Abstract
Background:
Barrett’s esophagus (BE) is the only known precursor of esophageal adenocarcinoma (EAC). Although endoscopy and biopsy are standard methods for BE diagnosis, their high cost and risk limit their use as a screening modality. Here, we sought to develop a BE detection method based on methylation status in cytology samples captured by EsophaCap using a streamlined sensitive technique, methylation on beads (MOB).
Experimental Design:
We conducted a prospective cohort study on 80 patients (52 in the training set; 28 in the test set). We employed MOB to extract and bisulfite-convert DNA, followed by qMSP to assess methylation levels of 8 previously selected candidate markers. Lasso regression was applied to establish a prediction model in the training set, which was then tested on the independent test set.
Results:
In the training set, 5 of 8 candidate methylation biomarkers (p16, HPP1, NELL1, TAC1, and AKAP12) were significantly higher in BE patients than in controls. We built a 4-biomarker-plus-age lasso regression model for BE diagnosis. The AUC was 0.894, with sensitivity 94.4% (95% CI 71%~99%) and specificity 62.2% (95% CI 44.6%~77.3%) in the training set. This model also performed with high accuracy for BE diagnosis in an independent test set: AUC= 0.929 (P<0.001, 95% CI 0.810~1), with sensitivity = 78.6% (95% CI 48.8%~94.3%) and specificity = 92.8% (95% CI 64.1%~99.6%).
Conclusions:
EsophaCap, in combination with an epigenetic biomarker panel and the MOB method, is a promising, well-tolerated, low-cost esophageal sampling strategy for BE diagnosis. This approach merits further prospective studies in larger populations.
Keywords: Barrett’s Esophagus, Cytologic sampling, methylation, biomarkers, methylation on beads, Lasso regression
Introduction
Esophageal adenocarcinoma (EAC) is the eighth-most common cancer and the sixth-most frequent worldwide cause of cancer-related death.(1) The incidence of EAC, particularly with regional or distant spread, is rapidly increasing in the Western World and confers very poor survival.(2) Barrett’s esophagus (BE), a replacement of the normal squamous esophageal epithelium by a metaplastic columnar lining, is the only known premalignant precursor of EAC. BE can progress to low-grade dysplasia (LGD), high-grade dysplasia (HGD), or EAC.(3) For this reason, endoscopic surveillance is recommended in BE patients, and EACs detected via surveillance have improved outcomes.(4) However, BE patients have a low absolute risk of progression to EAC, only 0.11% annually.(5) Moreover, the majority of patients with EAC were not previously diagnosed with BE and have advanced disease.(6) Thus, methods of BE screening that permit risk stratification as well as patient-tailored surveillance and therapy have been proposed(7).
An ideal screening program for BE and EAC should reduce the burden of disease and be cost-effective.(8) Screening for BE with subsequent surveillance for dysplasia and EAC seems feasible, given the long time interval to neoplastic progression in BE and the presence of intermediate stages (LGD, HGD) where endoscopic therapy has proven effective in preventing EAC.(9) However, the current screening paradigm for patients with chronic GERD and multiple risk factors for BE/EAC is problematic for several reasons. First, only a small proportion (<5%) of patients with EAC undergoing resection had a prior diagnosis of BE, suggesting that most BE cases are not detected with current screening.(10) Furthermore, only 35% of patients found to have BE report a history of GERD, suggesting that focusing screening solely on GERD patients will miss a significant proportion of BE patients.(11) Finally, while initial studies found that while endoscopy in high-risk patients was cost-effective, these studies dramatically overestimated EAC risk in BE patients and did not account for BE surveillance costs, suggesting that the current screening paradigm is likely not cost-effective.(12–14) Similarly, modeling studies examining BE screening using white light endoscopy in the general population have demonstrated poor cost-effectiveness.(12)
Given this lack of efficacy and cost-effectiveness, alternative methods of BE screening, including new endoscopic modalities as well as cytological methods, have been investigated.(15) Transnasal endoscopy is an alternative to standard endoscopy, which has been studied. This method increases patient tolerance by avoiding sedation although its smaller working channel limits the size of biopsy samples.(16) Studies have shown good agreement with standard endoscopy for detecting BE and 70% of patients prefer transnasal endoscopy.(17) However, its cost-effectiveness and efficacy in population-based screening have not been demonstrated.(18) Brush cytology sampling during endoscopy allows larger areas of the esophagus to be sampled at lower cost, but has a sensitivity of only 33-60% for diagnosing BE.(19–21) In addition, molecular marker assays in cytological samples have been studied to improve BE detection. Trefoil factor (TFF3) is a secretory protein expressed in the goblet cells of the intestinal mucosa that has shown significant promise for molecular BE diagnosis.(22) A swallowable sponge device, the Cytosponge, was used to sample esophageal mucosal cells with immunohistochemical analysis for TFF3, exhibiting 73.3% sensitivity and 93.8% specificity for diagnosing short-segment BE.(23) Moreover, Markowitz et al. recently reported detection of BE using an inflatable balloon in conjunction with epigenetic biomarkers (24). In the current prospective study, we examined the safety, feasibility, and efficacy of a sponge cytology-sampling device, EsophaCap, which Iyer et al. also recently studied in combination with a methylation biomarker panel to diagnose BE.(25) Our group identified 8 epigenetic biomarkers, viz., p16 (CDKN2A), RUNX3, HPP1, NELL1, TAC1, SST, AKAP12, and CDH13, which are methylated early in BE-associated neoplastic progression.(7, 26–31) A retrospective, multicenter, double-blinded validation study showed that these 8 methylation biomarkers assessed in biopsy specimens allowed for risk stratification in BE patients, accurately predicting future HGD and EAC that would not otherwise have been predicted.(7) In the current prospective study, we examined the safety, feasibility, and efficacy of a sponge cytology-sampling device, EsophaCap, in conjunction with our previously identified BE biomarkers. We applied a recently developed technology, methylation on beads (MOB), which permits the capture, retention, and bisulfite conversion of minute quantities of DNA,(32) thereby augmenting sensitivity for assessing DNA methylation in small sponge cytology samples.
