Abstract
In contrast to terminally differentiated cells, cancer cells and stem cells retain the ability to re-enter the cell cycle and proliferate. In order to proliferate, cells must increase the uptake and catabolism of nutrients to support anabolic cell growth. Intermediates of central metabolic pathways have emerged as key players that can influence cell differentiation ‘decisions’, processes relevant for both oncogenesis and normal development. Consequently, how cells rewire metabolic pathways to support proliferation may have profound consequences for cellular identity. Here, we discuss the metabolic programs that support proliferation and explore how metabolic states are intimately entwined with the cell fate decisions that characterize stem cells and cancer cells. By comparing the metabolism of pluripotent stem cells and cancer cells, we hope to illuminate common metabolic strategies as well as distinct metabolic features that may represent specialized adaptations to unique cellular demands.
Introduction
At their core, cell survival and growth are metabolic problems. Cells catabolize nutrients to generate the energy and reducing equivalents required to maintain basic cellular processes. Likewise, anabolic metabolic pathways convert nutrients into the macromolecules necessary for cell growth and proliferation. This intimate relationship between metabolism and cellular fitness is best exemplified by the observation that the growth of most unicellular organisms is directly tied to nutrient availability1. In contrast, cells of multicellular organisms must cooperate to share relatively constant nutrient supplies; consequently, metazoan cell proliferation is regulated by growth factors that license the acquisition of extracellular nutrients and activation of anabolic growth programs. During development, growth factor signaling pathways direct the proliferation, migration and death of selected populations to ensure proper organ size and function. These same pathways frequently become subverted in cancer: oncogenic activation of growth factor signaling or inhibition of cell death enables the pathological proliferation that drives tumor growth. Therefore, it is perhaps not surprising that cancer cells share many metabolic features with normal developmental programs. For example, just as folate deficiency is a major cause of early embryonic growth defects2, therapies interfering with folate metabolism are key components of many successful chemotherapeutic regimens3.
Metabolites play many roles beyond serving as substrates for energy generation and anabolic growth. Metabolites contribute to the regulation of intracellular redox balance4, directly alter the activity of intracellular signaling cascades5, and serve as co-substrates for enzymes that modify macromolecules such as DNA and proteins6. As a result, intracellular metabolic pathways may influence many cellular programs beyond proliferation. The dual role of metabolites as substrates for both anabolic and regulatory processes raises the possibility that the utilization of nutrients for cell proliferation inherently affects the availability of metabolites for other, non-anabolic roles. This metabolic convergence between proliferation and cell fate regulation may be particularly relevant in stem cells, which accomplish the dual feat of retaining the capacity to proliferate rapidly and differentiate into specialized cell types. As a result, there is great interest in elucidating the metabolic networks that sustain stem cell self-renewal and identifying metabolic nodes that influence lineage-specific differentiation.
Pluripotent stem cells (PSCs) provide an ideal model system to study the intersection between proliferation, metabolism and differentiation. While pluripotency—the capacity to give rise to cells of all three embryonic germ layers—exists only transiently during early mammalian development, the pluripotent state can be captured indefinitely in vitro, providing an invaluable resource to study principles of dynamic cell fate transitions, elucidate pathways critical for lineage-specific differentiation, and explore possibilities for regenerative medicine7. Pluripotent stem cells can be isolated from the inner cell mass of pre-implantation epiblast (embryonic stem cells, ESCs) or the early post-implantation epiblast (epiblast stem cells, EpiSCs); alternatively, somatic cells can be reprogrammed to pluripotency by expression of key pluripotency-associated transcription factors (iPSCs). Intriguingly, the functional identity of PSCs is largely determined not by the cell of origin but rather by the culture conditions under which the cells are generated and propagated7. Naïve PSCs share molecular characteristics with cells of the inner cell mass and can readily give rise to all germ layers and contribute to chimeras8. In contrast, primed PSCs exhibit similarities to post-implantation epiblast cells and, while retaining pluripotency, represent a more committed developmental state8. By and large, PSCs can be coaxed into either the naïve or primed state depending on the exogenous cues present in the culture medium7. Therefore, understanding how specific nutrients and growth factors present in PSC culture regulate cellular phenotypes such as growth and gene expression programs likely will yield critical insights into the regulation of cell identity.
A key question when comparing the metabolism of specialized cell types is how to distinguish the metabolic alterations accompanying differences in growth and proliferation from the metabolic alterations that support cell-type specific fate decisions. Therefore, it is useful to compare the emerging literature describing the metabolism of stem cells with the vast literature on the metabolism of cancer cells. Like stem cells, cancer cells are capable of rapid proliferation; unlike stem cells, cancer cells are relatively locked into a malignant identity. In this review, we will discuss emerging evidence that cancer cells and stem cells engage similar metabolic pathways to support anabolic proliferation. We will further examine how specific metabolic alterations influence the balance between self-renewal and differentiation in cancer cells and stem cells. As both stem cells and cancer cells face many similar metabolic challenges, lessons from one system may provide generalizable insight into the potential metabolic avenues that support growth, survival and differentiation across multiple cell types.
Metabolic requirements of proliferation
Glucose and glutamine are fundamental substrates for mammalian cells growing in culture
Regardless of the identity of the proliferating cell, proliferation requires a net increase in biomass. In order to generate the nucleic acids, lipids and proteins required for proliferation, cells increase the uptake and catabolism of the nutrients that provide the raw materials for macromolecular synthesis. Glucose and glutamine, two of the most abundant metabolites in serum and in common tissue culture media, serve as the major sources of energy and reducing equivalents required to assemble metabolic building blocks into macromolecules (Fig. 1)9. Beyond ATP and NAD(P)H, catabolism of glucose and glutamine generates precursors for the biogenesis of non-essential amino acids required to sustain protein and nucleic acid synthesis. Additionally, glucose provides the ribose and glutamine provides the obligate source of reduced nitrogen for nucleic acid biosynthesis. In cultured cells, glucose-derived acetyl-CoA represents the primary source of carbon for de novo lipid biosynthesis while protein biosynthesis, which makes up the majority of cellular biomass, is sustained by the combination of direct amino acid uptake and de novo synthesis of non-essential amino acids from glucose and glutamine10. Therefore, although the relative dependency on each of these metabolic precursors can vary according to cell line, culture conditions and nutrient availability9,10, a fundamental feature of mammalian cells proliferating in culture is the central role of glucose and glutamine supporting anabolic growth.
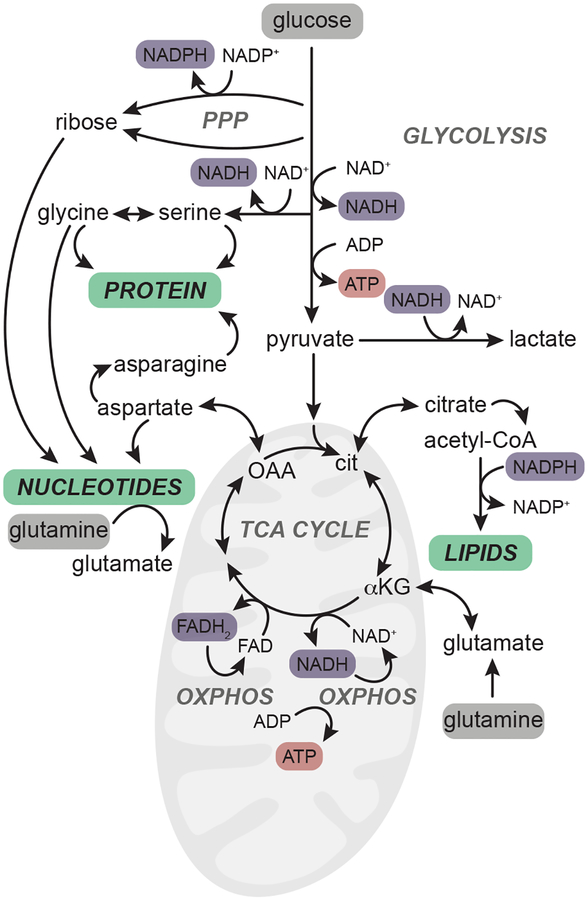
Figure 1.
Glucose and glutamine are critical inputs for major anabolic pathways. In proliferating cells, glucose and glutamine (highlighted in grey) are taken up from the extracellular environment and catabolized through major metabolic pathways including glycolysis, the pentose phosphate pathway (PPP) and the tricarboxylic acid (TCA) cycle to provide the reducing equivalents (purple) and high-energy carriers (ATP, red) required to synthesize major macromolecules (green). A subset of the non-essential amino acids that are synthesized from glucose and glutamine are shown. Reducing equivalents (NADH, FADH2) in the mitochondria fuel the electron transport chain and enable synthesis of ATP through oxidative phosphorylation (oxphos). TCA cycle intermediates such as citrate and oxaloacetate (OAA, converted to aspartate) likewise contribute to lipid and nucleotide biosynthesis, respectively.
The critical role of glucose and glutamine in cancer cell proliferation is well established and has been extensively reviewed elsewhere11–13. Like cancer cells, PSCs have the capacity to proliferate indefinitely in culture and are also heavily reliant on exogenous glucose and glutamine14,15. Although the inherent flexibility of metabolic networks ensures that cells have multiple mechanisms to cope with reduced abundance of either nutrient16, proliferation is maximized when both glucose and glutamine are abundant. Therefore, a major paradox of rapidly proliferating cells is the tendency to discard the overwhelming majority of glucose carbons as lactate. The conversion of glucose to lactate despite the presence of sufficient oxygen to sustain complete oxidation of glucose-derived carbons, commonly referred to as the “Warburg effect” or aerobic glycolysis, is a hallmark of all rapidly proliferating mammalian cells. Indeed, compared to their slower-growing differentiated counterparts, pluripotent stem cells—regardless of their cell of origin or stage of pluripotency—engage in aerobic glycolysis, consuming high levels of glucose and secreting large quantities of lactate17–20. This glycolytic flux is contingent upon cells maintaining their pluripotent identity and is rapidly reversible: glycolytic lactate production is reduced early during PSC differentiation19–22 and reprograming differentiated cells to the pluripotent state results in the re-acquisition of glycolytic phenotypes23–25. Accordingly, interfering with glycolytic metabolism impairs reprogramming and maintenance of the pluripotent state20–25. Mechanistically, transcription factors establishing the pluripotent state may directly regulate metabolic pathways: for example, the core pluripotency transcription factor Oct4 binds loci encoding glycolytic genes to promote glycolysis26. Not all differentiation results in downregulation of aerobic glycolysis: while human PSC differentiation to mesoderm and endoderm reduces glycolytic flux, differentiation to early neuroectoderm does not22. A key open question therefore is whether the links between aerobic glycolysis and pluripotency are the result of bioenergetic constraints of particular cell types or the consequences of altered accumulation of metabolites with the ability to regulate cell fate programs.
