Abstract
The World Health Organisation rotavirus surveillance networks have documented and shown eclectic geographic and temporal diversity in circulating G- and P- genotypes identified in children <5 years of age. To effectively monitor vaccine performance and effectiveness, robust molecular and phylogenetic techniques are essential to detect novel strain variants that might emerge due to vaccine pressure. This study inferred the phylogenetic history of the VP7 and VP4 genes of previously non-typeable strains and provided insight into the diversity of P[8] VP4 sequences which impacted the outcome of our routine VP4 genotyping method. Near-full-length VP7 gene and the VP8* fragment of the VP4 gene were obtained by Sanger sequencing and genotypes were determined using RotaC v2.0 web-based genotyping tool. The genotypes of the 57 rotavirus-positive samples with sufficient stool was determined. Forty-eight of the 57 (84.2%) had the P[8] specificity, of which 43 (89.6%) were characterized as P[8]a subtype and 5 (10.4%) as the rare OP354-like subtype. The VP7 gene of 27 samples were successfully sequenced and their G-genotypes confirmed as G1 (18/27) and G9 (9/27). Phylogenetic analysis of the P[8]a sequences placed them in subcluster IIIc within lineage III together with contemporary G1P[8], G3P[8], G8P[8], and G9P[8] strains detected globally from 2006–2016. The G1 VP7 sequences of the study strains formed a monophyletic cluster with African G1P[8] strains, previously detected in Ghana and Mali during the RotaTeq vaccine trial as well as Togo. The G9 VP7 sequences of the study strains formed a monophyletic cluster with contemporary African G9 sequences from neighbouring Burkina Faso within the major sub-cluster of lineage III. Mutations identified in the primer binding region of the VP8* sequence of the Ghanaian P[8]a strains may have resulted in the genotyping failure since the newly designed primer successfully genotyped the previously non-typeable P[8] strains. In summary, the G1, G9, and P[8]a sequences were highly similar to contemporary African strains at the lineage level. The study also resolved the methodological challenges of the standard genotyping techniques and highlighted the need for regular evaluation of the multiplex PCR-typing method especially in the post-vaccination era. The study further highlights the need for regions to start using sequencing data from local rotavirus strains to design and update genotyping primers.
Introduction
Diarrhoea is one of the leading causes of childhood mortality and was responsible for nearly 500, 000 childhood deaths in 2015 [1]. Rotavirus A (RVA), a member of the genus Rotavirus and family Reoviridae [2], is the leading cause of viral diarrhoea in children under five years. In 2016, rotavirus was responsible for an estimated 128, 500 deaths of children under the age of five, of which 104, 733 deaths occurred in sub-Saharan Africa [3]. Prior to the introduction of the monovalent rotavirus vaccine—Rotarix into Ghana’s Expanded Programme on Immunisation (EPI) in May 2012, rotaviruses killed about 2,090 children below five years of age accounting for 3.6% of annual diarrhoeal disease deaths in Ghana (https://path.azureedge.net/media/documents/VAD_rotavirus_ghana_fs.pdf).
Rotaviruses are non-enveloped viruses which possess a triple-layered capsid surrounding a genome of 11 segments of double-stranded RNA (dsRNA). The genome encodes six structural viral proteins (VP1—VP4, VP6 and VP7) and six non-structural proteins (NSP1—NSP6) [2]. The outer layer is made up of two proteins, VP7 and VP4 against which neutralizing antibodies are produced. The VP6 forms the middle layer whereas the VP2 forms the core capsid that surrounds the genome of the virus. The VP3 of rotavirus caps the viral mRNAs and helps combat cellular innate antiviral defenses. The non-structural proteins play various functional roles during rotavirus replication [2]. A binary classification of RVA strains is based on the VP7 (glycoprotein, G-genotype) and VP4 (protease-sensitive protein, P-genotype) proteins [4, 5]. Currently, 36 G genotypes and 51 P genotypes have been reported in humans and animals (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg). However, the majority of the global G and P genotypes of human rotaviruses detected are limited to G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] [6–8]. In addition, the G12 genotype, first detected in the Philippines in 1987 [9], emerged as a significant cause of rotavirus diarrhoea in children and have since persisted worldwide [10–19].
P[8] and P[4] RVAs have been identified as the first and second predominant P- genotypes, respectively, causing more than 90% of rotavirus-associated diarrhoea cases in many countries [20, 21]. Of note is the presence of P[8] in the world’s top six most frequently encountered RVA strains G1P[8], G3P[8], G4P[8], G9P[8], and G12P[8]. The P[8] genotype thus, accounts for 74% of the global prevalence of human VP4 rotavirus infections and hence its importance as an effective vaccine candidate [7]. It is therefore reasonable that the two widely used RVA vaccines, Rotarix a monovalent G1P[8] vaccine by GlaxoSmithKlein and RotaTeq a pentavalent vaccine containing the G1, G2, G3, G4, and P[8] genotypes by Merck, contain the P[8]-VP4 genotype as an antigenic component.
Phylogenetically, four distinct lineages of the P[8] genotype namely P[8]-Lineage I, P[8]-Lineage II, P[8]-Lineage III and P[8]-Lineage IV have been described [22, 23]. Current advances in molecular characterisation techniques have also distinguished the P[8] genotype into two antigenically distinct subtypes: P[8]a (P[8]-Lineage I–III) and P[8]b (OP354-like, i.e. P[8]-Lineage IV) [24]. Most of the world’s P[8] strains bear the P[8]a subtype; on the other hand, the P[8]b subtype is rare.