Methods
Study Design
We conducted a prospective case-control study at the Johns Hopkins University School of Medicine. All studies were conducted in accordance with the U.S. Common Rule. This study was approved by Institutional Review Boards of Johns Hopkins University School of Medicine. And all assays were performed at the Johns Hopkins University School of Medicine. We consecutively recruited patients undergoing upper endoscopy for the evaluation of symptoms in our gastroenterology outpatient services between February 1, 2016 and February 1, 2018 according to the following inclusion criteria: 1) undergoing esophagastroduodenoscopy at Johns Hopkins University between 2/0½016 and 2/0½018; 2) age 18 years or greater; 3) followed at Johns Hopkins Hospital or outpatient clinics; and 4) ability to swallow the EsophaCap capsule. We also applied the following exclusion criteria: 1) patients with extra-esophageal malignancies, including head and neck or gastric cancer; 2) patients who had undergone esophagectomy; 3) patients who had undergone radiation therapy to the chest; 4) patients younger than 18; 5) patients with esophageal stents; 6) patients with esophageal strictures preventing swallowing of the capsule; 7) patients with severe dysphagia or odynophagia. Clinic physicians and staff contacted patients or volunteers by email, telephone or face-to-face in their offices. They introduced the purpose of the study and explained the procedure and its risks and benefits. If the patient agreed to join the study, written informed consent was obtained. Then, participants completed a survey to capture sociodemographic and clinical information. Specifically, they were queried on the presence of symptoms including dysphagia, odynophagia, abdominal pain, nausea, vomiting, melena or hematochezia.
Eligible subjects were asked to swallow the EsophaCap sponge capsule. Each patient swallowed the EsophaCap immediately prior to undergoing upper endoscopy. Alternatively, patients were asked to swallow the EsophaCap during outpatient clinic visits occurring within 6 months before or after each patient’s upper endoscopy. In addition, it is worthily mentioned that some volunteers without any symptom also enrolled as control. These volunteers did not have any clinical indications for endoscopy and therefore did not undergo this procedure. There were 4 such volunteers in the training set and 0 volunteers in the test set.
The diagnosis of BE was made by gastrointestinal pathologists with expertise in BE. BE was defined by the consensus definition of ≥1 cm of salmon-colored mucosa proximal to the gastroesophageal junction with biopsy confirmation of specialized columnar-lined mucosa. Patients with an irregular Z-line or less than 1 cm of salmon-colored mucosa were not considered to have BE. Author S.I. collected clinicopathological data, while all other co-authors were blinded to this data, including those who performed methylation biomarker analyses on EsophaCap specimens.
Next, DNA was extracted from sponge cytology samples and treated with bisulfite using the MOB method. Methylation levels of 8 biomarkers and an unmethylatable beta-actin internal control were determined by quantitative methylation-specific PCR (qMSP). Then, the statistical analysis was performed.
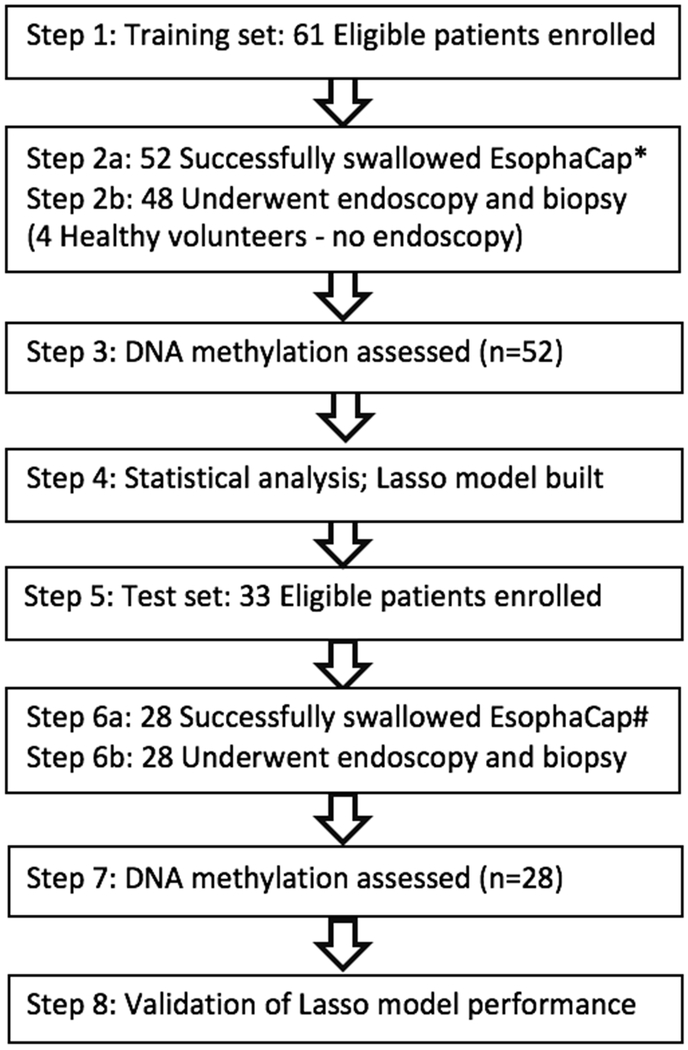
Participants were prospectively enrolled in two independent cohorts: training and test sets. The training set comprised patients enrolled from February 1, 2016 until May 1, 2017; the test set consisted of patients enrolled from May 1, 2017 until February 1, 2018. Author Z.W. performed methylation assays on the training set samples in blinded fashion, as above, while author Y.C. performed methylation assays in blinded fashion on the test set. Training set data were used to build the lasso model, after which this model was applied to the test set. All steps were performed strictly independently and blinded to the other steps. The summary flow of the study is displayed in Figure 1.
Figure 1:
Summary flow of the study. * 9 cases in training set were unable to swallow EsophaCap. # 5 cases in test set were unable to swallow EsophaCap.