Despite the ubiquity of the Warburg effect in proliferating cells, the potential benefits of this metabolic hallmark remain controversial27. Warburg’s initial hypothesis, that the aerobic conversion of pyruvate to lactate results from defective mitochondrial respiration, has since been disproven in both cancer cells and normal cells12. Mitochondria are notably distinct in PSCs relative to their differentiated counterparts: PSCs tend to harbor fewer mitochondria per cell and these mitochondria exhibit immature morphology marked by sparse cristae18,19,28,29. Nevertheless, PSCs are not defective in mitochondrial substrate oxidation. Respiratory complexes are fully functional in PSCs19 and as a result, PSCs have the flexibility to engage in oxidative or glycolytic programs depending on culture conditions18,30. Indeed, even under basal culture conditions, PSCs engage in significant glutamine oxidation to sustain bioenergetics14,15,30,31 and enhanced respiration may be a common feature of PSCs in the naïve state18,32,33. Therefore, as in other proliferating cell types, the aerobic glycolysis that characterizes PSCs does not appear to result from a defect in mitochondrial function.
Lacking an understanding of the origin or consequences of the Warburg effect in PSCs, it is difficult to ascribe any particular benefit or function to aerobic glycolysis in the regulation or maintenance of pluripotent cell growth. Intriguingly, it has recently been suggested that aerobic glycolysis might arise in part as a secondary consequence of the metabolic requirements of proliferating cells rather than as a primary driver of proliferation. For example, the conversion of pyruvate to lactate becomes favored when the cytosolic NADH/NAD+ ratio rises, which may occur in proliferating cells due to rapid consumption of NAD+ to drive the oxidative reactions required for nucleotide production13. PSCs are notable for their rapid proliferation (12–24h doubling time) and high nuclear to cytoplasmic ratio and therefore may be disproportionally reliant on nucleotide biosynthesis to sustain proliferation. Alternatively, since high mitochondrial substrate oxidation produces reactive oxygen species that promote differentiation and exert deleterious effects on cellular fitness, discarding reduced electrons in the form of lactate rather than transferring these reducing equivalents to the mitochondria for oxidation may be particularly beneficial to PSCs that must resist spontaneous differentiation and are primed for apoptosis34,35. In support of this hypothesis, the metabolome of ESCs becomes more oxidized upon differentiation36.
Taken together, the emerging literature on PSC metabolism demonstrates that cultured PSCs meet the anabolic demands of proliferation in a manner largely similar to cancer cells and other proliferating mammalian cells. This correspondence between the metabolic profiles of PSCs and cancer cells may arise simply as a consequence of the conserved metabolic requirements of proliferation. Nevertheless, emerging evidence suggests that there are subtle differences in how different types of PSCs use glucose and glutamine. For example, both mouse and human PSCs in the naïve state of pluripotency exhibit higher basal respiration than their primed counterparts18,32,33. At the same time, whereas glycolysis is increased in human naïve PSCs relative to primed PSCs, the opposite trend is true in mouse PSCs18,20. To what extent these differences arise from variations in cell size, proliferation rate, and culture conditions (including medium nutrient composition, growth factor availability, and choice of extracellular matrix) rather than functional variation between the cell types remains an important open question. Certainly, simply culturing the same cell type in different medium formulations can significantly alter how cells engage metabolic pathways to fulfill anabolic demands30. However, as PSC identity is intimately linked to culture conditions7, these metabolic differences could also result from—or contribute to—the establishment of different functional states. Therefore, a critical avenue of future work will be to unravel the metabolic phenotypes that are secondary to altered growth conditions from those metabolic phenotypes that are the specific result of variations in PSC identity.
Beyond glucose and glutamine: alternative metabolic strategies
Although aerobic glycolysis and glutamine oxidation represent nearly ubiquitous features of cancer cells proliferating in culture, recent studies examining tumor metabolism in vivo illuminate the diverse metabolic strategies and substrates that support cancer growth (Fig. 2a)12,37. For example, several types of brain and lung tumors tend to increase oxidation of glucose-derived carbons and rely on glucose for replenishment of TCA cycle intermediates38–40, while clear cell renal carcinomas retain profiles consistent with aerobic glycolysis41. Likewise, several tumor types are reported to have minimal reliance on glutamine oxidation to sustain TCA cycle metabolite pools and may even synthesize glutamine de novo from glucose-derived carbons38,39,42,43. Increasingly, substrates beyond glucose and glutamine are recognized to play critical roles as bioenergetic substrates in vivo. Lactate, produced in high quantities in the tumor microenvironment, may function as a major source of carbon for growing tumors40,44,45. Similarly, acetate has been reported to serve as a fuel for brain and liver tumors46,47 and circulating branched chain amino acids can contribute to biomass accumulation in non-small cell lung carcinoma48. Clearly, we are just beginning to understand the myriad metabolic strategies employed by tumors to sustain proliferation. Diverse factors such as local nutrient availability43, transforming oncogene49 and tissue of origin48 will likely converge to determine the unique metabolic preferences and liabilities of a given tumor.
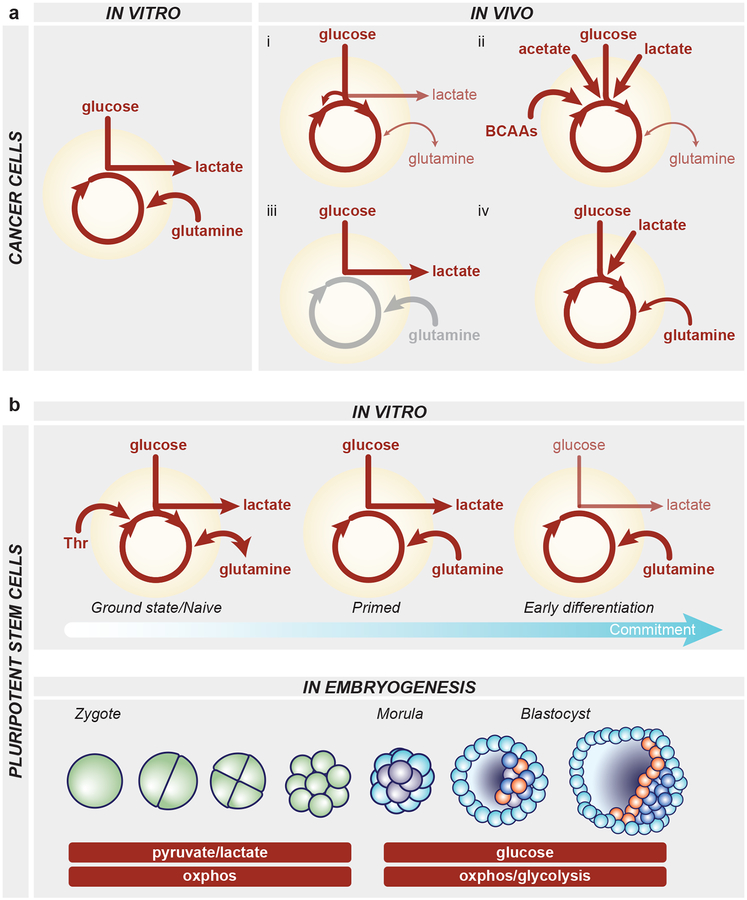
Figure 2.
Metabolic strategies utilized by cancer cells and pluripotent stem cells. a, Left, cancer cells in vitro take up high levels of glucose and glutamine. Much of the glucose is converted to lactate via aerobic glycolysis and glutamine-derived carbons provide tricarboxylic acid (TCA) cycle anaplerosis to fuel oxidative phosphorylation. Right, a sampling of the diverse metabolic strategies utilized by cancer cells in vivo. i) Many tumors including lung and brain tumors exhibit high glucose uptake and catabolism in the TCA cycle, often in addition to aerobic glycolysis. Glucose-derived carbons can fuel the TCA cycle through forward, pyruvate dehydrogenase flux or via pyruvate carboxylase-mediated anaplerotic entry as oxaloacetate. In this scenario, glutamine is a relatively minor contributor to TCA cycle metabolites and often tumors can net produce glutamine de novo from glucose-derived carbons. ii) Increasingly, substrates beyond glucose and glutamine are recognized to server as major substrates for anabolic pathways and the TCA cycle, including acetate, lactate and branched chain amino acids in lung, brain and liver tumors. iii) In human clear cell renal carcinoma, tumors exhibit high glycolysis and lactate production, consistent with the canonical Warburg effect. Glutamine metabolism has not been assessed and is therefore shaded in grey. iv) Studies in mouse pancreatic cancer reveal that glucose, lactate and glutamine all contribute to TCA cycle intermediates. b, Top, PSCs exhibit a variety of metabolic strategies depending on the culture condition or stage of differentiation. Naïve mouse PSCs in the ground state of pluripotency can synthesize glutamine de novo from glucose-derived carbons; naïve mouse PSCs also can catabolize threonine (Thr). Both naïve and primed PSCs in culture exhibit high consumption of glucose and glutamine as well as extensive production of lactate. Upon differentiation, the proliferative hallmark of aerobic glycolysis decreases along with proliferation rate. Bottom, while little is known about pluripotent cell metabolism in vivo, studies of embryos developing ex vivo have identified two major metabolic stages: from zygote to morula, embryos are dependent upon the moncarboxylates pyruvate and lactate and are highly oxidative; around the morula stage, embryos begin to use glycolysis to fuel metabolic pathways and the trophectoderm exhibits features consistent with oxidative phosphorylation while the inner cell mass may rely more on aerobic glycolysis. Green, blastomeres; light blue, trophectoderm; purple, inner cell mass; orange, primitive endoderm; dark blue, epiblast.
