Abstract
The typical abnormalities observed in the brain of Alzheimer’s disease (AD) patients include synaptic alterations, neuronal death, brain inflammation, and the accumulation of protein aggregates in the form of amyloid plaques and neurofibrillary tangles. Despite the development of many animal and in vitro models for AD, there is a lack of an experimental approach that fully recapitulates essential aspects of the disease in human cells. Here, we report the generation of a new model to study AD, consisting of cerebral organoids (COs) produced from human-induced pluripotent stem cells (iPSCs). Under our experimental conditions, COs grow to form three-dimensional (3D) structures containing neural areas with cortical-like organization. Analysis of COs by histological and biochemical methods revealed that organoids produced from iPSCs derived from patients affected by familial AD or Down syndrome (DS) spontaneously develop over time pathological features of AD, including accumulation of structures highly reminiscent to amyloid plaques and neurofibrillary tangles. These pathological abnormalities were not observed in COs generated from various controls, including human iPSCs from healthy individuals, human iPSCs from patients affected by Creutzfeldt–Jakob disease, mouse embryonic stem cells (ESCs), or mouse iPSCs. These findings enable modeling genetic AD in a human cellular context in a 3D cortical-like tissue developed in vitro from patient-specific stem cells. This system provides a more relevant disease model compared to pre-existing methods and offers a new platform for discovery of novel targets and screening of drugs for therapeutic intervention.
Introduction
Alzheimer’s disease (AD) is the most common cause of late-life dementia and a significant public health problem. Currently, over 35 million individuals worldwide and almost 6 million Americans suffer from AD, representing the sixth-leading cause of death. AD is characterized by progressive memory and cognitive dysfunction associated to various alterations in the brain, including synaptic abnormalities, neuronal death, brain inflammation, and the accumulation of protein aggregates in the form of extracellular amyloid-beta (Aβ) plaques and intracellular tau neurofibrillary tangles (NFTs) [1]. Despite the considerable progress in understanding the pathogenesis of AD, the knowledge has unfortunately not translated into the development of a cure or even efficient therapies. The current treatments are just palliative because they do not slow down or halt the disease progression. Many clinical studies have failed to demonstrate efficacy in human patients for compounds that were able to produce a large effect in pre-clinical disease models [2], suggesting that development of more predictable models is a top priority in the field.
Although extensive research has focused on the development of various cellular and animal models for AD [3], none of the existing models fully recapitulate all of the disease features. A possibility to better reproduce AD abnormalities in experimental conditions is to study human nerve cells derived from patients. Human somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) that are like embryonic stem cells (ESCs) because they divide and renew indefinitely and can be differentiated into any cell type, including neurons [4]. Several recent studies have supported the usefulness of patient-derived iPSCs to model diverse aspects of AD. These studies showed that cells derived from AD patients display some features associated to AD pathogenesis, including higher production of more amyloidogenic variants of the Aβ protein, increased tau phosphorylation, higher susceptibility to Aβ-mediated neurotoxicity, and/or presence of cellular stress markers [5–14]. However, the relevance of studies in cell cultures is compromised by the fact that AD pathology is intricate and involves diverse cell types, tissue structures and cellular pathways. Hence, to properly model AD pathogenesis, it is likely necessary to reproduce the complexity of the human brain tissue.
Recent publications have described methodologies to generate cultures of cerebral organoids (COs). COs are three-dimensional (3D) structures highly reminiscent of certain human brain regions [15, 16], including the cerebral cortex that is one of the areas heavily affected in AD. In this study, we generated COs starting from iPSCs derived from patients affected by AD, Down syndrome (DS), other neurodegenerative diseases and healthy controls. Our data show that “mini-brains” derived from DS or familial AD iPSCs recapitulate some of the abnormalities observed in the AD brain, including the accumulation of Aβ and tau aggregates and markers of cellular apoptosis.
Materials
Patient samples and fibroblast culture
To generate iPSC, some of the human fibroblasts were purchased from Coriell Cell Repositories (Coriell, Candem, NJ, USA). The following donors were used: (1) a 56-year-old male patient with familial AD (cell number: AG06840) carrying a missense mutation (A246E) in the presenilin 1 (PSN1) gene linked to early-onset AD [17]; (2) a 5-month-old male with DS (cell number: AG07096); and (3) a 66-year-old female (unaffected spouse of an AD family member) whose cells were used to generate control iPSCs (cell number: AG08517). Some human fibroblasts were obtained from skin biopsies and generously provided by Dr. Fabrizio Tagliavini (Istituto Neurologico Carlo Besta, Milan, Italy), including: (1) a 61-year-old CJD patient carrying a mutation (E200K) in the cellular prion protein gene and (2) a 49-year-old CJD patient with CJD expressing the cellular prion protein with 8 extra octarepeats. Upon arrival, the fibroblasts were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), 1:100 minimum essential media non-essential amino acids (MEM-NEAA) solution (Gibco), 1:100 Glutamax-I (Gibco), 4 ng/ml human basic fibroblast growth factor bFGF (R&D Systems) and 1:100 antibiotic-antimycotic (Gibco) at 37 °C with 5% CO2. Before generating the iPSC lines, the dermal fibroblasts were passaged 10–20 times.
Brain samples from an AD patient (79 years old, female, AD clinical diagnosis that was confirmed post-mortem) and aged control (59 years old, male, non-demented diagnosis) were obtained from the National Disease Research Interchange (Philadelphia, PA, USA).