EsophaCap Procedure
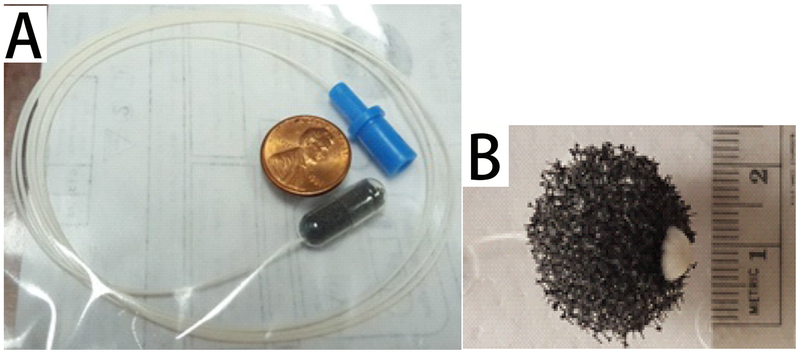
EsophaCap (EsophaCap, CapNostics, NJ, USA) is a swallowable cellular retrieval capsule sponge device. EsophaCap is composed of an open-cell graft polyether polyurethane foam sphere compressed within a soluble gelatin capsule and attached to a polyurethane filament (Figure 2). The end of the filament is held outside the mouth while the capsule is swallowed. Once inside the stomach, the capsule dissolves after several minutes, and the sponge expands. The sponge is then retrieved by pulling on the filament. Cytologic material attaches to the sponge during exit, including cells from the esophagus and proximal stomach. The cytosponge device provides a generous collection of exfoliated cells (approximately 500,000 cells) as previously study(33). Our data show that average DNA yield per EsophaCap device is 5730 ng. If we assume 6 pg of DNA per diploid cell, then the average number of cells captured per EsophaCap is 955,000. This EsophaCap procedure was executed by trained study staff. Sponge samples were collected, marked with consecutive numbers by authors S. I. or S.J.M., and delivered to researchers (authors Z.W. or Y.C.), who were kept blinded to clinical diagnoses.
Figure 2:
The EsophaCapTM Swallowable Sponge Device. The collapsible black plastic sponge is tethered to a white filament and compressed in a soluble gelatin capsule(A). The end of the filament is held outside the mouth while the capsule is swallowed. Once inside the stomach, the capsule dissolves after several minutes and the sponge expands(B). It is then retrieved by pulling on the filament. Cytologic material attaches during exit, including cells from BE and normal esophagus.
Methylation on Beads (MOB)
Because the quantity of cells captured via EsophaCap was considerably less than via biopsy samples, we employed the recently developed method, methylation on beads (MOB), which efficiently achieves capture of bisulfite-converted DNA from small specimens, as previously described.(34) MOB is a single-tube cellular processing technique that uses superparamagnetic beads (SSBs) as a DNA carrier for both DNA extraction and bisulfite conversion.(34) MOB yields, on average, 6-fold greater amplifiable converted DNA vs. commercial column-based techniques or phenolchloroform extraction.(34)
Real-Time Quantitative Methylation-Specific PCR (qMSP)
We measured target gene methylation levels and an internal unmethylatable control β-actin level in bisulfite-converted genomic DNAs obtained from EsophaCap samples by MOB. Methylation levels of 8 genes (p16, HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12, and SST) were determined by qMSP on an ABI 7900 Sequence Detection (Taqman) System, as previously described. (7) A standard curve was generated using serial dilutions of CpGenome Universal Methylated DNA (CHEMICON, Temecula, CA) to allow absolute quantification. The amplication level of β-actin was used for internal normalization. The normalized value of each gene’s methylation level was defined as fractional methylation of its matched β-actin reference value. Primer and probe sequences are listed in Supplementary Table S1.
Data Analysis and Statistics
The Mann-Whitney test was used to analyze methylation differences of the 8 above biomarkers between BE patients and control subjects without BE. Receiver-operating characteristic (ROC) curves and the area under the ROC curve (AUC) were applied to assess the diagnostic performance of each gene in univariate analyses. In addition, a lasso regression predictive model was built using R and glmnet packages. The Youden index (sensitivity + specificity-1) was employed to choose cutoff values. Hanley & McNeil test was used to ascertain whether there was any difference between AUCs of the two models.
Lasso Regression
Lasso is a popular regression method suitable for analyzing data with higher dimensionality and strongly relevant variables.(35) Here, we used lasso regression to build a BE diagnostic classifier, using the glmnet package(36). The function glmnet returned a sequence of lambdas (λs) and models. By using the function cv.glmnet, 10-fold cross-validation was conducted to select the best model. As shown in Supplementary Figure S1, we plotted the partial likelihood deviance versus log (λ), where λ was the tuning parameter. Herein, a value λ = 0.0500565 was chosen by 10-fold cross-validation via minimum criteria. Cross-validation was run 100 times (Figure S1). Next, we ran the function cv.glmnet again with the lambda.min and extracted variables with nonzero coefficients and their corresponding coefficients. We then derived a formula to calculate the diagnostic probability value of each case based on the selected variables and their corresponding coefficients. This formula was 1/[1+EXP (−(−3.91827023 + 6484.49114860 *p16 + 672.38584609 *NELL1 + 223.10561447 *AKAP12 + 7.71493793 *TAC1 + 0.03866346 *age))]. The cutoff value of 0.23 was decided by ROC curve and Youden index based on training set data. The predicted diagnosis of each case in the test set was decided according to its predicted value. Cases with predicted values greater than 0.23 were classified as BE, while all other cases were classified as non-BE.
Results
Participants and Clinical Characteristics
Total 94 participants were originally enrolled in our study. 9 patients in training set and 5 cases in test set who did not swallow the EsophaCap was excluded. No side effects were reported related to swallowing the device or retrieving the capsule.
In the training set, 52 participants were included. Baseline characteristics of these 52 patients are shown in Table 1. 61.5% were male, with age ranging from 34 to 83 years (mean 58.7 ±11.5). 18 patients had BE on subsequent endoscopy, including BE with no dysplasia (13), low-grade dysplasia (1), high-grade dysplasia (4), and 34 control patients without BE. These controls included 21 patients with GERD, 6 with gastritis, 1 with a gastric ulcer, 1 with functional dysphagia, 1 with functional odynophagia, and 4 patients without symptoms. Age in the BE group was significantly higher than in controls (p=0.022, Table 1). There were no significant differences between groups in other parameters, including gender, smoking history, BMI, and race in the training set (all p>0.05, Table 1).
