Abstract
During our studies on fungal diversity from plant substrates, a new species of Valsaria was isolated from dead branches of Ostrya carpinifolia. The taxon is morphologically similar to other taxa in Valsariaceae and is characterized by pseudostromata, apically free pseudoparaphyses, bitunicate asci, and dark brown, 2-celled ascospores. However, it differs from extant species in number of guttules and ornamentation of spore. It is introduced herein as Valsaria ostryae sp. nov. within the family Valsariaceae. Multigene phylogenies based on combined LSU, ITS and RPB2 DNA sequence data generated from maximum likelihood, maximum parsimony and MrBayes analyses indicate that V. ostryae is basal to V. lopadostomoides and V. rudis and its establishment as a new species is strongly supported. No discordance was found between our morphological and phylogenetic species boundaries as postulated by other researchers and our molecular data strongly supports ornamentation of spore as useful for species delineation. Valsaria species do not appear to be host specific. Full morphological details are provided herein and phylogenetic relationships of Valsaria species are also discussed in light with host association.
Introduction
Valsariaceae was introduced by Jaklitsch et al. [1] and classified in the order Valsariales. Species of this family have a worldwide distribution occurring on sun-exposed, corticated logs, branches of coniferous and broadleaf trees and on bamboo [1] and exist as saprobes, plant pathogens or necrotrophs [2,3]. The family comprises three genera namely Bambusaria Jaklitsch et al., Myrmaecium Nitschke ex Fuckel and Valsaria Ces. & De Not [1,4]. The asexual morphs are known to be coelomycetous [4]. Valsaria was introduced by Cesati and De Notaris [5] with Valsaria insitiva (Tode) Ces. & De Not. as the type species. Ju et al. [2] examined numerous species to provide an update of its taxonomy. Multigene phylogenetic analyses have also been performed to clarify species taxonomy [1]. In this paper, we introduce one new species of Valsaria, which was found on dead terrestrial branches of Ostrya carpinifolia (Betulaceae). Combined analyses of LSU, ITS and RPB2 DNA sequence data using maximum-likelihood (ML), maximum-parsimony (MP) and Bayesian inference analyses clearly indicate that this species belongs to the Valsariaceae with strong statistical support. Phylogeny also provides important clues on the taxonomic usefulness of morphological features that can be used to differentiate among Valsaria species and its allies. In this paper, the new species is described, illustrated and compared with similar taxa.
Materials and methods
Isolates and morphology
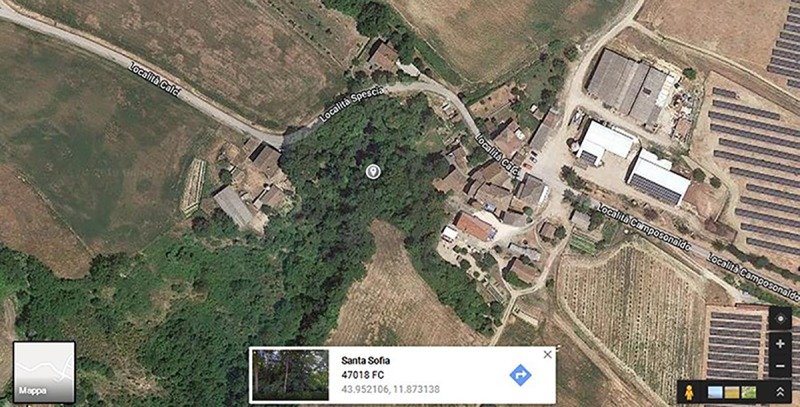
Specimens were collected from dead branches of Ostrya carpinifolia in the Province of Forlì-Cesena [FC], Santa Sofia, Camposonaldo, Italy and were pressed and dried individually between blotting papers and labelled. The geographic coordinates for the data collected is 43.952106, 11.873138 (Fig 1). Specimens were observed under a Motic SMZ 168 series dissecting stereo-microscope. Hand-cut sections of the fruiting structures were mounted in water for microscopic studies and photomicrography. Pure cultures were obtained from single spore isolation following the method designated in Chomnunti et al. [6]. Germinating ascospores were transferred aseptically to malt extract agar (MEA) plates and incubated at 16°C. Colony characters on media were observed and growth rates were measured after one week and at weekly intervals. No specific permissions were required for the location because permissions are standardized only for macro-fungi such as only 3 fruiting-bodies for each collection, the indication of the name of the species that it needs to collect and other things valid only for macrofungi. Also, the field studies did not involve endangered or protected species. The holotype specimen was deposited in the Mae Fah Luang University Herbarium (MFLU), Chiang Rai, Thailand. Ex-type living cultures are deposited in the Culture Collection at Mae Fah Luang University (MFLUCC) and at the International Collection of Microorganisms from Plants (ICMP). Faces of Fungi and Index Fungorum numbers are provided as in Jayasiri et al. [7] and Index Fungorum [8]. New taxa were established based on recommendations outlined by Jeewon and Hyde [9].
Fig 1. Geographical coordinates of sample collected.
DNA extraction, PCR amplification and sequencing
Fungal isolates were grown on MEA media at 16 ± 2°C for 4 weeks. Fungal mycelium was extracted with Biospin Fungus Genomic DNA Extraction Kit-BSC14S1 (BioFlux, P.R. China). Amplification reactions were performed using known primer pairs, LR0R/LR5 for the 28S large subunit rDNA (LSU) [10], ITS5/ITS4 for 5.8S nrRNA gene with the two flanking internal transcribed spacers (ITS) [11] and fRPB2-5F/ fRPB2-7cR for RNA polymerase II second largest subunit (RPB2) [12]. The ultimate volume of the PCR reaction were 25 μl, containing 1 μl of DNA template (40 μg), 1 μl of each forward and reverse primer (0.2 μM) and 12.5 μl of 2xMaster Mix and 9.5 μl of ddH2O. The PCR condition was as follows: initial denaturation 95°C for 3 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 48 s, elongation at 72°C for 1 min, and final extension at 72°C for 10 min for LSU, ITS and RPB2. PCR products were sequenced with the same primers mentioned above at Sangon Biotech, Shanghai, China.