Widespread use of rotavirus vaccines could result in strain selection due to vaccine-induced selective pressure as demonstrated by Roczo-Farkas et al. [25] who noted changes in the diversity and distribution of circulating rotavirus genotypes in children less than 5 years who were hospitalized with rotavirus gastroenteritis after vaccine introduction in Australia. Since Ghana has introduced the Rotarix vaccine, the emergence of novel variants of circulating strains due to vaccine pressure may occur; therefore monitoring of changes in rotavirus genotype prevalence as well as identification of potential vaccine escape strains are important to improve vaccination strategies and the development of new rotavirus vaccines.
There is an abundance of molecular epidemiological data on the prevalence of both common and unusual G/P types determined by RT-PCR in Ghana. These studies described RVA strains with unusual G and P combinations and revealed the G9 genotype as an important emerging RVA in Ghana. In addition, it was also shown that G1P[8] RVA was replaced as the major circulating strain during the pre-vaccination era with G12P[8] as the most predominant strain post vaccine introduction. [26–29]. However, in-depth genetic comparisons at both the nucleotide and phylogenetic level has been rarely performed to detect if any the emergence of novel variants of the commonly detected strains in the country. Enweronu-Laryea et al. [30] employed a RT-PCR based genotyping method to characterize the G and P genotypes of RVA strains from stool samples collected from children with diarrhoea during the WHO sponsored surveillance study in Ghana. Their study recorded a substantial number of uncharacterized G (135/876) and P (167/876) genotypes. In this study, we determined the genotypes of previously non-typeable strains based on nucleotide sequence data and provided insight into the evolutionary history of the VP7 and VP4 genes by phylogenetic analysis. We also examined the effect of VP8* P[8] genotype diversity on the routinely used RT-PCR based genotyping techniques and resolved challenges associated with P[8]-genotyping failure.
Materials and methods
Ethical approval and background of study samples
The study protocol was approved by the Institutional Review Board, Noguchi Memorial Institute for Medical Research, Legon, Accra, Ghana. A total of 57 RVA positive samples analysed in this study were collected from children <5years who were hospitalized with a primary diagnosis of acute gastroenteritis between 2007 and 2011. The G-genotypes of these 57 samples were previously reported in studies carried out by Enweronu-Laryea et al. [30], Damanka et al. [31], [32] and Binka et al. [33]. The distribution of the G-genotypes were as follows: G1 (38), G2 (3), G6 (1), and G9 (15). Their P-types could however not be determined using the routine genotyping primers.
RNA extraction, reverse transcription polymerase chain reaction and Sanger sequencing
Total RNA was extracted from 10% stool suspension by the phenol/chloroform method as described by Steele and Alexander [34] and the RNA purified with an RNaid kit (Bio101, Inc., California, USA) according to the manufacturer’s protocol. The RNA was reverse transcribed and amplified using the VP4 gene consensus primer set VP4F/VP4R [35]. The reaction was carried out with an initial reverse transcription step at 42°C for 30 minutes, followed by initial denaturation at 94°C for 2 minutes, 30 cycles of amplification (denaturation at 94°C for 1 minute, annealing at 42°C for 2 minutes and extension at 72°C for 3 minutes) and a final extension at 72°C for 7 minutes in a thermal cycler (Eppendorf, Hamburg, AG). The corresponding VP7 gene of the samples were amplified using the VP7-F/VP7-R consensus primers [35] and the same PCR cycle conditions.
The amplicons were purified with the QIAquick PCR purification kit (Qiagen/Westburg) according to the manufacturer’s protocol and sequenced in the forward and reverse directions using the respective amplification primers and the BigDye Terminator cycle sequencing kit v.3.1 (Applied Biosystems). The cycle sequenced products were purified, using ethanol-sodium acetate precipitation method, dried under vacuum and reconstituted in Hi-Di formamide. DNA sequences were determined using ABI 3130xl genetic analyser (Applied Biosystems, Foster City, CA).
Sequence and phylogenetic analyses
Near-full-length VP7 and VP8* portion of the VP4 gene were obtained by combining the sequences obtained from the forward and reverse reactions. The genotypes were determined for the VP8* portion and confirmed for the G-types using the online RotaC v2.0 RVA genotyping tool [36]. Using the Basic Local Alignment Search Tool (BLAST) and the rotavirus resource in virus variation available at the NCBI website [37, 38], reference sequences for phylogenetic analysis were retrieved. Multiple sequence alignment was carried out on the datasets and the best fit nucleotide sequence substitution models were determined using MEGA version 6.06 [39].
Based on the best fit nucleotide substitution models with the lowest Bayesian Information Criterion scores [40], i.e. Tamura 3-parameter model with gamma distribution (T92+G) for the VP8* and G9 VP7 datasets and T92 + G + Invariant sites for the G1 VP7 dataset, maximum likelihood phylogenetic trees for the VP8* and the VP7 gene sequences were constructed using with 1000 bootstrap replicate trials. Lineages defined in the G9 VP7 phylogenetic tree were based on designations by Doan et al. [41] whereas those identified in the G1 VP7 and P[8] VP4 trees were based on designations by Cunliffe et al. [23] and Do et al. [42].