Table 1:
Clinical characteristics of patients in study.
| Characteristic | Training set | Test set | ||||
|---|---|---|---|---|---|---|
| Control | BE | P value | Control | BE | P value | |
| Age | 0.022* | 0.010* | ||||
| <=60 | 23 | 6 | 11 | 2 | ||
| >60 | 11 | 12 | 3 | 12 | ||
| Gender | 0.133 | 0.705 | ||||
| Male | 18 | 14 | 8 | 7 | ||
| Female | 16 | 4 | 6 | 7 | ||
| BMI | 0.551 | 0.663 | ||||
| <=26 | 14 | 6 | 3 | 4 | ||
| >26 | 17 | 12 | 11 | 10 | ||
| NA | 3 | 0 | 0 | 0 | ||
| Smoking | 0.130 | 0.058 | ||||
| Former/current | 11 | 11 | 5 | 10 | ||
| Never | 17 | 6 | 9 | 4 | ||
| NA | 6 | 1 | 0 | 0 | ||
| Alcohol | 0.180 | 0.842 | ||||
| Former/current | 8 | 8 | 6 | 7 | ||
| Never | 23 | 10 | 7 | 7 | ||
| NA | 3 | 0 | 1 | 0 | ||
| Race | 0.280 | 0.309 | ||||
| White | 22 | 16 | 13 | 14 | ||
| Other | 8 | 2 | 1 | 0 | ||
| NA | 4 | 0 | 0 | 0 | ||
In the test set, a total of 28 patients were enrolled. These individuals comprised 14 BE cases and 14 non-BE patients with gastrointestinal symptoms. 13 BE cases were nondysplastic and 1 had intramucosal adenocarcinoma. Except for age, there were no significant differences between the case and control groups in clinical parameters, including gender, smoking history, BMI, and race in the test set (all p>0.05, Table 1).
Potential DNA Methylation Markers in Cytology Samples for Detecting BE
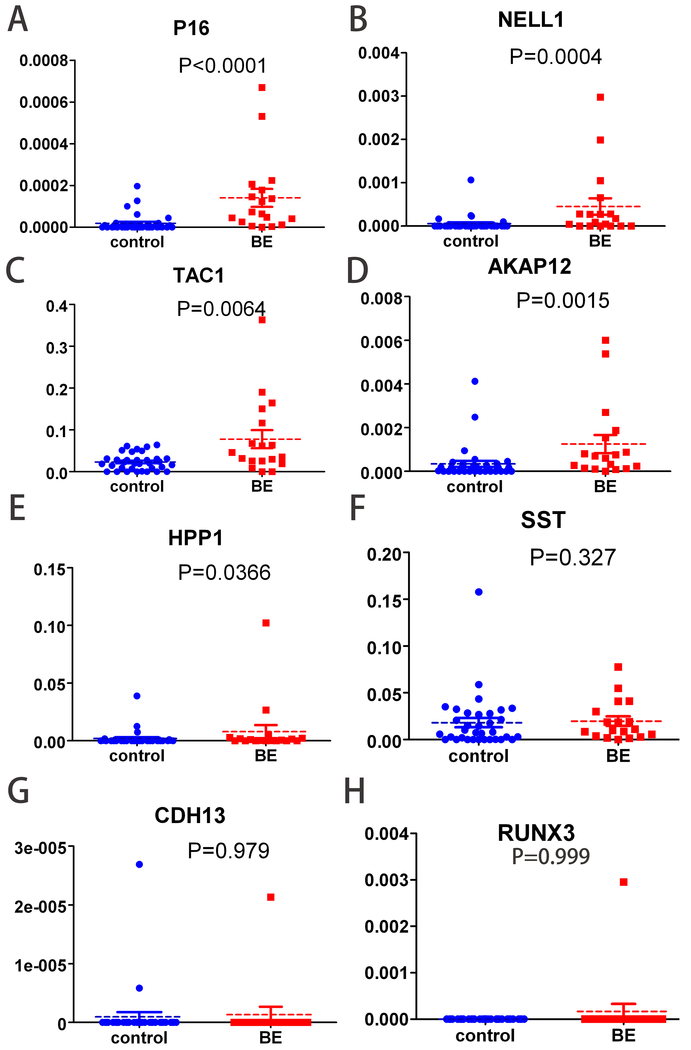
We studied 8 genes, CDKN2A (p16), HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12 and SST, which had been extensively proven to become methylated in the vast majority of BE tissue biopsies. We compared methylation levels of these 8 genes in sponge sample-derived DNAs between BE cases and controls. Methylation levels of p16, NELL1, TAC1, AKAP12 and HPP1(all P<0.05, Figure 3A, B, C, D, E, and F), but not SST, CDH13, or RUNX3 (all P>0.05, Figure 3G, H, and I), were significantly higher in sponge DNAs from BE patients than from control subjects. In addition, methylation frequencies of CDH13 and RUNX3 in sponge samples were very low (2/52 and 1/52, respectively), in contrast to previous findings in biopsies(7). The CDH13 and RUNX outliers shown in Figure 3 were not the same individuals. There were no associations between methylation status or levels and clinical parameters, including gender, BMI, age, and smoking (Supplementary Table S2).
Figure 3:
Methylation values of 8 genes in sponge DNAs from BE cases vs. control subjects. Methylation values of p16(A), NELL1(B), TAC1(C), AKAP12(D) and HPP1(E) were significantly higher in BE than controls (p<0.0001, p=0.0004, p=0.0064, p=0.0015, and p=0.0366, respectively), but differences for SST(F), RUNX3(G), and CDH13(H) were not significant (p=0.327, p=0.979, and p=0.999).