Early evidence suggests that PSCs may likewise exhibit substantial metabolic diversity, even under routine culture conditions (Fig. 2b). Indeed, many of the unique metabolic phenotypes of cancer cells that become revealed only upon tumor growth in vivo are readily apparent in PSCs in vitro. For example, driving the normally metastable and heterogeneous population of naïve mouse ESCs into homogenous ground state of pluripotency substantially increases the contribution of glucose-derived carbons to TCA cycle metabolites14,50. As a result, naïve, ground state ESCs can generate glutamine de novo from glucose-derived carbons and therefore possess the unique capacity to proliferate in the absence of exogenous glutamine14. Analogous to the distinct utilization of branched chain amino acids by certain tumor in vivo48, ESCs may also be able to catabolize amino acids in unusual ways. Mouse ESCs express an active version of the enzyme threonine dehydrogenase that enables robust catabolism of threonine to glycine and acetyl-CoA; consequently, mouse ESCs are exquisitely reliant upon exogenous threonine to sustain viability and cell identity51,52. Moreover, just as tumor cells may efficiently scavenge macromolecular nutrients from the microenvironment53, PSCs appear to preferentially utilize exogenously-supplied lipids over those synthesized de novo30. Furthermore, acetate is one of the most changed metabolites during early differentiation, suggesting that alterations in acetate production or consumption may follow changes in pluripotent identity21.
Because the state of pluripotent self-renewal is inherently intertwined with culture conditions, whether these metabolic phenotypes of PSCs will translate to the transient pluripotent state in vivo remains an open question. Relative to their inner cell mass counterparts, PSCs have undergone significant adaptation to cell culture and acquired the ability to sustain continuous self-renewal; consequently, it is unclear to what extent the metabolism of cultured PSCs reflects the growth of the inner cell mass or epiblast cells in vivo54,55. Very little is known about metabolism during early mammalian development. As embryos progress from zygote to blastocyst, there is no substantial increase in cell mass: rather, these early cell divisions are “reductive,” marked by an increase in nuclei at the expense of cytoplasm56. Therefore, the metabolic demands of proliferation during early development are likely to be quite different from those of most proliferating cells which must generate net biomass. While it is technically difficult to assess how embryos developing in situ meet their metabolic needs, studies on embryos developing in culture revealed that pyruvate and lactate serve as the primary sources of energy in early cleavage embryos (Fig. 2b)57. At the morula stage, cells become competent to utilize glucose as an energy source and studies suggest that the cells of the inner cell mass (ICM) may be more glycolytic relative to the oxidative cells of the trophectoderm58. Intriguingly, studies examining amino acid turnover in murine blastocysts and dissociated ICM cells did not identify glutamine as a highly-consumed amino acid58, suggesting that early blastocyst development may impose very different requirements for amino acids compared to cultured cells. Indeed, perhaps because of a reduced requirement for net protein synthesis, mouse blastocysts can develop in vitro with just the presence of single, select amino acids or even without any free amino acids at all59,60. Future studies, perhaps utilizing tools such as single-cell metabolite sensors, heterologous enzymes and genetic modification of metabolic pathways combined with linage tracing, may begin to unravel the metabolic pathways required for early mammalian development.
Regulation of anabolic metabolic pathways
Taken together, these studies suggest that cultured cancer cells and PSCs engage a core set of common metabolic pathways to support rapid proliferation. In normal cells, growth factor-mediated activation of receptor tyrosine kinases engage signaling pathways such as Ras and phosphatidylinositol 3-kinase (PI3K) that in turn activate downstream effectors including MEK/ERK, AKT and mTOR. Collectively, these pathways enhance nutrient uptake and promote anabolic metabolic pathways that drive lipid, protein and nucleic acid synthesis53. Mutations that drive aberrant activation of these gatekeepers of anabolic metabolism are among the most common alterations in human cancer, underscoring the deep mechanistic links between metabolic reprogramming and malignant transformation. Other commonly mutated genes can also activate anabolic pathways. MYC, a transcription factor that positively regulates glucose and glutamine metabolism and coordinates anabolic gene expression programs, undergoes frequent amplification in human cancer61. Similarly, loss of wild type p53, the most common event in human cancer, results in a constitutive shift from nutrient catabolism to anabolism, increased glycolysis and altered redox regulation62.
Many of these same pathways may regulate pluripotent cell growth. Hypoxia-inducible factor-1 (HIF1), which induces glycolysis and is frequently activated by a combination of genetic and environmental factors in tumors, triggers a glycolytic switch that promotes the metabolic transition from naïve to primed pluripotency and provides a selective advantage during early reprogramming18,25. Likewise, naïve human PSCs exhibit signatures of elevated MYC activity and accumulation of N-MYC in the nucleus correlates with enhanced glycolytic programs20. Accordingly, sustained N-MYC activity promotes glycolysis during hPSC self-renewal and early ectoderm differentiation while loss of MYC activity coincides with reduced glycolysis during endoderm and mesoderm differentiation22. Deletion of both c- and N-Myc from mouse ESCs decreases anabolic programs without affecting markers of pluripotency and triggers a state of growth arrest reminiscent of embryonic diapause, a reversible state of arrest during pre-implantation embryonic development63. Similarly, partial inhibition of mTOR induces a reversible “paused” state of low anabolic activity but sustained pluripotency in cultured ESCs and is sufficient to induce diapause in cultured mouse embryos64. Together, these studies raise the intriguing possibility that cellular capacity to engage in anabolic metabolic programs is a prerequisite for exit from pluripotency and successful development of peri-implantation embryos.
Beyond these exciting studies, however, comparatively little is known about regulation of metabolism in PSCs by signal transduction pathways despite the fact that many of the pathways that regulate tumor cell metabolism have been implicated in aspects of pluripotent cell biology. For example, loss of wild-type p53 function accelerates reprogramming and iPSCs tend to acquire p53 mutations upon prolonged culture65,66, although this may simply be the result of the proliferative advantage conferred by loss of p53-mediated cell cycle regulation rather than any specific metabolic effect per se. Similarly, Akt activation promotes pluripotent self-renewal67,68, but whether this results from any metabolic consequences of Akt signaling remains unexplored. Nevertheless, it seems likely that growth factor-mediated metabolic regulation may indeed contribute to PSC cell fitness. Leukemia inhibitory factor (LIF) sustains the undifferentiated state of naïve PSCs, and JAK/STAT3 activation downstream of LIF signaling can induce a subset of STAT3 to translocate to the mitochondria. Mitochondrial STAT3 enhances oxidative phosphorylation that supports cell proliferation but does not appear to influence self-renewal33. Likewise, fibroblast growth factor (FGF)-mediated activation of MEK/ERK signaling provides a critical trigger for exit from naïve pluripotency and thus inhibition of MEK signaling is a common component of formulations that capture PSCs in the naïve pluripotent state7. However, MEK inhibition may impose a metabolic limitation on cells that can be rescued by combined inhibition of MEK and GSK3β69. Although there are clearly key signaling differences between cancer cells and stem cells—notably, the ability to maintain proliferation despite impaired MEK/ERK signaling appears to be a unique and defining feature of naïve ESCs8—further study into how these networks regulate metabolism in stem cells and cancer cells will continue to reveal the strategies cells employ to wire metabolic pathways to meet cell-type and context-specific demands.
Metabolic regulation of chromatin and differentiation
The influence of metabolites on gene expression programs
In addition to serving as critical substrates that fuel cell growth and proliferation, metabolites can also actively influence differentiation in both stem cells and cancer cells6,11. Several key metabolic intermediates function as obligate substrates or co-factors for enzymes that deposit and remove chemical modifications on DNA and histones. This biochemical link between metabolites and chromatin-modifying enzymes has led to the intriguing hypothesis that metabolites exert effects on cell fate decisions primarily through altering chromatin modifications, which in turn regulate gene expression programs involved in self-renewal and lineage differentiation70. Acetylation of histones and methylation of both histones and DNA may be particularly responsive to metabolic inputs and will therefore be the focus of this discussion (Fig. 3)71.
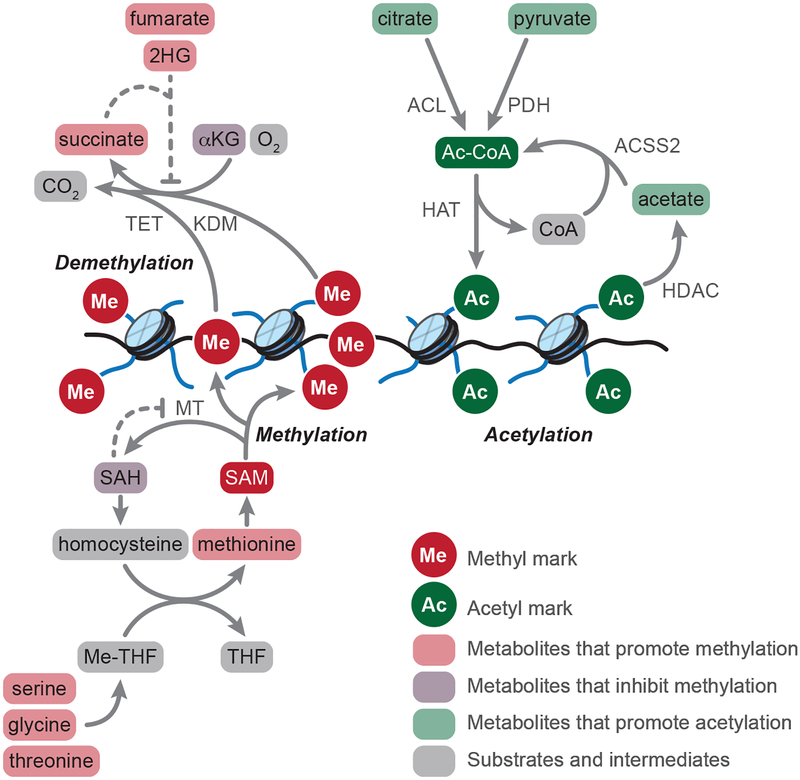
Figure 3.