iPSC generation
To generate human iPSCs from adult fibroblasts, we followed the protocol described by Nethercott and colleagues [18] with minor modifications. Lentiviral plasmids carrying human cDNAs for OCT4, SOX2, C-MYC, KLF4, NANOG, and LIN28 were cloned in DH5α Competent Cells (Invitrogen). The plasmids were recovered from bacterial cultures using a Qiagen EndoFree Plasmid MegaKit following the manufacturer’s instructions. To produce the viral particles, 293FT packaging cell line (Gibco) were transfected with the lentiviral plasmids pMD2.G (contains VSV-G envelope protein gene) and psPAX2 (contains gagpol genes) using Lipofectamine 2000 (Invitrogen). Briefly, 7.5 μg psPAX2 plasmid, 2.5 μg pMD2.G, and 6 μg of one of the target clone vectors (pSIN4-EF2-O2S, pSIN4-EF2-N2L, or pSIN4-CMV-K2M) were combined with 36 μl of Lipofectamine 2000 (Invitrogen). All plasmids and vectors were purchased from Addgene. After 48 and 72 h, virus-containing supernatants derived from these 293FT cultures were collected and filtered through a 0.45-μm cellulose acetate filter (Millipore). The virus titer was determined using the HIV-1 p24 Antigen Capture Assay (Advanced BioScience Laboratories). The day before transduction, human dermal fibroblasts were seeded at 2 × 105 cells/well in six-well gelatin-coated plates. The day after, the cells were transduced three times (on day 0, day 1, and day 2) with 5 multiplicity of infection (MOI) of the virus/polybrene-containing supernatants. On day 7, the cells were transferred to 6-well plates (1 × 105 cells/well) previously seeded with irradiated mouse embryonic fibroblasts (MEFs) and cultured in standard human ESC culture medium containing KnockOut DMEM/F12 supplemented with 20% KnockOut serum replacement (Gibco), 1:100 MEM-NEAA solution, 1:100 Glutamax-I 100 μM β-mercaptoethanol (Sigma), 30 μg/ml bFGF and 1:100 antibiotic-antimycotic solution. The culture medium was further supplemented with small molecules (0.5 μM thiazovivin, 0.5 μM Y-27632, and 0.5 μM PD0325901, R&D Systems) to increase the survival of generated iPSCs. The culture medium was replaced every day. On day 30, the potential iPSCs colonies (selected by morphological features) were selected from the wells and expanded on irradiated MEFs feeder layers in ESC standard culture medium as described above. In addition, Dr. Ying Liu (University of Texas Health Science Center, Houston, TX, USA) kindly provided an iPSC line derived from a different healthy patient’s urine that was reprogrammed with non-integrating Sendai viral vectors, as described [19]. Mouse ESCs and iPSCs were purchased from Gibco and System Bioscience, respectively.
RT-PCR analysis
Total RNA was extracted from iPSCs using RNeasy Mini kit (Qiagen) and stored at −20 °C. One microgram of total RNA was used to generate cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. RT-PCR was performed for 32 cycles for all markers as follows: denaturing for 45 s at 94 °C, annealing temperature of 55 °C for 20 s and extending at 72 °C for 30 s. PCR products were resolved on a 2% agarose gel.
Alkaline phosphatase (AP) staining
The iPSCs were stained with an AP staining kit (Stemgent). Briefly, the cells were washed twice with PBS and fixed at room temperature for 1–2 min. After removing the fixative solution, the cells were washed with PBS containing 0.5% Tween 20 and incubated for 30 min with freshly prepared AP staining solution. Finally, the cells were covered with mounting medium and stored at 4 °C.
Embryoid body assay
iPSCs were harvested by trypsinization and transferred to ultra-low attachment plates in human ESC medium without bFGF. After 3 days, the embryoid bodies were collected and plated onto gelatin-coated covers and cultured for three additional weeks. Finally, the samples were fixed and processed for immunofluorescence.
Generation of cerebral organoids
Cerebral organoids (COs) were generated following the protocol described by Lancaster and Knoblich [16] with minor modifications. Briefly, on day 0 of the culture procedure, the iPSCs colonies were detached from the culture dishes by treating them with collagenase IV (Sigma) and dispase (StemCell), re-suspended in DMEM/F12 medium (Gibco) and spun down at 200 × g for 5 min. Thereafter, the recovered material was treated with accutase (Millipore) and mechanically disaggregated to obtain a single cell suspension. The cells were plated in 96-well plates with a concave bottom (ultra-low attachment plates, Sigma) at 9000 cells/well in human ESC culture medium with a high concentration (50 μM) of Rock inhibitor (Y-27632, R&D Systems) and 4 ng/ml of bFGF. 70% of the culture medium was completely replaced every day during 6 consecutive days. At day 6, the culture medium was replaced with neural induction medium containing DMEM/F12 medium supplemented with 1:100 N2 supplement (Gibco), 1:100 Glutamax-I, 1:100 MEM-NEAA, 1 μg/ml heparin (Sigma) and 1:100 antibiotic-antimycotic solution (Gibco). On days 11 and 12 of the protocol, COs were embedded in Geltrex LDEV-Free drops (Gibco) and cultured in ultra-low attachment six-well plates (16 COs/well) in differentiation medium containing DMEM/F12 and Neurobasal medium (Gibco) (1:1 ratio) supplemented with 1:200 N2 supplement (Gibco), 1:100 B27 supplement without vitamin A (Gibco), 3.5 μl/l 2-mercaptoethanol, 1:4000 human insulin solution (Sigma), 1:100 Glutamax-I, 1:200 MEM-NEAA and 1:100 antibiotic-antimycotic solution. After 4–5 days, COs were transferred to differentiation medium (same as described above with B27 supplement with vitamin A (Gibco)) and the plates were placed in an orbital shaker (Boekel) set at 90 rpm. The culture medium was replaced every 5 days.
Microscopy
The iPSCs or the differentiated embryoid bodies, both cultured on covers, were fixed in Bouin’s fluid for 20 min at 4 °C and then washed in 50 mM TRIS HCl buffer pH 7.8 and stored at 4 °C until further processing. The COs, kept 30 and 110 days in vitro (DIV), were fixed in Carnoy’s fixative at 4 °C O/N and embedded in paraffin. 5 μm thick sections were obtained for histology analysis. The samples (covers and histological sections) were incubated in the primary antibody diluted in Tris-carrageenan-triton solution containing TRIS HCl buffer, λ-Carrageenan 0.7% and triton X-100 0.5% (O/N at R/T). The primary antibodies were revealed with AlexaFluor labeled secondary antibodies (Life Technologies) diluted 1:500 in TCT. We use the following primary antibodies for the immunostaining procedure: Oct4 (09–0023, Stemgent, 1:200), SOX2 (ab97959, abcam, 1:100), SSEA4 (MC-813–70, DSHB, 1:10), TRA-1–60 (ab16288, abcam, 1:200), brachyury (ab20680, abcam, 1:100), α-fetoprotein (ab3980, abcam, 1:100), human nestin (mab5326, Millipore, 1:100), H3 (ab5176, abcam, 1:1000), DCX (ab18723, abcam, 1:200), MAP-2 (AB5622, Millipore, 1:200), Tuj1 (MMS-435P, Covance, 1:1000), NeuN (MAB377, Millipore, 1:100), CNPase (ab6319, abcam, 1:200), anti-amyloid β 4G8 (SIG-39220, Covance, 1:500), anti-amyloid β 6E10 (SIG-39320, Covance, 1:500), anti-phospho-tau AT8 (MN1020, Thermo Fisher Scientific, 1:100), anti-phospho-tau PHF-1 (kindly provided by Dr. Peter Davies, 1:250), SATB2 (ab34735, abcam, 1:200), Tbr1 (AB10554, Millipore, 1:50), GFAP (MAB360, Millipore, 1:200), Aquaporin 4 (ab46182, abcam, 1:100) and cleaved caspase-3 (9664, Cell Signaling, 1:200). BTA-1, a fluorescent Thioflavin-T derivative, staining was performed by incubating tissue slices with a BTA-1 (Sigma) solution (500 nM in DMSO) for 30 min after deparaffinization. For Hematoxylin staining, samples were incubated for 10 min in hematoxylin and rinsed in water for 10 min. For Gallyas silver staining, samples were treated for 10 min with Potassium permanganate solution (0.3%), 1 min with Oxalic acid solution (1%), 1 min with Alkali silver iodide solution, 1 min with gold chloride solution (0.5%) and 1 min with an aqueous solution of sodium thiosulfate (1%). Samples were dehydrated in graded ethanol, cleared in xylene and cover-slipped with DPX mounting medium (Innogenex).