Univariate and Multivariate Logistic Regression Analyses
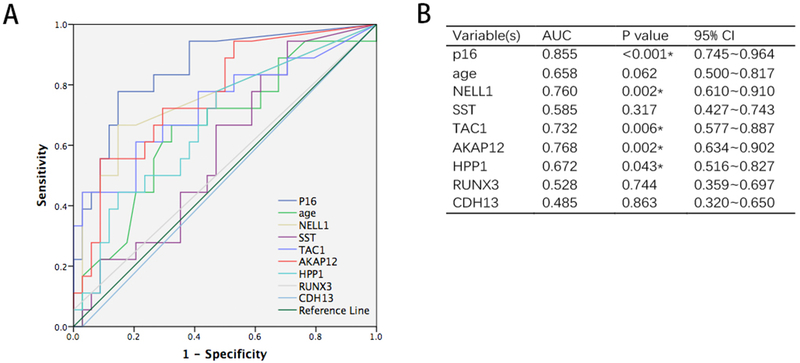
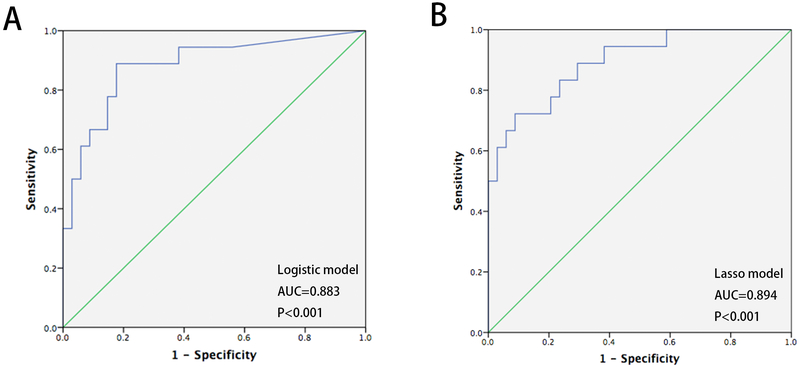
To assess the diagnostic value of single biomarkers, we measured classification accuracy using receiver-operating characteristic (ROC) curves and area under curve (AUC) calculations. As shown in figure 4, AUCs for p16, NELL1, TAC1, AKAP12, and HPP1 were all significantly greater than 0.500 (p16, AUC=0.855, P<0.001, NELL1, AUC=0.760, P=0.002, TAC1, AUC=0.732, P=0.006, AKAP12, AUC=0.768, P=0.002, HPP1, AUC=0.672, P=0.043). The Youden index was used to assess the sensitivity and specificity of each biomarker. To develop a multimarker panel for the diagnosis of BE, we first applied logistic regression to the 8 biomarkers plus age. Only p16 and NELL1 were found to be significant in this logistic regression model (P=0.003 and 0.041, respectively). The AUC of this 2-biomarker regression model was higher than that of any single biomarker (AUC=0.883, 95% CI 0.781~0.986, P<0.001, Figure 5A). Cutoff value was 0.21, decided by highest Youden index. The sensitivity and specificity of the 2-biomarker logistic regression model were 88.9% and 82.4%, respectively.
Figure 4:
ROC curves showing individual diagnostic performance of p16, NELL1, AKAP12, TAC1, HPP1, SST, RUNX3, CDH13 and age in sponge DNAs from BE cases vs. controls in the training set (A and B). AUCs = 0.855 (p16), 0.760 (NELL1), 0.768 (AKAP12), 0.732 (TAC1), 0.672 (HPP1), 0.585 (SST), 0.528 (RUNX3), 0.485 (CDH13) and 0.658 (age); P=0.000, 0.002, 0.002, 0.006, 0.043, 0.317, 0.774, 0.863 and 0.062, respectively.
Figure 5:
ROC curves showing diagnostic performance of logistic regression and LASSO models in sponge samples from BE cases and controls in the training set. (A) AUC for logistic regression model based on p16 (CDKN2A) and NELL1 = 0.883, P<0.001; (B) AUC for LASSO model based on 4 biomarkers plus age = 0.894, P<0.001.
Lasso Regression Analysis
In order to obtain a higher-dimensionality predictive model, we next applied lasso regression to select the most useful prognostic markers from the 8 biomarkers and age. Five variables, namely p16, NELL1, AKAP12, TAC1 and age, were chosen by the model (Figure S2). A formula containing 4 biomarkers plus age was established based on data from the training set, where diagnostic probability score = 1/[1+EXP (−(−3.91827023 + 6484.49114860 *P16 + 672.38584609 *NELL1 + 223.10561447 *AKAP12 + 7.71493793 *TAC1 + 0.03866346 *age))]. The AUC of the 4-biomarker-plus-age lasso model was 0.894 (95% CI 0.804~0.984) (Figure 5B). Next, the cutoff value of 0.230 was chosen via Youden index. The diagnostic sensitivity and specificity of this cutoff value in the training set were 94.4% (95%CI 71%~99%) and 62.2% (95% CI 44.6%~77.3%), respectively.
In addition, we performed Hanley & McNeil testing to ascertain whether there was any difference between AUCs of the two models. This testing showed no significant difference between AUCs of the two models (P= 0.855; AUC=0.883 vs. 0.894).
Performance of The Lasso Regression Model in The Test Set
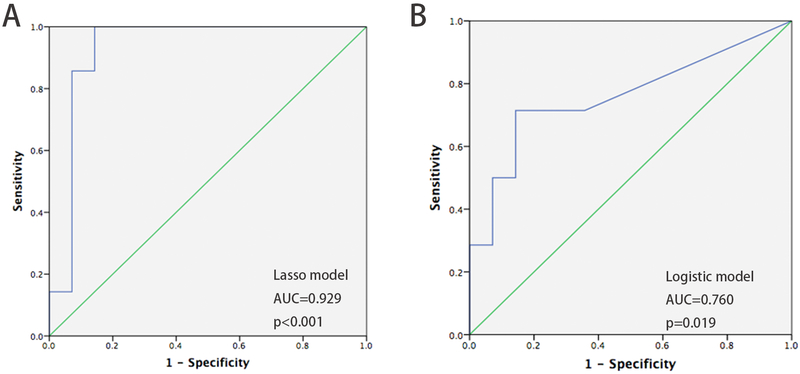
To validate the performance of lasso model, we enrolled an independent second cohort of 28 individuals as a test set. We measured methylation levels of the 4 biomarkers in the lasso model, then calculated the probability value of each case according to the above model formula. The predicted diagnosis of each case was decided using the same cutoff values as in the training set. These individuals comprised 14 BE cases and 14 non-BE patients after unblinding. In the test set, the diagnostic accuracy of the lasso model was 85.7%, with a AUC of 0.929 (P<0.001, 95% CI 0.810~1) (Figure 6A). The sensitivity and specificity were 78.6% (95% CI 48.8%~94.3%) and 92.8% (95% CI 64.1%~99.6%), respectively. In addition, we tested the diagnostic accuracy of a logistic regression model, and its accuracy was only 71.4%, with an AUC of 0.760 (P=0.019, 95% CI 0.575~0.946) (Figure 6B).