Sequence alignment and phylogenetic analyses
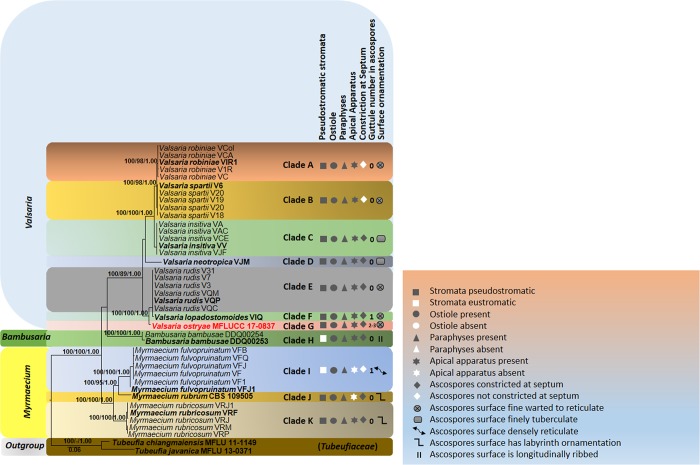
Phylogenetic analyses were conducted on a combined dataset of LSU, ITS and RPB2 sequence data. Taxa used in the analyses were obtained through recent publications [1, 13]. Separate LSU, ITS and RPB2 DNA sequences were also subjected to BLAST search engine tool of NCBI for verification and selection of taxa for subsequent phylogenetic analyses. Maximum likelihood analysis was performed by using raxmlGUIv.0.9b2 [14]. The search strategy was set to rapid bootstrapping and the analysis carried out using the GTRGAMMAI model of nucleotide substitution. The number of replicates was inferred using the stopping criterion [15]. Maximum Likelihood bootstrap values equal or greater than 90% are given as the first set of numbers above the nodes (Fig 2). PAUPv4.0b10 was used to conduct the parsimony analysis to obtain the phylogenetic trees. Trees were inferred using the heuristic search option with 1000 random sequence additions. Maxtrees were setup to 500 and branches of zero length were collapsed and all multiple parsimonious trees were saved. Descriptive tree statistics for parsimony (Tree Length [TL], Consistency Index [CI], Retention Index [RI], Relative Consistency Index [RC] and Homoplasy Index [HI] were calculated for trees generated under different optimality criteria. Kishino-Hasegawa tests (KHT) [16] were performed in order to determine whether trees were significantly different. Maximum-parsimony bootstrap values equal or greater than 90% are given as the second set of numbers above the nodes (Fig 2). Bayesian Inference (BI) analysis was conducted with MrBayes v. 3.1.2 [17] to evaluate Posterior probabilities (BYPP) (Rannala and Yang 1996, Zhaxybayeva and Gogarten 2002) by Markov Chain Monte Carlo sampling (BMCMC). Two parallel runs were conducted, using the default settings, but with the following adjustments: Six simultaneous Markov chains were run for 5,000,000 generations and trees were sampled every 1000 generation. The distribution of log-likelihood scores was examined to determine stationary phase for each search and to decide if extra runs were required to achieve convergence, using the program Tracer 1.5 [18]. First 20% of generated trees were discarded and remaining 80% of trees were used to calculate posterior probabilities (PP) of the majority rule consensus tree. Phylograms were visualized with FigTree v1.4.0 program [19] and reorganized in Microsoft power point (2016) and Adobe Illustrator CS5 (Version 15.0.0, Adobe, San Jose, CA). The finalized alignment and tree is deposited in TreeBASE, submission ID: 22749 (http://purl.org/phylo/treebase/phylows/study/TB2:S22749).
Fig 2. RAxML tree based on DNA sequence analyses of a combined dataset of LSU, ITS and RPB2 gene regions.
Bootstrap support values for ML and MP equal to or greater than 90%, Bayesian posterior probabilities (PP) equal to or greater than 0.90 are defined as ML/MP/PP above or below the nodes. Taxonomic novelty is indicated in red and ex-type strains are in bold text. GenBank accession numbers are indicated at the end of the species name. The tree was rooted to Tubeufia chiangmaiensis MFLU 11–1149, Tubeufia javanica MFLU 13–0371.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to Index Fungorum from where they will be made available to the Global Names Index. The unique Index Fungorum number can be resolved and the associated information viewed through any standard web browser by appending the Index Fungorum number contained in this publication to the prefix www.indexfungorum.org/. The online version of this work is archived and available from the following digital repositories: PubMed Central, LOCKSS.
Compliance with ethical standards
There is no conflict of interest (financial or non-financial) and all authors have agreed to submission of paper. The authors also declare that they have no conflict of interest and confirm that the field studies did not involve endangered or protected species.
Results
Phylogenetic analyses
The combined LSU, ITS, RPB2 dataset which comprises 38 strains of Valsariaceae species was analyzed to determine the placement of our new taxon and infer relationships at the intrageneric level. The phylogenetic trees obtained from maximum likelihood and parsimony analyses yielded trees with similar overall topology and in agreement with previous study based on maximum likelihood analysis [1]. The maximum parsimony dataset consists of 2414 characters with 1707 characters as constant information, 52 characters as variable characters are parsimony-uninformative, and 655 characters were counted as parsimony informative character. The most parsimonious tree had the following tree scores: TL = 1666, CI = 0.744, RI = 0.930, RC = 0.692, HI = 0.256 values. The best scoring RAxML tree is presented in Fig 2. For better phylogenetic inferences derived from our analyses, we divided the ingroup taxa in the phylogram (Fig 2) into eleven clades (A–K) which corresponds to different Valsaria species, including our new taxon. All V. robiniae strains clustered together in one clade (clade A) with high statistical support (100% ML/ 99% MP/ 1.00 PP). Clade B appearing in 100% ML/ 98% MP/ 1.00 PP of the bootstrap replicates comprises V. spartii while V. insitiva strains are basal (clade C) and all these taxa constitute a strongly supported monophyletic clade (100% ML/ 100% MP/ 1.00 PP). Valsaria neotropica (clade D) is basal to V. robiniae, V. spartii and V. insitiva, but its relationships to these species receive weak support while V. rudis (clade E) and V. lopadostomoides (clade F) cluster together in a monophyletic group but with no support. Our new taxon, V. ostryae (clade G) is basal to V. lopadostomoides and V. rudis. Individual LSU, ITS and RPB2 single gene trees were generated from individual datasets and phylogenies recovered were essentially topologically similar with no conflicting phylogenetic signals (data not shown). The new sequence data are deposited in GenBank (Table 1).