Results
G/P types determined based on sequence data
All 57 rotavirus-positive samples had enough bulk stool specimens for further analysis in this study. The VP8* fragment of the VP4 genes of these samples were successfully sequenced and genotypes were determined. Forty-eight of the 57 (84.2%) had the P[8] specificity, of which 43 (89.6%) were characterized as P[8]a subtype and 5 (10.4%) as the rare P[8]b (OP354-like) subtype. The P-genotypes of the remaining 9/57 samples were P[6]: 7(12.3%), P[4]: 1(1.75%), and P[14]: 1(1.75%). Out of the 43 P[8]a sequences, 39 (Table 1, S1 Table) were of appreciable length (nucleotide position 178–787 of the VP8* portion of the VP4 gene) and were included for further sequence comparison and phylogenetic analysis. Of these 39 successfully sequenced P[8]a samples, the corresponding VP7 gene of only 27 of them could be sequenced even though their G-genotypes were known through the routine PCR genotyping method. These G-genotypes were confirmed as G1 (18/27) and G9 (9/27) upon sequencing (Table 1, S1 Table).
Table 1. Genotypes and lineages determined for the VP7 and VP4 of previously non-typeable strains.
| G/P type | Sample | Year of collection | VP7 gene Lineage | VP4 gene Lineage |
|---|---|---|---|---|
| G1P[8]a | GHA-00240/DC | 2007 | G1; sequence ND | P[8]-Lineage III |
| GHA-00319/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00328/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00378/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00532/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00324/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00495/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00141/PML | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-00329/DC | 2008 | G1; sequence ND | P[8]-Lineage III | |
| GHA-5059/EB | 2009 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00592/DC | 2009 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00850/DC | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-0099/M | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-0123/P | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-0139/P | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-0028/K | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-0021/K | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00892/DC | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00840/DC | 2010 | G1; sequence ND | P[8]-Lineage III | |
| GHA-009I9/DC | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-474/AG | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00845/DC | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00702/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00759/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00800/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00789/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00796/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00793/PML | 2010 | G1-Lineage I | P[8]-Lineage III | |
| GHA-00713/PML | 2010 | G1; sequence ND | P[8]-Lineage III | |
| GHA-0093/M | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00293/LA | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00886/DC | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00894/DC | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| G9P[8] | GHA-0176/P | 2010 | G9-sequence ND | P[8]-Lineage III |
| GHA-00710/PML | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00802/PML | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00801/PML | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00810/PML | 2010 | G9-Major sub-lineage III | P[8]-Lineage III | |
| GHA-00784/PML | 2010 | G9-Major sub-lineage III | P[8]-Lineage III |
ND: Sequence Not determined
Sequence analyses and phylogenetic inference
Sequence analyses
The nucleotide and amino acid sequence diversity of the VP8* fragment of the VP4 gene among the Ghanaian P[8]a strains was relatively high even between strains detected from the same place and year, ranging from 0% - 5% and 0% - 9.4%, respectively. Specifically, strains from DC/2008 varied from 0%– 2.9%, those from DC/2010 varied from 0.2% - 3.2%, those from PML/2010 varied from 0.7% - 3.9%, those from EB/M/P/2010 varied from 0% - 2.9% (S2 and S3 Tables). Similarly, the G9 VP7 genes were 0% - 3.6% and 0% - 3.8% diverse at the nucleotide and amino acid levels respectively. On the other hand, the G1 VP7 genes were relatively closely related with nucleotide and amino acid diversity ranging from 0% - 1.8% and 0% - 2.9% respectively.
Phylogenetic inference of the P[8] VP8* sequences
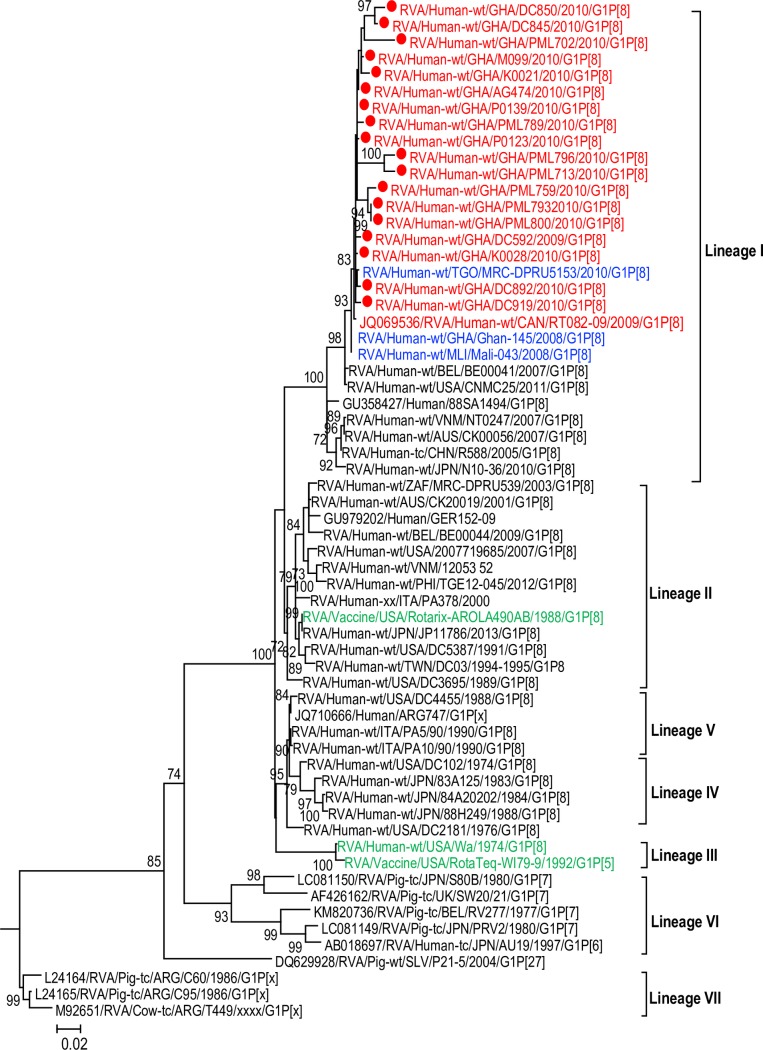
Phylogenetic analysis of the VP8* fragment of the VP4 gene using sequences available in the DNA databases maintained the previously established four distinct lineages designated lineage I–lineage IV (Fig 1). The P[8]a sequences of the study strains belonged to sub-cluster IIIc within lineage III (Fig 1). This sub-cluster also included contemporary G1P[8], G3P[8], G8P[8], and G9P[8] strains detected from across the world from 2006–2016. Within sub-cluster IIIc, strain GHA-00532/DC/2008/G1P[8]a (indicated with a red dot) clustered separately from the rest of the Ghanaian P[8]a sequences but was closely related to the VP4 of two Malawian G8P[8] detected in 2006 (indicated in blue font). The phylogenetic analysis clearly showed that the study strains have a different evolutionary pathway from the P[8] sequences of historical strains such as the prototype G1P[8] strain Wa, and the vaccine strains of Rotarix and RotaTeq-WI79-4 (indicated in green font).