Figure 6:
Diagnostic performance of the 4-biomarker-plus-age LASSO model and logistic model in the test set (28 cases). (A) AUC of LASSO model based on 4 biomarkers plus age = 0.929, P<0.001; (B) AUC of logistic model = 0.760, P=0.019.
We also compared the accuracy of the two models. the accuracy of the lasso model was significantly higher than that of the logistic model (P=0.001, AUC=0.760 vs. 0.929). Notably, the logistic model excluded age, which is itself an important risk factor for BE. Therefore, we chose the lasso model as our final predictive tool.
In order to explore whether abnormalities of the GEJ (such as intestinal metaplasia or dysplasia) influenced the predictive power of our panel, we checked detailed pathology reports of false positives and false negatives in both training and test sets. No false positives in the training or test sets showed intestinal metaplasia at the GEJ. In the training set, 17 of 18 BE cases were true positives; the only false negative result occurred in a non-dysplastic case. In the test set, there were 11 true positives; all 3 false negative results occurred in non-dysplastic cases.
Discussion
Here, we demonstrate excellent safety and feasibility of a novel non-endoscopic esophageal cytologic sampling device, the EsophaCap. Usage of esophageal balloons for cytological analysis has a long history, when it was shown that it exhibited discordance with standard endoscopy for BE detection as well as with dysplasia detection.(37) Our device is smaller and safer than inflatable balloons, and contains an open-cell graft polyether polyurethane foam sphere, making it much easier and safer to swallow, with rapid spontaneous deployment (by dissolution of the gelatin capsule) in the stomach and comfortable retrieval. Thus, 85% (80/94) of our patients who attempted to swallow the EsophaCap successfully swallowed it, with 100% successful sponge retrieval. There were no instances of bleeding, severe or persistent pain, endoscopic evidence of trauma, or any other adverse events in any subjects. Similarly, the safety and feasibility of EsophaCap were previously established in Iyer’s study, which showed a 98% successful swallowing rate, no mucosal injury in 68%, and minimal abrasion without bleeding in 32% of patients who swallowed the EsophaCap(25). Recently, a new balloon device sampling the distal esophagus was described by Markowitz et al. (24). These researchers described a compliance (successful swallowing) rate of 82%. Administration of this balloon device may have been more complicated than that of EsophaCap, since it requires expert inflation by specialized medical personnel in order to be safely deployed without causing esophageal damage.
Our study was also novel in utilizing Methylation-on-Beads (MOB). Cytologic sampling limits the amount of material for analysis of DNA; MOB permits the extraction and bisulfite modification of minuscule amounts of methylated DNA from limited samples, such as peripheral blood or sputum(38). The sensitivity of MOB is 25-fold greater than traditional DNA extraction and bisulfite convertion.(39) Using MOB, we measured 8 biomarkers and 1 internal control gene in EsophaCap cytology samples. The presence of food material in the stomach resulted in failed DNA extraction, leading us to perform the study only on patients with an empty stomach.
Prior studies utilizing cytologic brushings for BE detection demonstrated limited sensitivity and specificity, limiting their usage in clinical practice.(21, 40) Biomarkers have been combined with cytologic specimen recovery to improve BE detection accuracy. Trefoil factor 3 (TFF3), a secretory protein expressed by goblet cells in intestinal metaplasia, is one such biomarker.(22) The Cytosponge, a swallowable sponge used in England in conjunction with immunohistochemical staining for TFF3, showed 73.3% sensitivity and 93.8% specificity in detecting BE.(23, 41) However, the TFF3 protein marker requires interpretation by expert pathologists and may be affected by inter-observer variation. Recently, several methylation biomarkers combined with cytosponge sampling were introduced by the same research group; these biomarkers showed high accuracy in discriminating BE(33). Iyer et al. (25) reported a two-biomarker panel (VAV3 and ZNF682) achieving a combined AUC of 1.0 (100% specificity and 100% sensitivity); notably, their study enrolled EsophaCap specimens from only 19 BE patients and 20 controls. Efficacies of testing DNA methylation levels of Cytosponge specimens in predicting BE diagnosis vary among data from different clinical centers. For example, Chettouh et al.(33) assessed methylation levels of four biomarkers (TFPI2, TWIST1, ZNF345 and ZNF569) on Cytosponge specimens, finding AUCs between 78.7% and 87.7%. Markowitz et al. (24) employed bisulfite sequencing to assess methylation levels of two genes, CCNA1 and VIM, in esophageal balloon specimens, reporting 95% sensitivity and 91% specificity for BE and EAC detection. However, these researchers reported that their biomarkers were positive in normal squamous esophageal epithelium from tobacco smokers, as well as in intestinal metaplasia of the stomach. Nevertheless, taken together, data from our own and others’ studies are exciting and suggest substantial potential of EsophaCap-based biomarker assays in the clinic.
In our study, we created a lasso model containing four DNA methylation markers, viz., p16, NELL1, AKAP12, and TAC1, as well as age. This high-dimensionality lasso model performed better than did any individual biomarkers or the two-biomarker logistic model in diagnosing BE from EsophaCap samples, with a sensitivity of 94.4% (95% CI 71%~99%), a specificity 62.2% (95% CI 44.6%~77.3%), and AUC = 0.894 in the training set. The performance of this model was also excellent in an independent test set, with AUC= 0.929 (P<0.001, 95% CI 0.810~1), sensitivity = 78.6% (95% CI 48.8%~94.3%), and specificity = 92.8% (95% CI 64.1%~99.6%). Notably, we combined a novel and highly sensitive DNA extraction and bisulfite treatment method, MOB, with the EsophaCap sampling device, which should translate easily to starndard clinical laboratories.