Table 1. Culture collection code and GenBank accession numbers of fungal strains used in this study.
| Genbank Accession No | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species | Isolate No. | Code | Herbarium No. | Substrate/Host | Country | LSU | ITS | Rpb2 |
| Bambusaria bambusae | MFLUCC 12–0851 | DDQ00253 | MFLU 15–0050 | Thyrsostachys siamensis | Thailand | KP687812 | KP687812 | KP687890 |
| Bambusaria bambusae | CBS 139763 | DDQ00254 | MFLU 15–0051 | Thyrsostachys siamensis | Thailand | KP687813 | KP687813 | KP687891 |
| Myrmaecium fulvopruinatum | CBS 139057 | VF | WU 33433 | Fagus sylvatica | Austria | KP687858 | KP687858 | KP687933 |
| Myrmaecium fulvopruinatum | VF1 | WU 33434 | Fraxinus excelsior, Alnus glutinosa | Austria | KP687859 | KP687859 | KP687934 | |
| Myrmaecium fulvopruinatum | VFB | WU 33436 | Betula pendula | Austria | KP687860 | KP687860 | KP687935 | |
| Myrmaecium fulvopruinatum | CBS 139058 | VFJ | NY (M.E.B.B. 6905)(E) | Fagus grandifolia | U.S.A. | KP687861 | KP687861 | KP687936 |
| Myrmaecium fulvopruinatum | VFJ1 | WU 33437 | unidentified corticated twigs | Taiwan | KP687862 | KP687862 | KP687937 | |
| Myrmaecium fulvopruinatum | CBS 139059 | VFQ | WU 33438 | Quercus cerris | Austria | KP687863 | KP687863 | KP687938 |
| Myrmaecium rubricosum | CBS 139067 | VRF | WU 33447 | unidentified bark | France | KP687881 | KP687881 | KP687955 |
| Myrmaecium rubricosum | VRJ | Quercus robur | France | KP687882 | KP687882 | KP687956 | ||
| Myrmaecium rubricosum | VRJI | WU 33448 | unidentified bark | U.S.A. | KP687883 | KP687883 | KP687957 | |
| Myrmaecium rubricosum | CBS 139069 | VRM | WU 33449 | Picea abies | Austria | KP687884 | KP687884 | |
| Myrmaecium rubricosum | CBS 139068 | VRP | WU 33450 | Quercus pubescens | Croatia | KP687885 | KP687885 | KP687958 |
| Myrmaecium rubrum | CBS 109505 | Quercus sp. | Italy | GU456324 | - | GU456344 | ||
| Tubeufia chiangmaiensis | MFLUCC 11–0514 | - | MFLU 11–1149 | dead wood of an unidentified tree | Thailand | KF301538 | KF301530 | - |
| Tubeufia javanica | MFLUCC 12–0545 | - | MFLU 13–0371, | dead culm-sheath of bamboo | Thailand | KJ880036 | KJ880034 | - |
| Valsaria insitiva | CBS 139056 | VA | WU 33453 | Acer monspessulanum | Croatia | KP687847 | - | KP687922 |
| Valsaria insitiva | VAC | WU 33454 | Acer campestre | Austria | KP687849 | KP687849 | KP687924 | |
| Valsaria insitiva | VAF | WU 33455 | Ficus carica | France | KP687850 | KP687850 | KP687925 | |
| Valsaria insitiva | VCE | WU 33456 | Cercis siliquastrum | Greece | KP687854 | KP687854 | ||
| Valsaria insitiva | CBS 127882 | VV | WU 33462 (E) | Vitis vinifera | Croatia | KP687886 | KP687886 | KP687959 |
| Valsaria lopadostomoides | CBS 139062 | VIQ | WU 33470 (H) | Quercus ilex | Greece | KP687868 | KP687868 | |
| Valsaria neotropica | CBS 139064 | VJM | WU 33471 (H) | unidentified corticated twig | France | KP687874 | KP687874 | KP687948 |
| Valsaria ostryae | MFLUCC 18–1123 | MFLU 17–0837 | Ostrya carpinifolia | Italy | MH337873 | MH337874 | MH337875 | |
| Valsaria robiniae | CBS 121890 | VC | WU 33472 | Hippocrepis emerus | Slovenia | KP687851 | KP687851 | KP687926 |
| Valsaria robiniae | CBS 128015 | VCA | WU 33474 | Caragana arborescens | Austria | KP687853 | KP687853 | KP687928 |
| Valsaria robiniae | CBS 125583 | VCo1 | WU 33475 | Colutea arborescens | Austria | KP687855 | KP687855 | KP687930 |
| Valsaria robiniae | V1R | WU 33477 | Robinia pseudacacia | Italy | KP687869 | KP687869 | KP687944 | |
| Valsaria robiniae | CBS 139063 | VIR1 | WU 33478 | Robinia pseudacacia | Italy | KP687870 | KP687870 | KP687945 |
| Valsaria rudis | VQC | WU 33483 | Quercus cerris | Croatia | KP687877 | KP687877 | KP687951 | |
| Valsaria rudis | CBS 139065 | VQM | WU 33484 | Quercus macrolepis | Greece | KP687878 | KP687878 | KP687952 |
| Valsaria rudis | CBS 139066 | VQP | WU 33485 (E) | Quercus pubescens | Austria | KP687879 | KP687879 | KP687953 |
| Valsaria rudis | V3 | WU 33486 | Quercus cerris | Italy | KP687836 | KP687836 | KP687914 | |
| Valsaria rudis | V7 | WU 33487 | Quercus petraea | Austria | KP687844 | KP687844 | KP687920 | |
| Valsaria rudis | V31 | WU 33488 | Quercus petraea | Austria | KP687838 | KP687838 | KP687916 | |
| Valsaria spartii | CBS 139070 | V6 | WU 33505 (E) | Spartium junceum | Italy | KP687843 | KP687843 | KP687919 |
| Valsaria spartii | V18 | WU 33515 | Ulex parviflorus | Spain | KP687823 | KP687823 | KP687901 | |
| Valsaria spartii | V19 | WU 33516 | Teline linifolia | Spain | KP687824 | KP687824 | KP687902 | |
| Valsaria spartii | V20 | WU 33517 | Cytisus baeticus | Spain | KP687826 | KP687826 | KP687904 | |
| Valsaria spartii | V21 | WU 33518 | Retama monosperma | Spain | KP687827 | KP687827 | KP687905 | |
Taxonomy
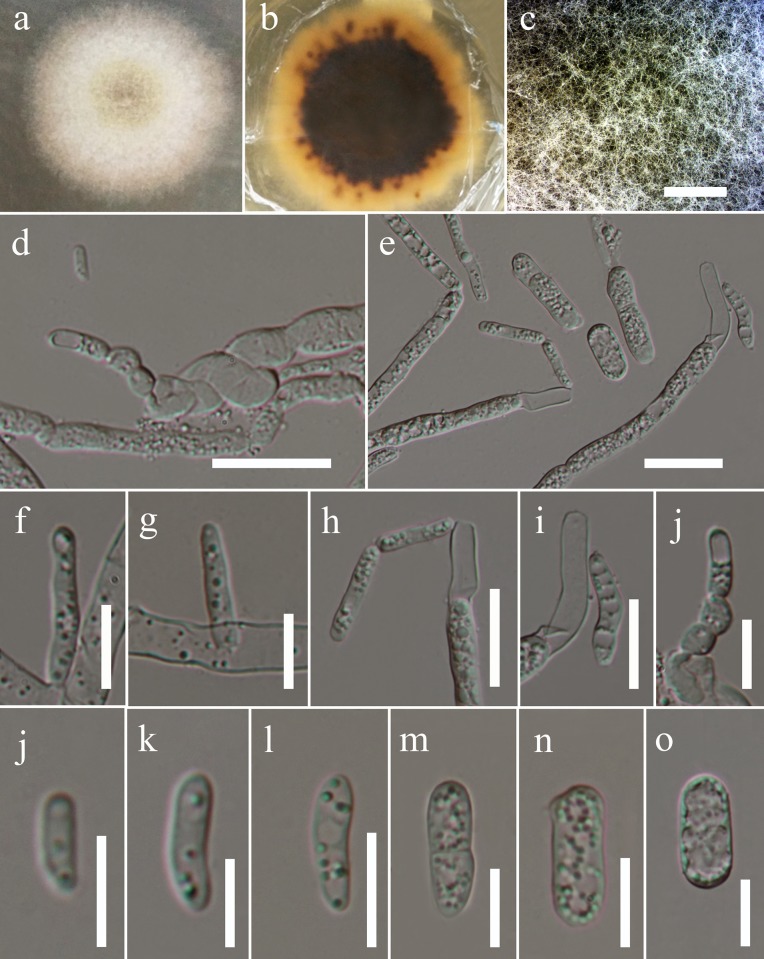
Valsaria ostryae D. Pem, R. Jeewon, Camporesi & K.D. Hyde, sp. nov. Fig 3
Fig 3. Valsaria ostryae (MFLU 17–0837).