Metabolic regulation of chromatin marks. Methyltransferase (MT) enzymes transfer methyl (Me) groups from S-adenosylmethionine (SAM) to histones and DNA. Methylation reactions generate S-adenosylhomocysteine (SAH) as a product; SAH inhibits methylation reactions, making MT activity responsive to the relative ratio of SAM:SAH. Methionine serves as the direct precursor for SAM, and methionine pools are regulated both by cellular uptake (not shown) and the methionine cycle, which regenerates methionine from homocysteine. The methionine cycle is intimately connected to the folate cycles, in which serine, glycine, and (in mouse PSCs) threonine provide one-carbon units to tetrahydrofolate (THF) for transfer to homocysteine. Lysine demethylases (KDM) and ten-eleven translocation (TET) enzymes catalyze demethylation of histones and DNA, respectively, using alpha-ketoglutarate (αKG), oxygen, and ferrous iron (not shown) as substrates. Succinate, fumarate, and 2-hydroxyglutarate (2HG) function as competitive inhibitors of KDM and TET enzymes, thereby promoting accumulation of methyl marks. Histone acetyltransferases (HAT) transfer acetyl (Ac) groups from acetyl-coenzyme A (Ac-CoA) to histones. Ac-CoA derives from cytosolic and/or nuclear citrate, pyruvate, and acetate, which serve as substrates for the Ac-CoA generating enzymes ATP citrate lyase (ACL), pyruvate dehydrogenase (PDH), or acyl-coenzyme A synthetase short-chain family member 2 (ACSS2). Moreover, class I, II, and IV histone deacetylase (HDAC) reactions generate acetate as a product, which can be captured by nuclear ACSS2 to regenerate Ac-CoA.
Acetylation of histones
Histone acetylation generally functions to increase chromatin accessibility and activate gene transcription72. Histone acetyltransferases catalyze the transfer of acetyl groups from acetyl-CoA to histones, whereas histone deacetylases remove acetyl marks (Fig. 3). Despite its abundant production in the mitochondria to fuel the TCA cycle, acetyl-CoA cannot diffuse across the mitochondrial membrane and is thus unavailable for histone acetylation reactions73. Instead, mitochondrial acetyl-CoA (derived from pyruvate, acetate, fatty acids, or amino acids) condenses with oxaloacetate to produce citrate, which can undergo transport out of the mitochondria. In the cytosol, ATP-citrate lyase (ACL) cleaves citrate into acetyl-CoA and oxaloacetate providing a major source of acetyl-CoA for acetylation reactions and lipid biosynthesis. In addition to cytosolic ACL, nuclear localization of ACL and the pyruvate dehydrogenase complex provide localized sources of acetyl-CoA production to facilitate histone acetylation74–77. Activation of oncogenic signaling, most notably the PI3K/AKT pathway, increases generation of glucose-derived citrate and enhances activity of ACL through AKT-mediated phosphorylation74,78. This results in increased cytosolic acetyl-CoA pools and globally enhanced histone acetylation, which helps to maintain oncogenic programs of gene expression74,78,79. Histone acetylation may in turn further influence the metabolic state of the cell by regulating the expression of metabolic enzymes in both cancer cells and stem cells80–82.
Under conditions of hypoxic stress, cellular pools of citrate become limiting and acyl-CoA synthetase short chain family member 2 (ACSS2) serves as an important source of cytosolic acetyl-CoA through the capture of free acetate83. Accordingly, tumors growing in vivo exhibit high consumption of acetate47 and dependence on ACSS2/acetate for the maintenance of acetyl-CoA production, lipid synthesis, and histone acetylation46,84. In particular, nuclear translocation of ACSS2 in response to oxygen and nutrient limitation may play a key role in maintaining histone acetylation46,85. Hypoxia and defective mitochondrial respiration also promote reductive carboxylation of glutamine-derived αKG to citrate, which can serve as an additional source of cytosolic acetyl-CoA for lipid production in cancer cells cultured in vitro86–88. The potential contribution of reductive carboxylation to histone acetylation remains unknown, but isotopic labeling experiments suggest that utilization of this pathway may be limited in tumors growing in vivo38,47.
While histone acetylation therefore likely responds to defined metabolic inputs that regulate local acetyl-CoA pools, the effects of histone acetylation on stem cell and cancer cell differentiation vary depending on the cellular context. During early embryonic development, several TCA cycle enzymes including pyruvate dehydrogenase localize to the nucleus where they may provide the acetyl-CoA and other metabolites that collectively help to establish the permissive chromatin landscape that facilitates zygotic genome activation (Fig. 4)77. Likewise, metabolic changes may initiate decreases in histone acetylation that allow exit from the pluripotent state. PSCs induced to differentiate experience a rapid decline in histone acetylation due to decreased glycolytic flux and reduced generation of glucose-derived acetyl-CoA; forced restoration of histone acetylation through supplementation with exogenous acetate impairs the onset of differentiation21. In contrast, when multipotent progenitor cells from adult tissues differentiate into specialized lineages, increased histone acetylation drives expression of lineage-specific genetic programs. Consequently, disruption of acetyl-CoA synthesis through ACL or ACSS2 can impair differentiation of progenitor cells into adipocytes or neurons, respectively74,89. It remains uncertain to what degree metabolic regulation of histone acetylation influences cancer cell differentiation. However, histone deacetylase inhibitors exhibit particular efficacy in hematologic malignancies, at least in part through promoting terminal differentiation of transformed stem/progenitor cells90. Interestingly, histone acetylation may be intimately linked with the regulation of intracellular pH; intracellular acidification decreases global histone acetylation, while inhibition of histone deacetylation increases intracellular pH91. It remains to be determined whether coordinate regulation of histone acetylation and pH might contribute to phenotypic heterogeneity within tumors. Likewise, it will be important to determine if metabolic manipulation of histone acetylation can serve as a more broadly applicable strategy to target cancer cell differentiation.
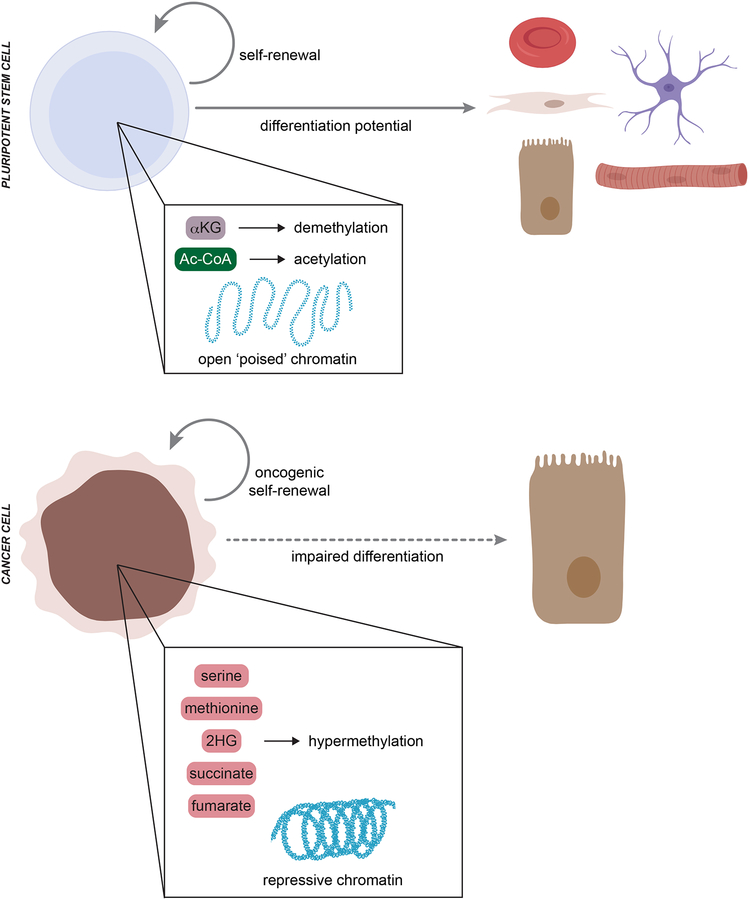
Figure 4.
Metabolic control of differentiation in stem cells and cancer cells. Naïve pluripotent stem cell (PSC) self-renewal and differentiation potential depend on a characteristic open (‘poised’) chromatin structure. Metabolic inputs from alpha-ketoglutarate (αKG) and acetyl-CoA (Ac-CoA) drive active demethylation and histone acetylation to maintain naïve PSC identity. In contrast, cancer cells generally exhibit a hypermethylated chromatin landscape that represses expression of both tumor suppressors and gene expression programs required for normal lineage differentiation. The hypermethylated state in cancer cells can be driven, at least in part, by metabolic rewiring that results in accumulation of serine, methionine, 2-hydroxyglutarate (2HG), succinate, and/or fumarate.
Methylation of histones and DNA
Methylation of specific histone residues can function either to activate or repress transcription, whereas DNA methylation results in transcriptional repression92. The net balance of histone and DNA methylation depends on the relative rates of deposition and removal of methyl marks, a metabolically responsive process that will be discussed in further detail below. Hypermethylation of DNA and repressive histone marks represents a well described hallmark of cancer, which contributes to oncogenesis by silencing tumor suppressors and repressing gene expression programs required for normal differentiation93. Methylation changes in tumors can be heterogeneous: for example, while cancer-associated hypermethylation generally localizes to promoter elements, cancer cells often exhibit hypomethylation of repetitive DNA elements and gene bodies94–99. In contrast to the often hypermethylated phenotype of cancer, early mammalian development is marked by extremely dynamic chromatin methylation100. Naïve PSCs exhibit a globally hypomethylated landscape similar to pre-implantation blastocysts whereas primed PSCs exhibit levels of DNA methylation consistent with post-implantation development7.
