilage pathology is well established. They not only suppress the expression of genes related to the cartilage phenotype but also the expression of proteinases such as MMP21, thereby disrupting the balance between anabolism and catabolism. The interleukin family includes important inflammatory factors. IL‐1β is one of the most intensely studied cytokines in cartilage degeneration. IL‐1β and its receptor IL‐1RI have been detected in OA chondrocytes22, 23. The destructive effect of IL‐1β has been proven in an OA model, an ex vivo articular cartilage model, and a protective experiment of IL‐1 receptor agonist24. The actions of IL‐1α and IL‐1β are similar. Their receptors share the same ligand binding chain and signal transducing chain25. However, different from IL‐1β, IL‐1α activity is independent of the inflammasome caspase‐1 pathway26. Regarding inflammation‐induced arthritis and cartilage degeneration, IL‐1β has been mentioned considerably more than IL‐1α. IL‐6 may modulate the destructive cytokines instead of destroying cartilage itself. It has been reported that IL‐6 suppresses the expression of anabolic genes, although not as strongly as IL‐127. Some experts propose that IL‐6 has both protective and destructive roles28. In our study, IL‐1β and IL‐1α were differentially expressed in degenerated cartilage endplates, which is in accordance with the results of studies on articular cartilage. Expression of IL‐1α was correlated with MMP‐3, which suggests that IL‐1α plays a role in cartilage turnover. Expression of IL‐6 was not changed in degenerated tissue and this may imply cartilage endplate degrading without the effect of IL‐6.
For the first time, our study examined the expression pattern of MMP, TIMP, and IL in human degenerated cartilage endplates and elucidated the difference between articular cartilage and cartilage endplate. MMP‐3, MMP‐9, and TIMP‐3 may be the major MMP and TIMP, and IL‐1α and IL‐1β but not IL‐6 may be effective IL in the process of cartilage endplate degeneration. MMP‐3 may be regulated by IL‐1α, and TIMP‐3 may be associated with MMP‐9 levels and be regulated by IL‐1α and IL‐1β.
However, this a primary study involving only the transcriptional levels of these genes. Further studies are planning to knock down the possible upstream factors (inflammatory factors) and to evaluate the expression of possible downstream catabolism genes to confirm the relationship between them.
Conclusion
In this study, the transcriptional levels of MMP, TIMP, and IL, and the correlations between them were evaluated. We found that MMP‐3, MMP‐9, TIMP‐3, IL‐1α, and IL‐1β may play a role in the process of cartilage endplate degeneration. MMP‐3 may be regulated by IL‐1α, and TIMP‐3 may be associated with MMP‐9 and be regulated by IL‐1α and IL‐1β.
Acknowledgments
The authors did not receive any outside funding or grants in support of their research or for preparation of this work.
Disclosure: This study was supported by the Chinese National Natural Science Foundation (Grant Nos. 81601924 and 81472064), the Natural Science Foundation of Zhejiang Province (Grant No. LZ15H060002), the Platform Major Project of Health and Family Planning Commission of Zhejiang Province (Grant No. 2016145597, 2015KYA133, and 2017KY087), and the Project of Education Department of Zhejiang Province (Grant No. Y201017108).
References
- 1. Deyo RA, Weinstein JN. Low back pain. N Engl J Med, 2001, 344: 363–370. [DOI] [PubMed] [Google Scholar]
- 2. Samartzis D, Karppinen J, Mok F, Fong DY, Luk KD, Cheung KM. A population‐based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J Bone Joint Surg Am, 2011, 93: 662–670. [DOI] [PubMed] [Google Scholar]
- 3. Battié MC, Videman T, Levalahti E, Gill K, Kaprio J. Heritability of low back pain and the role of disc degeneration. Pain, 2007, 131: 272–280. [DOI] [PubMed] [Google Scholar]
- 4. Ogata K, Whiteside LA. Volvo award winner in basic science. Nutritional pathways of the intervertebral disc. An experimental study using hydrogen washout technique. Spine (Phila Pa 1976), 1980, 1981: 211–216. [PubMed] [Google Scholar]
- 5. Neidlinger‐Wilke C, Boldt A, et al Molecular interactions between human cartilaginous endplates and nucleus pulposus cells: a preliminary investigation. Spine (Phila Pa 1976), 2014, 39: 1355–1364. [DOI] [PubMed] [Google Scholar]
- 6. Yurube T, Takada T, Suzuki T, et al Rat tail static compression model mimics extracellular matrix metabolic imbalances of matrix metalloproteinases, aggrecanases, and tissue inhibitors of metalloproteinases in intervertebral disc degeneration. Arthritis Res Ther, 2012, 14: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen S, Huang Y, Zhou ZJ, et al Upregulation of tumor necrosis factor alpha and ADAMTS‐5, but not ADAMTS‐4, in human intervertebral cartilage endplate with modic changes. Spine (Phila Pa 1976), 2014, 39: E817–E825. [DOI] [PubMed] [Google Scholar]