Each group of samples, analyzed by bright field and immunofluorescence (IF), were collected from at least three independent cultures containing a total number of 50–200 COs. Samples were mounted in VECTASHIELD HardSet Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (H-1500, Vector laboratories). Human samples were treated with 1 mg/ml Propidium iodide (PI) for 20 min and mounted in Prolong Antifade Mountant medium (Gibco). Images were acquired with a Leica DMI 6000B microscope. Confocal imaging was performed on Nikon A1R Confocal Laser Microscope at the Center for Advanced Microscopy, Department of Integrative Biology & Pharmacology at McGovern Medical School, UTHealth.
Western blot analysis
COs were collected at 110 DIV. They were washed two times with PBS, snap-frozen in liquid nitrogen and stored at −80 °C. To prepare the tissue homogenates, the samples were thawed partially on ice, resuspended (20% v/v) in ice-cold PBS containing protease (complete cocktail, Roche) and phosphatase inhibitors (50 mM NaF, 0.2 mM Na3VO4) and lysed using 20 gauge needles. The protein concentration was estimated by the bicinchoninic acid method (BCA, Pierce). A volume of each sample, containing 40–150 μg protein, was mixed with a 5 times volume of methanol, in order to precipitate the proteins. The supernatant was discarded and the pellet was resuspended in SDS sample buffer (containing 33 mM dithiothreitol), boiled for 10 min, and electrophoresed through 4–12% Bis-Tris precast gels (Invitrogen). Proteins were transferred onto a PVDF (0.2 μm, Bio-Rad) membrane. Membranes were incubated with 5% non-fat milk for blocking. Phosphorylated tau (p-tau) was recognized using the PHF-1 antibody (1:1000, kindly provided by Dr. Peter Davies). The blot was subsequently incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (1:1000, Sigma). The blots were developed with ECL Plus reagents (Amersham GE Healthcare), and the immunoreactive bands were visualized by scanning with a Bio-Rad image analysis system. Each group of samples analyzed by western blot was collected from at least three independent cultures containing a total number of 50–200 organoids.
Amyloid β and tau ELISA
The Aβ peptide and tau protein were quantified using commercially available ELISA kits. The following kits were purchased from Invitrogen: human Aβ40 (KHB3482), human Aβ42 (KHB3441), total human tau protein (KHB0041), p-tau [pS396] (KHB7031), p-tau [pT181] (KHO0631) and p-tau [pT231] (KHB8051). The protein concentration was estimated in the 20% organoid homogenates using the BCA method. Two hundred micrograms of protein (in 400 μl) were centrifuged in a L100K ultracentrifuge (Beckman-Coulter, Brea,CA) at 100,000 × g, for 1 h at 4 °C. Supernatants (soluble fraction) were collected and the pellet was resuspended in 50 μl of 70% formic acid (FA), sonicated and neutralized with Tris buffer 2.5 M pH 8.5 (insoluble fraction). Aβ40, Aβ42, total tau, and p-tau levels were measured according the manufacturer’s instructions in soluble and insoluble fractions. Samples were read on an ELISA reader (EL800 BIOTEK, BioTek, Winooski, VT) at 450 nm. Each group of samples analyzed by ELISA was collected from at least three independent cultures containing a total number of 80–220 organoids.
Statistical analysis
The data on Aβ and tau aggregates in different COs was analyzed by one-way ANOVA, followed by Tukey’s multiple comparison post-test, using the Graph Pad software.
Results
Human iPSCs were produced from adult fibroblasts obtained from skin biopsies of a patient affected by familial early-onset Alzheimer’s disease (fAD) carrying a missense mutation (A246E) in presenilin 1 and a patient affected by DS. Reprogramming was done by infection with lentivirus expressing OCT4, SOX2, C-MYC, KLF4, NANOG, and LIN28 as previously described [18] (Supplementary Fig. 1a). All the colonies generated showed the expected morphology (Supplementary Fig. 1b), as well as high levels of alkaline phosphatase (AP) and the expression of pluripotent markers including Oct4, TRA-1–60, Sox2 and SSEA4 (Supplementary Fig. 1c). RT-PCR analysis confirmed the expression of genes typically expressed in iPSCs (Supplementary Fig. 1d). Embryoid body assays revealed that the iPSC lines were able to generate cells derived from the three primitive germ layers of the embryo (Supplementary Fig. 1e). Taken together, these results indicate that iPSC lines produced from control and patient fibroblasts meet essential characteristics of pluripotent stem cells, including gene expression profile, the presence of multiple stem cells markers and pluripotency.