S-adenosylmethionine
Methyltransferase enzymes catalyze the transfer of methyl groups from S-adenosylmethionine (SAM) to lysine and arginine residues on histones and (primarily) cytosine nucleotides in DNA. Analogous to histone acetylation, the methylation status of cancer cells and stem cells may depend on the metabolic inputs that produce and recycle the methyl donor SAM (Fig. 3)71. Intracellular SAM pools are regulated by both cellular import of methionine through amino acid transporters and the methionine cycle, which salvages the homocysteine produced by methyltransferase reactions to regenerate methionine. Moreover, re-methylation of homocysteine occurs via one-carbon donation from the folate cycle, which is intimately related to serine and glycine metabolism71. Physiologic or pathologic alterations to any of these metabolic inputs may influence histone and DNA methylation with consequent effects on the differentiation of cancer cells and stem cells.
Many oncogenic lesions lead to enhanced uptake and/or synthesis of amino acids involved in SAM metabolism101–105, which in turn can influence methylation patterns and dependencies in cancer cells and in some cases affect differentiation status106,107. In cultured cancer cells, methionine restriction causes global decreases in histone methylation with decreased expression of genes implicated in cancer cell proliferation and lineage identity108,109. In lung cancer models, increased expression of the methionine transporter LAT1 enhances intracellular SAM pools to maintain the histone methyltransferase activity of EZH2110. Restriction of methionine or targeting of the SAM-generating enzyme MAT2A de-represses expression of the differentiation-promoting transcription factor RXRα110. Oncogenic cooperativity between loss of liver kinase B 1 (LKB1) and mutant KRAS results in enhanced serine biosynthesis to maintain DNA methylation, conferring a therapeutic vulnerability to inhibitors of the methionine salvage pathway106. Acute myeloid leukemias (AML) overexpress the one-carbon folate pathway enzyme methylenetetrahydrofolate dehydrogenase-cyclohydrolase 2 (MTHFD2), and inhibiting MTHFD2 promotes myeloid differentiation of AML blasts111. These examples suggest that perturbations in SAM homeostasis may contribute to cancer progression, and it will be important for future work to elucidate the combination of factors that enable specific phenotypes to emerge from general perturbations in the availability of the required methyl donor.
Mouse PSCs derive much of their methyl-donating SAM pools from the catabolism of threonine, a metabolic feature which is absent in humans51,52. Threonine deprivation or inhibition of threonine dehydrogenase in mouse ESCs impairs self-renewal and potentiates differentiation. These effects appear to result from depletion of SAM pools and loss of H3K4 methylation, a modification important to the maintenance of the transcriptionally ‘poised’ state of pluripotency112. Analogously, human PSCs critically depend on methionine uptake and recycling for the maintenance of intracellular SAM levels. Interference with methionine metabolism causes a loss of methylation on histones (primarily H3K4) and DNA and potentiates ESC differentiation113. To date these studies have relied on removal of essential amino acids from PSC culture. It will be important for future work to determine whether dynamic regulation of SAM metabolic pathways and/or physiologic changes in nutrient availability in the developing embryo are sufficient to alter gene expression programs by modulating SAM availability.
Alpha-ketoglutarate
Although passive loss of methylation on histones and DNA can occur during cell division or as a result of nucleosome turnover114, the enzymatic removal of methyl marks plays a critical role in regulating the methylation status of cancer cells and stem cells93. The lysine-specific demethylase (LSD) enzymes catalyze removal of some histone marks in a flavin-dependent oxidative reaction. However, most demethylation reactions are catalyzed by the large family of αKG-dependent dioxygenases, which includes the Jumonji-domain containing histone demethylases (JHDM) and the ten-eleven-translocation (TET) enzymes that mediate the iterative oxidation and demethylation of methylated cytosine in DNA. The activity of these enzymes depends on the availability of the co-substrates—αKG, ferrous iron, oxygen—as well as the presence of competitive inhibitors with structural similarity to αKG such as 2-hydroxyglutarate, succinate, and fumarate115. Thus, the dynamic interplay between the metabolic pathways that regulate αKG, succinate, fumarate, and 2-hydroxyglutarate production and consumption collectively influences the activity of αKG-dependent dioxygenases with potential implications for cancer cells and stem cells (Fig. 3).
In myeloid leukemia stem cells, overexpression of the enzyme branched chain amino acid transaminase 1 (BCAT1) contributes to oncogenesis through direct effects on αKG levels. Specifically, BCAT1 depletes intracellular αKG resulting in impaired TET enzyme activity, DNA hypermethylation, and blockade of normal myeloid differentiation116. Ascorbate (vitamin C) enhances the activity of αKG-dependent dioxygenases by facilitating regeneration of ferrous iron117. Several recent studies demonstrated that ascorbate supplementation enhances TET-mediated DNA demethylation, which promotes differentiation of leukemia cells118,119. Enhanced ascorbate uptake was further implicated in the self-renewal of normal hematopoietic stem cells via regulation of TET-mediated demethylation118. In human melanoma tumors, hypoxic central regions exhibit significant depletion of extracellular glutamine, with a concomitant decrease in intracellular αKG levels and increase in repressive histone methylation120. Importantly, the hypermethylated phenotype promotes cancer cell de-differentiation and therapeutic resistance, effects that can be reversed by inhibition of the histone methyltransferase EZH2.
In contrast to the pro-differentiation effects observed in cancer cells, metabolic activation of αKG-dependent dioxygenases has notably context-specific roles in PSCs, likely as a consequence of the vastly different chromatin states in naïve and primed PSCs. Metabolic pathways in naïve mouse PSCs are wired to promote accumulation of intracellular αKG, perhaps in part to promote activity of the chromatin-modifying enzymes that sustain the open chromatin landscape characteristic of naïve pluripotency14,121. Interventions that increase αKG drive loss of repressive chromatin modifications and favor self-renewal over differentiation14,121. In another recent example, the protein prohibitin was identified as a key regulator of human ESC self-renewal. Mechanistic studies demonstrated that prohibitin interacts with histone H3.3 to regulate transcription of the isocitrate dehydrogenase enzymes, which produce the αKG required to maintain demethylation and pluripotency122. In contrast to the preceding examples, αKG accelerates differentiation of the more heavily methylated primed human PSCs, consistent with the requirement to demethylate and de-repress lineage-specific regions as part of the differentiation process31. These data argue that metabolic pathways may facilitate major transitions in the global chromatin landscape. In support of this hypothesis, ascorbic-acid mediated activation of αKG-dependent dioxygenases, including both JHDM and TET enzymes, is well-established to enhance reprogramming to pluripotency, a process that requires erasure of somatic methylation patterns123.
Antagonists of alpha-ketoglutarate
Perhaps the best examples of metabolic regulation of chromatin come from the seminal discovery of cancer-associated mutations in the isocitrate dehydrogenase (IDH) enzymes. IDH1/2 enzymes normally catalyze the interconversion of isocitrate and αKG in an NADP(H)-dependent manner. Oncogenic mutations in IDH1/2 occur at specific arginine residues in the active site of the enzyme, conferring a neomorphic enzymatic activity that efficiently reduces αKG to the structurally similar metabolite D-2-hydroxyglutarate (D-2HG)124,125. D-2HG functions as a competitive inhibitor of αKG-dependent dioxygenases126–128, and exogenous D-2HG is sufficient to induce many of the oncogenic effects of IDH mutations129. Although the relative importance of each enzyme inhibited by D-2HG is still being elucidated, inhibition of the TET and JHDM enzymes appears to play a key role in mutant IDH/D-2HG-mediated oncogenesis130,131. By poisoning the function of the TET and JHDM enzymes, D-2HG promotes hypermethylation of DNA and repressive histone marks; this locks IDH-mutant cancer cells in an undifferentiated stem cell-like state, conferring a differentiation ‘hit’ that contributes to oncogenesis130–133. In both preclinical models and patients with AML, targeted inhibition of mutant IDH enzyme function suppresses D-2HG production leading to reversal of repressive methylation marks and differentiation of IDH-mutant stem/progenitors134,135. Ongoing efforts seek to define physiologic sources and biologic effects of D-2HG and its enantiomer L-2HG, which will determine if targeted inhibition of 2HG metabolism might represent a more broadly applicable strategy to modulate chromatin structure, cell differentiation, and oncogenesis136–138.
Similar to the effects of 2-hydroxyglutarate, succinate and fumarate function as competitive inhibitors of αKG-dependent dioxygenases139. Succinate and fumarate catabolism depends on the activity of the TCA cycle enzymes succinate dehydrogenase (SDH) and fumarate hydratase (FH), respectively. Inactivating lesions in components of the SDH complex or FH have been implicated in the pathogenesis of a variety of rare cancers and inherited cancer predisposition syndromes140. Loss of SDH or FH activity results in cellular accumulation of succinate or fumarate, leading to stabilization of hypoxia-inducible factor 1 (HIF-1) via inhibition of αKG-dependent prolyl hydroxylases that normally target HIF-1 for degradation in the presence of oxygen141,142. This ‘pseudo-hypoxic’ state of SDH- and FH-deficient tumors, contributes to oncogenesis by driving glycolytic metabolism and tumor angiogenesis140. More recent evidence indicates that fumarate and succinate accumulation also drive widespread hypermethylation of DNA and repressive histone marks143,144, which alter cancer cell differentiation, at least in part, by inducing genetic programs that promote an invasive mesenchymal phenotype145. Importantly, restoration of the balance of αKG relative to succinate/fumarate reverses the hypermethylated phenotype and promotes HIF-1 degradation143,146.