Fig 1. Phylogenetic tree of the VP8* fragment of the VP4 genes of Ghanaian P[8]a strains and published P[8]a strains constructed using maximum likelihood method with Mega 6.06 software.
P[8]a strains analysed in the present study are indicated in red font. The prototype G1P[8] strain Wa and the P[8] vaccine strains in Rotarix and RotaTeq are in green font. Phylogenetic tree was supported statistically by bootstrapping with 1000 replicates. The scale bar at the bottom of the tree indicates genetic distance expressed as nucleotide substitutions per site. Percentage bootstrap support values ≥70% are shown.
Phylogenetic inference of the G1 and G9 VP7 sequences
Phylogenetic analysis of the G1 and G9 VP7 sequences of both human and porcine origin from the global rotavirus database resulted in previously established seven distinct lineages designated; lineage I–lineage VII (Figs 2 and 3). The G1 VP7 sequences of the study strains belonged to lineage I which is made of global G1 sequences of only contemporary human RVA strains detected from 2006–2016. Within lineage I, our study strains formed a monophyletic cluster with African G1P[8] strains detected previously in Ghana and Mali during Rotateq vaccine trial [43] and neighbouring Togo (indicated in blue font).
Fig 2. Phylogenetic tree of the VP7 sequences of G1P[8] strains and reference G1 strains retrieved from the GenBank database.
The tree was constructed using maximum likelihood method with Mega 6.06 software. G1 strains analysed in the present study are indicated in red font. The prototype G1P[8] strain Wa and the P[8] vaccine strains in Rotarix and RotaTeq are in green font. The closest G1 sequences from African rotaviruses to our study strains are indicated in blue font. The phylogenetic tree was supported statistically by bootstrapping with 1000 replicates. The scale bar at the bottom of the tree indicates genetic distance expressed as nucleotide substitutions per site. Percentage bootstrap support values ≥70% are shown.
Fig 3. Phylogenetic tree of the VP7 sequences G9P[8] strains and reference G9 strains retrieved from the GenBank database.
The tree was constructed using maximum likelihood method with Mega 6.06 software. G9 strains analysed in the present study are indicated in red font. The historical G9 strains reported from Ghana by Armah et al. [28] are indicated in green font. The closest G9 sequences from African rotaviruses to our study strains are indicated in blue font. The phylogenetic tree was supported statistically by bootstrapping with 1000 replicates. The scale bar at the bottom of the tree indicates genetic distance expressed as nucleotide substitutions per site. Percentage bootstrap support values ≥70% are shown.
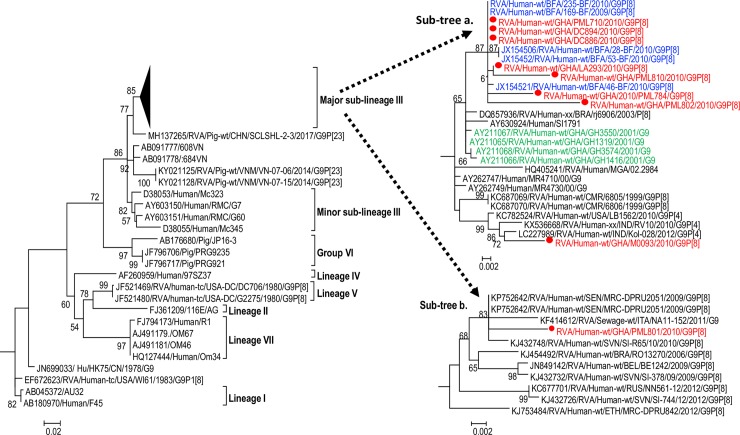
The G9 VP7 sequences of the study strains belonged to the major sub-cluster of lineage III which consists of global G9 sequences of both historical and contemporary human RVA strains. Within the major sub-lineage III, the study strains formed a monophyletic cluster (Fig 3 sub-tree a.) with African G9 sequences from neighbouring Burkina Faso (indicated in blue front) away from G9 sequences of Ghanaian G9 strains detected in 2001 (indicated in green front). It was noted that the VP7 sequence of the Ghanaian G9 study strain RVA/Human-wt/GHA/PML801/2010/G9P[8] had a different evolutionary history since it belonged to a sub-cluster consisting of strains from Africa, South America and Europe (Fig 3 sub-tree b).