a-d Habit and appearance of ectostromata on host surface. e Vertical section of stroma. f Peridium wall. g Apically free pseudoparaphyses. h-j Immature asci k Mature asci. l, m Apical rings in Congo red. n-p Immature ascospores q Mature ascospore r Germinating ascospores s, t Culture characters on MEA (s above view, t back view). Scale bars: a = 500 μm, b = 200 μm, c,d = 500 μm, e = 200 μm, f = 10 μm, g = 5 μm, h-l = 50 μm, n-r = 5 μm.
[urn:lsid:indexfungorum.org:names:554758]
Facesoffungi number: FoF 04614
Etymology–Name reflects the host from which the fungus is isolated.
Holotype–MFLU 17–0837
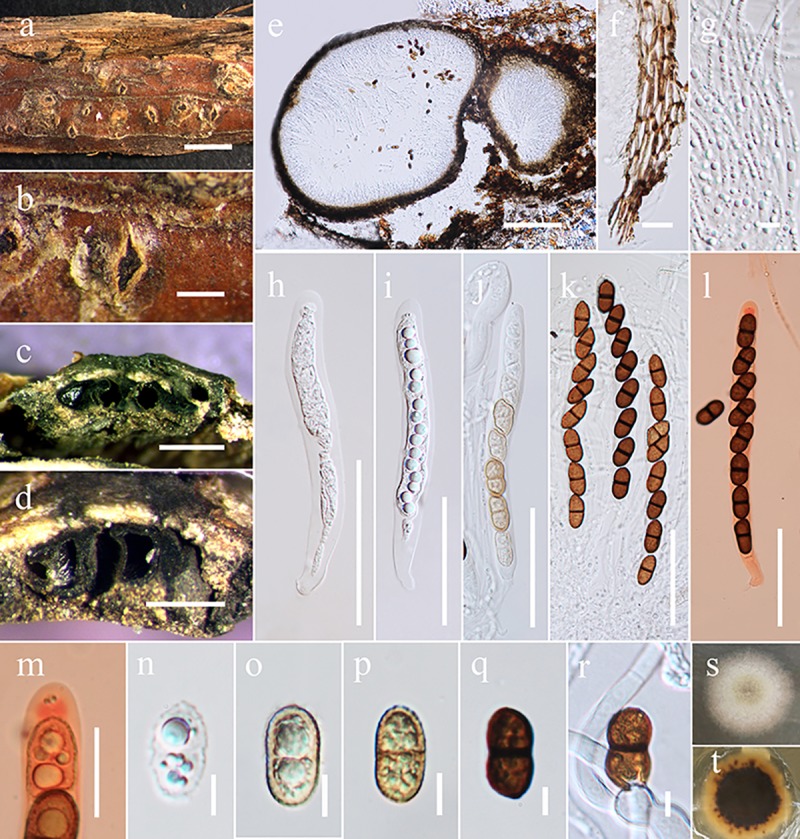
Saprobic on dead branch of Ostrya carpinifolia. Sexual morph: Stromata pseudostromatic, erumpent from bark, scattered, sometimes aggregating in groups of 2–3; pustular, conical to subglobose with flattened base, 500–800 μm high, 1000–1500 μm diam., enclosed on top and/or at sides by a black, 20–40 μm thick pseudoparenchymatous crust. Ectostroma often forming sub- or inversely stellate structures of 3–5 greyish, brown to black, tubercular segments; tissue beneath crusty pseudoparenchymatous; tissue between necks and at stromatal base prosenchymatous, grey, soft, mixed with bark cells. Ostiolar openings inconspicuous at the surface or indistinctly papillate. Ascomata arranged in valsoid configuration, monostichous, subglobose to flask-shaped, 277–479 μm high, 284–603 μm diam., peridium 25–40 μm thick, of pale brown compressed cells of textura angularis. Paraphyses numerous, unbranched, tapering upwards, apically free, 2–3 μm wide. Asci 82–131 × 9–12 μm ( = 110 × 11 μm, n = 15), 8-spored, bitunicate, fissitunicate, cylindrical, apex containing an ocular chamber and a short cylindrical to barrel-shaped ring, 2–3 μm wide, 3–4 μm high ( = 2 × 3 μm, n = 10), staining in Congo Red. Ascospores 16–17 × 7–9 μm, ( = 16 × 7 μm, n = 20), ellipsoid, 2-celled, dark brown, with a dark, thick, central, slightly constricted septum; surface finely warted to reticulate. Asexual morph: Pycnidia 0.1–0.4 mm diam, globose, multiloculate, black, irregularly surrounded by white mycelium, wall pseudoparenchymatous, thick-walled, globose to clavate cells, giving rise to short, cylindrical to ellipsoid hyaline cells. Phialides 7–18 × 2–5 μm ( = 13 × 3 μm, n = 20) produced in more or less parallel, densely packed in palisades, narrow, lageniform, cylindrical or conical. Pycnoconidia 14–18 × 2–9 μm ( = 16 × 5 μm, n = 20) oblong, cylindrical to ellipsoid, sometimes subglobose, 1-celled, hyaline, smooth, lower end sometimes truncate (Fig 4).