To generate cerebral organoids (COs), we followed the protocol described by Lancaster and Knoblich [16]. Cultures initially containing 9000 iPSCs per well (96-well plates) underwent a progressive compaction over time to form a cellular aggregate with sharp edges by 7 days (Supplementary Fig. 2a). After transferring the cellular aggregates into the neural induction medium, bright and compact areas emerged from their surfaces (Supplementary Fig. 2a). Histological examination of these areas at 30 DIV reveals that they have high cellular density and small cavities lined by neuroepithelial-like cells (Supplementary Fig. 2b). The immunoreactivity of these cells to phosphohistone H3 (Supplementary Fig. 2b, left inset) and the presence of numerous mitotic figures observed at their apical pole (Supplementary Fig. 2b, right inset) indicate that they are proliferative. Additionally, the neuroepithelial-like cells are nestin- (Supplementary Fig. 2c) and SOX2-positive (Supplementary Fig. 2d), confirming their neuroepithelial nature. At this stage (30 DIV), these cells were surrounded by doublecortin (DCX) positive cells, a marker of young neurons (Supplementary Fig. 2e). The neural areas composed by neuroepithelial cells and young neurons grew and matured over time. We closely monitored the maturation of the organoids and their cortical-like organization by daily surveillance. We performed more detailed quantitative studies at 65 DIV and 110 DIV. COs reaching 110 DIV exhibited increased cellular density (Fig. 1a–c), larger neural areas (MAP2+ and nestin+ cells) (Fig. 1d) and more mature cortical architecture. Indeed, cells expressed the neuronal markers Tuj1 and NeuN (Fig. 2a, b) and the cortical neuronal markers SABT2 and Tbr1 (Fig. 2c, d). At this stage (110 DIV), young neurons (DCX+ cells) were no longer visible. Astrocytes (GFAP+ and Aquaporin 4 (AQP4)+ cells) were observed in some COs (Fig. 2e), but no oligodendrocytes (CNPase+ cells) were present (Fig. 2f). Taken together, these results suggest that we were able to generate COs with neural tissue having a cortical-like organization, confirming previously published observations [15, 16, 20].
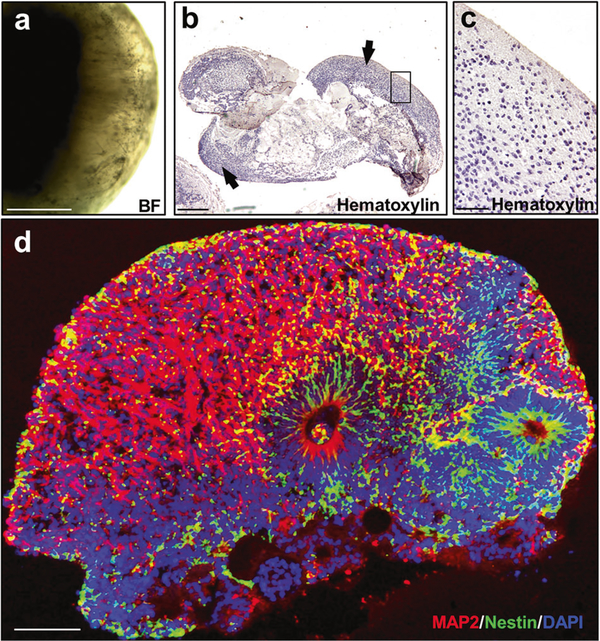
Fig. 1.
Histological analysis of COs at 110 days in culture. COs were generated from human healthy control (HC) iPSCs according to the Lancaster and Knoblich procedure, as described in Methods. a Bright field (BF) representative image of a CO maintained for 110 days in vitro (DIV). b Low-magnification image of one representative CO stained with hematoxylin. Peripheral structures with high cellular density were identified (arrows). c This panel shows the framed area in panel b at higher magnification. d A representative full-sized organoid stained with MAP2 (red) Nestin (green) and DAPI (blue). BF bright field. Scale bars: a, b: 400 μm; c: 50 μm; d: 100 μm
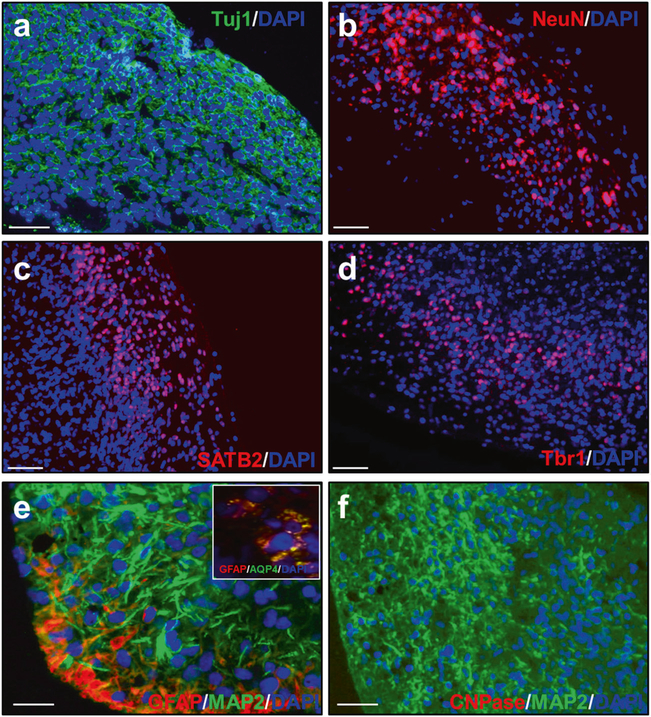
Fig. 2.
Cell types in COs at 110 days in culture. COs prepared from HC iPSCs showed that most of the cells express the neuronal markers Tuj1 (a) and NeuN (b). Additionally, the cells express the cortical markers SATB2 (c) and Tbr1 (d). e Some cells were positive for the astrocyte markers GFAP and aquaporin 4 (AQP4, inset), but no cells were positive for the oligodendrocyte marker CNPAse (f). DAPI in blue. Scale bar 50 μm in all panels
The development of AD neuropathological features was studied in COs at 110 DIV. Amyloid-β (Aβ) deposition was studied in 50–200 independently generated COs at 110 DIV, with 3–5 repeats for each method. Staining using an antibody specific for the sequence 17–24 of Aβ (4G8) revealed the presence of extracellular Aβ deposits of ~25–30 μm in COs derived from fAD and DS (Fig. 3a, center and bottom panels). Conversely, no Aβ reactive deposits were detectable in COs produced from healthy controls (HC; Fig. 3a, top panels). The use of an additional anti-Aβ antibody, 6E10 (recognizing residues 1–16 of human Aβ), showed similar immunoreactive structures (Fig. 3 a, right panels) in neuronal regions (confocal microscopy, Supplementary Fig. 3a) further confirming that these deposits are composed of the Aβ peptide. The Aβ deposits observed in fAD and DS are highly reminiscent in size and morphology to amyloid plaques observed in the brain of humans affected by AD (Fig. 3b, bottom panels), but absent in HC patients (Fig. 3b, top panels). To analyze the specificity of these results for fAD and DS COs, we studied whether Aβ deposition was observed in COs generated from several other iPSC lines, including cells coming from two patients affected by another neurodegenerative disease (familial Creutzfeldt-Jakob disease; Fig. 3c) as well as mouse iPSCs and ESCs (Supplementary Fig. 4). Pathology was not detected in any of the COs produced and analyzed from these cells, suggesting that amyloid deposition was specific for fAD and DS COs.