- 8. Wang X, Zhao X, Tang S. Inhibitory effects of EGb761 on the expression of matrix metalloproteinases (MMPs) and cartilage matrix destruction. Cell Stress Chaperones, 2015, 20: 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kullich W, Fagerer N, Schwann H. Effect of the NSAID nimesulide on the radical scavenger glutathione S‐transferase in patients with osteoarthritis of the knee. Curr Med Res Opin, 2007, 23: 1981–1986. [DOI] [PubMed] [Google Scholar]
- 10. Cawston TE, Galloway WA, Mercer E, Murphy G, Reynolds JJ. Purification of rabbit bone inhibitor of collagenase. Biochem J, 1981, 195: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou ZJ, Zhang JF, Xia P, et al Selection of suitable reference genes for normalization of quantitative real‐time polymerase chain reaction in human cartilage endplate of the lumbar spine. PLoS One, 2014, 9: e88892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagase H, Woessner JF Jr. Matrix metalloproteinases. J Biol Chem, 1999, 274: 21491–21494. [DOI] [PubMed] [Google Scholar]
- 13. Burrage PS, Mix KS, Brinckerhoff CE. Matrix metalloproteinases: role in arthritis. Front Biosci, 2006, 11: 529–543. [DOI] [PubMed] [Google Scholar]
- 14. Vincenti MP, White LA, Schroen DJ, Benbow U, Brinckerhoff CE. Regulating expression of the gene for matrix metalloproteinase‐1 (collagenase): mechanisms that control enzyme activity, transcription, and mRNA stability. Crit Rev Eukaryot Gene Expr, 1996, 6: 391–411. [DOI] [PubMed] [Google Scholar]
- 15. Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol, 2002, 3: 207–214. [DOI] [PubMed] [Google Scholar]
- 16. Bonassar LJ, Frank EH, Murray JC, et al Changes in cartilage composition and physical properties due to stromelysin degradation. Arthritis Rheum, 1995, 38: 173–183. [DOI] [PubMed] [Google Scholar]
- 17. Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta, 2000, 1477: 267–283. [DOI] [PubMed] [Google Scholar]
- 18. Murphy G, Willenbrock F. Tissue inhibitors of matrix metalloendopeptidases. Methods Enzymol, 1995, 248: 496–510. [DOI] [PubMed] [Google Scholar]
- 19. Dean DD, Martel‐Pelletier J, Pelletier JP, Howell DS, Woessner JF Jr. Evidence for metalloproteinase and metalloproteinase inhibitor imbalance in human osteoarthritic cartilage. J Clin Invest, 1989, 84: 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kashiwagi M, Enghild JJ, Gendron C, et al Altered proteolytic activities of ADAMTS‐4 expressed by C‐terminal processing. J Biol Chem, 2004, 279: 10109–10119. [DOI] [PubMed] [Google Scholar]
- 21. Reginato AM, Sanz‐Rodriguez C, Diaz A, Dharmavaram RM, Jimenez SA. Transcriptional modulation of cartilage‐specific collagen gene expression by interferon gamma and tumour necrosis factor alpha in cultured human chondrocytes. Biochem J, 1993, 294: 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moos V, Fickert S, Müller B, Weber U, Sieper J. Immunohistological analysis of cytokine expression in human osteoarthritic and healthy cartilage. J Rheumatol, 1999, 26: 870–879. [PubMed] [Google Scholar]
- 23. Martel‐Pelletier J, Alaaeddine N, Pelletier JP. Cytokines and their role in the pathophysiology of osteoarthritis. Front Biosci, 1999, 4: D694–D703. [DOI] [PubMed] [Google Scholar]
- 24. Pelletier JP, Caron JP, Evans C, et al In vivo suppression of early experimental osteoarthritis by interleukin‐1 receptor antagonist using gene therapy. Arthritis Rheum, 1997, 40: 1012–1019. [DOI] [PubMed] [Google Scholar]
- 25. Wesche H, Korherr C, Kracht M, Falk W, Resch K, Martin MU. The interleukin‐1 receptor accessory protein (IL‐1RAcP) is essential for IL‐1‐induced activation of interleukin‐1 receptor‐associated kinase (IRAK) and stress‐activated protein kinases (SAP kinases). J Biol Chem, 1997, 272: 7727–7731. [DOI] [PubMed] [Google Scholar]
- 26. Kwak A, Lee Y, Kim H, Kim S. Intracellular interleukin (IL)‐1 family cytokine processing enzyme. Arch Pharm Res, 2016, 39: 1556–1564. [DOI] [PubMed] [Google Scholar]
- 27. Guerne PA, Desgeorges A, Jaspar JM, et al Effects of IL‐6 and its soluble receptor on proteoglycan synthesis and NO release by human articular chondrocytes: comparison with IL‐1. Modulation by dexamethasone. Matrix Biol, 1999, 18: 253–260. [DOI] [PubMed] [Google Scholar]
- 28. Westacott CI, Sharif M. Cytokines in osteoarthritis: mediators or markers of joint destruction?. Semin Arthritis Rheum, 1996, 25: 254–272. [DOI] [PubMed] [Google Scholar]