Fig 4. Valsaria ostryae: Asexual morph (MFLU 17–0837).
a-c Culture characteristics and mycelium. d,e hyphae with denticles and conidia (partly budding). f-j Phialides. j-o Conidia. Scale bars: c = 1000 μm, d = 20 μm, e = 10 μm, f-j = 10 μm, j-o = 10 μm. Notes–Valsaria ostryae resembles V. insitiva (type species), in having pseudostromatic stromata, pseudoparenchymatous ectostroma, bitunicate asci containing short cylindrical ring and ellipsoid, dark brown ascospores. However, V. ostryae differs from V. insitiva in having narrower pseudoparaphyses (2–3 μm vs. 1.5–5 μm), guttule number in each ascospore cell (0 vs. 2–3) and in ornamentation and constriction of the ascospores. The ascospores are deeply constricted at septum and finely warted to reticulate in V. ostryae while those of V. insitiva are not or hardly constricted at the septum and finely tuberculate. Our multi-gene phylogeny suggests that V. ostryae is related to V. lopadostomoides and V. rudis but phylogenetically segregated from them with strong bootstrap support (100% ML, 100% MP, 1.00 PP). Valsaria ostryae has smaller stromata (500–800 μm high, 1000–1500 μm diam v.s 1200–1400 μm high, 1700–2500 μm diam), shorter asci (82–131 × 9–12 μm v.s (112–)115–128(–136)×(10.5–)11.0–14(–15.5) and have different ascospore ornamentation (finely warted to reticulate v.s distinctly warted, reticulate or spotted). Our new taxon is also morphologically different from V. rudis in having smaller stromata1000–1500 μm diam v.s 1200–1700 μm high, 1200–2500 μm diam) and smaller ascomata (277–479 μm high, 284–603 μm diam, v.s 3000–6000 μm high, 2000–5000 μm diam), Furthermore, a comparison of 600 nucleotides across the ITS (+5.8S) gene regions between V. ostryae and V. rudis reveals 5.31% base pair differences and that of V. ostryae and V. lopadostomoides reveals 6.60% base pair differences respectively. Hence, V. ostryae is introduced as a new species in the genus Valsaria.
Culture characteristics: Ascospores germinating on MEA within 24 h. Colonies growing on MEA, reaching 2 cm diam. in 1 week at 16°C. Mycelium medium dense, circular, slightly raised, surface smooth, edge slightly crenate, thinly hairy, slightly irregular margin, above whitish to pale yellow, reverse dark brown in middle and yellowish at the edge.
Material examined: ITALY, Province of Forlì-Cesena [FC], Santa Sofia, Camposonaldo, on dead branches of Ostrya carpinifolia (Betulaceae), 18 March 2017, E. Camporesi, IT 3290 (MFLU 17–0837, holotype; HKAS 97468, isotype) ex-type living culture MFLUCC 18–1123, ICMP 22558.
Discussion
Species of Valsaria occur primarily on woody parts of ornamental plants but is also distributed on a wide range of hosts. Our new taxon was isolated from Ostrya carpinifolia which belongs to Eudicots and Betulaceae family [20]. Substrates colonised by members of Valsaria recorded until now are all dicotyledons of the Fabaceae, Fagaceae and Vitaceae. Valsaria lopadostomoides can be found on evergreen oaks, such as Quercus ilex (Fagaceae) while V. rudis occur on deciduous oaks, such as Quercus cerris and Q. petrae (Fagaceae). Woody plant hosts of V. lopadostomoides and V. rudis are closely related to each other as both are accommodated in the Fagaceae which is a monophyletic family close to Quercoideae [21]. Valsaria neotropica has been isolated from unidentified hosts in the Neotropics and seems to be distributed in the tropical terrestrial ecoregions of the Americas and the South American temperate zone. Valsaria insitiva was described mainly from non-fabaceous hosts and Vitis (Vitaceae) which is accommodated in its own order Vitales and closely related to Rhamnaceae in the order Rhamnales [22]. Valsaria robiniae have been reported from Amorpha, Caragana, Colutea, Hippocrepis and Robinia species in Central Europe, submediterranean regions and Eastern USA while V. spartii occur on various species of woody Fabaceae in the Sub-Mediterranean. Host distribution of V. robiniae and V. spartii seems to be related as most of the species are from Fabaceae which is the third largest family of angiosperms after Orchidaceae and Asteraceae [23]. Based on available literature, it is apparent that Valsaria has a wide host range and species are less likely to be host specific. As far as we are aware, no Valsaria species are known from Ostrya carpinifolia. We compare the new species with V. insitiva, the type species of Valsaria and they share similar characters. However, they differ in the number of guttules in each ascospore cell, as well as in the surface ornamentation of the ascospores (Fig 3). Our new taxon can also be compared to V. sparti and V. robiniae and differs in ascospores that are not constricted. Valsaria ostryae also shares similar morphological characters to V. rudis and has 2–3 guttules in each ascospore cell, while V. rudis has no guttule in its ascospore cells. Previous morphological studies and phylogenetic investigations based on SSU, ITS-LSU RPB2 and TEF genes data have provided insights into the taxonomy and classification of Valsaria and its allies [1]. The most recent taxonomic work dealt with more than 100 collections of Valsaria sensu lato, including two new species, which has helped to resolve generic delimitation, circumscription and classification of Valsaria [1]. In this study, a new species with a unique morphological character is illustrated and its relationships with other species are investigated based on multi-gene phylogenetic analyses. Our molecular data analyzed herein generated phylogenies that are congruent with those of Jaklitsch et al. [1] and helps to resolve intergeneric relationships between Bambusaria, Myrmaecium and Valsaria. Three well supported monophyletic groups are recovered which support Bambusaria, Myrmaecium and Valsaria as phylogenetically distinct genera (Fig 2). Nevertheless, we observe that several morphological characters such as ostiole, apical ring, and pseudoparaphyses are interspersed in several clades which could be examples of homoplasy in morphological characters within the Valsariaceae. Our molecular phylogeny provides an overview of the usefulness of specific morphs for species and generic delineation.