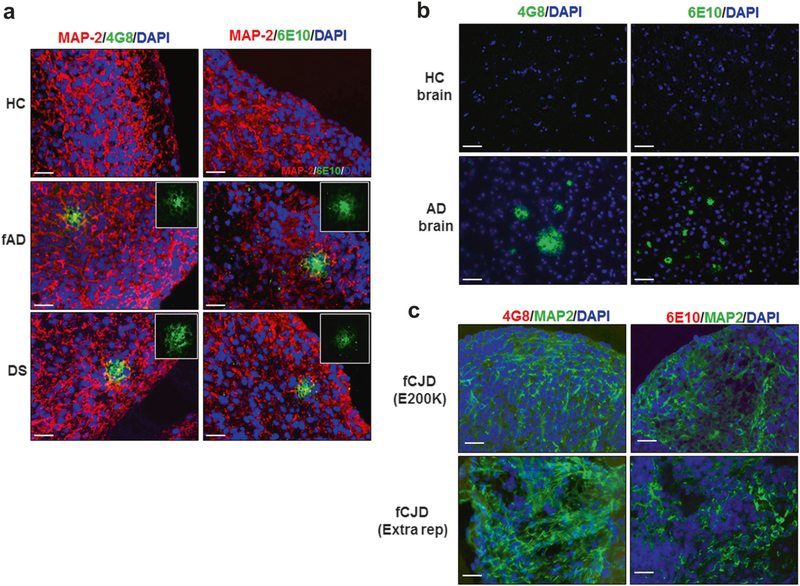
Fig. 3.
Aβ deposition in COs. a COs prepared from iPSCs derived from healthy control (HC, top panels) familiar Alzheimer’s disease (fAD, center panels), and Down syndrome (DS, bottom panels) were cultivated for 110 days and stained with the Aβ specific antibodies 4G8 (left panels) and 6E10 (right panels). Slides were co-stained with an antibody against MAP-2 (red) and DAPI (blue) to label neurons and the cell nuclei, respectively. To better appreciate the morphology of the aggregates, the inset in each panel show zoomed sections of the deposits stained only with the anti-Aβ antibodies. b Staining of Aβ plaques with the 4G8 (left) or 6E10 (right) antibodies (green) in the brain of HC and AD patients, counterstained with DAPI (blue). c To study the specificity of Aβ deposition, COs prepared from iPSCs derived from patients affected by two different forms of familial Creutzfeldt-Jakob disease (fCJD) were stained with 4G8 or 6E10 (red), in parallel with MAP-2 (green) and DAPI (blue). Scale bars: 25 μm
The thioflavin analog BTA-1, a dye that stains amyloid aggregates with high affinity [21], also labeled the amyloid deposits observed in fAD and DS. These results indicate that deposits are organized into amyloid-like aggregates (Fig. 4b, c) similar to the ones detected in the brain of humans affected by AD (Fig. 4e). As expected, BTA-1 positive aggregates were not present in COs or brain slides from healthy controls (Fig. 4a, d). To demonstrate by an independent technique the formation of Aβ aggregates in COs, we performed biochemical studies employing a fractionation procedure commonly used to study brain amyloid deposits, which utilizes formic acid to solubilize large aggregates [22, 23]. ELISA measurements of formic acid extracted Aβ1–42 and Aβ1–40, normalized by the total protein concentration, showed that DS and fAD COs have higher levels of Aβ42/Aβ40 ratio in the insoluble fraction compared with control samples (Fig. 4f).
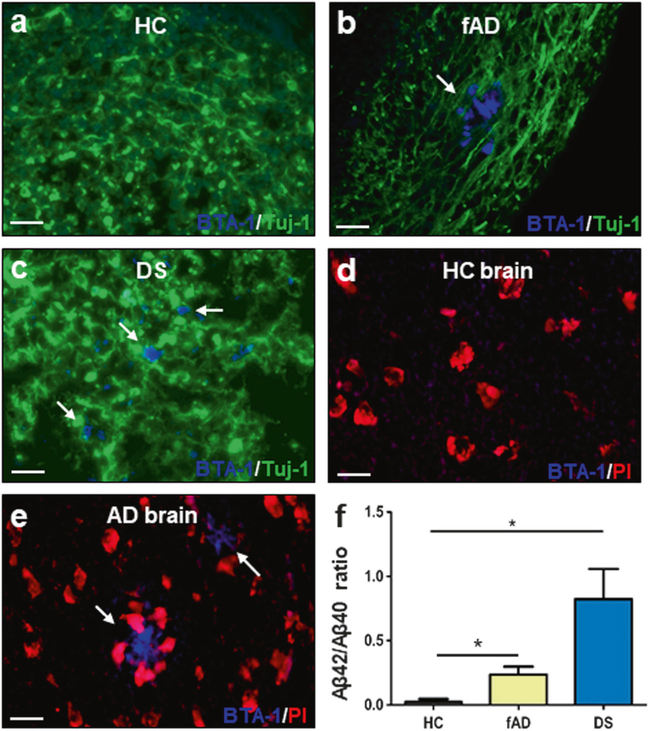
Fig. 4.