Investigation of VP4 genotyping failure
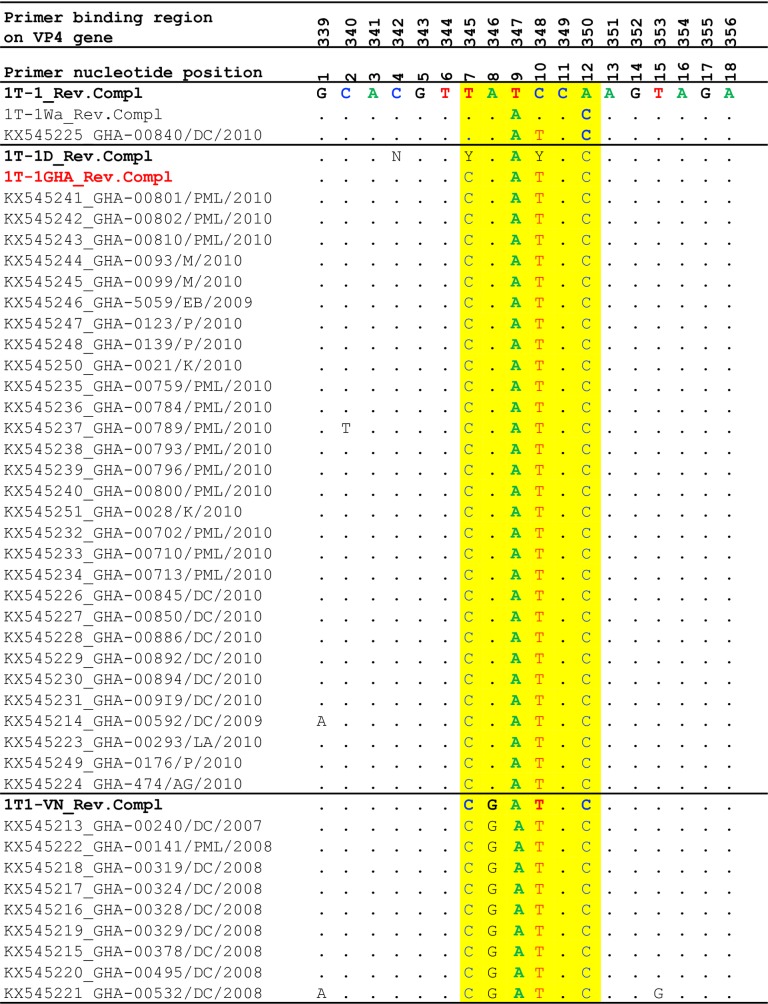
To investigate possible primer mismatches that led to the previously reported genotyping failure [30], 1T-1 as well as alternative P[8] genotyping primers namely 1T-1Wa, 1T1-VN, 1T-1D developed by WHO Rotavirus Collaborating Centers and Regional Laboratories were aligned with the P[8]a sequences. The following nucleotide mismatches were observed. First, strain GHA-00840 showed two mismatches when aligned with the routinely used P[8]-genotype specific primer 1T-1 at nt 10 (C→T) and 12 (A→C) and one mismatch at nt 10 (C→T) when aligned with the alternative primer 1T-1Wa primer (Fig 4).
Fig 4. Alignment of VP8* fragments of the VP4 gene sequences of the Ghanaian P[8]a rotavirus strains and the reverse complementary sequences of the routinely used P[8] primers, 1T-1, 1T-Wa, 1T1-VN and the degenerate primer, 1T-1D.
Dots indicate consensus with the primer and the yellow highlight indicates the region of variability of the primer binding site. Abbreviations: N: A or C or G or T; Y: C or T and R: A or G.
Twenty-nine of the P[8]a sequences consistently showed three mismatches at nucleotides 7 (T→C), 10 (C→T) and 12 (A→C) (Fig 4) upon alignment with 1T-1. These twenty-nine samples also showed—two consistent mismatches at nucleotides; 7 (T→C),—and 10 (C→T) upon comparison with 1T-1Wa (Fig 4). Also, a mismatch at nt 8 (G→A) was identified when aligned with the 1T1-VN primer. However, an alignment with the 1T-1D degenerate primer (CAN GTY AAY CCA GTA GA) showed 100% complementarity with the Ghanaian P[8]a when the ambiguous codes N, Y, and Y were respectively substituted with C, C, and T (Fig 4). The new primer 1T-1GHA was ordered and tested by PCR against the non-typeable strains which were successfully genotyped as P[8].
Additionally, nine of the strains showed five consistent mismatches at nucleotide (nt) 7 (T→C), 8 (A→G), 9 (T→A), 10 (C→T) and 12 (A→C) when aligned with 1T-1 primer (Fig 4). These nine strains also showed three nucleotide mismatches at nt 7 (T→C), 8 (A→G) and 10 (C→T) when aligned with the 1T-1Wa primer (Fig 4) and 100% complementarity when aligned with the 1T1-VN primer except for strain GHA-00532/DC/2008 which had additional mismatches at position 1 (G→A) and 15 (T→G). Upon comparison with the degenerate primer 1T-1D (CAN GTY AAY CCA GTA GA), a single nucleotide mismatch at nt 8 (A→G) was detected.
Discussion
Reverse transcription polymerase chain reaction genotyping methods used in rotavirus surveillance studies, have been useful in monitoring circulating strains and identifying rare and emerging strains worldwide. Advances in rotavirus molecular assays such as sequence analysis of rotavirus genes provide us the opportunity to i) genotype non-typeable rotavirus strains that have mutated hence might be recognized by the routine genotyping primers and ii) understand their evolutionary histories using molecular phylogenetic inferences. Our previous study on Ghanaian rotavirus strains revealed that a considerable proportion of the RVAs could not be G and/or P genotyped by the routine multiplex RT-PCR strategy by Gouvea et al. [4] and Gentsch et al., [5]. In this study, sequence data obtained by Sanger sequencing showed that the selected P-non-typeable strains possessed the G1P[8] and G9P[8] genotypes.