By coupling chromatin modifications to the metabolic state, cellular differentiation decisions may be influenced by nutrient availability and microenvironmental conditions (e.g. oxygen availability and pH). It will be interesting for future work to examine how competition for scarce resources in the tumor microenvironment and the adult stem cell niche affect chromatin regulation and gene expression programs. The consequences of metabolic regulation of chromatin structure often result in divergent effects on differentiation state of cancer cells and stem cells (Fig. 4). This context-specificity likely arises as a consequence of the many factors that collectively combine to determine gene expression programs, including local metabolite levels, the relative sensitivity of chromatin-modifying enzymes to metabolic fluctuations and the expression and recruitment of relevant transcription factors and co-activators to key genetic loci6. Understanding how global shifts in nutrient availability translate into specific gene expression programs is therefore a major area for future investigation.
Conclusions and future directions
Metabolites that are key intermediates of central metabolic pathways supporting bioenergetics and anabolic growth can also influence the regulation of gene expression programs and cell fate decisions. Therefore, the particular bioenergetic requirements of a given cell—and the pathways engaged to meet those requirements—may have important consequences for the regulation of cell identity. Indeed, while all cells proliferating in vitro appear to share several canonical hallmarks of proliferative metabolism, accumulating evidence demonstrates that there is no single metabolic profile of a cancer cell, just as there is no single profile of a stem cell. We are likely just beginning to understand the diversity of metabolic strategies that support cell proliferation and the factors that regulate these myriad metabolic profiles. Clearly, both cell-intrinsic and cell-extrinsic factors play critical roles determining cellular metabolic phenotypes.
It is interesting to note that to date, the metabolic plasticity of cancer cells is largely revealed in vivo, while PSCs may engage a wider array of metabolic strategies in vitro. Because many oncogenes and tumor suppressors regulate cellular metabolism, the very oncogenic lesions that drive malignant identity may also lock cultured cancer cells into a relatively inflexible metabolic identity. In contrast, the diversity of extracellular stimuli experienced in vivo may enable rewiring of both intracellular signaling and metabolic pathways even in malignant cells. Similarly, although several key signaling pathways sustain the growth of PSCs, these pathways are dynamic and largely tied to the regulation of cell identity. It is therefore tempting to speculate that the inherent metabolic plasticity of PSCs may arise as a consequence of their functional plasticity.
Several avenues of research may provide valuable insight into the connections between metabolic pathways, gene regulation and cell fate decisions in cancer and development. First, inborn errors of metabolism represent a largely untapped opportunity for elucidating how defined changes in cellular metabolite levels influence cellular phenotypes and disease susceptibility147. Some intriguing examples are beginning to emerge. Heterozygous mutations in genes encoding FH or subunits of SDH are associated with susceptibility to specific cancer syndromes and widespread changes in chromatin modifications consistent with inhibition of αKG-dependent dioxygenases mediated by high levels of fumarate or succinate, respectively140,143. Similarly, biallelic mutation of the gene encoding the L-2HG dehydrogenase, which converts L-2HG to αKG, drives high levels of L-2HG and predisposes patients to central nervous system malignancies in addition to other adverse metabolic consequences138,148–150. Intriguingly, inborn errors of metabolism that result in elevated D-2HG, including mutations in the D-2HG dehydrogenase and gain-of-function mutations in IDH2, induce phenotypes including developmental delays and cardiomyopathy but do not appear to be associated with an increased risk of cancer148,151,152. How specific metabolic alterations contribute to developmental phenotypes, why certain metabolic perturbations predispose to tumorigenesis and why some tissues are more or less sensitive to transformation in the background of these germline mutations remain key open questions that may be illuminated by the continued study of inborn errors of metabolism.
Second, cancer stem cells (or tumor initiating cells) represent an intriguing example of cells that can exhibit features of both normal stem cells and aberrantly proliferating tumor cells153. To date, the metabolism of these tumor initiating cells remains poorly understood—indeed, it seems unlikely that there will be one single metabolic signature of a cancer stem cell, just as there is no single metabolic phenotype of adult stem cells from which these cancer stem cells may arise153,154. Some studies suggest that cancer stem cells exhibit decreased mitochondrial function and increased dependence on glycolytic flux155–160, while other studies report enhanced dependence on mitochondrial function and oxidative phosphorylation161–166. Lipid metabolism also appears to play an important role in cancer stem cell maintenance by supplying bioenergetic substrates and potentially influencing cell fate decisions156,167–171. Cell lineage, environmental niche and dynamic cellular phenotypes—including the switch from self-renewal to transient amplification and differentiation—are likely to result in a heterogeneous metabolic profile. Nevertheless, continued study of metabolic requirements of adult stem cells and their transformed counterparts may yield critical insights into how metabolism supports both normal and aberrant stem cell phenotypes and provide opportunities for targeted therapeutic intervention.
We are just beginning to understand the intersection between metabolic activity and the regulation of cell fate decisions and several outstanding questions will shape future research in this area. First, the mechanisms that link metabolic profiles to cellular phenotypes remain largely unidentified. While the most compelling hypothesis is that metabolic regulation of chromatin mechanistically links metabolic profiles to gene expression programs, careful genetic experiments will be required to determine to what extent individual metabolites influence cell fate through specific chromatin modifications. Second, the aspects of cellular metabolic phenotypes that result from bioenergetic demands as opposed to intrinsically determined metabolic preferences linked to cell identity remain an open question. Third, it will be important to determine whether physiological changes in nutrient abundance contribute to altered cell fate decisions during cancer progression or embryonic development. Addressing these questions will require new, sensitive techniques to assess the metabolism of rare cell populations in vivo. Together, these studies will enable a better understanding of whether individual metabolic features are cause or consequence of changing cellular programs and reveal opportunities for targeted manipulation of cellular metabolism that hold promise for anti-cancer therapies and regenerative medicine approaches.
Acknowledgements
We thank members of the Intlekofer and Finley labs for discussion and Juanma Schvartzman for critical reading of the manuscript. A.I. is supported by the NIH/NCI (K08 CA201483), Damon Runyon Cancer Research Foundation (CI 95–18), Burroughs Wellcome Fund (CAMS 1015584), Leukemia & Lymphoma Society (SCOR 7011–16), Susan & Peter Solomon Divisional Genomics Program, Steven A. Greenberg Fund, and Cycle for Survival. L.F is a Dale F. Frey-William Raveis Charitable Fund Scientist supported by the Damon Runyon Cancer Research Foundation (DFS-23–17). This work was additionally supported by the Searle Scholars program (to L.F.), The Starr Foundation (I11–0039 to L.F.) and the Memorial Sloan Kettering Cancer Center Support Grant P30 CA008748.
Footnotes
Competing Interests. A.I. has previously consulted for Foundation Medicine, Inc.
References
- 1.Vander Heiden MG, Cantley LC & Thompson CB Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033, doi: 10.1126/science.1160809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudin AE & Stover PJ Insights into metabolic mechanisms underlying folate-responsive neural tube defects: a minireview. Birth Defects Res A Clin Mol Teratol 85, 274–284, doi: 10.1002/bdra.20553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luengo A, Gui DY & Vander Heiden MG Targeting Metabolism for Cancer Therapy. Cell Chem Biol 24, 1161–1180, doi: 10.1016/j.chembiol.2017.08.028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong H & Chandel NS Regulation of redox balance in cancer and T cells. The Journal of biological chemistry 293, 7499–7507, doi: 10.1074/jbc.TM117.000257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chantranupong L, Wolfson RL & Sabatini DM Nutrient-sensing mechanisms across evolution. Cell 161, 67–83, doi: 10.1016/j.cell.2015.02.041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schvartzman JM, Thompson CB & Finley LWS Metabolic regulation of chromatin modifications and gene expression. The Journal of cell biology 217, 2247–2259, doi: 10.1083/jcb.201803061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberger L, Ayyash M, Novershtern N & Hanna JH Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nature reviews. Molecular cell biology 17, 155–169, doi: 10.1038/nrm.2015.28 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Martello G & Smith A The nature of embryonic stem cells. Annual review of cell and developmental biology 30, 647–675, doi: 10.1146/annurev-cellbio-100913-013116 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Fan J et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol 9, 712, doi: 10.1038/msb.2013.65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hosios AM et al. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Developmental cell 36, 540–549, doi: 10.1016/j.devcel.2016.02.012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell metabolism 23, 27–47, doi: 10.1016/j.cmet.2015.12.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ & Chandel NS Fundamentals of cancer metabolism. Sci Adv 2, e1600200, doi: 10.1126/sciadv.1600200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Heiden MG & DeBerardinis RJ Understanding the Intersections between Metabolism and Cancer Biology. Cell 168, 657–669, doi: 10.1016/j.cell.2016.12.039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey BW, Finley LW, Cross JR, Allis CD & Thompson CB Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416, doi: 10.1038/nature13981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tohyama S et al. Glutamine Oxidation Is Indispensable for Survival of Human Pluripotent Stem Cells. Cell metabolism 23, 663–674, doi: 10.1016/j.cmet.2016.03.001 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Boroughs LK & DeBerardinis RJ Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17, 351–359, doi: 10.1038/ncb3124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung S et al. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 4 Suppl 1, S60–67, doi: 10.1038/ncpcardio0766 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J 31, 2103–2116, doi: 10.1038/emboj.2012.71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 30, 4860–4873, doi: 10.1038/emboj.2011.401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu W et al. Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell stem cell 19, 476–490, doi: 10.1016/j.stem.2016.08.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moussaieff A et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell metabolism 21, 392–402, doi: 10.1016/j.cmet.2015.02.002 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Cliff TS et al. MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell stem cell 21, 502–516 e509, doi: 10.1016/j.stem.2017.08.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folmes CD et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell metabolism 14, 264–271, doi: 10.1016/j.cmet.2011.06.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panopoulos AD et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell research 22, 168–177, doi: 10.1038/cr.2011.177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathieu J et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell stem cell 14, 592–605, doi: 10.1016/j.stem.2014.02.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H et al. Core Pluripotency Factors Directly Regulate Metabolism in Embryonic Stem Cell to Maintain Pluripotency. Stem Cells 33, 2699–2711, doi: 10.1002/stem.2073 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Liberti MV & Locasale JW The Warburg Effect: How Does it Benefit Cancer Cells? Trends in biochemical sciences 41, 211–218, doi: 10.1016/j.tibs.2015.12.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prigione A, Fauler B, Lurz R, Lehrach H & Adjaye J The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28, 721–733, doi: 10.1002/stem.404 (2010). [DOI] [PubMed] [Google Scholar]