Characterization of Aβ deposition by staining with an amyloid-binding dye and by biochemical studies of insoluble Aβ. a–c COs prepared from iPSCs derived from familiar Alzheimer’s disease (fAD), Down syndrome (DS) and healthy control (HC) patients were cultivated for 110 days. Amyloid deposits were stained with the BTA-1 dye (blue) and Tuj-1 (green). d, e Staining of Aβ plaques with the BTA-1 dye (blue) and propidium iodide (PI, red) in brain slides from HC and AD patients. Scale bars in panels a–e: 25 μm. f ELISA assay measuring the levels of total insoluble Aβ peptide (Aβ40 and Aβ42) in COs homogenates. Protein concentration in the COs homogenates was measured by BCA and equal amounts of total protein were used. Extraction was done with formic acid as described in Methods. The figure shows the ratio Aβ42/Aβ40 in the insoluble fraction. Each value corresponds to the average ± SEM of six replicates. For this analysis, 80–220 organoids were used for each replicate. Data was analyzed by one-way ANOVA, and unpaired Student’s t-test. *P <0.05; **P<0.01; ***P<0.001
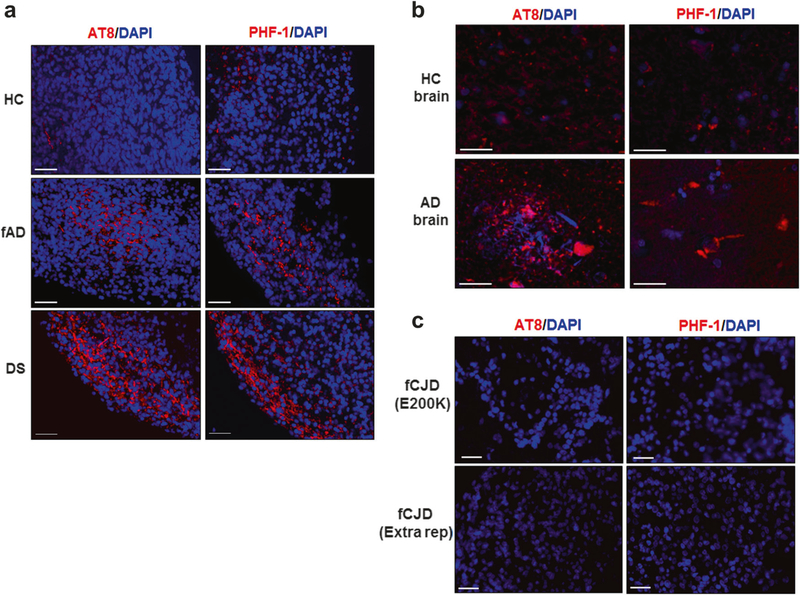
The same group of COs was analyzed for the presence of tau pathology by immunofluorescence, utilizing two widely used antibodies that recognize p-tau: AT8 and PHF-1. Despite the detection of a variable number of neurons containing p-tau, cells positive for AT8 and PHF-1 staining were the highest in DS derived COs, followed by fAD, and much lower for HC samples (Fig. 5a). The reactivity with AT8 and PHF-1 were similar to the staining observed in AD patient brains (Fig. 5b), which were much less intense in the HC brains (Fig. 5b). The accumulation of p-tau appears to come both from cytoplasmic tangle-like deposits and neurite staining of p-tau, which co-localized with the neuron-specific cytoskeletal protein, MAP-2 (confocal microscopy; Supplementary Fig. 3b). To evaluate the specificity of these findings, we studied whether tau pathological abnormalities were also observed in COs generated from non-AD iPSC lines, including iPSCs coming from 2 patients affected by another neurodegenerative disease (familial Creutzfeldt-Jakob disease; Fig. 5c) as well as mouse iPSCs and ESCs (Supplementary Fig. 4). Pathology was not detected in any of the COs analyzed from these cells, suggesting that staining for abnormal tau was specific for the fAD and DS COs.
Fig. 5.
Accumulation of hyper-phosphorylated tau in COs. a COs prepared from iPSCs derived from healthy control (HC, top), familiar Alzheimer’s disease (fAD, center), and Down syndrome (DS, bottom) patients were cultivated for 110 days and processed for histological analysis as described in the Methods. Representative images of COs stained with AT8 (left) and PHF-1 (right), two antibodies that recognize p-tau protein. b Staining of p-tau with the same antibodies was done in slides coming from the brain of HC (top) and AD (bottom) patients. c To evaluate the specificity of p-tau accumulation, COs prepared from iPSCs derived from patients affected by two different forms of familial Creutzfeldt-Jakob disease (fCJD) were stained with AT8 (left) and PHF-1 (right). DAPI in blue. Scale bars: 25 μm
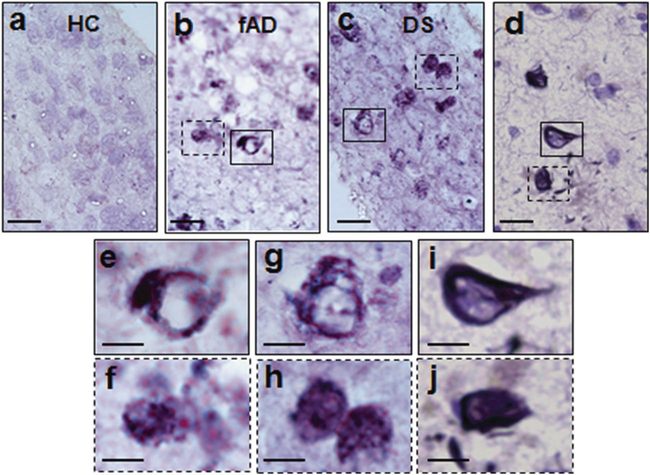
In further support of the tau pathological abnormalities, Gallyas silver staining revealed many positive cells bearing NFTs-like structures only in COs derived from fAD or DS (Fig. 6a–c). These structures showed very similar morphology and size compared to NFTs present in an AD brain (Fig. 6d), as appreciated better in the zoomed pictures showing individual aggregates (Fig. 6e–j).
Fig. 6.
Gallyas silver staining of COs. a–c Gallyas silver staining of representative sections of COs generated with healthy control (HC), familiar Alzheimer’s disease (fAD), and Down syndrome (DS) iPSCs. Cells with NFT-like structures were only observed in fAD and DS COs. d Gallyas silver staining of NFT in an AD human brain. Magnified versions of NFT-like structures observed in fAD COs and DS COs are shown in panels e, f and g, h, respectively. These images are shown to display the morphology of the aggregates and were magnified from the boxes in panels a and b. i, j Magnification of the NFT-like structures observed in the brain of an AD patient (magnified from the boxes in panel d). Scale bars are 50 μm in a-d and 10 μm in e–j
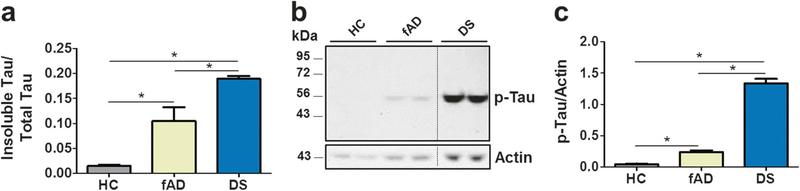
To confirm biochemically the presence of aggregated and abnormally phosphorylated tau in COs, we performed measurements using ELISA and western blots. To evaluate the extent of tau aggregation, we determined the amount of tau protein in the soluble and insoluble fractions prepared from COs homogenates using a fractionation procedure with aqueous and formic acid extraction. The results were expressed as the ratio of total insoluble tau over total tau (Fig. 7a). The relative amount of insoluble tau was significantly higher in patients than control COs. The highest amount was found in DS, followed by fAD and little (if any) in HC COs. Moreover, there was a strong correlation between the levels of p-tau231 and tau protein phosphorylated at Thr181 and Ser396, two nonpathological tau phospho-epitopes, suggesting that tau is hyper-phosphorylated in fAD COs (Supplementary Fig. 5). Western blot analysis using PHF-1 antibodies confirmed the presence p-tau protein in the samples analyzed (Fig. 7b). The densitometric analysis of the blots revealed significant differences among the groups (Fig. 7c). Confirming the observations done by immunofluorescence with p-tau antibodies, the highest amount was found in DS followed by fAD, with no signal observed in HC COs.