Circa 1999–2000, G1P[8] strains were rarely reported in Ghana while the G9P[8] strains emerged and predominated in the 4th quarter of 1999 [28, 44]. From 2005 however, G1P[8] strains became the most prevalent strains accounting for 80% of strains from the Kasena Nankana district [45]. The G1 strains consistently remained prevalent while the G9 strains became rarely detected in Ghana [26, 46]. We inferred the phylogenetic history of the G1 and G9 VP7 sequences and noted that the strains characterized in this study were not very different from previously reported ones by Armah et al. (G9) [28] and Heylen et al. (G1P[8]) [43] from Ghana. There was evidence that our strains and those reported in the past diverged from a common ancestor. The little diversity observed can be considered typical taking into consideration the evolutionary rates of G1 VP7 gene (0.93 x10-3 nucleotide substitutions/site/year (Highest posterior density interval: 0.68 x10-3–1.18 x10-3) [47] and G9 VP7 gene (1.87 x10-3 nucleotide substitutions/site/year (Highest posterior density interval: 1.45 x10-3–2.27 x10-3) [11].
Phylogenetic analysis of P[8]—VP4 sequences from the global rotavirus database maintained that P[8] rotaviruses could be divided into the four previously established distinct phylogenetic lineages: P[8]-lineage I, P[8]-lineage II, P[8]-lineage III and P[8]-lineage IV. All the Ghanaian strains were grouped into lineage III and seemed to be related to African, Australian and European strains detected between 2006 and 2014, indicating that they shared common ancestral strains (Fig 1). In the phylogenetic tree, the Ghanaian strains clustered in P[8]-lineage III which differed from the lineages of vaccine strains (Rotarix and RotaTeq) which clustered in P[8]-lineage I and II respectively). This indicated that the evolutionary relationship between the P[8]-lineage of human RVAs detected in Ghana and the two vaccine strains are different.
Sequence comparison of the VP4 genes revealed mismatches at the P[8] primer (1T-1) binding site which we considered to be responsible for the genotyping failure. The substantial numbers of incompletely genotyped strains could be attributed to genetic drift due to the accumulation of point mutations or the lack of specific oligonucleotide primers for amplifying novel strains [48]. Majority of genotype specific primers used in multiplex PCR were designed from the sequences of strains detected in the 1980s and may have sequence mismatches in the primer binding region of the target gene of more recent rotavirus strains. Series of mutations were detected at the primer binding sites of the P[8] templates (our strains) thus decreased the primer binding probability, which explains the initial failure of the primers to bind to the P[8]a sites. Iturriza-Gomara and colleagues previously reported mismatches in the primer binding site of P[8] strains that resulted in genotyping failure [48]. Sequence based genotyping has helped determine the genotypes of more than 70% of non-typeable strains detected during the surveillance period thus reducing the percentage of non-typeable strains. It is interesting to note that majority of the VP4 strains characterized in this study belonged to the P[8]a subtype, an essential component of both Rotarix and RotaTeq vaccines. Failure to determine the P[8]a subtypes in our previous study indicated extreme simplification of the previous RT-PCR based genotyping system employed in our Regional Reference laboratory. The successful determination of the P[8]a genotype by sequencing showed that the prevalence of these strains were underestimated during the WHO sponsored rotavirus surveillance study in Ghana. A study in the United Kingdom showed that a degenerate version (1T-1D) of the P[8]-specific primer (1T-1) allowed strains previously non-typeable due to point mutations at the primer binding site to be P- typed by RT-PCR [33]. This study met with methodological challenges such as inter-primer suppression when employing a standard multiplex RT–PCR procedure using the commonly used P[8]-specific primer (1T-1) [20] and VP4 genotyping strategy developed by WHO Rotavirus Collaborating Centers and Regional Laboratories using 1T-1Wa, 1T1-VN (http://apps.who.int/iris/bitstream/handle/10665/70122/WHO_IVB_08.17_eng.pdf;jsessionid=8C5703F323752AD2934C58EDEAE338AA?sequence=1). The observed difficulty in P-typing was solved by including a new primer 1T-1GHA—designed based on the 1T-1D degenerate typing primer in our PCR reactions and this additional primer helped genotype the non-typeable strains reported previously in the reference laboratory in Ghana.
Our analysis provided information on how Ghanaian pre-vaccine introduction period G1P[8] and G9P[8] strains relate to a global collection of G1 and G9 strains at the lineage level. The study also resolved the methodological limitations of the standard genotyping techniques employed in the Rotavirus Regional Reference laboratory in Ghana. Genotyping failure attributable to natural variation in primer binding sites could be cited among problems encountered in large surveillance studies. The study highlights the importance of close monitoring of rotavirus genotyping methods as well as regularly updating the primer sequences employed for molecular typing of rotaviruses. The findings also suggest that regions should start to use sequencing data from local rotavirus strains to design and update genotyping primers.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors acknowledge with gratitude the commitment of the staff of the Department of Electron Microscopy and Histopathology, Noguchi Memorial Institute for Medical Research, for their technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files. GenBank Accession numbers: KX545213 - KX545251 and LC456060 - LC456086.
Funding Statement
The authors received no specific funding for this work.