We also correlated methylation levels of 8 candidate genes (p16, HPP1, RUNX3, CDH13, TAC1, NELL1, AKAP12 and SST) in sponge samples with clinical characteristics associated with BE. Methylation of these 8 genes in biopsy samples had already been proven to associate strongly with BE (7, 26–31). In the current study, methylation levels of these genes were not associated with any clinical parameters, including smoking history. The finding contrasts with Markowitz et al., whose biomarkers were increased in esophageal mucosa from tobacco smokers (24, 42). In our study, only Caucasian race was associated with higher rates of p16 methylation (Supplementary Table S2).
Notably, most of our enrolled cases described gastrointestinal symptoms, which may explain why the rate of BE in our study was higher than in the general population. Moreover, the majority of our BE patients had short-segment, nondysplastic BE, in contrast to the cohorts studied by Iyer (25) and Markowitz (24) , both of which were enriched for long-segment, dysplastic BE and esophageal adenocarcinoma patients: these conditions likely increased both rates and levels of methylation for the markers they tested.
In conclusion, the EsophaCap is a safe and tolerable minimally invasive esophageal sampling device. A lasso prediction model that incorporated 4 DNA methylation markers and age exhibited excellent sensitivity and very good specificity for BE diagnosis in the training set, with comparable results in the test set. These findings suggest that this approach is a promising low-cost strategy for the early detection of BE. Future large-scale studies are indicated to consolidate and further validate this strategy.
Supplementary Material
Translational Relevance.
Non-endoscopic swallowable balloon-based esophageal sampling devices, used as minimal invasive and cost-effective tools for Barrett’s esophagus screening, have attracted considerable attention recently. The current study sought to diagnose Barrett’s esophagus (BE) based on methylated DNA biomarkers in specimens captured by a sponge-on-a-string device, EsophaCap. Here, we employed a recently introduced technique, methylation on beads (MOB), to extract, bisulfite-modify, and concentrate limited DNA available from EsophaCap-collected samples for downstream methylation analysis. We demonstrated that EsophaCap, in combination with an epigenetic biomarker panel and the MOB method, is a promising, low-cost esophageal sampling strategy for BE early diagnosis.
Acknowledgments
The authors would also like to acknowledge the generous support of Drs. Alan, Rosaline Barron, and Sariat Ibrahim.
Funding: This work was supported by the National Institutes of Health (Grants CA211457 and DK118250), the Emerson Cancer Research Fund, and a Discovery Award from The Johns Hopkins University School of Medicine. Dr. Meltzer is the Harry and Betty Myerberg-Thomas R. Hendrix Professor and an American Cancer Society Clinical Research Professor. Zhixiong Wang was supported by a Scholarship from the China Scholarship Council (CSC) and the 3×3 Fund from the First Affiliated Hospital of Sun Yat-sen University.
Footnotes
Declarations of interests: All authors state that there are no conflicts of interest.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spechler SJ. Barrett esophagus and risk of esophageal cancer: a clinical review. JAMA. 2013;310:627–36. [DOI] [PubMed] [Google Scholar]
- 4.El-Serag HB, Naik AD, Duan Z, Shakhatreh M, Helm A, Pathak A, et al. Surveillance endoscopy is associated with improved outcomes of oesophageal adenocarcinoma detected in patients with Barrett’s oesophagus. Gut. 2015. [DOI] [PubMed] [Google Scholar]
- 5.Hvid-Jensen F, Pedersen L, Drewes AM, Sorensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. The New England journal of medicine. 2011;365:1375–83. [DOI] [PubMed] [Google Scholar]
- 6.Bhat SK, McManus DT, Coleman HG, Johnston BT, Cardwell CR, McMenamin U, et al. Oesophageal adenocarcinoma and prior diagnosis of Barrett’s oesophagus: a population-based study. Gut. 2015;64:20–5. [DOI] [PubMed] [Google Scholar]
- 7.Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, Mori Y, et al. A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res. 2009;69:4112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberg AJ, Park JW, Hager BW, Brock MV, Diener-West M. The use of “overall accuracy” to evaluate the validity of screening or diagnostic tests. J Gen Intern Med. 2004;19:460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett’s esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26–33. [DOI] [PubMed] [Google Scholar]
- 11.Ward EM, Wolfsen HC, Achem SR, Loeb DS, Krishna M, Hemminger LL, et al. Barrett’s esophagus is common in older men and women undergoing screening colonoscopy regardless of reflux symptoms. Am J Gastroenterol. 2006;101:12–7. [DOI] [PubMed] [Google Scholar]
- 12.Barbiere JM, Lyratzopoulos G. Cost-effectiveness of endoscopic screening followed by surveillance for Barrett’s esophagus: a review. Gastroenterology. 2009;137:1869–76. [DOI] [PubMed] [Google Scholar]
- 13.Inadomi JM, Sampliner R, Lagergren J, Lieberman D, Fendrick AM, Vakil N. Screening and surveillance for Barrett esophagus in high-risk groups: a cost-utility analysis. Ann Intern Med. 2003;138:176–86. [DOI] [PubMed] [Google Scholar]
- 14.Soni A, Sampliner RE, Sonnenberg A. Screening for high-grade dysplasia in gastroesophageal reflux disease: is it cost-effective? Am J Gastroenterol. 2000;95:2086–93. [DOI] [PubMed] [Google Scholar]
- 15.di Pietro M, Chan D, Fitzgerald RC, Wang KK. Screening for Barrett’s Esophagus. Gastroenterology. 2015;148:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shariff MK, Bird-Lieberman EL, O’Donovan M, Abdullahi Z, Liu X, Blazeby J, et al. Randomized crossover study comparing efficacy of transnasal endoscopy with that of standard endoscopy to detect Barrett’s esophagus. Gastrointest Endosc. 2012;75:954–61. [DOI] [PubMed] [Google Scholar]