Fig. 7.
Biochemical analysis of tau pathology in COs. Groups of COs produced from healthy control (HC), familiar Alzheimer’s disease (fAD), or Down syndrome (DS) iPSCs were analyzed by ELISA and western blotting. Before treatment, total protein concentration was measured by BCA, and analysis was done using samples containing the same amount of total protein. a Total insoluble tau protein found in the COs homogenates was measured after fractionation with formic acid using an ELISA assay. Results are shown as the ratio of the amount of insoluble tau over the total tau protein (soluble + insoluble). b Western blot analysis of total COs homogenates using PHF-1 antibody revealed the presence of p-tau at the expected molecular weight. Duplicate samples were analyzed for each group of COs. c Densitometric analysis of the blots revealed significant differences among all groups. For data in panels a and c, each value corresponds to the average ± SEM of three and four replicates, respectively. For this analysis, 80–220 organoids were used for each replicate. Data was analyzed by one-way ANOVA, followed by Tukey’s multiple comparison post-test. *P<0.05; **P<0.01; ***P<0.001
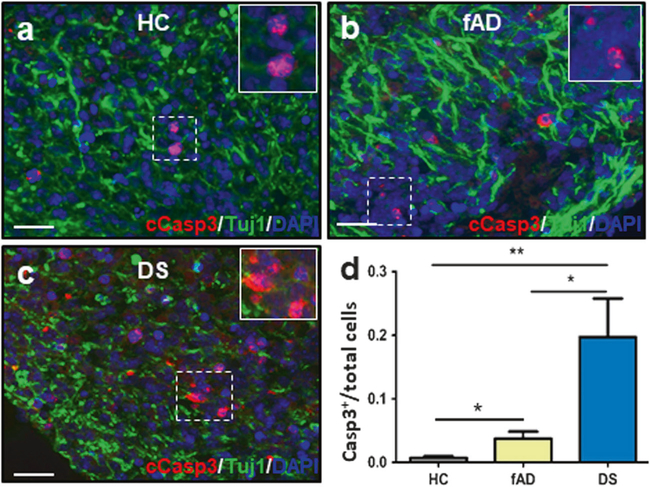
To study whether COs develop some neurodegenerative features as those observed in AD brain, we performed staining for cleaved caspase-3, the active form of this enzyme. Active caspase-3 plays a major role as one of the main executors of programmed cell death and is an established marker of apoptosis [24]. Cells positive for cleaved caspase-3 staining (relative the total number cells, measured by DAPI staining) were higher in DS derived COs than fAD- and HC-derived COs (Fig. 8). These results suggest a higher extent of cell death in the COs containing the largest quantities of Aβ and tau pathology.
Fig. 8.
Analysis of cellular apoptosis in COs. a–c Cleaved caspase-3 (cCasp3, red) and Tuj-1 (green) staining for representative sections of COs made from a healthy control (HC), b familiar Alzheimer’s disease (fAD), or c Down syndrome (DS) iPSCs. Scale bars: 25 μm. D Immunofluorescence analysis of the ratio of cCasp3+ cells per total number of cells, as measured by DAPI staining. Data was analyzed by one-way ANOVA, and unpaired Student’s t-test. *P <0.05; **P <0.01
Discussion
In the past years, various in vitro AD models have been developed with cell lines and iPSCs. Interesting results have been reported with monolayer cultures of iPSC-derived cells [5–14], predominantly neurons. These observations are interesting because they recapitulate some aspects of AD pathology, but are far from reproducing the hallmark pathological features of the disease, most likely because this would require the brain tissue organization. For example, amyloid plaques (likely the most pathognomonic feature of AD) are deposited in the extracellular space of the brain parenchyma and disrupt synaptic connections among cells. In a monolayer system, there is no extracellular organization or establishment of an ordered connection between neurons. Recently, various types of second generation 3D neurospheres have been developed, which displayed significant improvements as AD models. Wild-type iPSC-derived neuroepithelial stem cells grown in a self-assembling peptide scaffold were shown to form 3D cultures that reproduce some of the in vivo-like responses related to AD, not recapitulated with conventional 2D cultures [25]. Kim, Tanzi and colleagues showed that a matrigel matrix supporting immortalized, ESC-derived neural progenitor cells overexpressing multiple fAD mutations developed both mature plaques and tangles after 90 days in culture [26]. Although this model represents a substantial step forward from monolayer cultures, it utilizes artificially manipulated cell lines engineered to express various mutations. In addition, the 3D cultures produced correspond to a mass of cells held together by matrigel, but do not recapitulate the organization of the human brain. More recently, fAD-derived iPSCs were grown in a scaffold-free 3D model which began with an embryoid body-like aggregate that progressively matured into a cerebral organoid. Using this technology, the organoids spontaneously developed amyloid and tau pathology over time [27].
COs are a superior in vitro model to the other available 3D systems because they uniquely permit the spontaneous generation of organized cortical-like structures with striking resemblance to the human cortex. In addition to their potential to model developmental disorders and diseases [15, 28], our study indicates that COs might be useful to model also an age-related neurodegenerative disease. However, despite the fact that COs mimic the structure of the human cortex, they do not contain the same proportion of different type of nerve cells as the human brain, since they are mostly composed by neurons, with a smaller proportion of glial cells and virtually no oligodendrocytes. Another limitation is that COs do not establish mature synaptic connections (at least at the time in which our experiments were done), since no markers of synapsis were observed by either biochemical or immunohistochemical techniques (data not shown). Nevertheless, it has been reported that COs exhibit some form of electrical activity when maintained for longer periods in culture [15, 16]. Finally, another problem is the lack of vascularization, which limit the growing and survival of the tissue and may complicate modeling some important aspects of the brain (e.g. the blood brain barrier) [29].