- 29.St John JC et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells 7, 141–153, doi: 10.1089/clo.2005.7.141 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Zhang H et al. Distinct Metabolic States Can Support Self-Renewal and Lipogenesis in Human Pluripotent Stem Cells under Different Culture Conditions. Cell Rep 16, 1536–1547, doi: 10.1016/j.celrep.2016.06.102 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.TeSlaa T et al. alpha-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell metabolism 24, 485–493, doi: 10.1016/j.cmet.2016.07.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takashima Y et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 162, 452–453, doi: 10.1016/j.cell.2015.06.052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carbognin E, Betto RM, Soriano ME, Smith AG & Martello G Stat3 promotes mitochondrial transcription and oxidative respiration during maintenance and induction of naive pluripotency. EMBO J 35, 618–634, doi: 10.15252/embj.201592629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Setoguchi K, TeSlaa T, Koehler CM & Teitell MA P53 Regulates Rapid Apoptosis in Human Pluripotent Stem Cells. J Mol Biol 428, 1465–1475, doi: 10.1016/j.jmb.2015.07.019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madden DT, Davila-Kruger D, Melov S & Bredesen DE Human embryonic stem cells express elevated levels of multiple pro-apoptotic BCL-2 family members. PloS one 6, e28530, doi: 10.1371/journal.pone.0028530 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanes O et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nature chemical biology 6, 411–417, doi: 10.1038/nchembio.364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muir A, Danai LV & Vander Heiden MG Microenvironmental regulation of cancer cell metabolism: implications for experimental design and translational studies. Dis Model Mech 11, doi: 10.1242/dmm.035758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marin-Valencia I et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell metabolism 15, 827–837, doi: 10.1016/j.cmet.2012.05.001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson SM et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell metabolism 23, 517–528, doi: 10.1016/j.cmet.2016.01.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hensley CT et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 164, 681–694, doi: 10.1016/j.cell.2015.12.034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Courtney KD et al. Isotope Tracing of Human Clear Cell Renal Cell Carcinomas Demonstrates Suppressed Glucose Oxidation In Vivo. Cell metabolism, doi: 10.1016/j.cmet.2018.07.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tardito S et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol 17, 1556–1568, doi: 10.1038/ncb3272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir A et al. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. Elife 6, doi: 10.7554/eLife.27713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hui S et al. Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118, doi: 10.1038/nature24057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faubert B et al. Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371 e359, doi: 10.1016/j.cell.2017.09.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comerford SA et al. Acetate dependence of tumors. Cell 159, 1591–1602, doi: 10.1016/j.cell.2014.11.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mashimo T et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell 159, 1603–1614, doi: 10.1016/j.cell.2014.11.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayers JR et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165, doi: 10.1126/science.aaf5171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuneva MO et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism 15, 157–170, doi: 10.1016/j.cmet.2011.12.015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell stem cell 19, 66–80, doi: 10.1016/j.stem.2016.05.009 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Shyh-Chang N et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226, doi: 10.1126/science.1226603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J et al. Dependence of mouse embryonic stem cells on threonine catabolism. Science 325, 435–439, doi: 10.1126/science.1173288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palm W & Thompson CB Nutrient acquisition strategies of mammalian cells. Nature 546, 234–242, doi: 10.1038/nature22379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J et al. Metabolism in Pluripotent Stem Cells and Early Mammalian Development. Cell metabolism 27, 332–338, doi: 10.1016/j.cmet.2018.01.008 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Tang F et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell stem cell 6, 468–478, doi: 10.1016/j.stem.2010.03.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaneko KJ Metabolism of Preimplantation Embryo Development: A Bystander or an Active Participant? Curr Top Dev Biol 120, 259–310, doi: 10.1016/bs.ctdb.2016.04.010 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Houghton FD, Thompson JG, Kennedy CJ & Leese HJ Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev 44, 476–485, doi: (1996). [DOI] [PubMed] [Google Scholar]
- 58.Houghton FD Energy metabolism of the inner cell mass and trophectoderm of the mouse blastocyst. Differentiation 74, 11–18, doi: 10.1111/j.1432-0436.2006.00052.x (2006). [DOI] [PubMed] [Google Scholar]
- 59.Brinster RL Effect of glutathione on the development of two-cell mouse embryos in vitro. J Reprod Fertil 17, 521–525 (1968). [DOI] [PubMed] [Google Scholar]
- 60.Cholewa JA & Whitten WK Development of two-cell mouse embryos in the absence of a fixed-nitrogen source. J Reprod Fertil 22, 553–555 (1970). [DOI] [PubMed] [Google Scholar]
- 61.Stine ZE, Walton ZE, Altman BJ, Hsieh AL & Dang CV MYC, Metabolism, and Cancer. Cancer discovery 5, 1024–1039, doi: 10.1158/2159-8290.CD-15-0507 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kruiswijk F, Labuschagne CF & Vousden KH p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nature reviews. Molecular cell biology 16, 393–405, doi: 10.1038/nrm4007 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Scognamiglio R et al. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 164, 668–680, doi: 10.1016/j.cell.2015.12.033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulut-Karslioglu A et al. Inhibition of mTOR induces a paused pluripotent state. Nature 540, 119–123, doi: 10.1038/nature20578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merkle FT et al. Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233, doi: 10.1038/nature22312 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hanna J et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595–601, doi: 10.1038/nature08592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paling NR, Wheadon H, Bone HK & Welham MJ Regulation of embryonic stem cell self-renewal by phosphoinositide 3-kinase-dependent signaling. The Journal of biological chemistry 279, 48063–48070, doi: 10.1074/jbc.M406467200 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Watanabe S et al. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 25, 2697–2707, doi: 10.1038/sj.onc.1209307 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Ying QL et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523, doi: 10.1038/nature06968 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reid MA, Dai Z & Locasale JW The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 19, 1298–1306, doi: 10.1038/ncb3629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su X, Wellen KE & Rabinowitz JD Metabolic control of methylation and acetylation. Curr Opin Chem Biol 30, 52–60, doi: 10.1016/j.cbpa.2015.10.030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tessarz P & Kouzarides T Histone core modifications regulating nucleosome structure and dynamics. Nature reviews. Molecular cell biology 15, 703–708, doi: 10.1038/nrm3890 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Sivanand S, Viney I & Wellen KE Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends in biochemical sciences 43, 61–74, doi: 10.1016/j.tibs.2017.11.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wellen KE et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080, doi: 10.1126/science.1164097 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sutendra G et al. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97, doi: 10.1016/j.cell.2014.04.046 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Sivanand S et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Molecular cell 67, 252–265 e256, doi: 10.1016/j.molcel.2017.06.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagaraj R et al. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell 168, 210–223 e211, doi: 10.1016/j.cell.2016.12.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JV et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell metabolism 20, 306–319, doi: 10.1016/j.cmet.2014.06.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai L, Sutter BM, Li B & Tu BP Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell 42, 426–437, doi: 10.1016/j.molcel.2011.05.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sebastian C et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199, doi: 10.1016/j.cell.2012.10.047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong L et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293, doi: 10.1016/j.cell.2009.12.041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang J et al. Inhibiting histone deacetylases suppresses glucose metabolism and hepatocellular carcinoma growth by restoring FBP1 expression. Sci Rep 7, 43864, doi: 10.1038/srep43864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamphorst JJ, Chung MK, Fan J & Rabinowitz JD Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab 2, 23, doi: 10.1186/2049-3002-2-23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schug ZT et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer cell 27, 57–71, doi: 10.1016/j.ccell.2014.12.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bulusu V et al. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep 18, 647–658, doi: 10.1016/j.celrep.2016.12.055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Metallo CM et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384, doi: 10.1038/nature10602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mullen AR et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388, doi: 10.1038/nature10642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wise DR et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America 108, 19611–19616, doi: 10.1073/pnas.1117773108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mews P et al. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386, doi: 10.1038/nature22405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de The H Differentiation therapy revisited. Nature reviews. Cancer 18, 117–127, doi: 10.1038/nrc.2017.103 (2018). [DOI] [PubMed] [Google Scholar]
- 91.McBrian MA et al. Histone acetylation regulates intracellular pH. Molecular cell 49, 310–321, doi: 10.1016/j.molcel.2012.10.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berger SL The complex language of chromatin regulation during transcription. Nature 447, 407–412, doi: 10.1038/nature05915 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Plass C et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 14, 765–780, doi: 10.1038/nrg3554 (2013). [DOI] [PubMed] [Google Scholar]
- 94.Feinberg AP & Vogelstein B Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983). [DOI] [PubMed] [Google Scholar]
- 95.Kulis M et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nature genetics 44, 1236–1242, doi: 10.1038/ng.2443 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Ehrlich M & Lacey M DNA hypomethylation and hemimethylation in cancer. Adv Exp Med Biol 754, 31–56, doi: 10.1007/978-1-4419-9967-2_2 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Jakel C et al. Genome-wide genetic and epigenetic analyses of pancreatic acinar cell carcinomas reveal aberrations in genome stability. Nature communications 8, 1323, doi: 10.1038/s41467-017-01118-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ehrlich M DNA methylation in cancer: too much, but also too little. Oncogene 21, 5400–5413, doi: 10.1038/sj.onc.1205651 (2002). [DOI] [PubMed] [Google Scholar]