- 17.Jobe BA, Hunter JG, Chang EY, Kim CY, Eisen GM, Robinson JD, et al. Office-based unsedated small-caliber endoscopy is equivalent to conventional sedated endoscopy in screening and surveillance for Barrett’s esophagus: a randomized and blinded comparison. Am J Gastroenterol. 2006;101:2693–703. [DOI] [PubMed] [Google Scholar]
- 18.Peery AF, Hoppo T, Garman KS, Dellon ES, Daugherty N, Bream S, et al. Feasibility, safety, acceptability, and yield of office-based, screening transnasal esophagoscopy (with video). Gastrointest Endosc. 2012;75:945–53 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk GW. Cytology in Barrett’s esophagus. Gastrointest Endosc Clin N Am. 2003;13:335–48. [DOI] [PubMed] [Google Scholar]
- 20.Kumaravel A, Lopez R, Brainard J, Falk GW. Brush cytology vs. endoscopic biopsy for the surveillance of Barrett’s esophagus. Endoscopy. 2010;42:800–5. [DOI] [PubMed] [Google Scholar]
- 21.Robey SS, Hamilton SR, Gupta PK, Erozan YS. Diagnostic value of cytopathology in Barrett esophagus and associated carcinoma. Am J Clin Pathol. 1988;89:493–8. [DOI] [PubMed] [Google Scholar]
- 22.Dunn LJ, Jankowski JA, Griffin SM. Trefoil Factor Expression in a Human Model of the Early Stages of Barrett’s Esophagus. Dig Dis Sci. 2015;60:1187–94. [DOI] [PubMed] [Google Scholar]
- 23.Kadri SR, Lao-Sirieix P, O’Donovan M, Debiram I, Das M, Blazeby JM, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett’s oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moinova HR, LaFramboise T, Lutterbaugh JD, Chandar AK, Dumot J, Faulx A, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett’s esophagus. Sci Transl Med. 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer PG, Taylor WR, Johnson ML, Lansing RL, Maixner KA, Yab TC, et al. Highly Discriminant Methylated DNA Markers for the Non-endoscopic Detection of Barrett’s Esophagus. Am J Gastroenterol. 2018;113:1156–66. [DOI] [PubMed] [Google Scholar]
- 26.Jin Z, Cheng Y, Olaru A, Kan T, Yang J, Paun B, et al. Promoter hypermethylation of CDH13 is a common, early event in human esophageal adenocarcinogenesis and correlates with clinical risk factors. Int J Cancer. 2008;123:2331–6. [DOI] [PubMed] [Google Scholar]
- 27.Jin Z, Hamilton JP, Yang J, Mori Y, Olaru A, Sato F, et al. Hypermethylation of the AKAP12 promoter is a biomarker of Barrett’s-associated esophageal neoplastic progression. Cancer Epidemiol Biomarkers Prev. 2008;17:111–7. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z, Mori Y, Hamilton JP, Olaru A, Sato F, Yang J, et al. Hypermethylation of the somatostatin promoter is a common, early event in human esophageal carcinogenesis. Cancer. 2008;112:43–9. [DOI] [PubMed] [Google Scholar]
- 29.Jin Z, Mori Y, Yang J, Sato F, Ito T, Cheng Y, et al. Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene. 2007;26:6332–40. [DOI] [PubMed] [Google Scholar]
- 30.Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, et al. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin Cancer Res. 2007;13:6293–300. [DOI] [PubMed] [Google Scholar]
- 31.Schulmann K, Sterian A, Berki A, Yin J, Sato F, Xu Y, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–48. [DOI] [PubMed] [Google Scholar]
- 32.Guzzetta AA, Pisanic Ii TR, Sharma P, Yi JM, Stark A, Wang TH, et al. The promise of methylation on beads for cancer detection and treatment. Expert Rev Mol Diagn. 2014;14:845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chettouh H, Mowforth O, Galeano-Dalmau N, Bezawada N, Ross-Innes C, MacRae S, et al. Methylation panel is a diagnostic biomarker for Barrett’s oesophagus in endoscopic biopsies and non-endoscopic cytology specimens. Gut. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey VJ, Zhang Y, Keeley BP, Yin C, Pelosky KL, Brock M, et al. Single-tube analysis of DNA methylation with silica superparamagnetic beads. Clin Chem. 2010;56:1022–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibshirani R The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95. [DOI] [PubMed] [Google Scholar]
- 36.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J Stat Softw. 2011;39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witte S [Cytologic diagnosis of the upper gastrointestinal tract]. Leber Magen Darm. 1984;14:8–17. [PubMed] [Google Scholar]
- 38.Hulbert A, Jusue-Torres I, Stark A, Chen C, Rodgers K, Lee B, et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin Cancer Res. 2017;23:1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeley B, Stark A, Pisanic TR 2nd, Kwak R, Zhang Y, Wrangle J, et al. Extraction and processing of circulating DNA from large sample volumes using methylation on beads for the detection of rare epigenetic events. Clin Chim Acta. 2013;425:169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rygiel AM, Milano F, Ten Kate FJ, de Groot JG, Peppelenbosch MP, Bergman JJ, et al. Assessment of chromosomal gains as compared to DNA content changes is more useful to detect dysplasia in Barrett’s esophagus brush cytology specimens. Genes Chromosomes Cancer. 2008;47:396–404. [DOI] [PubMed] [Google Scholar]
- 41.Ross-Innes CS, Debiram-Beecham I, O’Donovan M, Walker E, Varghese S, Lao-Sirieix P, et al. Evaluation of a minimally invasive cell sampling device coupled with assessment of trefoil factor 3 expression for diagnosing Barrett’s esophagus: a multi-center case-control study. PLoS Med. 2015;12:e1001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaz AM, Wong CJ, Varadan V, Willis JE, Chak A, Grady WM. Global DNA methylation patterns in Barrett’s esophagus, dysplastic Barrett’s, and esophageal adenocarcinoma are associated with BMI, gender, and tobacco use. Clin Epigenetics. 2016;8:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.