In this study, we demonstrated that fAD and DS patient-derived COs with endogenous levels of protein expression developed the two main pathological features of AD, Aβ and p-tau protein aggregates. COs also exhibited some degree of cellular apoptosis in a manner proportional to the accumulation of protein aggregates. It is noteworthy that our system allowed aggregates to mature over time into structures highly reminiscent to the extracellular senile amyloid plaques and intracellular NFTs associated with late-onset AD. COs produced from iPSCs derived from two HC patients, patients affected by another neurodegenerative disorder (genetic forms of Creutzfeldt–Jakob disease) or mouse iPSCs or ESCs did not show any significant amount of Aß aggregates or tau pathology. These results suggest that the development of AD pathology in COs is specific for cells coming from patients genetically predisposed to the disease (fAD and DS). Nevertheless, studies with a much larger number of patient-derived cells, including different types of mutations in distinct genes, are needed to firmly support this conclusion. fAD and DS iPSCs express naturally occurring genetic alterations that have been shown to increase the production of either total Aβ or the more amyloidogenic Aβ42 peptide [6, 8]. In contrast, as also observed in AD patients, tau obtained its pathological phenotype in both types of COs without any mutations in the tau gene. Further supporting a link between these pathological features of AD, the fAD and the DS COs with the largest amount of Aβ aggregates had also the highest levels of tau abnormalities. Together, these findings support the hypothesis that Aβ aggregation and tau pathology are closely related events in the pathogenesis of AD [30].
In summary, our findings indicate that COs may represent an excellent model of genetic forms of AD that may enable studying the pathology at more physiological levels of gene expression in cortical tissue derived from stem cells with the same genetic background of the patients. The facility and large number in which organoids can be generated, added to the flexibility of in vitro experiments, may enable to study the underlying disease mechanisms and to screen novel drug candidates for efficacy [31]. Future experiments using this technology may allow studying in real-time the cellular reaction to the accumulation of protein aggregates, the inter-relationship between amyloid and tangles, the potential toxicity of the aggregates, the proteins and signaling pathways altered by the appearance of protein deposits, etc. Many of these findings may bring potentially new therapeutic targets. In addition, COs may offer a great alternative to animal models for screening of drugs, with the added advantage that all the components are of human origin, as opposed to just one protein as in transgenic animals. Also, thousands of organoids can be generated and maintained in a small space, which could be incubated with large chemical libraries. Indeed, miniaturized spinning bioreactors were recently designed to aid in scaling-up the production of COs for such purposes [32]. Furthermore, this technology may be extended to produce relevant models for other neurodegenerative diseases.
Supplementary Material
Acknowledgements
We are grateful to Dr. Brian Davis (University of Texas Medical School at Houston) for his valuable help in establishing the iPSC technology in our lab, Dr. Fabrizio Tagliavini (Istituto Carlo Besta, Italy) for kindly providing human fibroblasts from patients affected by inherited Creutzfeldt-Jakob disease. We are also grateful to Dr. Ying Liu (Institute of Molecular Medicine, University of Texas at Houston) for providing an iPSC line obtained from healthy controls. This project was partially funded by a pilot grant from the University of Texas Brain Initiative Program.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41380-018-0229-8) contains supplementary material, which is available to authorized users.
References
- 1.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362:329–44. [DOI] [PubMed] [Google Scholar]
- 2.Laurijssens B, Aujard F, Rahman A. Animal models of Alzheimer’s disease and drug development. Drug Discov Today Technol. 2013;10:e319–27. [DOI] [PubMed] [Google Scholar]
- 3.Duyckaerts C, Potier MC, Delatour B. Alzheimer disease models and human neuropathology: similarities and differences. Acta Neuropathol. 2008;115:5–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T. et al. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530–9. [DOI] [PubMed] [Google Scholar]
- 6.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C. et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y. et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–96. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Sci Transl Med. 2012;4:124ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN. et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB. et al. APP metabolism regulates tau proteostasis in human cerebral cortex neurons. Cell Rep. 2015;11:689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koch P, Tamboli IY, Mertens J, Wunderlich P, Ladewig J, Stuber K. et al. Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of gamma-secretase activity in endogenous amyloid-beta generation. Am J Pathol. 2012;180:2404–16. [DOI] [PubMed] [Google Scholar]
- 12.Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M. et al. Characterization and molecular profiling of PSEN1 familial Alzheimer’s disease iPSC-derived neural progenitors. PLoS ONE. 2014;9:e84547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahairaki V, Ryu J, Peters A, Chang Q, Li T, Park TS. et al. Induced pluripotent stem cells from familial Alzheimer’s disease patients differentiate into mature neurons with amyloidogenic properties. Stem Cells Dev. 2014;23:2996–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armijo E, Gonzalez C, Shahnawaz M, Flores A, Davis B, Soto C. Increased susceptibility to Abeta toxicity in neuronal cultures derived from familial Alzheimer’s disease (PSEN1-A246E) induced pluripotent stem cells. Neurosci Lett. 2017;639:74–81. [DOI] [PubMed] [Google Scholar]
- 15.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME. et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M. et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–60. [DOI] [PubMed] [Google Scholar]
- 18.Nethercott HE, Brick DJ, Schwartz PH. Derivation of induced pluripotent stem cells by lentiviral transduction. Methods Mol Biol. 2011;767:67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Zheng Y, Li S, Xue H, Schmitt K, Hergenroeder GW. et al. Human neural progenitors derived from integration-free iPSCs for SCI therapy. Stem Cell Res. 2017;19:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A. et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klunk WE, Wang Y, Huang GF, Debnath ML, Holt DP, Mathis CA. Uncharged thioflavin-T derivatives bind to amyloid-beta protein with high affinity and readily enter the brain. Life Sci. 2001;69:1471–84. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayashi T, Younkin LH, Saido TC, Shoji M, Ashe KH, Younkin SG. Age-dependent changes in brain, CSF, and plasma amyloid (beta) protein in the Tg2576 transgenic mouse model of Alzheimer’s disease. J Neurosci. 2001;21:372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rostagno A, Ghiso J. Isolation and biochemical characterization of amyloid plaques and paired helical filaments. Curr Protoc Cell Biol. 2009;Chapter 3:Unit 3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter AG, Janicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A. A 3D Alzheimer’s disease culture model and the induction of P21-activated kinase mediated sensing in iPSC derived neurons. Biomaterials. 2014;35:1420–8. [DOI] [PubMed] [Google Scholar]
- 26.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C. et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515:274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J. et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE. 2016;11:e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcez PP, Loiola EC, Madeiro da CR, Higa LM, Trindade P, Delvecchio R. et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–8. [DOI] [PubMed] [Google Scholar]
- 29.Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The hope and the hype of organoid research. Development. 2017;144:938–41. [DOI] [PubMed] [Google Scholar]
- 30.Musiek ES, Holtzman DM. Three dimensions of the amyloid hypothesis: time, space and ‘wingmen’. Nat Neurosci. 2015;18:800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. [DOI] [PubMed] [Google Scholar]
- 32.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.