References
- 1.GBD_Diarrhoeal_Diseases_Collaborators. (2017) Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:909–948. 10.1016/S1473-3099(17)30276-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estes MK, Greenberg HB (2013) Rotaviruses In: Knipe D. M., Howley P.M (Eds.), Fields Virology. Wolters Kluwer Health/Lippincott, Williams and Wilkins Philadelphia; pp. 1347–1401. [Google Scholar]
- 3.Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T, et al. (2018) Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr 10.1001/jamapediatrics.2018.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. (1990) Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol 28:276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, et al. (1992) Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol 30:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, et al. (2005) Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 192 Suppl 1:S146–159. [DOI] [PubMed] [Google Scholar]
- 7.Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, et al. (2012) Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 30 Suppl 1:A122–130. [DOI] [PubMed] [Google Scholar]
- 8.Santos N, Hoshino Y. (2005) Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. 10.1002/rmv.448 [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi N, Lintag IC, Urasawa T, Taniguchi K, Saniel MC, Urasawa S. (1989) Unusual human rotavirus strains having subgroup I specificity and "long" RNA electropherotype. Arch Virol 109:11–23. [DOI] [PubMed] [Google Scholar]
- 10.Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, et al. (2007) Evolutionary history and global spread of the emerging g12 human rotaviruses. J Virol 81:2382–2390. 10.1128/JVI.01622-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. (2010) Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 27:2431–2436. 10.1093/molbev/msq137 [DOI] [PubMed] [Google Scholar]
- 12.Pun SB, Nakagomi T, Sherchand JB, Pandey BD, Cuevas LE, Cunliffe NA, et al. (2007) Detection of G12 human rotaviruses in Nepal. Emerg Infect Dis 13:482–484. 10.3201/eid1303.061367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castello AA, Arguelles MH, Rota RP, Olthoff A, Jiang B, Glass RI, et al. (2006) Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J Clin Microbiol 44:2046–2050. 10.1128/JCM.02436-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagomi T, Do LP, Agbemabiese CA, Kaneko M, Gauchan P, Doan YH, et al. (2017) Whole-genome characterisation of G12P[6] rotavirus strains possessing two distinct genotype constellations co-circulating in Blantyre, Malawi, 2008. Arch Virol 162:213–226. 10.1007/s00705-016-3103-5 [DOI] [PubMed] [Google Scholar]
- 15.Luchs A, Cilli A, Morillo SG, de Souza Gregorio D, Farias de Souza KA, Vieira HR, et al. (2016) Detection of the emerging rotavirus G12P[8] genotype at high frequency in brazil in 2014: Successive replacement of predominant strains after vaccine introduction. Acta Trop 156:87–94. 10.1016/j.actatropica.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 16.Delogu R, Ianiro G, Camilloni B, Fiore L, Ruggeri FM. (2015) Unexpected spreading of G12P[8] rotavirus strains among young children in a small area of central Italy. J Med Virol 87:1292–1302. 10.1002/jmv.24180 [DOI] [PubMed] [Google Scholar]
- 17.Mijatovic-Rustempasic S, Teel EN, Kerin TK, Hull JJ, Roy S, Weinberg GA, et al. (2014) Genetic analysis of G12P[8] rotaviruses detected in the largest U.S. G12 genotype outbreak on record. Infect Genet Evol 21:214–219. 10.1016/j.meegid.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pietsch C, Liebert UG. (2009) Human infection with G12 rotaviruses, Germany. Emerg Infect Dis 15:1512–1515. 10.3201/eid1509.090497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S, Varghese V, Chaudhury S, Barman P, Mahapatra S, Kojima K, et al. (2003) Emergence of novel human group A rotavirus G12 strains in India. J Clin Microbiol 41:2760–2762. 10.1128/JCM.41.6.2760-2762.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sai L, Sun J, Shao L, Chen S, Liu H, Ma L. (2013) Epidemiology and clinical features of rotavirus and norovirus infection among children in Ji'nan, China. Virol J 10:302 10.1186/1743-422X-10-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayindou G, Ngokana B, Sidibe A, Moundele V, Koukouikila-Koussounda F, Christevy Vouvoungui J, et al. (2016) Molecular epidemiology and surveillance of circulating rotavirus and adenovirus in Congolese children with gastroenteritis. J Med Virol 88:596–605. 10.1002/jmv.24382 [DOI] [PubMed] [Google Scholar]
- 22.Arista S, Giammanco GM, De Grazia S, Ramirez S, Lo Biundo C, Colomba C, et al. (2006) Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J Virol 80:10724–10733. 10.1128/JVI.00340-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunliffe NA, Gondwe JS, Graham SM, Thindwa BD, Dove W, Broadhead RL, et al. (2001) Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J Clin Microbiol 39:836–843. 10.1128/JCM.39.3.836-843.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagashima S, Kobayashi N, Paul SK, Alam MM, Chawla-Sarkar M, Krishnan T. (2009) Characterization of full-length VP4 genes of OP354-like P[8] human rotavirus strains detected in Bangladesh representing a novel P[8] subtype. Arch Virol 154:1223–1231. 10.1007/s00705-009-0436-3 [DOI] [PubMed] [Google Scholar]