One interesting finding herein is the significance of spore surface ornamentation in taxonomy. Ornamentation is a character commonly used to delineate species [24–28]. The taxonomy of fungi is based to a large extent on spore characteristics including spore size, shape, color, surface ornamentation and ontogeny [29–34]. Among asexual fungi, spore pigmentation and ornamentation have been crucial in the taxonomy of appendaged coelomycetes. Jeewon et al. [35, 36], Maharachchikumbura et al. [37] and Liu et al. [38] have demonstrated that versicolourous and nonversicolourous spore can be used to segregate species. Among Pleosporalean fungi, Punithalingam [39] used spore surface ornamentation as a diagnostic feature to differentiate Coniothyrium species. Among other sexual fungi, twenty-one species of Eupenicillium were successfully segregated into two groups (with v/s without distinct equatorial ridges) based on ornamentation of ascospores [40]. Spore septation and colour have also been used as characters for distinguishing genera and species of the family Lophiostomataceae by many mycologists such as Saccardo, who established 9 separate genera using these criteria [41]. Species of the genus Aspergillus Section Nigri, which are taxonomically confused and complex have also been identified and delineated based on spore ornamentation [42]. The latter has also been reported to be useful to demarcate sexual and asexual species at the familial and generic level [43–47]. Surface ornamentation and wall development have also been examined to segregate species of the Pachyphlodes (Pezizaceae, Pezizales) as well as to differentiate between species, genera and families of the Basidiomycetes [48].Villegas et al. [49] reported spore ornamentation as a useful character in differentiating various genera within Gomphales. Clitopilus reticulosporus was described as a new species (Entolomataceae, Agaricales) based on its unique spore ornamentation and the spore evolution theory has been well discussed by Morgado et al. [50]. Interestingly, ornamentation of ascospores as well as guttule number have also been significant to establish new genera and demarcate taxa within Helvellaceae (Pezizales, Ascomycota) such as Gyromitra subgenus Melaleucoides (warted, non apiculate, biguttulate ascospores), Gyromitra subgenus Caroliniana (coarsely reticulate ascospores with multiple spicules at the poles) and Helvella subgenus Cerebriformae (globose, echinate ascospores) [51].
A similar phylogenetic scenario is reported herein. The three clades corresponding to different genera can be easily distinguished based on spore ornamentation. Valsaria is characterized by fine warted to reticulate or finely tuberculate ascospores surface, whereas Bambusaria and Myrmaecium species possess densely reticulate and longitudinally ribbed ornamentation respectively. Even at the species level, ornamentation can be used to a certain extent to segregate species. For example, V. insitiva the type species, can easily be delineated from other Valsaria species herein, except V.neotropica, based on finely tuberculate spore ornamentation. Valsaria rudis and Valsaria lopadostomoides are similar in having ascospores surface ‘warted to reticulate’ but additionally the ascospores of V. lopadostomoides is described as ‘spotted’ [1]. In addition, M. fulvopruinatum, characterized by densely reticulate spores can be distinguished from M. rubrum and M. rubricosum, whose spore surface has labyrinth ornamentation. We can therefore rely mostly on such morphology to circumscribe species at this taxonomic level. However, relationships based on other morphs, such as stromatal characteristics, presence or absence of ostioles/ pseudoparaphyses/apical ring, septum constriction and number of guttules show little congruence with findings from molecular data and are not phylogenetically significant for intergeneric segregation. Nevertheless some of these characters, especially the latter two, if used judiciously can be used to circumscribe species. For instance, V. lopadostomoides and M. fulvopruinatum can be distinguished from other Valsaria and Myrmaecium species on the basis of number of guttules. The number of guttules per spore has also been considered as a diagnostic feature to delineate taxa at the intraspecific level. For example, guttule character was found as important to separate species and varieties recognized in the genus Phoma [52] and helped to provide a key to differentiate among thirty taxa. Recently, Southworth et al. [53] demonstrated that number of guttules in each spore has been useful to delineate species of Balsamia when other characters overlap. Vander [54] reported that the occurrence of large guttules was useful to distinguish species of Phyllosticta, especially when used in combination with other characters. Presence of guttule is especially useful when confirming diagnosis of herbarium material from fresh specimen and this has helped to validate Lophiostoma diminuens (Pers.) Fuckel [55]. The diagnostic microscopical characters supporting ranks of different taxa of the family Sarcoscyphaceae were mainly based upon differences in the guttule (lipid) number and pattern of living ascospores [41]. To differentiate genera within Gnomoniaceae, Monod [56] used characters such as presence or absence of small guttules. The latter has also been considered as a diagnostic feature to separate Diaporthe species from grapevine [57] as well as freshwater fungal species [58, 59]. Guttule characters can also be used to distinguish our new taxon, V. ostryae from others. Phylogeny herein indicates V. ostryae to be more closely related to V. lopadostomoides and V. rudis than any other species. Although these three species share several common morphological features (pseudostromatic stromata, apically free pseudoparaphyses, apical ring and dark brown ascospores), our new taxa is clearly distinct in number of guttules and surface ornamentation.
To further substantiate establishment of Valsaria ostryae as a new species, the complete ITS and RPB2 gene regions were compared. Nucleotides differences reveal clear cut differences that warrant new species status. For example, V. ostryae differs from V. lopadostomoides by 5.31% bp difference in the ITS gene regions. Considering both molecular and morphological data we regard V. ostryae as a new taxon that warrants species rank. The genus Valsaria is known to comprise taxa with minor morphological variations that result in taxonomic confusion. For example, the ascomata of Valsaria insitiva, V. lopadostomoides and V.neotropica are monostichously arranged in valsoid configuration measuring 250–450 μm high, 180–400 μm diam, peridium 14–25 μm thick, cylindrical asci with a short cylindrical to barrel-shaped ring measuring in between 96–112 μm × 11–18.5 μm and 2-celled, ellipsoid dark brown ascospores with the size ranging from 12–19 μm × 6.5–11.7 μm. However, when analyzing the gene regions, they are all genetically different. ITS base pair difference between Valsaria insitiva to V. lopadostomoides is 2.4%, V insitiva to V. neotropica is 11.8%, and V. lopadostomoides to V. neotropica is 14.3%. Likewise, other Valsaria species such as V. robiniae, V. rudis, V. spartii and our new taxon share similar morphological characters which are not really meaningful in species delineation.
This study provides delightful taxonomic insights across Valsaria species. We anticipate that taxonomists will pay particular attention to these characters that can potentially aid in species identification of other genera. Our study also reveals that Valsaria could be a speciose genus and there is a need for more collections targeting a wide variety of hosts which can help to answer pertinent questions with regards to fungal diversity issues.