- 99.Kulis M & Esteller M DNA methylation and cancer. Adv Genet 70, 27–56, doi: 10.1016/B978-0-12-380866-0.60002-2 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Smith ZD et al. DNA methylation dynamics of the human preimplantation embryo. Nature 511, 611–615, doi: 10.1038/nature13581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeNicola GM et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nature genetics 47, 1475–1481, doi: 10.1038/ng.3421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Locasale JW et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics 43, 869–874, doi: 10.1038/ng.890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Possemato R et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350, doi: 10.1038/nature10350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maddocks OD et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546, doi: 10.1038/nature11743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edinger AL & Thompson CB Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell 13, 2276–2288, doi: 10.1091/mbc.01-12-0584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kottakis F et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539, 390–395, doi: 10.1038/nature20132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maddocks OD, Labuschagne CF, Adams PD & Vousden KH Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Molecular cell 61, 210–221, doi: 10.1016/j.molcel.2015.12.014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mentch SJ et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell metabolism 22, 861–873, doi: 10.1016/j.cmet.2015.08.024 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dai Z, Mentch SJ, Gao X, Nichenametla SN & Locasale JW Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nature communications 9, 1955, doi: 10.1038/s41467-018-04426-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dann SG et al. Reciprocal regulation of amino acid import and epigenetic state through Lat1 and EZH2. EMBO J 34, 1773–1785, doi: 10.15252/embj.201488166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pikman Y et al. Targeting MTHFD2 in acute myeloid leukemia. The Journal of experimental medicine 213, 1285–1306, doi: 10.1084/jem.20151574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bernstein BE et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326, doi: 10.1016/j.cell.2006.02.041 (2006). [DOI] [PubMed] [Google Scholar]
- 113.Shiraki N et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell metabolism 19, 780–794, doi: 10.1016/j.cmet.2014.03.017 (2014). [DOI] [PubMed] [Google Scholar]
- 114.Chory EJ et al. Nucleosome Turnover Regulates Histone Methylation Patterns over the Genome. Molecular cell, doi: 10.1016/j.molcel.2018.10.028 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Losman JA & Kaelin WG Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes & development 27, 836–852, doi: 10.1101/gad.217406.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Raffel S et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551, 384–388, doi: 10.1038/nature24294 (2017). [DOI] [PubMed] [Google Scholar]
- 117.Knowles HJ, Raval RR, Harris AL & Ratcliffe PJ Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer research 63, 1764–1768 (2003). [PubMed] [Google Scholar]
- 118.Agathocleous M et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481, doi: 10.1038/nature23876 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cimmino L et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 170, 1079–1095 e1020, doi: 10.1016/j.cell.2017.07.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pan M et al. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol 18, 1090–1101, doi: 10.1038/ncb3410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hwang IY et al. Psat1-Dependent Fluctuations in alpha-Ketoglutarate Affect the Timing of ESC Differentiation. Cell metabolism 24, 494–501, doi: 10.1016/j.cmet.2016.06.014 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Zhu Z et al. PHB Associates with the HIRA Complex to Control an Epigenetic-Metabolic Circuit in Human ESCs. Cell stem cell 20, 274–289 e277, doi: 10.1016/j.stem.2016.11.002 (2017). [DOI] [PubMed] [Google Scholar]
- 123.Cimmino L, Neel BG & Aifantis I Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol 28, 698–708, doi: 10.1016/j.tcb.2018.04.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dang L et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744, doi: 10.1038/nature08617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ward PS et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell 17, 225–234, doi: 10.1016/j.ccr.2010.01.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu W et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell 19, 17–30, doi: 10.1016/j.ccr.2010.12.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chowdhury R et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports 12, 463–469, doi: 10.1038/embor.2011.43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koivunen P et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488, doi: 10.1038/nature10898 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Losman JA et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625, doi: 10.1126/science.1231677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu C et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478, doi: 10.1038/nature10860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Figueroa ME et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell 18, 553–567, doi: 10.1016/j.ccr.2010.11.015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saha SK et al. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature 513, 110–114, doi: 10.1038/nature13441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sasaki M et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488, 656–659, doi: 10.1038/nature11323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Amatangelo MD et al. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood 130, 732–741, doi: 10.1182/blood-2017-04-779447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen C et al. Cancer-associated IDH2 mutants drive an acute myeloid leukemia that is susceptible to Brd4 inhibition. Genes & development 27, 1974–1985, doi: 10.1101/gad.226613.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fan J et al. Human phosphoglycerate dehydrogenase produces the oncometabolite D-2-hydroxyglutarate. ACS chemical biology 10, 510–516, doi: 10.1021/cb500683c (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Intlekofer AM et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell metabolism 22, 304–311, doi: 10.1016/j.cmet.2015.06.023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ye D, Guan KL & Xiong Y Metabolism, Activity, and Targeting of D- and L-2-Hydroxyglutarates. Trends Cancer 4, 151–165, doi: 10.1016/j.trecan.2017.12.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kaelin WG Jr. & McKnight SL Influence of metabolism on epigenetics and disease. Cell 153, 56–69, doi: 10.1016/j.cell.2013.03.004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gottlieb E & Tomlinson IP Mitochondrial tumour suppressors: a genetic and biochemical update. Nature reviews. Cancer 5, 857–866, doi: 10.1038/nrc1737 (2005). [DOI] [PubMed] [Google Scholar]
- 141.Selak MA et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer cell 7, 77–85, doi: 10.1016/j.ccr.2004.11.022 (2005). [DOI] [PubMed] [Google Scholar]
- 142.Isaacs JS et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer cell 8, 143–153, doi: 10.1016/j.ccr.2005.06.017 (2005). [DOI] [PubMed] [Google Scholar]
- 143.Xiao M et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes & development 26, 1326–1338, doi: 10.1101/gad.191056.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Killian JK et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer discovery 3, 648–657, doi: 10.1158/2159-8290.CD-13-0092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sciacovelli M et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 537, 544–547, doi: 10.1038/nature19353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.MacKenzie ED et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Molecular and cellular biology 27, 3282–3289, doi: 10.1128/MCB.01927-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Erez A & DeBerardinis RJ Metabolic dysregulation in monogenic disorders and cancer - finding method in madness. Nature reviews. Cancer 15, 440–448, doi: 10.1038/nrc3949 (2015). [DOI] [PubMed] [Google Scholar]
- 148.Kranendijk M, Struys EA, Salomons GS, Van der Knaap MS & Jakobs C Progress in understanding 2-hydroxyglutaric acidurias. J Inherit Metab Dis 35, 571–587, doi: 10.1007/s10545-012-9462-5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ma S et al. L2hgdh Deficiency Accumulates l-2-Hydroxyglutarate with Progressive Leukoencephalopathy and Neurodegeneration. Molecular and cellular biology 37, doi: 10.1128/MCB.00492-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rzem R et al. A mouse model of L-2-hydroxyglutaric aciduria, a disorder of metabolite repair. PloS one 10, e0119540, doi: 10.1371/journal.pone.0119540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kranendijk M et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science 330, 336, doi: 10.1126/science.1192632 (2010). [DOI] [PubMed] [Google Scholar]
- 152.Akbay EA et al. D-2-hydroxyglutarate produced by mutant IDH2 causes cardiomyopathy and neurodegeneration in mice. Genes & development 28, 479–490, doi: 10.1101/gad.231233.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Batlle E & Clevers H Cancer stem cells revisited. Nature medicine 23, 1124–1134, doi: 10.1038/nm.4409 (2017). [DOI] [PubMed] [Google Scholar]
- 154.Snyder V, Reed-Newman TC, Arnold L, Thomas SM & Anant S Cancer Stem Cell Metabolism and Potential Therapeutic Targets. Front Oncol 8, 203, doi: 10.3389/fonc.2018.00203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou Y et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. The Journal of biological chemistry 286, 32843–32853, doi: 10.1074/jbc.M111.260935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen CL et al. NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell metabolism 23, 206–219, doi: 10.1016/j.cmet.2015.12.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li Z et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell 15, 501–513, doi: 10.1016/j.ccr.2009.03.018 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dong C et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer cell 23, 316–331, doi: 10.1016/j.ccr.2013.01.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang YH et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell 158, 1309–1323, doi: 10.1016/j.cell.2014.07.048 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Saito Y, Chapple RH, Lin A, Kitano A & Nakada D AMPK Protects Leukemia-Initiating Cells in Myeloid Leukemias from Metabolic Stress in the Bone Marrow. Cell stem cell 17, 585–596, doi: 10.1016/j.stem.2015.08.019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Janiszewska M et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes & development 26, 1926–1944, doi: 10.1101/gad.188292.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lagadinou ED et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell stem cell 12, 329–341, doi: 10.1016/j.stem.2012.12.013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sancho P et al. MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell metabolism 22, 590–605, doi: 10.1016/j.cmet.2015.08.015 (2015). [DOI] [PubMed] [Google Scholar]
- 164.Jones CL et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer cell 34, 724–740 e724, doi: 10.1016/j.ccell.2018.10.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vlashi E et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences of the United States of America 108, 16062–16067, doi: 10.1073/pnas.1106704108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kuntz EM et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nature medicine 23, 1234–1240, doi: 10.1038/nm.4399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ye H et al. Leukemic Stem Cells Evade Chemotherapy by Metabolic Adaptation to an Adipose Tissue Niche. Cell stem cell 19, 23–37, doi: 10.1016/j.stem.2016.06.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pascual G et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45, doi: 10.1038/nature20791 (2017). [DOI] [PubMed] [Google Scholar]
- 169.Samudio I et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. The Journal of clinical investigation 120, 142–156, doi: 10.1172/JCI38942 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li J et al. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell stem cell 20, 303–314 e305, doi: 10.1016/j.stem.2016.11.004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Helou El R. et al. miR-600 Acts as a Bimodal Switch that Regulates Breast Cancer Stem Cell Fate through WNT Signaling. Cell Rep 18, 2256–2268, doi: 10.1016/j.celrep.2017.02.016 (2017). [DOI] [PubMed] [Google Scholar]