- 25.Roczo-Farkas S, Kirkwood CD, Cowley D, Barnes GL, Bishop RF, Bogdanovic-Sakran N, et al. (2018) The Impact of Rotavirus Vaccines on Genotype Diversity: A Comprehensive Analysis of 2 Decades of Australian Surveillance Data. J Infect Dis 10.1093/infdis/jiy197 [DOI] [PubMed] [Google Scholar]
- 26.Lartey BL, Damanka S, Dennis FE, Enweronu-Laryea CC, Addo-Yobo E, Ansong D, et al. (2018) Rotavirus strain distribution in Ghana pre- and post- rotavirus vaccine introduction. Vaccine 10.1016/j.vaccine.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armah GE, Pager CT, Asmah RH, Anto FR, Oduro AR, Binka F, et al. (2001) Prevalence of unusual human rotavirus strains in Ghanaian children. J Med Virol 63:67–71. [PubMed] [Google Scholar]
- 28.Armah GE, Steele AD, Binka FN, Esona MD, Asmah RH, Anto F, et al. (2003) Changing patterns of rotavirus genotypes in ghana: emergence of human rotavirus G9 as a major cause of diarrhea in children. J Clin Microbiol 41:2317–2322. 10.1128/JCM.41.6.2317-2322.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asmah RH, Green J, Armah GE, Gallimore CI, Gray JJ, Iturriza-Gomara M, et al. (2001) Rotavirus G and P genotypes in rural Ghana. J Clin Microbiol 39:1981–1984. 10.1128/JCM.39.5.1981-1984.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enweronu-Laryea CC, Sagoe KW, Damanka S, Lartey B, Armah GE. (2013) Rotavirus genotypes associated with childhood severe acute diarrhoea in southern Ghana: a cross-sectional study. Virol J 10:287 10.1186/1743-422X-10-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damanka S, Lartey B, Agbemabiese C, Dennis FE, Adiku T, Nyarko K, et al. (2016) Detection of the first G6P[14] human rotavirus strain in an infant with diarrhoea in Ghana. Virol J 13:183 10.1186/s12985-016-0643-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damanka S, Dennis FE, Agbemabiese C, Lartey B, Adiku T, Nyarko K, et al. (2016) Identification of OP354-like human rotavirus strains with subtype P[8]b in Ghanaian children with diarrhoea. Virol J 13:69 10.1186/s12985-016-0523-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binka E, Vermund SH, Armah GE. (2012) Rotavirus as a cause of diarrhea among children under 5 years of age in urban Ghana: Prevalence and serotypes/genotypes. Paediatr Infect Dis J 30:718–720. [Google Scholar]
- 34.Steele AD, Alexander JJ. (1987) Molecular epidemiology of rotavirus in black infants in South Africa. J Clin Microbiol 25:2384–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, et al. (2008) New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol 42:368–373. 10.1016/j.jcv.2008.02.011 [DOI] [PubMed] [Google Scholar]
- 36.Maes P, Matthijnssens J, Rahman M, Van Ranst M. (2009) RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol 9:238 10.1186/1471-2180-9-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, Ostapchuck Y, et al. (2017) Virus Variation Resource—improved response to emergent viral outbreaks. Nucleic Acids Res 45:D482–D490. 10.1093/nar/gkw1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brister JR, Bao Y, Zhdanov SA, Ostapchuck Y, Chetvernin V, Kiryutin B, et al. (2014) Virus Variation Resource—recent updates and future directions. Nucleic Acids Res 42:D660–665. 10.1093/nar/gkt1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarz G. (1978) Estimating the dimension of a model. Ann Stat 6:461–464. [Google Scholar]
- 41.Doan YH, Nakagomi T, Agbemabiese CA, Nakagomi O. (2015) Changes in the distribution of lineage constellations of G2P[4] Rotavirus A strains detected in Japan over 32 years (1980–2011). Infect Genet Evol 34:423–433. 10.1016/j.meegid.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 42.Do LP, Doan YH, Nakagomi T, Kaneko M, Gauchan P, Ngo CT, et al. (2015) Molecular characterisation of wild-type G1P[8] and G3P[8] rotaviruses isolated in Vietnam 2008 during a vaccine trial. Arch Virol 10.1007/s00705-015-2706-6 [DOI] [PubMed] [Google Scholar]
- 43.Heylen E, Zeller M, Ciarlet M, Lawrence J, Steele D, Van Ranst M, et al. (2015) Comparative analysis of pentavalent rotavirus vaccine strains and G8 rotaviruses identified during vaccine trial in Africa. Sci Rep 5:14658 10.1038/srep14658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Binka FN, Anto FK, Oduro AR, Awini EA, Nazzar AK, Armah GE, et al. (2003) Incidence and risk factors of paediatric rotavirus diarrhoea in northern Ghana. Trop Med Int Health 8:840–846. [DOI] [PubMed] [Google Scholar]
- 45.Silva PA, Stark K, Mockenhaupt FP, Reither K, Weitzel T, Ignatius R, et al. (2008) Molecular characterization of enteric viral agents from children in northern region of Ghana. J Med Virol 80:1790–1798. 10.1002/jmv.21231 [DOI] [PubMed] [Google Scholar]
- 46.Enweronu-Laryea CC, Sagoe KW, Glover-Addy H, Asmah RH, Mingle JA, Armah GE. (2012) Prevalence of severe acute rotavirus gastroenteritis and intussusceptions in Ghanaian children under 5 years of age. J Infect Dev Ctries 6:148–155. [DOI] [PubMed] [Google Scholar]
- 47.Afrad MH, Matthijnssens J, Afroz SF, Rudra P, Nahar L, Rahman R, et al. (2014) Differences in lineage replacement dynamics of G1 and G2 rotavirus strains versus G9 strain over a period of 22 years in Bangladesh. Infect Genet Evol 28:214–222. 10.1016/j.meegid.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 48.Iturriza-Gomara M, Green J, Brown DW, Desselberger U, Gray JJ. (2000) Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J Clin Microbiol 38:898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. GenBank Accession numbers: KX545213 - KX545251 and LC456060 - LC456086.