Data Availability
We have submitted our data on online resources: 1. DNA sequences to Genbank (sequences will be released upon acceptance); Accession number: MH337873; MH337874; MH337875. 2. Alignment of DNA data to Treebase: submission ID: 22749 (http://purl.org/phylo/treebase/phylows/study/TB2:S22749). 3. All our figures are available in a public repository such as Figshare: https://figshare.com/account/articles/8167580#).
Funding Statement
The authors received no specific funding for this work.
References
- 1.Jaklitsch WM, Fournier J, Dai DQ, Hyde KD, Voglmayr H. Valsaria and the Valsariales. Fungal Divers. 2015; 73:159–202. 10.1007/s13225-015-0330-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ju YM, Rogers JD, Huhndorf SM. Valsaria and notes on Endoxylina, Pseudothyridaria, Pseudovalsaria, and Roussoella. Mycotaxon. 1996; 58:419–481. [Google Scholar]
- 3.Hyde KD, Jones EBG, Liu JK, Ariyawansa H, Boehm E, Boonmee S et al. Families of Dothideomycetes. 2013; Fungal Divers 63:1–313 [Google Scholar]
- 4.Wijayawardene NN, Hyde KD, Lumbsch TH, Liu JK, Maharachchikumbura SN, Ekanayaka AH et al. Outline of Ascomycota: 2017. Fungal Divers. 2018; 88(1):167–263. [Google Scholar]
- 5.Cesati V de, Notaris G. de. Schema di classificazione degli sferiacei Italici aschigeri piu o meno appartenenti al genere Sphaeria nell’antico significato attribuitogli de Persoon. Comm. Soc. crittog. 1863; Ital.1 (fasc. 4): 205 (1863). [Google Scholar]
- 6.Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Peršoh D, Dhami MK et al. The sooty moulds. Fungal Divers. 2014; 66:1–36. [Google Scholar]
- 7.Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L et al. The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015; 74(1):3–18. [Google Scholar]
- 8.Index Fungorum. 2018 http://www.indexfungorum.org/names/Names.asp (Accessed 26 February 2018)
- 9.Jeewon R, Hyde KD. Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere. 2016; 7:1669–1677. [Google Scholar]
- 10.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990; 172:4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White TJ, Burns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics PCR protocols: A Guide to Methods and Applications (Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds). Academic Press, San Diego, California, USA; 1990. pp. 315–322. [Google Scholar]
- 12.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Bio Evo. 1999; 16(12):1799–1808. [DOI] [PubMed] [Google Scholar]
- 13.Liu JK, Hyde KD, Jeewon R, Phillips AJL, Maharachchikumbura SSN, Ryberg M et al. Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal divers. 2017; 84:75–99. [Google Scholar]
- 14.Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012; 12:335–337. [Google Scholar]
- 15.Pattengale ND, Alipour M, Bininda-Emonds OR, Moret BM, Stamatakis A. How many bootstrap replicates are necessary? J Comput Biol. 2010; 17(3):337–54. 10.1089/cmb.2009.0179 [DOI] [PubMed] [Google Scholar]
- 16.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989; 29:170–179. [DOI] [PubMed] [Google Scholar]
- 17.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001; 294:2310–2314. 10.1126/science.1065889 [DOI] [PubMed] [Google Scholar]
- 18.Rambaut A, Drummond AJ. 2004 Tracer v1.3http://evolve.zoo.ox.ac.uk/software.html (Accessed: 22 January 2018)
- 19.Rambaut A, Drummond AJ. FigTree: Tree figure drawing tool, version 1.4. 0. Institute of Evolutionary Biology, University of Edinburgh; 2012. [Google Scholar]
- 20.Taylor TN, Taylor EL, Krings M. Flowering plants In: Paleobotany. The Biology and Evolution of Fossil Plants, 2nd edn Academic Press, 2009, pp 873–997. [Google Scholar]
- 21.Crepet WL, Nixon KC. Earliest Megafossil Evidence of Fagaceae: Phylogenetic and Biogeographic Implications. Am J Botany. 1989; 76:842–855. [Google Scholar]
- 22.Doweld AB. The systematic relevance of fruit and seed structure in Bersama and Melianthus (Melianthaceae). Plant Syst Evol. 2001; 227:75–103. [Google Scholar]
- 23.Doyle JJ. Leguminosae In: Brenner S. & Miller J. H. (Eds.), Encyclopedia of Genetics (pp. 1081–1085). New York: Academic Press; 2001. [Google Scholar]
- 24.Jeewon R, Cai L, Zhang K, Hyde KD. Dyrithiopsis lakefuxianensis gen et sp. nov. from Fuxian Lake, Yunnan, China and notes on the taxonomic confusion surrounding Dyrithium. Mycologia. 2003a; 95:911–920. [PubMed] [Google Scholar]
- 25.Liu AR, Chen SC, Wu SY, Xu T, Guo LD, Jeewon R et al. Cultural studies coupled with DNA based sequence analyses and its implication on pigmentation as a phylogenetic marker in Pestalotiopsis taxonomy. Mol Phylogenetics Evol. 2010; 57:528–535. [DOI] [PubMed] [Google Scholar]
- 26.Liu JK, Phookamsak R, Dai DQ, Tanaka K, Jones EBG, Xu JC et al. Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa. 2014; 181 (1):001–033. [Google Scholar]
- 27.Phookamsak R, Liu JK, McKenzie E, Manamgoda D, Ariyawansa H, Thambugala K et al. Revision of Phaeosphaeriaceae. Fungal divers. 2014; 68 (1):159–238. [Google Scholar]
- 28.Wijayawardene NN, Hyde KD, Wanasinghe DN, Papizadeh M, Goonasekara ID, Camporesi E et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 2016; 77:1–316. [Google Scholar]
- 29.Pegler DN, Young TWK. The gasteroid Russulales. Trans Br Mycol Soc. 1979; 72:353–388. [Google Scholar]
- 30.Pegler DN, Young TWK. A natural arragement of the Boletales, with reference to spore morphology. Trans Br Mycol Soc. 1981. 76: 103–146. [Google Scholar]
- 31.Petersen R, Pegler DN, Young TWK. Basidiospora morphology in the Agaricales. Mycologia. 1973; 65(1):257. [Google Scholar]
- 32.Kühner R. La notion d’espece chez les champignons superieurs. Bull Mens Soc Linn Lyon. 1976, 45:19–48. [Google Scholar]
- 33.Cai L, Jeewon R, Hyde KD. Phylogenetic evaluation and taxonomic revision of Schizothecium based on ribosomal DNA and protein coding genes. Fungal Divers. 2005; 19:1–17. [Google Scholar]
- 34.Pinnoi A, Jeewon R, Sakayaroj J, Hyde KD, Jones EBG. Berkleasmium crunisia sp. nov. and its teleomorphic affinities to the Pleosporales based on 18S, 28S and ITS-5.8S rDNA sequence analyses. Mycologia. 2007: 99:378–384. [DOI] [PubMed] [Google Scholar]
- 35.Jeewon R, Liew ECY, Hyde KD. Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol Phylogenet Evol. 2002; 25:378–392. [DOI] [PubMed] [Google Scholar]
- 36.Jeewon R, Liew ECY, Hyde KD. Phylogenetic evaluation of species nomenclature of Pestalotiopsis in relation to host association. Fungal Divers. 2003b; 17:39–55. [Google Scholar]
- 37.Maharachchikumbura SSN, Hyde KD, Groenewald JZ, Xu J, Crous PW. Pestalotiopsis revisited. Stud. Myco. 2014; 79:121–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu AR, Chen SC, Wu SY, Xu T, Guo LD, Jeewon R et al. Cultural studies coupled with DNA based sequence analyses and its implication on pigmentation as a phylogenetic marker in Pestalotiopsis taxonomy. Mol Phylogenetics Evol. 2010; 57:528–535. [DOI] [PubMed] [Google Scholar]
- 39.Punithalingam E. A new species of Coniothyrium. Trans Br Mycol Soc. 1970; 55: 497–499. [Google Scholar]
- 40.Udagawa SI, Horie Y. Some Eupenicillium from soils of New Guinea. Trans Br Mycol Soc Japan. 1973; 14:370–387. [Google Scholar]
- 41.Chesters CGC, Bell ANN. Studies in the Lophiostomataceae Sacc. Mycol pap. 1970; 120–175. [Google Scholar]
- 42.Silva DM, Batista LR, Rezende EF, Fungaro MH, Sartori D, Alves E. Identification of fungi of the genus Aspergillus section nigri using polyphasic taxonomy. Braz J Microbiol. 2011; 42(2):761–773. 10.1590/S1517-838220110002000044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai L, Jeewon R, Hyde KD. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Fungal Biol. 2006a; 110:137–150. [DOI] [PubMed] [Google Scholar]
- 44.Cai L, Jeewon R, Hyde KD. Molecular systematics of Zopfiella and allied genera: evidence from multigene sequence analyses. Fungal Biol. 2006b. 110:359–368. [DOI] [PubMed] [Google Scholar]
- 45.Kodsueb R, McKenzie EHC, Lumyong S, Hyde KD, Jeewon R. Molecular phylogeny of Aquaticheirospora broccolii; a new synnematous hyphomycete taxon from Thailand and its teleomorphic affinities to Massarinaceae. Bot J Linean Soc. 2007; 155:283–296. [Google Scholar]
- 46.Zhang Y, Jeewon R, Fournier J, Hyde KD. Multi-gene phylogeny and morphotaxonomy of Amniculicola lignicola: novel freshwater fungus from France and its relationships to the Pleosporales. Fungal Biol. 2008; 112:1186–1194. [DOI] [PubMed] [Google Scholar]
- 47.Thongkantha S, Jeewon R, Vijaykrishna D, Lumyong S, McKenzie EC, Hyde KD. Molecular phylogeny of Magnaporthaceae (Sordariomycetes) with a new species, Ophioceras chiangdaoense from Dracaena loureiroi in Thailand. Fungal Divers. 2009; 34:157–173. [Google Scholar]
- 48.Larsson E, Larsson KH. Phylogenetic relationships of russuloid basidiomycetes with emphasis on aphyllophoralean taxa. Mycologia. 2003; 95(6):1037–1065. [DOI] [PubMed] [Google Scholar]
- 49.Villegas M, Cifuentes J, Estrada-Torres A. Sporal characters in Gomphales and their significance for phylogenetics. Fungal Divers. 2005; 18:157–175. [Google Scholar]
- 50.Morgado L, Noordeloos ME, Hausknecht A. Clitopilus reticulosporus, a new species with unique spore ornamentation, its phylogenetic affinities and implications on the spore evolution theory. Mycol Progress. 2016; 15:1–8. [Google Scholar]
- 51.Abbott SP, Currah RS. The Helvellaceae: Systematic revision and Occurrence in Northern and Northwestern North America. Mycotaxon. 1997; 61:1–125. [Google Scholar]
- 52.Gruyter De. Contributions towards a monograph of Phoma (Coelomycetes)–IX. Section Macrospora. Persoonia. 2002; 18(1):85–102. [Google Scholar]
- 53.Southworth D, Frank JL, Castellano MA, Smith ME, Trappe JM. Balsamia (Sequestrate Helvellaceae, Ascomycota) in western North America. Fungal Sys Evol. 2018; 2:11–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van der Aa HA. Studies in Phyllosticta I. Stud Mycol. 1973; 5:1–110. [Google Scholar]
- 55.Baral HO. The European and North-American species of Sarcoscypha. In: Vital taxonomy and ecology of Ascomycetes with special regard to inoperculate discomycetes. Available at: http://www.gbif-mycology.de/HostedSites/Baral/index.html (Accessed 31 May 2018)
- 56.Monod M. Monographie taxonomique des Gnomoniaceae (Ascomycètes de l'ordre des Diaporthales). I. Beihefte zur Sydowia. 1983; 9:1–314. [Google Scholar]
- 57.Lawrence DP, Travadon R, Baumgartner K. Diversity of Diaporthe species associated with wood cankers of fruit and nut crops in northern California. Mycologia. 2015; 107(5):926–940. 10.3852/14-353 [DOI] [PubMed] [Google Scholar]
- 58.Vijaykrishna D, Jeewon R, Hyde KD. Fusoidispora aquatica: New freshwater ascomycetes from Hong Kong based on morphology and molecules. Sydowia. 2005; 57:267–280. [Google Scholar]
- 59.Vijaykrishna D, Mostert L, Jeewon R, Hyde KD, Crous PW. Pleurostomosphora, an anamorph of Pleurostoma (Calosphaeriales), a new anamorph genus morphologically similar to Philaphora. Stud Mycol. 2004; 50:387–398. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have submitted our data on online resources: 1. DNA sequences to Genbank (sequences will be released upon acceptance); Accession number: MH337873; MH337874; MH337875. 2. Alignment of DNA data to Treebase: submission ID: 22749 (http://purl.org/phylo/treebase/phylows/study/TB2:S22749). 3. All our figures are available in a public repository such as Figshare: https://figshare.com/account/articles/8167580#).